Abstract
Background
Choline is essential for healthy cognitive development. Single nucleotide polymorphisms (SNPs; rs3199966(G), rs2771040(G)) within the choline transporter SLC44A1 increase risk for choline deficiency. In a choline intervention trial of children who experienced prenatal alcohol exposure (PAE), these alleles are associated with improved cognition.
Objective
This study aimed to determine if SNPs within SLC44A1 are differentially associated with cognition in children with PAE compared with normotypic controls (genotype × exposure). A secondary objective tested for an association of these SNPs and cognition in controls (genotype-only).
Design
This is a secondary analysis of data from the Collaborative Initiative on Fetal Alcohol Spectrum Disorders. Participants (163 normotypic controls, 162 PAE) underwent psychological assessments and were genotyped within SLC44A1. Choline status was not assessed. Association analysis between genotype × exposure was performed using an additive genetic model and linear regression to identify the allelic effect. The primary outcome was the interaction between SLC44A1 genotype × exposure status with respect to cognition. The secondary outcome was the cognitive–genotype association in normotypic controls.
Results
Genotype × exposure analysis identified 7 SNPs in SLC44A1, including rs3199966(G) and rs2771040(G), and in strong linkage (D′ ≥ 0.87), that were associated (adjusted P ≤ 0.05) with reduced performance in measures of general cognition, nonverbal and quantitative reasoning, memory, and executive function (β, 1.92–3.91). In controls, carriers of rs3199966(GT or GG) had worsened cognitive performance than rs3199966(TT) carriers (β, 0.46–0.83; P < 0.0001), whereas cognitive performance did not differ by rs3199966 genotype in those with PAE.
Conclusions
Two functional alleles that increase vulnerability to choline deficiency, rs3199966(G) (Ser644Ala) and rs2771040(G) (3′ untranslated region), are associated with worsened cognition in otherwise normotypic children. These alleles were previously associated with greater cognitive improvement in children with PAE who received supplemental choline. The findings endorse that choline benefits cognitive development in normotypic children and those with PAE.
Keywords: choline, cognition, CTL1, fetal alcohol spectrum disorder, memory, nutrigenomics, rs3199966, rs2771040, single nucleotide polymorphism (SNP), SLC44A1
Introduction
Prenatal alcohol exposure (PAE) causes life-long deficits in cognition, learning, memory, and executive function [1,2]. These can be accompanied by growth restrictions and physical anomalies and are collectively described under the umbrella term Fetal Alcohol Spectrum Disorders (FASD). FASD is more common than generally appreciated; in the United States, 5.2% of pregnant females self-report binge drinking (>4 drinks per occasion) in the prior 30 d [3], and 8.4% of newborn bloodspots test positive for alcohol exposure [4]. These frequencies are consistent with prevalence studies in which 1% to 5% of children meet the diagnostic criteria for FASD [5]. There is considerable interest in identifying factors that modulate the impact of PAE, and a major focus of these intervention trials is the micronutrient choline [6,7].
Although choline is essential for healthy brain development [8,9], its intake is often limiting during the high-demand periods of pregnancy and lactation [10,11]. Supplemental choline significantly improves offspring cognition and memory [8,9,12], prompting parallel investigations for FASD. Preclinical studies report that supplemental choline improves memory, learning, and cognition in alcohol-exposed offspring [6,[13], [14], [15]], whether provided during the fetal and/or postnatal periods. Clinical trials report similar improvements. Infants born to females who received prenatal choline (740 mg/d choline) and a multivitamin/mineral supplement had significantly improved measures of visual memory in a habituation–dishabituation memory test [16] and better performance in a reaction time attentional task at preschool follow-up [17]. Similarly, infants born to mothers having greater adherence to a gestational choline intervention (2 g/d) had better learning in an eyeblink conditioning task at age 6 mo and a visual recognition memory task at 12 mo [18]. In young children (ages 2–5 y), a 9-mo choline intervention (500 mg/d) conferred improvements in tests of delayed memory [19], and nonverbal intelligence quotient, working memory, and executive function at 4- and 7-y follow up [20,21], suggesting these benefits persist.
However, choline’s nuanced benefits in these clinical studies suggest the presence of additional modifying factors. An important modulator of choline’s action is genetics. Numerous genetic variants have been described for choline-related genes that govern choline synthesis, absorption, or utilization [[22], [23], [24]]. Carriers of these effect alleles may be at greater risk for deficiency under marginal choline intakes. We recently identified single nucleotide polymorphisms (SNPs) within the ubiquitous choline transport SLC44A1 (CTL1, choline-like transporter-1) that are associated with cognitive performance. Specifically, in children with FASD who received a 9-mo choline intervention [19], we identified effect alleles in SLC44A1, including the functional SNPs rs3199966(G) and rs2771040(G), that were associated with greater pre/postintervention improvement in a sequential memory task [25]. Because these minor alleles are also associated with greater vulnerability to choline deficiency [22,24], we speculated that those carriers had unmet choline needs and thus derived greater benefit from the supplement. These findings suggest that variants affecting choline metabolism could modulate cognition in those with PAE. Here, we tested the hypothesis that these same variants in SLC44A1 were associated with poorer cognitive performance in individuals with heavy PAE who were not part of a choline intervention trial. A secondary objective tested for associations between these SNPs and cognitive performance of normotypic controls. These results highlight the potential influence of these functional SNPs for healthy brain function.
Participants and methods
Study design and participants
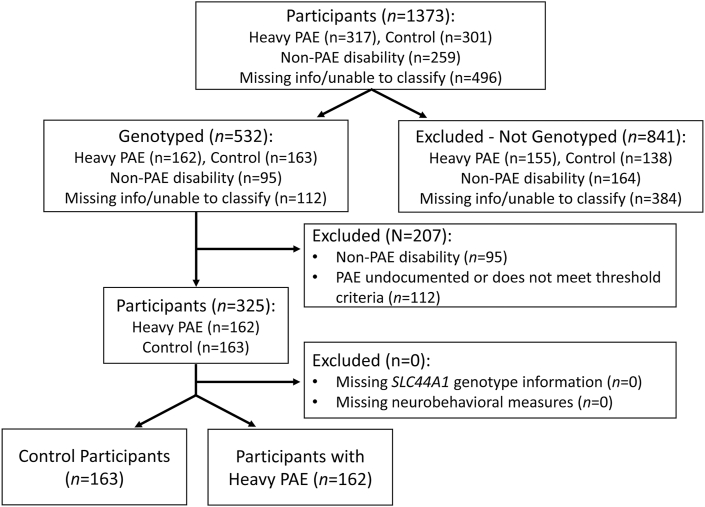
This study is a secondary analysis of preexisting cognitive and genetic data [26]. The primary outcome tested whether alleles in SLC44A1 that are associated with increased vulnerability to choline deficiency are also associated with worsened cognitive performance in children who experienced heavy PAE (genotype × exposure). A secondary outcome tested whether these alleles were associated with cognition in normotypic controls (genotype-only). The study design is shown in Figure 1. Participants (N = 1373) were recruited as part of the Collaborative Initiative on FASD (CIFASD), a multisite consortium focused on understanding the range of neurodevelopmental effects of PAE, including the clinical effects on children. Participants were recruited during CIFASD Phases 2 (2007–2012) and 3 (2013–2017) from sites in Atlanta, GA, Los Angeles, CA, Minneapolis, MN, and San Diego, CA [27,28]. Children (aged 5–16 y) either experienced heavy PAE with or without a diagnosis of FASD (N = 317) or were normotypic controls with either minimal (<1 drink/wk or <2 drinks/occasion) or no PAE history (N = 301). Heavy alcohol exposure was strictly defined as >4 drinks/occasion at least weekly or >13 drinks/wk. Children were excluded who had exposures that exceeded the control criteria but were below the heavy criteria, or whose exposure could not be documented (n = 496). Children were also excluded who had a known (non-FASD) cause of mental disability, significant head injury with loss of consciousness, or significant psychiatric or physical disability that prevented participation (n = 259) [27,28]. Information regarding nutrient intake, nutrient status, or diet patterns was not gathered; none of these participants were known to have been enrolled in a choline intervention trial.
FIGURE 1.
CONSORT flow diagram of study participants. CONSORT, Consolidated Standards of Reporting Trials; PAE, prenatal alcohol exposure.
Neurobehavioral measurements
Children underwent a series of standardized psychological assessments as described in [2,28]. The selection of assessment tools for Phase 3 was informed by the prior findings from Phase 2, and thus the test batteries did not wholly overlap. This analysis focused on the measures from Phase 3 that were also assessed in Phase 2. The behavioral instruments spanned measures of general intellectual ability, attention and concentration, executive function, memory and working memory, motor reaction time, and psychological symptomatology, and are detailed in Table 1 [[29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40]]. Tests were age-appropriate, age-normalized within each test instrument, and were not administered when an age-appropriate version was unavailable for that participant. Tests were administered on a single day by trained personnel blinded as to diagnosis. Results were expressed as Z- or T-scores as defined by the respective test. Informed assent or consent was obtained from each participant or their legal caregiver, respectively. Protocols for Phases 2 and 3 were approved by Institutional Review Boards at the lead site (San Diego State University) and all recruitment sites. A total of 398 neurobehavioral metrics were included in the association analysis.
TABLE 1.
Neurobehavioral test battery for CIFASD-2 and CIFASD-31
| Domain | Instrument and subtests | Phase | Ref. |
|---|---|---|---|
| General intellectual ability | Wechsler Intelligence Scale for Children – 4th Edition (Full Scale IQ) | 2 | [29] |
| Differential Ability Scales – 2nd Edition (DAS-II) | 3 | [30] | |
| Attention and concentration | Developmental NEuroPSYchological Assessment (Speeded Naming; Arrows) | 3 | [31] |
| Executive function | Delis-Kaplan Executive Function System (Verbal Fluency, Trail Making; Color-Word Interference; Tower; 20 Questions; Design Fluency) | 2 | [32] |
| Cambridge Neuropsychological Test Automated Battery (Intra/Extradimensional Shift) | 2, 3 | [33] | |
| Behavior Rating Inventory of Executive Function (Parent Version) | 2, 3 | [34] | |
| Developmental NEuroPSYchological Assessment (Word Generation; Inhibition) | 3 | [31] | |
| Working memory/memory | Cambridge Neuropsychological Test Automated Battery (Spatial Working Memory; Delayed Match-to-Sample) | 2 | [33] |
| California Verbal Learning Test – Children’s Version | 3 | [35] | |
| Developmental NEuroPSYchological Assessment (Memory for Names; Memory for Designs: Memory for Faces; Narrative Memory) | 3 | [31] | |
| Motor/reaction time | Cambridge Neuropsychological Test Automated Battery (Motor Screening; Big/Little Circle; Simple Reaction Time; Choice Reaction Time) | 2 | [33] |
| Psychological symptomatology | Achenbach System of Empirically Based Assessment (Child Behavior Checklist; Teacher Report Form; Youth Self Report) | 2, 3 | [36] |
| Disruptive Behavior Rating Scale | 2, 3 | [37] | |
| Sluggish Cognitive Tempo Scale | 2, 3 | [38] | |
| Vineland Adaptive Behavior Scales, 2nd Edition (Parent/Caregiver Rating Form; Teacher Rating Form) | 2, 3 | [39] | |
| Conners Comprehensive Behavior Rating Scale | 2, 3 | [40] |
IQ, intelligence quotient; Ref., reference.
Adapted from [2,27,28] and www.cifasd.org. CIFASD, Collaborative Initiative on Fetal Alcohol Spectrum Disorders, Phases 2 and 3.
Genetic analysis
Of the 1373 participants, 532 contributed saliva samples, including 162 who had heavy PAE and 163 normotypic controls. Participant sex as reported by the caregiver was checked against X-linked SNPs, and discordant samples were excluded due to the possibility of sample mix-up or misidentification. Genome-wide SNP data were genotyped on the OmniExpress genome array (Illumina) and on the Multi-Ethnic Genotyping Array at the Johns Hopkins Genetic Resources Core Facility [26]. Following a previously published genome-wide association study cleaning pipeline [41], the 2 datasets were imputed separately using the Michigan Imputation Server [42] to the 1000 Genomes Phase 3, b37 reference panel [43] and then combined. The final dataset consisted of 4,000,362 directly genotyped and imputed SNPs with minor allele frequency (MAF) ≥0.01, genotype rate ≥0.99, and Hardy–Weinberg equilibrium P ≥ 0.000001. In SLC44A1 (chr9:108,006,932–108,200,785), 7 SNPs were directly genotyped and 113 imputed with R2 values ≥0.4. Thirteen SNPs had alleles with a MAF <0.05 and were removed. Most of the 107 variants were SNPs, although some were insertion/deletions (Supplementary Table 1). These variants were distributed across the entire 179 kb length of the gene; 2 variants (rs3199966, rs2771040) were functional SNPs located in exon 16 (Ser644Ala) and the 3′ untranslated region, respectively; the remainder were intronic. Principal component analysis (PCA) of each participant’s SNP profile for chromosomes 1 to 22, including individuals in the 1000 Genomes database [43] as reference genetic ancestry groups, was computed using SNPRelate [44]. The resulting eigenvalues were used to group samples with similar genetic ancestry, using 1000 Genomes as the reference population to estimate linkage disequilibrium (LD) values and MAFs in the participant population.
Association analyses
Analysis of SNP associations was performed using R (version 4.2.0; R Foundation for Statistical Computing; http://cran.r-project.org/) and PLINK v2.00a3 64-bit (8 June, 2021). The 398 neurobehavioral measures were analyzed against each of the 107 SNPs employing a linear regression model to test for genotype × exposure associations. In addition to the main effects of genotype and exposure, sex, age at time of behavioral assessment, the first 2 principal components computed from all 22 autosomes (to account for differences in allele frequency due to ancestry), and site (coded with 4 levels for 4 sites) were utilized as covariates. The primary outcome of interest was the association of genotype with neurobehavioral performance in the genotype × exposure interaction, extracting the β coefficient, P values, and effect allele from PLINK for each SNP. Analyses used an additive genetic model as prior work on SLC44A1 found that this model best explained the associations [25]. P values were adjusted for false discovery across the entire analysis using the Bonferroni correction (P adjusted [Padj]), which is a conservative approach that treats each SNP as independent; a Padj value <0.05 was considered significant. For variants showing significant associations, we computed LD stratified by European ancestry and African ancestry using the “LDlinkR” (version 1.2.2) package in R. Associations were visualized with scatter plots created in the R package “ggplot” (version 3.36). To visualize the genotype × exposure interaction at rs3199966 and its effect size within the affected cognitive measures, we calculated both the mean performance of these 325 participants for that cognitive measure and each individual’s Z-score normalized against the group mean, followed by generalized linear regression using the “glm” package in R, followed by 2-way analysis of variance with Tukey’s post hoc analysis and stratified by exposure and genotype.
PCA from “FactoMineR” package (version 2.4) and “MICE” package (version 3.14.0) were used in combination to explore the participants’ separation with respect to the 107 variants in SLC44A1 based on sex, age, ancestry assignment from the PCA, and alcohol exposure status. Eigenvalues from the PCA were computed using the R package “Factoextra” (version 1.0.7) to produce scree plots to evaluate the variation defining each component. PC1 by PC2 plots were produced to visualize how strongly each variable influenced, if at all, a principal component in plots that catered to the individual as well as the variable.
Results
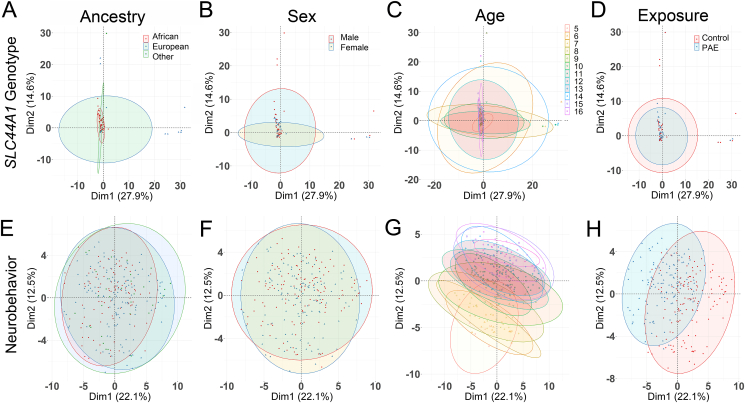
There were no differences in the study participants’ characteristics with respect to mean age, sex, and genetic ancestry representation between the heavy PAE and control groups (Table 2). PCA of SLC44A1 genotype against participant characteristics revealed that PC1 explained 27.9% of the genotype variance, and PC2 accounted for 14.6% of that variance. Neither genetic ancestry (Figure 2A), sex (Figure 2B), age at enrollment (Figure 2C), or PAE status (Figure 2D) contributed to the variance of genotypes. The PCA loadings revealed 2 sets of variants that contributed to PC1 and PC2, respectively (Supplementary Figure 1A, B), but their removal did not change the loadings (Supplementary Figure 1C) and indicated they did not drive the variance. PCA of the neurobehavioral endpoints against these same participant characteristics again revealed no influence of genetic ancestry (Figure 2E) and sex (Figure 2F), whereas exposure status had a modest influence upon PC1 (21.9%, Figure 2H), and participant age upon PC2 (12.2%, Figure 2G). Components beyond PC2 contributed ≤7.2% to this variance (Supplementary Figure 1D), and the top contributors to PC1 were all cognitive measures including general cognition; verbal, nonverbal, and sequential quantitative reasoning; and recall of designs (Supplementary Figure 1E). These same cognitive domains were the top contributors to PC2 (Supplementary Figure 1F). The half that were age-adjusted may reflect that those with heavy PAE performed below their age normative.
TABLE 2.
Participant characteristics
| Characteristic1 | Controls (N = 163) | Heavy PAE (N = 162) | P2 |
|---|---|---|---|
| Age at assessment3, y | 12.7 ± 2.4 | 12.7 ± 2.2 | 0.80 |
| Sex4 | |||
| Male | 82 (50.3%) | 96 (59.3%) | 0.29 |
| Female | 81 (49.7%) | 66 (40.7%) | 0.22 |
| Genetic ancestry5 | |||
| European | 88 (54.0%) | 78 (48.1%) | 0.44 |
| African | 49 (30.1%) | 62 (38.3%) | 0.22 |
| Other | 26 (16.0%) | 22 (13.1%) | 0.56 |
| Ethnic ancestry6 | |||
| Hispanic or Latino | 27 (16.6%) | 26 (16.0%) | 0.89 |
| Not Hispanic or Latino | 133 (81.6%) | 126 (77.8%) | 0.66 |
| Unknown | 3 (1.8%) | 10 (6.2%) | 0.052 |
| FAS diagnostic category7 | |||
| Not FAS | 109 (66.9%) | 49 (30.2%) | <0.0001 |
| FAS | 0 (0.0%) | 28 (17.3%) | N.D. |
| Deferred | 41 (25.2%) | 69 (42.6%) | <0.0001 |
FAS, fetal alcohol syndrome; N.D., not determined; PAE, prenatal alcohol exposure; SNP, single nucleotide polymorphism.
Values are n (%) or mean ± SD.
All P values are derived from chi-square tests except for age, which was an unpaired t test.
Age at assessment for their most recent interview.
Provided by caregiver completing the surveys.
Genetic ancestry assigned using principal component analysis and subsequent eigenvalues of an individual’s SNP distribution against the 1000 Genomes database [43].
Ethnic ancestry based on self-report.
FAS status based on evaluation of dysmorphological features according to the diagnostic criteria in [1]. Deferred status was assigned to individuals who met some but not all the physical diagnostic criteria for FAS.
FIGURE 2.
Principal component analysis of SLC44A1 genotypes and neurobehaviors against population characteristics. (A–D) Principal component analysis (PCA; FactoMineR, v2.3) was used to examine the genomic separation of all participants based on ancestry, sex, age, and exposure status. (A) Genetic ancestry based on comparison against 1000 Genomes vs. genotype. Red, African; Blue, European; Green, Other. (B) Sex vs. genotype. Red, males. Blue, females. (C) Age vs. genotype, stratified in 1-y intervals. (D) Exposure status vs. genotype. Red, low-to-no prenatal exposure (PAE); Blue, heavy PAE. No significant associations between genotype and these population characteristics were observed in the PCA. (E–H) PCA to examine the separation of neurobehaviors in all participants based on ancestry, sex, age, and exposure status. All color symbols as described for (A–D). (E) Genetic ancestry vs. neurobehavior. (F) Sex vs. neurobehavior. (G) Age vs. neurobehavior. (H) Exposure status vs. neurobehavior. There was no influence of ancestry and sex on neurobehavior. PAE status contributed to the behavioral variance in PC1. Age contributed to the behavioral variance in PC2. The 95% confidence interval surrounding each group mean is indicated by the ellipseing.
Association analysis of genotype × exposure identified 7 variants within SLC44A1 that were associated with any behavioral outcome (Table 3). Five of these variants were intronic SNPs, 4 in intron 1 and 1 in intron 3, and all were single nucleotide variants having an unknown potential function. Two additional variants, rs3199966 and rs2771040, are functional and were previously associated with risk for choline deficiency [[22], [23], [24]]. The intronic variants were in strong LD (D′ ≥ 0.90) with rs3199966 and rs2771040 in this population, and rs3199966 and rs2771040 were in complete LD with each other (D′ = 1.00; Supplementary Table 2).
TABLE 3.
All SNPs in SLC44A1 associated with any neurobehavioral measure in the genotype × exposure interaction1
| ID | Position, GRCh38 | Location | Type | Ref/Alt2 | MAF, this cohort3 | MAF, Europeans | MAF, Afr. Am. | Alternate allele phenotype |
|---|---|---|---|---|---|---|---|---|
| rs75106836 | 105252720 | Intron 1 | SNV | T > C | 2.9% | 0.04% | 5.5% | unknown |
| rs143438338 | 105285891 | Intron 1 | SNV | A > G | 3.0% | 0.04% | 5.5% | unknown |
| rs59370172 | 105291402 | Intron 1 | SNV | C > T | 3.0% | 0.04% | 5.6% | unknown |
| rs12347364 | 105297774 | Intron 1 | SNV | T > A | 5.6% | 5.0% | 0.7% | unknown |
| rs10991629 | 105331975 | Intron 3 | SNV | C > T | 18.7% | 11.8% | 36.3% | unknown |
| rs3199966 | 105385482 | Exon 15 (Ser644Ala) | SNV | T > G | 19.1% | 9.0% | 41.5% | increases risk for choline deficiency |
| rs2771040 | 105389918 | Exon 16 (3′ UTR) | SNV | A > G | 21.3% | 12.0% | 43.8% | increases risk for choline deficiency |
MAF, minor allele frequency; SNP, single nucleotide polymorphism; SNV, single nucleotide variant; UTR, untranslated region.
Only SNPs with adjusted P values ≤ 0.05 are included.
Ref/Alt, Reference allele/Alternate (minor) allele.
Frequencies for participants using principal component analysis. Frequencies for European and African American (Afr. Am.) populations as reported in dbSNP [45].
In the analysis of genotype × exposure, these 7 SNPs were associated with 6 behavioral measures including assessments of intellectual ability, working memory, and executive function (Table 4; Supplementary Table 3). These included general cognitive ability, nonverbal reasoning, recall of designs, sequential and quantitative reasoning, executive function, and approach to learning. The strongest associations (Padj = 0.006–0.046) were with the functional SNPs rs3199966 and rs2771040. For all but the Conners Executive Function, an increasing β value reflects a better performance. For those measures, the βs indicated that the effect alleles rs3199966(T) and rs2771040(A) were associated with improved performance with effect sizes of 1.92 to 3.91 (β, 0.46–0.86; SE, 0.17–0.24). For the Conners, an increasing score represents worsening executive function, and the negative β value (−0.37 to −0.43) indicated improved performance. No associations emerged in the secondary analysis stratified by exposure status, that is, normotypic-only and PAE-only (all Padj > 0.64; Table 4).
TABLE 4.
Associations between functional SNPs rs3199966 and rs2771040 and behavioral measures in the models of genotype-only and genotype × exposure1
| Cognitive assessment | SNP | Genotype-only, normotypic (N=163) |
Genotype-only, PAE (N=162) |
Genotype × exposure (N=325) |
Effect allele | ||||
|---|---|---|---|---|---|---|---|---|---|
| Padj | β ± SE | Padj | β ± SE | Padj | β ± SE | Effect size | |||
| DAS-II, General Cognitive Abilities2 | rs3199966 | 0.88 | –0.14 ± 0.20 | 0.93 | 0.06 ± 0.16 | 0.0056 | 0.72 ± 0.21 | 3.43 | T(T>G) |
| rs2771040 | 0.88 | –0.07 ± 0.18 | 0.90 | 0.12 ± 0.15 | 0.0058 | 0.70 ± 0.21 | 3.33 | A(A>G) | |
| DAS-II, Nonverbal Reasoning Cluster2 | rs3199966 | 0.88 | –0.12 ± 0.21 | 0.93 | 0.17 ± 0.19 | 0.0074 | 0.74 ± 0.23 | 3.22 | T(T>G) |
| rs2771040 | 0.88 | –0.06 ± 0.19 | 0.93 | 0.18 ± 0.18 | 0.010 | 0.70 ± 0.22 | 3.18 | A(A>G) | |
| DAS-II, Recall of Designs2 | rs3199966 | 0.88 | –0.05 ± 0.22 | 0.93 | –0.04 ± 0.17 | 0.046 | 0.52 ± 0.23 | 2.26 | T(T>G) |
| rs2771040 | 0.88 | –0.07 ± 0.20 | 0.98 | –0.01 ± 0.16 | 0.043 | 0.53 ± 0.22 | 2.41 | A(A>G) | |
| DAS-II, Sequential and Quantitative Reasoning2 | rs3199966 | 0.88 | –0.25 ± 0.19 | 0.87 | 0.24 ± 0.19 | 0.0030 | 0.86 ± 0.22 | 3.91 | T(T>G) |
| rs2771040 | 0.88 | –0.12 ± 0.18 | 0.87 | 0.23 ± 0.18 | 0.0056 | 0.75 ± 0.22 | 3.41 | A(A>G) | |
| Conners, Executive Function Difficulties3 | rs3199966 | 0.88 | 0.25 ± 0.15 | 0.93 | –0.07 ± 0.13 | 0.057 | –0.43 ± 0.17 | –2.53 | T(T>G) |
| rs2771040 | 0.88 | 0.21 ± 0.17 | 0.90 | –0.09 ± 0.12 | 0.035 | –0.37 ± 0.17 | –2.18 | A(A>G) | |
| CVLT, Serial Cluster Ratio3 | rs3199966 | 0.91 | 0.03 ± 0.26 | 0.64 | 0.31 ± 0.15 | 0.080 | 0.46 ± 0.24 | 1.92 | T(T>G) |
| rs2771040 | 0.88 | –0.14 ± 0.24 | 0.64 | 0.32 ± 0.14 | 0.043 | 0.54 ± 0.23 | 2.35 | A(A>G) | |
CVLT, California Verbal Learning Test; DAS-II, Differential Abilities Scales, 2nd edition; Padj, adjusted P value; PAE, prenatal alcohol exposure; SE, standard error; SNP, single nucleotide polymorphism.
Cognitive assessments are standardized to age-norms.
Value is the normalized T-score calculated within the test instrument.
Value is the normalized Z-score calculated within the test instrument.
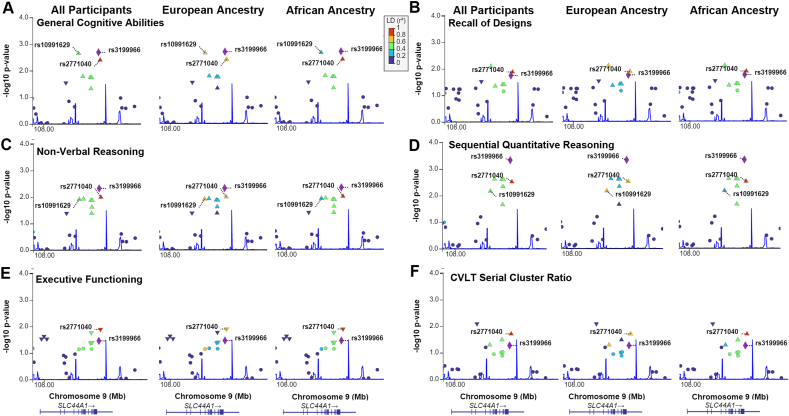
These genotype × exposure associations were explored utilizing linkage maps to assess whether additional SNPs in SLC44A1 that were in linkage with rs3199966 were also associated with these 6 cognitive measures, analyzed both in the overall cohort and separately by genetic ancestry, because ancestry affects LD. For each cognitive measure, rs3199966 was in LD with 3 to 6 additional SNPs in SLC44A1 that demonstrated similar or less significant associations with those cognitive measures (Figure 3). These included rs4538947 (D′ = 0.83; intron 10) and rs11506819 (D′ = 1.00; intron 12), as well as the previously identified rs2771040 (D′ = 1.00; 3′ UTR) and rs10991629 (D′ = 0.78; intron 3); none of these are known to have a functional influence on SLC44A1 or choline status. SNPs that were in LD with rs3199966 were flanked by recombination hotspots located within intron 2 and downstream of exon 16 and its 3′ untranslated region.
FIGURE 3.
Gene-wide linkage plots reveal SNPs in linkage with rs3199966 are also associated with cognitive measures. LD plots were generated for the 6 cognitive measures from Table 4: (A) General Cognitive Abilities, (B) Nonverbal Reasoning, (C) Recall of Designs, (D) Sequential Quantitative Reasoning, (E) Conners Executive Functioning, and (F) CVLT Serial Cluster Ratio. The x-axis is the position on the chromosome in kilobase pairs. The y-axis is −log10(P value). Each SNP is represented by a triangle, with the purple diamond representing rs3199966. The warmth of the color indicates the strength of LD of a SNP with rs3199966 as per the scale bar at the bottom right. Because linkage strength varies with genetic ancestry, LD was calculated based on all participants (ALL), and separately for participants having European (EUR) and African (AFR) ancestries. The blue line indicates recombination hot spots. LD, linkage disequilibrium; SNP, single nucleotide polymorphism.
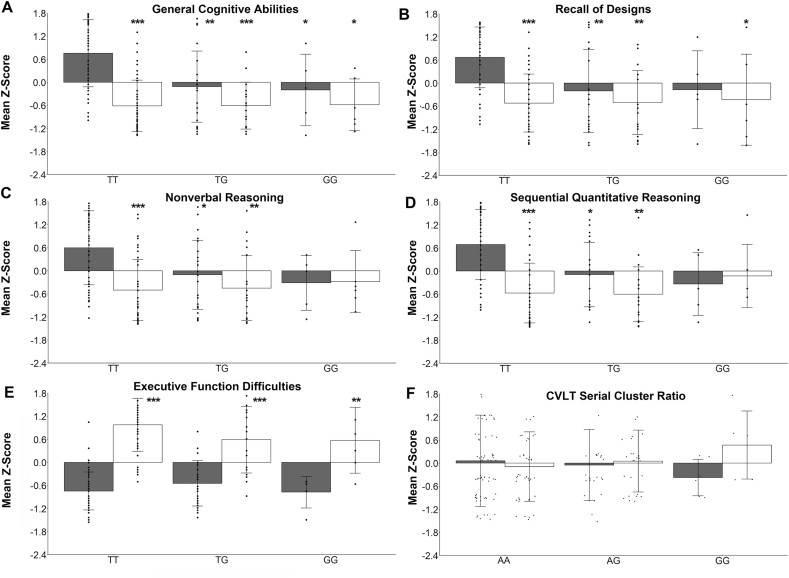
Regression analysis of each participant’s genotype at rs3199966 (TT compared with TG compared with GG) and exposure status revealed that the association of cognitive measures with genotype differed by exposure status. Specifically, in the measures of cognition (Figure 4A), there were effects of exposure (F(1,162) = 67.41; P = 6.57×10−14), genotype (F(2,162) = 8.96; P = 2.03×10−4), and genotype × exposure (F(2,162)=6.42; P = 0.0021), such that rs3199966(TT) controls had better assessment scores than did other controls (compared with GT, P = 1.33×10−4; compared with GG, P = 0.041) or those with PAE (compared with TT or GT, P < 1.0×10−8; compared with GG, P = 0.0033). Similar results were obtained for the assessments of memory (Figure 4B), and nonverbal and quantitative reasoning (Figure 4C, D; Supplementary Table 4). Genotype at rs3199966 did not affect these measures in those with PAE. For the Executive Function Difficulties (Figure 4E), there was an effect of exposure (F(1,169) = 257; P < 2×10−16), exposure × genotype (F(2,169) = 3.64; P = 0.028), and no genotype effect, and those with PAE had greater difficulties. For the California Verbal Learning Test Serial Cluster Ratio, there were no effects of exposure, genotype, or genotype × exposure at rs2771040 (Figure 4F) or rs3199966 (Table 4); although GG appeared to influence the learning style used by controls (grouped by meaning) compared with those with PAE (grouped by order) to recall a list of words, the sample size was too small (n = 6 and 5, respectively) to test for such an effect. Although the influence of rs3199966(T) upon measures of cognition, memory, and reasoning appeared to be dominant, there were too few homozygous GG carriers to distinguish between the additive and dominant models.
FIGURE 4.
Regression analysis by genotype reveals that rs3199966(G) or rs2771040(G) is associated with reduced cognitive performance regardless of PAE history. Analyses are shown for the 6 cognitive measures from Table 4: (A) General Cognitive Abilities, (B) Nonverbal Reasoning, (C) Recall of Designs, (D) Sequential Quantitative Reasoning, (E) Executive Function Difficulties, and (F) CVLT Serial Cluster Ratio (learning approach). Shown are the mean z-scores ± SD for the cohort’s performance in that measure, stratified by exposure (control vs. PAE) and genotype at rs3199966 (TT vs. GT vs. GG; A–E); for Serial Cluster Ratio, results for the association with rs277104 (AA vs. AG vs. GG) are shown because that for rs3199966 was not significant. Dots represent the individual z-scores for that exposure/genotype subgroup with respect to the cohort’s mean performance in that measure. For measures in (A-D), 1 or 2 copies of rs3199966(G) reduced the scores in the controls to levels not different from those with PAE. That is, rs3199966(TT) is associated with better cognitive measures for controls, but not those with PAE. ∗P < 0.05, ∗∗P < 0.001, ∗∗∗P < 10−4 differs from Control rs3199966(TT) or Control rs2771040(AA) by Tukey multiple comparison of means. CVLT, California Verbal Learning Test; PAE, prenatal alcohol exposure.
Discussion
To our knowledge, this is the first demonstration that choline-related SNPs, and specifically, several previously implicated in risk for choline deficiency, may be associated with intellectual ability. We identified SNPs within the ubiquitous choline transporter SLC44A1 that were associated with the variance in cognitive performance in a population of US children. None of these children were part of a choline intervention trial, and they therefore likely experienced conventional dietary choline intakes during their prenatal and postnatal development. Two of these alleles, rs3199966(G) and rs2771040(G), are associated with greater risk of muscle damage under a low-choline diet [22,23], which suggests those carriers are at a greater risk for choline insufficiency. Here we show those alleles are also associated with worsened cognitive performance. Under supraphysiological choline intakes, such carriers have increased conversion of choline to betaine and methionine [24] and have greater improvements in a memory task [25], suggesting that increasing their choline supply may overcome the potentially detrimental effects of this transporter variant. Significantly, only ∼8% of pregnant females consume the adequate intake for choline, and prenatal supplements are recommended but not standard-of-care [9,46,47]. Although this study could not determine when or where these alleles in CTL1 exerted their influence—maternal choline uptake, placental/lactational transfer, or offspring utilization—this association between choline-related SNPs and cognitive performance highlights that current dietary practices during gestational and childhood life stages might not fully address the choline needs of those who carry these risk alleles.
It is not unexpected that these genotype associations would emerge for measures of cognition and executive function including memory and reasoning; persistent deficits in these neurocognitive domains are among the most consequential effects of PAE, and choline supplements have been shown to improve performance in these domains in numerous preclinical studies [6,[13], [14], [15]] and several human intervention trials of PAE [7,[16], [17], [18], [19], [20], [21]]. These findings are also consistent with our prior study of young children with PAE, wherein those who carried rs3199966(G) and rs2771040(G) had the greatest pre/postintervention improvements in imitative memory following a 9-mo daily choline intervention [25]. We interpreted that result to mean that those carriers entered the trial with the poorest choline status and thus benefited most from the extra choline intake. That those same alleles are associated here with poorer cognitive measures is consistent with that interpretation. However, what was unexpected was that this association transcended PAE history and was present across the cohort. Normotypic children who carried rs3199966(GT or GG) had performance in these measures no different from that of children of any rs3199966 genotype who also experienced PAE, and all had worse performance than normotypic children who carried TT. That is, although rs3199966 genotype did not protect those who experienced PAE, it did confer a benefit to otherwise normotypic children who carried TT. These findings are exploratory and require independent validation. However, they are consistent with suggestions that attainment of adequate choline intake should be a priority for pregnancy, lactation, and childhood, especially because rs3199966(G) and rs2771040(G) are present in 11.8% of the US population [22,45].
How might SLC44A1 affect choline needs and brain development? The protein encoded by SLC44A1, CTL1, is ubiquitous and the primary choline transporter for nearly all cells; it controls dietary choline uptake and its entry into tissues and subcellular compartments to affect cellular choline status. It is distinct from the transporter (SLC5A7) that transports choline for acetylcholine synthesis in cholinergic neurons [48]. Choline transported by CTL1 supports diverse biochemical reactions, including the synthesis of phosphatidylcholine, sphingomyelin, and ceramide lipids, and provides 1-carbon groups for purine and pyrimidine synthesis and the methyl modifications of DNA, RNAs, and histones [12,23,24,48,49]. CTL1 loss-of-function impairs myelination [50] and causes early neurodegeneration and cognitive decline [51], emphasizing the transporter’s importance for healthy brain development.
The CTL1 protein sequence (Q8WWI5) is highly conserved, and the most significant variant identified here, rs3199966, is the only exonal coding SNP having an abundance greater than 0.01% [45]; it converts serine 644 to alanine (Ser644Ala) and is predicted to reside in an extracellular domain of this integral membrane protein [48,52]. rs3199966(G) is associated with increased conversion of choline to betaine and methionine under supraphysiological intake [22,23], suggesting this variant might influence choline flux between the Kennedy and methylation pathways. Because alanine 644 is present in other homologs including rat CTL1, this variant is likely functional. Interestingly, it is in complete LD (D′ = 1.00) with rs2771040(G), which resides within the 3′ untranslated region of the plasma membrane isoform; it is absent from the mitochondrial splice variant [48]. This suggests the relevant mechanism might involve cytosolic rather than mitochondrial choline pools, although reduction in the former could limit the latter. Although the impact of these alleles upon CTL1 expression, activity, or stability is unknown, their high LD suggests a strong evolutionary pressure may maintain both.
This study has several limitations. Children with PAE are often adopted, and information was scant in this database about potential confounders such as maternal age and diet, age of adoption, other substance exposures, home environment, adverse childhood experience, socioeconomic status, and other external factors. These could substantially influence diet, choline intake, and these behaviors. However, we note that these children with PAE were brought to the CIFASD clinics to receive help, suggesting a supportive environment. Second, genetic association studies are strengthened by the use of a validation cohort; however, such cohorts for PAE with both genotype and cognitive phenotyping are rare. No information was collected about pre- or postnatal choline intake, choline supplement use, or choline-related biomarkers, rendering the choline status of these participants as unknown. The sample size was relatively modest and there were few homozygous carriers of rs3199966(G), and thus we could not determine if the allele’s influence was additive or dominant. We also could not distinguish if the association was due to maternal versus offspring genotype, which would inform if future interventions should target the prenatal or postnatal periods or both. However, because choline positively affects cognition across the lifespan, it is likely that both periods would benefit from choline [8,9,12].
In summary, we identified alleles within the choline transporter SLC44A1 that were associated with worsened cognitive measures in otherwise normotypic children. These same alleles were also associated with greater vulnerability to choline deficiency when choline intake is limiting [22], suggesting they might reduce the transporter’s functionality. Increasing choline through supplementation may counteract deleterious consequences of this transporter variant [[23], [24], [25]]. These findings highlight that genetic variance in nutrient transporters may be an important influence on tissue-level nutrient status. Because choline insufficiency is common during pregnancy, lactation, and early childhood [[9], [10], [11],46], these data support the importance of choline adequacy for healthy brain development through dietary and/or supplement approaches. The interaction between PAE status and genotype was associated with reduced cognitive performance in normotypic carriers to levels that did not differ from those with PAE under either genotype. This finding does not contradict prior demonstrations of choline’s cognitive benefits for those with PAE; rather, it further supports choline’s beneficial impact for those at risk for or who previously experienced PAE.
Acknowledgments
This work was done in conjunction with the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD), which is funded by grants from the National Institute on Alcohol Abuse and Alcoholism (NIAAA). Additional information about CIFASD can be found at www.cifasd.org.
Author contributions
The authors’ contributions were as follows—SMS: designed research; SMS, MSV, SV, LW, TDW: conducted research; LW, TSA, SNM, ES, JRW, CDC, JAK: provided data; SMS, TDW, LW: analyzed data; SMS: wrote the paper and had primary responsibility for final content; and all authors: read and approved the final manuscript.
Conflict of interest
The authors report no conflicts of interest.
Funding
Supported by National Institutes of Health grants UH2 AA029056 (SMS); U01 AA026103 and U24 AA030169 (LW); U01 AA014834 (SNM, CDC, JAK); and U01 AA0017122 (ES, JRW).
Data availability
Data described in the manuscript and data dictionaries are available by application to CIFASD (cifasd.org).
Declaration of interests
Susan Smith reports financial support was provided by National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajcnut.2023.10.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hoyme H.E., Kalberg W.O., Elliott A.J., Blankenship J., Buckley D., Marais A.S., et al. Updated clinical guidelines for diagnosing fetal alcohol spectrum disorders. Pediatrics. 2016;138(2) doi: 10.1542/peds.2015-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattson S.N., Roesch S.C., Glass L., Deweese B.N., Coles C.D., Kable J.A., et al. Further development of a neurobehavioral profile of fetal alcohol spectrum disorders. Alcohol Clin. Exp. Res. 2013;37(3):517–528. doi: 10.1111/j.1530-0277.2012.01952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gosdin L.K., Deputy N.P., Kim S.Y., Dang E.P., Denny C.H. Alcohol consumption and binge drinking during pregnancy among adults aged 18-49 years - United States, 2018-2020. MMWR Morb. Mortal. Wkly. Rep. 2022;71(1):10–13. doi: 10.15585/mmwr.mm7101a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakhireva L.N., Sharkis J., Shrestha S., Miranda-Sohrabji T.J., Williams S., Miranda R.C. Prevalence of prenatal alcohol exposure in the state of Texas as assessed by phosphatidylethanol in newborn dried blood spot specimens. Alcohol Clin. Exp. Res. 2017;41(5):1004–1011. doi: 10.1111/acer.13375. [DOI] [PubMed] [Google Scholar]
- 5.May P.A., Chambers C.D., Kalberg W.O., Zellner J., Feldman H., Buckley D., et al. Prevalence of fetal alcohol spectrum disorders in 4 US communities. JAMA. 2018;319(5):474–482. doi: 10.1001/jama.2017.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akison L.K., Kuo J., Reid N., Boyd R.N., Moritz K.M. Effect of choline supplementation on neurological, cognitive, and behavioral outcomes in offspring arising from alcohol exposure during development: a quantitative systematic review of clinical and preclinical studies. Alcohol Clin. Exp. Res. 2018;42(9):1591–1611. doi: 10.1111/acer.13817. [DOI] [PubMed] [Google Scholar]
- 7.Ernst A.M., Gimbel B.A., de Water E., Eckerle J.K., Radke J.P., Georgieff M.K., et al. Prenatal and postnatal choline supplementation in Fetal Alcohol Spectrum Disorder. Nutrients. 2022;14(3):688. doi: 10.3390/nu14030688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbyshire E., Obeid R. Choline, neurological development and brain function: a systematic review focusing on the first 1000 days. Nutrients. 2020;12(6):1731. doi: 10.3390/nu12061731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace T.C., Blusztajn J.K., Caudill M.A., Klatt K.C., Zeisel S.H. Choline: the neurocognitive essential nutrient of interest to obstetricians and gynecologists. J. Diet. Suppl. 2020;17(6):733–752. doi: 10.1080/19390211.2019.1639875. [DOI] [PubMed] [Google Scholar]
- 10.Food and Nutrition Board, Institute of Medicine . National Academy Press; Washington, DC: 1998. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. [PubMed] [Google Scholar]
- 11.Wallace T.C., McBurney M., Fulgoni V.L., 3rd Multivitamin/mineral supplement contribution to micronutrient intakes in the United States, 2007-2010. J. Am. Coll. Nutr. 2014;33(2):94–102. doi: 10.1080/07315724.2013.846806. [DOI] [PubMed] [Google Scholar]
- 12.Blusztajn J.K., Slack B.E., Mellott T.J. Neuroprotective actions of dietary choline. Nutrients. 2017;9(8):815. doi: 10.3390/nu9080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan S.H., Williams J.K., Thomas J.D. Choline supplementation attenuates learning deficits associated with neonatal alcohol exposure in the rat: effects of varying the timing of choline administration. Brain Res. 2008;1237:91–100. doi: 10.1016/j.brainres.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider R.D., Thomas J.D. Adolescent choline supplementation attenuates working memory deficits in rats exposed to alcohol during the third trimester equivalent. Alcohol Clin. Exp. Res. 2016;40(4):897–905. doi: 10.1111/acer.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waddell J., Mooney S.M. Choline and working memory training improve cognitive deficits caused by prenatal exposure to ethanol. Nutrients. 2017;9(10):1080. doi: 10.3390/nu9101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kable J.A., Coles C.D., Keen C.L., Uriu-Adams J.Y., Jones K.L., Yevtushok L., et al. The impact of micronutrient supplementation in alcohol-exposed pregnancies on information processing skills in Ukrainian infants. Alcohol. 2015;49(7):647–656. doi: 10.1016/j.alcohol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kable J.A., Coles C.D., Keen C.L., Uriu-Adams J.Y., Jones K.L., Yevtushok L., et al. The impact of micronutrient supplementation in alcohol-exposed pregnancies on reaction time responses of preschoolers in Ukraine. Alcohol. 2022;99:49–58. doi: 10.1016/j.alcohol.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson S.W., Carter R.C., Molteno C.D., Stanton M.E., Herbert J.S., Lindinger N.M., et al. Efficacy of maternal choline supplementation during pregnancy in mitigating adverse effects of prenatal alcohol exposure on growth and cognitive function: a randomized, double-blind, placebo-controlled clinical trial. Alcohol Clin. Exp. Res. 2018;42(7):1327–1341. doi: 10.1111/acer.13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wozniak J.R., Fuglestad A.J., Eckerle J.K., Fink B.A., Hoecker H.L., Boys C.J., et al. Choline supplementation in children with fetal alcohol spectrum disorders: a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 2015;102(5):1113–1125. doi: 10.3945/ajcn.114.099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wozniak J.R., Fink B.A., Fuglestad A.J., Eckerle J.K., Boys C.J., Sandness K.E., et al. Four-year follow-up of a randomized controlled trial of choline for neurodevelopment in fetal alcohol spectrum disorder. J. Neurodev. Disord. 2020;12(1):9. doi: 10.1186/s11689-020-09312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimbel B.A., Anthony M.E., Ernst A.M., Roediger D.J., de Water E., Eckerle J.K., et al. Long-term follow-up of a randomized controlled trial of choline for neurodevelopment in fetal alcohol spectrum disorder: corpus callosum white matter microstructure and neurocognitive outcomes. J. Neurodev. Disord. 2022;14(1):59. doi: 10.1186/s11689-022-09470-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Costa K.A., Corbin K.D., Niculescu M.D., Galanko J.A., Zeisel S.H. Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups. FASEB J. 2014;28(7):2970–2978. doi: 10.1096/fj.14-249557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganz A.B., Klatt K.C., Caudill M.A. Common genetic variants alter metabolism and influence dietary choline requirements. Nutrients. 2017;9(8):E837. doi: 10.3390/nu9080837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganz A.B., Cohen V.V., Swersky C.C., Stover J., Vitiello G.A., Lovesky J., et al. Genetic variation in choline-metabolizing enzymes alters choline metabolism in young women consuming choline intakes meeting current recommendations. Int. J. Mol. Sci. 2017;18(2):E252. doi: 10.3390/ijms18020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith S.M., Virdee M.S., Eckerle J.K., Sandness K.E., Georgieff M.K., Boys C.J., et al. Polymorphisms in SLC44A1 are associated with cognitive improvement in children diagnosed with fetal alcohol spectrum disorder: an exploratory study of oral choline supplementation. Am. J. Clin. Nutr. 2021;114(2):617–627. doi: 10.1093/ajcn/nqab081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou X., Menkari C., Mitsuyama R., Foroud T., Wetherill L., Hammond P., et al. L1 coupling to ankyrin and the spectrin-actin cytoskeleton modulates ethanol inhibition of L1 adhesion and ethanol teratogenesis. FASEB J. 2018;32(3):1364–1374. doi: 10.1096/fj.201700970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattson S.N., Foroud T., Sowell E.R., Jones K.L., Coles C.D., Fagerlund A., et al. Collaborative Initiative on Fetal Alcohol Spectrum Disorders: methodology of clinical projects. Alcohol. 2010;44(7–8):635–641. doi: 10.1016/j.alcohol.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattson S.N., Roesch S.C., Fagerlund A., Autti-Rämö I., Jones K.L., May P.A., et al. Toward a neurobehavioral profile of fetal alcohol spectrum disorders. Alcohol Clin. Exp. Res. 2010;34(9):1640–1650. doi: 10.1111/j.1530-0277.2010.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wechsler D. 4th ed. Psychological Corporation; San Antonio, TX: 2004. Manual for the Wechsler Intelligence Scale for Children. [Google Scholar]
- 30.Elliott C.D. Second Edition. Harcourt Assessment; San Antonio, TX: 2007. Differential Ability Scales –. DAS-II. [Google Scholar]
- 31.Korkman M., Kirk U., Kemp S. 2nd ed. Harcourt Assessment; San Antonio, TX: 2007. NEPSY-II Administration Manual. [Google Scholar]
- 32.Delis D.C., Kaplan E., Kramer J.H. Psychological Corporation; San Antonio, TX: 2001. Manual for the Delis-Kaplan Executive Function System. [Google Scholar]
- 33.Cambridge Cognition Limited . Cambridge Cognition Ltd.; Cambridge, UK: 2006. CANTABeclipse Version 3.0.0: Test Administration Guide. [Google Scholar]
- 34.Gioia G.A., Isquith P.K., Guy S.C., Kenworthy L. Psychological Assessment Resources, Inc; Lutz, FL: 2000. Behavior Rating Inventory of Executive Function: Professional Manual. [Google Scholar]
- 35.Delis D.C., Kramer J.H., Kaplan E., Ober B.A. Psychological Corporation; San Antonio, TX: 1994. Manual for the California Verbal Learning Test – Children’s Version. [Google Scholar]
- 36.Achenbach T.M. University of Vermont Department of Psychiatry; Burlington, VT: 1991. Manual for the Child Behavior Checklist/4-18 and 1991 Profile. [Google Scholar]
- 37.Pelham W.E., Jr., Gnagy E.M., KE Greenslade, Milich R. Teacher ratings of DSM-III-R symptoms for the disruptive behavior disorders. J. Am. Acad. Child Adolesc. Psychiatry. 1992;31(2):210–218. doi: 10.1097/00004583-199203000-00006. [DOI] [PubMed] [Google Scholar]
- 38.McBurnett K., Pfiffner L. University of California; San Francisco: 2005. Sluggish Cognitive Tempo (SCT) Scale.lindap@lppi.ucsf.edu Available at. [Google Scholar]
- 39.Sparrow S.S., Cicchetti D.V., Balla D.A. Survey Forms Manual. 2nd ed. AGS Publishing; Minnesota City, MN: 2005. Vineland Adaptive Behavior Scales. [Google Scholar]
- 40.Conners C.K., Sitarenios G., Parker J.D., Epstein J.N. The revised Conners’ Parent Rating Scale (CPRS-R): factor structure, reliability, and criterion validity. J. Abnorm. Child Psychol. 1998;26(4):257–268. doi: 10.1023/a:1022602400621. [DOI] [PubMed] [Google Scholar]
- 41.Schwantes-An T.H., Darlay R., Mathurin P., Masson S., Liangpunsakul S., Mueller S., et al. Genome-wide association study and meta-analysis on alcohol-associated liver cirrhosis identifies genetic risk factors. Hepatology. 2021;73(5):1920–1931. doi: 10.1002/hep.31535. [DOI] [PubMed] [Google Scholar]
- 42.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fairley S., Lowy-Gallego E., Perry E., Flicek P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020;48(D1):D941–D947. doi: 10.1093/nar/gkz836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., Weir B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Library of Medicine . 2023. dbSNP Short Genetic Variations.https://www.ncbi.nlm.nih.gov/snp/rs3199966 [Internet]. [cited 23 February. Available from: [Google Scholar]
- 46.Korsmo H.W., Jiang X., Caudill M.A. Choline: exploring the growing science on its benefits for moms and babies. Nutrients. 2019;11(8):182. doi: 10.3390/nu110818233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace T.C., Fulgoni V.L., 3rd Assessment of total choline intakes in the United States. J. Am. Coll. Nutr. 2016;35(2):108–112. doi: 10.1080/07315724.2015.1080127. [DOI] [PubMed] [Google Scholar]
- 48.Michel V., Bakovic M. The ubiquitous choline transporter SLC44A1. Cent. Nerv. Syst. Agents Med. Chem. 2012;12(2):70–81. doi: 10.2174/187152412800792733. [DOI] [PubMed] [Google Scholar]
- 49.Traiffort E., O’Regan S., Ruat M. The choline transporter-like family SLC44: properties and roles in human diseases. Mol. Aspects Med. 2013;34(2–3):646–654. doi: 10.1016/j.mam.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 50.Heffernan C., Jain M.R., Liu T., Kim H., Barretto K., Li H., et al. Nectin-like 4 complexes with choline transporter-like protein-1 and regulates Schwann cell choline homeostasis and lipid biogenesis in vitro. J. Biol. Chem. 2017;292(11):4484–4498. doi: 10.1074/jbc.M116.747816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fagerberg C.R., Taylor A., Distelmaier F., Schrøder H.D., Kibæk M., Wieczorek D., et al. Choline transporter-like 1 deficiency causes a new type of childhood-onset neurodegeneration. Brain. 2020;143(1):94–111. doi: 10.1093/brain/awz376. [DOI] [PubMed] [Google Scholar]
- 52.Yuan Z., Tie A., Tarnopolsky M., Bakovic M. Genomic organization, promoter activity, and expression of the human choline transporter-like protein 1. Physiol. Genomics. 2006;26(1):76–90. doi: 10.1152/physiolgenomics.00107.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript and data dictionaries are available by application to CIFASD (cifasd.org).