Abstract
Genomic imprinting results in parent-specific monoallelic expression of a small number of genes in mammals. The identity of imprints is unknown, but much evidence points to a role for DNA methylation. The maternal alleles of the imprinted H19 gene are active and hypomethylated; the paternal alleles are inactive and hypermethylated. Roles for other epigenetic modifications are suggested by allele-specific differences in nuclease hypersensitivity at particular sites. To further analyze the possible epigenetic mechanisms determining monoallelic expression of H19, we have conducted in vivo dimethylsulfate and DNase I footprinting of regions upstream of the coding sequence in parthenogenetic and androgenetic embryonic stem cells. These cells carry only maternally and paternally derived alleles, respectively. We observed the presence of maternal-allele-specific dimethylsulfate and DNase I footprints at the promoter indicative of protein-DNA interactions at a CCAAT box and at binding sites for transcription factors Sp1 and AP-2. Also, at the boundary of a region further upstream for which existent differential methylation has been suggested to constitute an imprint, we observed a number of strand-specific dimethylsulfate reactivity differences specific to the maternal allele, along with an unusual chromatin structure in that both strands of maternally derived DNA were strongly hypersensitive to DNase I cutting over a distance of 100 nucleotides. We therefore reveal the existence of novel parent-specific epigenetic modifications, which in addition to DNA methylation, could constitute imprints or maintain monoallelic expression of H19.
Imprinted genes are expressed from only one parental allele (6). While imprints are laid down in the germ line, for some imprinted genes additional modifications appear to be required after fertilization for the induction of monoallelic expression (21, 42). The epigenetic modifications that constitute imprints and which regulate monoallelic expression are yet to be determined, but there is much evidence that DNA methylation plays a role in at least maintaining monoallelic expression (23). Also, at least one model proposes a role for transcription factor DNA binding. It has been suggested that such factors could be imprint “readers” in that their imprint-dependent binding constitutes an additional epigenetic modification which is necessary for monoallelic expression, or protein factor-DNA binding could constitute the imprint itself (12). However, differential allele-specific transcription factor binding has not been described for any autosomal imprinted gene.
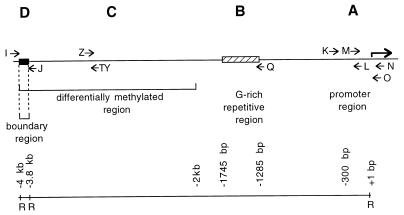
One of the best-studied imprinted genes is H19, which is expressed from only the maternal allele in somatic cells. Two lines of evidence suggest that a 4-kb domain upstream of the coding sequence is important in conferring imprinting on this gene. First, a number of CpGs, between kb −2 and −4 in this domain, are methylated in sperm but not in oocytes. This methylation persists paternally throughout preimplantation development; and thereby could constitute an imprint which leads to monoallelic expression (45). Second, in transgenic mice, a DNA fragment comprised of this domain, the H19 coding sequence, and two downstream enhancers is expressed like the endogenous gene, that is, only when inherited maternally, although this occurs only when multiple copies are integrated (3, 13, 36). Also consistent with the possibility that this domain determines H19 imprinting is that in H19 knockout mice, imprinting of a neomycin selection cassette is not conferred when the coding sequence is deleted along with 10 kb of upstream sequence (22) but is conferred when only the coding sequence is deleted (38). In differentiated tissues, it is of interest that the promoter contains nuclease-hypersensitive sites when maternally inherited (3, 16). Also, additional CpGs in the paternally inherited domain are methylated (3, 44), and this pronounced difference in parent-specific methylation in differentiated tissues is also observed at other sites, including the promoter region (3, 15). While the differential methylation at the promoter is imparted after fertilization and therefore does not constitute the imprint, it is not known when in development the parent-specific differences in nuclease hypersensitivity are conferred, and it remains possible that they are already present in the gametes. In addition, it has been noted that simple sequence repeats are often associated with regions of imprinted genes that exhibit differential DNA methylation, and these repeats may facilitate the inactivation of one of the parental alleles (30). For the mouse H19 gene, one such repeat is located at kb −1.3 to −1.7 from the transcription start site (45).
To shed further light on the possible epigenetic mechanisms determining monoallelic expression of H19, we have used a ligation-mediated PCR (LMPCR) genomic sequencing technique (29, 35) to conduct footprinting and methylation analyses of various regions upstream of the coding sequence, including the putative imprinting region, in parthenogenetic (PG) and androgenetic (AG) embryonic stem (ES) cells. These cells were used because they (i) are diploid and contain only maternally and paternally derived genomes, respectively, (ii) retain imprints as assessed by the developmental potential of chimeras (4, 25–27), (iii) are representative of an early undifferentiated stage of mouse development, being derived from, and similar in developmental potency to, the primitive ectoderm of peri-implantation-stage blastocysts (5, 10), (iv) exhibit parent-specific differential methylation of at least some sites upstream of the H19 coding region that are differentially methylated in differentiated tissues, while retaining a relatively very low level of expression of H19 as exists in the primitive ectoderm (1, 41, 42), and (v) can be obtained in sufficient quantity for conducting LMPCR.
The analyses reveal significant differences in binding of transcription factors, chromatin structure, and methylation between the maternally and paternally derived upstream regions of the H19 gene that have not been previously described and implicate these epigenetic modifications in imprinting and regulation of monoallelic expression of H19.
MATERIALS AND METHODS
ES cells.
The type, name, and passage number at DNA collection of the ES cell lines used were as follows: PG (two maternal genomes), LG.1, passage 13; wild-type, WT (one maternal and one paternal genome), W9.5, passage 11; and AG (two paternal genomes), LB.4, passage 10. The derivation and culture conditions for these cell lines have been previously described (26, 41). For each type of cell line, the methylation and expression of imprinted genes were found to be consistent between independently derived cell lines (41).
Treatment of ES cells with reagents for in vivo footprinting.
In preparing ES cells for treatment, STO feeder cells were removed from trypsinized cell suspensions by differential adherence to tissue culture plates. This results in an ES purity of at least 97% by cell number (41). The following descriptions are for a washed ES cell pellet of ∼15 × 106 cells obtained from one semiconfluent 10-cm-diameter dish. Treatments and isolation of DNA were performed as previously described (33), with the following modifications.
(i) DMS treatment.
The ES cell pellet was resuspended in 6 ml of serum-free medium containing 0.2% dimethylsulfate (DMS) and was incubated for 5 min at room temperature (RT). The action of DMS was stopped by adding ice-cold Dulbecco’s phosphate-buffered saline with calcium and magnesium (DPBS) to a volume of 50 ml, followed by centrifugation at 250 × g for 3 min to pellet the cells. After two washes with 20 ml of ice-cold DPBS, the pellet was dissolved in 10 ml of buffer A containing 1% Nonidet P-40 (33), and then the DNA was isolated. As a control, genomic DNA was isolated from untreated ES cells (33), and then 80 to 100 μg was treated with DMS (28).
(ii) DNase I treatment.
The ES cell pellet was resuspended in 5 ml of ice-cold solution I containing lysolecithin (0.25 mg/ml) to permeabilize cells (32). The cells were immediately pelleted, then resuspended in solution II containing DNase I (1 μg/ml; Boehringer Mannheim catalog no. 104132), and incubated for 1 to 6 min at RT to obtain an optimal nicking frequency of one nick every 200 to 800 nucleotides, as assessed by alkaline gel electrophoresis after DNA purification and before LMPCR. Cells were pelleted and resuspended in 2 ml of buffer C containing proteinase K (600 μg/ml), then 2 ml of buffer B was added, and the sample was incubated at 37°C overnight (33). As a control, 40 μg of genomic DNA isolated from untreated ES cells as described above was mixed with 10 μg of DNase I per ml in a 200-μl reaction volume and incubated at RT for 5 min.
(iii) KMnO4 treatment.
The ES cell pellet was resuspended in 2 ml of PBS containing 20 mM KMnO4. After 2 min at RT, the reaction was stopped by addition of 50 ml of ice-cold DPBS. The cells were pelleted and resuspended in 10 ml of buffer B containing 1.0 M β-mercaptoethanol, then 10 ml of buffer C containing proteinase K (600 μg/ml) was added, and the sample was incubated at 37°C for 3 h (33). As an in vitro control, KMnO4 treatment of 20 μg of DNA was performed in the presence of fresh 20 mM KMnO4 for 2 min at RT in a reaction volume of 100 μl. The reaction was stopped by adding 2-mercaptoethanol to a concentration of 1 M. After ethanol precipitation, 1 M piperidine was used to cleave the DNA at sites of KMnO4 modifications, and the samples were used in LMPCR.
Other treatments.
For methylation analysis at C residues, genomic DNA was isolated from ES cells as described above, and 80 to 100 μg was treated with hydrazine in the presence of NaCl (28). As a control, a reaction was performed with 0.5 μg of H19 lambda clone DNA mixed with 80 μg of tRNA. The amount of lambda DNA was adjusted before LMPCR to match the copy number of H19 sequences in genomic DNA. For analysis of C+T and G+A modifications, 80 to 100 μg of DNA was treated with hydrazine and formic acid, respectively (28).
LMPCR.
LMPCRs were performed as previously described (33), with the following modifications. The Sequenase reaction with the first primer was carried out at 49°C for 15 min. To increase reaction specificity, DNase I-treated samples were isolated on paramagnetic beads by using biotinylated primers before LMPCR (43). The PCR in which the second primer was included was carried out at 95, 65, and 74°C for 23 cycles. Following a booster step with an additional aliquot of Taq polymerase, the reaction was extracted with phenol-chloroform, and then the PCR product was precipitated. The product was run on a denaturing 8% polyacrylamide gel, then electroblotted to a nylon membrane, and hybridized at 65°C overnight with a runoff probe made on an isolated PCR fragment by using the third primer.
The primers (the first primer in each series was biotinylated) and their sequences (in parentheses; 5′-3′) were K1 (GGGGGTTAGCCACTCGTAG), K2 (CCACTCGTAGGCTGTTCATACTCCG), K3 (GTATGAGACCCATGCCCTCAAATTCC), L1 (CCACCTCCACCCTGTATCG), L2 (CCCTGTATCGTTCCAGCGCACGTT), L3 (GCCTCACCCCACACCCGCA), M1 (TCTAAGGGATTCCAAAGTGG), M2 (TGTGGTGAGGCTGTCTTTGGAGAATT), M3 (TTTGGAGAATTTCAGGACGGGTGCG), N1 (CCAGACTCCAGATGCCGA), N2 (GGTGCTCCTCGGACCCCACG), N3 (ACCCCACGACTCTCCTCCAGCTCTC), I1 (TGCCTACAGTTCCCGAATC), I2 (CGAATCACCACAAGGAAAGAAAAAGG), I3 (GAAAGAAAAAGGTTGGTGAGAAAAATAGAG), J1 (GTGACCCCCCTGAGGTACT), J2 (TGAGGTACTGAACTTGGGTGACCCAC), J3 (GTGACCCACAGCATTGCCATTTG), O1 (GCTCTCCCATCTTCCCCA), O2 (TCCCATCTTCCCCGGTTTCCCCG), O3 (CCCCCCACCCCCCTCCCACA), Q1 (GTGGTGGCAGTTGGTCTCT), Q2 (TGTCTCCCATCACCCCCCACAT), Q3 (CCCACATCACCCTTGCTATACTCCC), TY1 (GCTGACACCCAAGGCTTG), TY2 (GACACCCAAGGCTTGATGTAGGATTC), TY3 (CTATCCGAAGCTGGCAGCTGAGC), Z1 (CGGCTCAGGGCTGCAA), Z2 (GCTGCAAACAATTCTGAAACTGCATTC), and Z3 (GCATTCTCTCTCAATGGGGCTCAGC).
RESULTS
Chromatin and methylation analysis.
Intact ES cells were treated with two agents that are sensitive to chromatin structure and protein-DNA interactions. DNase I nicks DNA inside permeabilized cells with a specificity similar to that in isolated DNA, but this nicking is inhibited at positions where protein factors are bound (32). DMS reacts with protein-bound DNA with either a lower or higher rate of N-7 methylation of G residues (14). After DNA isolation, piperidine was used to cleave DNA at the modified G bases. LMPCR was then used to amplify gene-specific sequences and visualize, at single-nucleotide resolution, cuts made by DNase I or derived from DMS modification (29, 35). DNase I footprints usually appear as areas of protection on sequencing gels, while DMS footprints are stronger or weaker single-G bands compared to isolated DNA controls. Methylation of C bases in the whole population of chromosomes was also investigated at single-nucleotide resolution. DNA was isolated from PG ES cells and AG ES cells, modified with hydrazine in the presence of sodium chloride, and cleaved with piperidine. Methylation inhibits modification; therefore, bands corresponding to methylated cytosines are missing from the sequencing gels while unmethylated cytosines are present.
Analysis of the H19 promoter.
An approximately 350-bp-long region containing the H19 promoter (segment A [Fig. 1]) was analyzed by using five primer sets, K, L, M, N, and O. The proximal promoter fragment contains several CpG dinucleotides. Compared to unmethylated cloned DNA, AG ES cells show heavy methylation at all of these sites, while PG ES cells show bands at these positions, suggesting no or partial methylation (Fig. 2 and 3). Using Southern blot analysis (41), we obtained similar data for one of these sites with the methylation-sensitive restriction enzyme SmaI. The data obtained by the genomic sequencing method indicate that this differential methylation extends to all CpG sites in the promoter region that we examined.
FIG. 1.
Map of the 4-kb region upstream of the H19 gene. The A, B, C, and D segments have been analyzed by in vivo footprinting. Approximate locations of LMPCR primer sets K, L, M, N, O, Q, TY, Z, I, and J are indicated by horizontal arrows. The scale underneath shows the distances from the transcription start site. R, EcoRI sites.
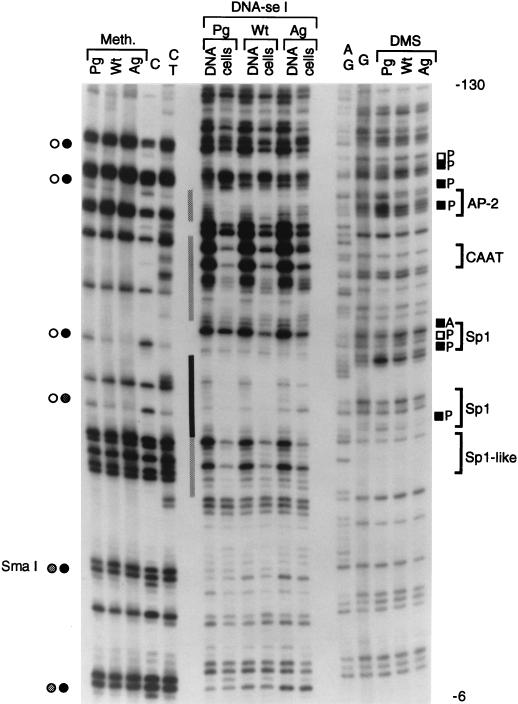
FIG. 2.
In vivo footprinting of the proximal H19 promoter in PG ES cells, WT ES cells, and AG ES cells. Methylation (Meth.), genomic DNase I footprinting (DNA-se I), and in vivo DMS footprinting (DMS) analyses were conducted. The N primer set was used for LMPCR analysis of the upper strand. Pg, PG ES cells; Wt, WT ES cells; Ag, AG ES cells. Open and filled circles indicate unmethylated and fully methylated CpGs, respectively, representing PG ES cells on the left and AG ES cells on the right; grey bars indicate partial DNase I footprints in PG ES cells; black bars indicate complete protection; black and open squares indicate increases and decreases, respectively, in DMS reactivity; A, AG ES cells; P, PG ES cells; brackets on the right side show consensus binding sites for transcription factors.
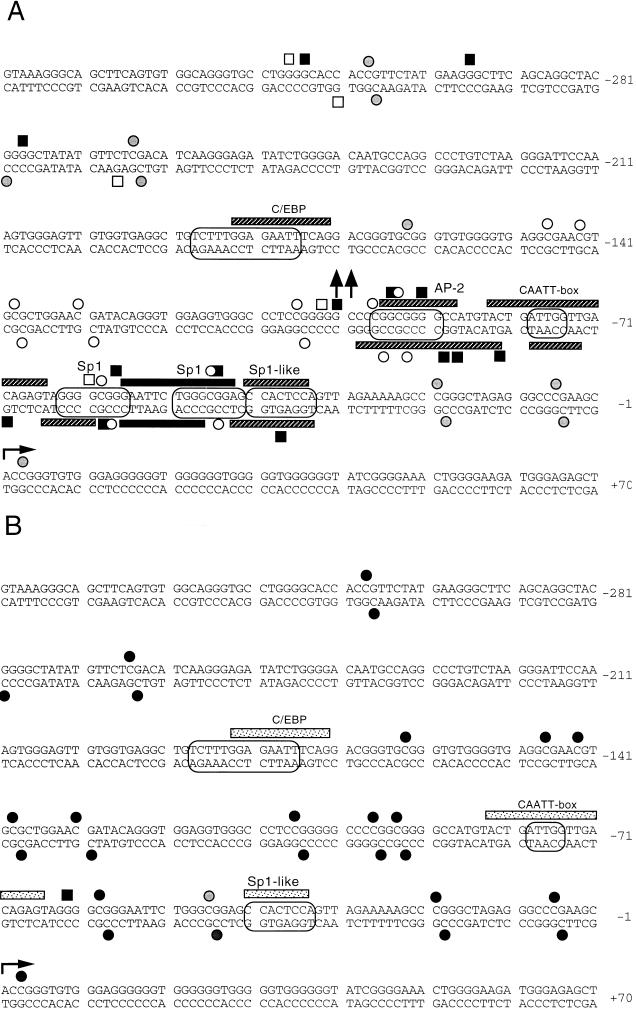
FIG. 3.
Summary of the promoter footprinting data. Primary data were taken from experiments with the K, L, M, N, and O primer sets. (A) Maternal chromosome, PG ES cells; (B) paternal chromosome, AG ES cells. Circles around the DNA sequences indicate consensus sequences for transcription factors. Striped bars indicate partial DNase I footprints; black bars indicate complete protection; arrows pointing up or down indicate increased DNase I sensitivity of the in vivo-treated versus the in vitro-treated samples; filled or open squares indicate increases or decreases, respectively, in DMS reactivity; open or closed circles stand for hypomethylated or hypermethylated CpGs, respectively; grey circles stand for partially methylated sites.
DNase I footprinting revealed binding of protein factors in the H19 promoter at three Sp1 sites, one CCAAT box sequence, and one AP-2 site in PG ES cells (Fig. 2 and 3). The second Sp1 site (nucleotides [nt] −49 to −44) exhibited complete protection by the factor, while all other sites were partially protected. DMS footprinting results supported occupation of two Sp1 sites and the AP-2 consensus site by protein factors. In AG ES cells, there was a weak protection near the first Sp1-like site and the CCAAT box. LMPCR performed on the lower strand of the proximal promoter region showed similar methylation differences and confirmed the footprints (data not shown and Fig. 3).
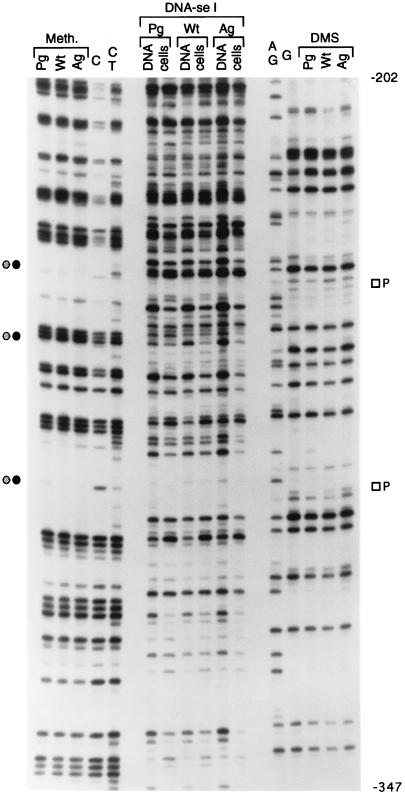
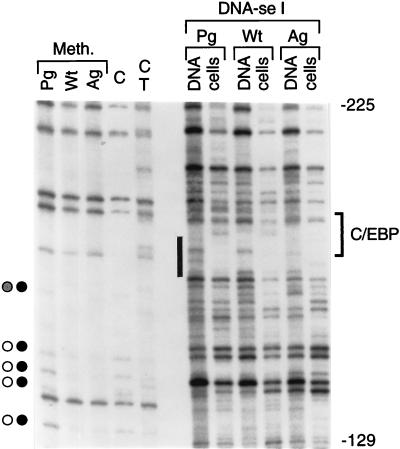
The more distal promoter region had a similar hypomethylation of CpGs in PG ES cells and methylation in AG ES cells (Fig. 3 to 5). A C/EBP consensus site near bp −180 was partially protected from DNase I cleavage (Fig. 3 and 4). Binding of two additional protein factors in PG ES cells is suggested by in the vivo DMS footprinting data (Fig. 3A and 5).
FIG. 5.
In vivo footprinting of the distal H19 promoter in PG ES cells and AG ES cells. The lower strand of the region between nt −202 and −347 was analyzed. The K primer set was used in the LMPCR. Open squares indicate protection from DMS modification in PG ES cells. For other details, see the legend to Fig. 2.
FIG. 4.
In vivo footprinting of the H19 promoter in PG ES cells and AG ES cells. The upper strand of the region between nt −129 and −225 was analyzed. The O primer set was used in LMPCR. The bracket on the right indicates the C/EBP recognition sequence, and the black rectangle indicates the protection at this site by a protein factor in PG, AG, and WT ES cells. For other details, see the legend to Fig. 2.
The WT ES cells showed an intermediate protection/reactivity in the methylation analysis and the DNase I or DMS footprinting compared to PG ES cells and AG ES cells. This is the expected result in cells containing both maternal and paternal chromosomes (Fig. 2 and 5).
Analysis of the G-rich repetitive region.
Analysis of the upper strand of segment B (Fig. 1), which carries the short tandem repeat sequence [(G)GGGGTATA]32 (45), is shown in Fig. 6A. This area does not contain CpG dinucleotides, which is reflected in the identical hydrazine modification pattern of C’s between the cell lines. A very similar modification pattern was obtained with DMS. DNase I generated a pattern in vivo which was different from the in vitro cleavage pattern but identical between all three cell lines. The T and A nucleotides at the 5′ ends of the repeats usually have decreased DNase I cleavage intensities in vivo compared to the rest of the sequence (Fig. 6B).
FIG. 6.
Footprinting of the G-rich repetitive region located at kb −1.3 to −1.4. (A) Autoradiogram. The Q primer set was used for LMPCR. Filled bars on the right show perfect repeats; open bars show partially homologous repeats. For other details, see legend to Fig. 2. (B) Summary of the analysis. Arrows pointing up or down indicate increased or decreased DNase I sensitivity of the in vivo-treated versus the in vitro-treated samples. No major differences in DNase I patterns were observed between PG, WT, and AG ES cells.
Analysis of the far upstream paternally methylated region.
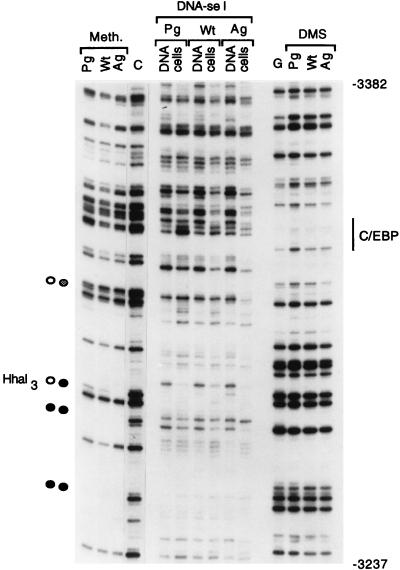
We analyzed a portion of the ∼2-kb upstream paternally methylated region with primer sets TY and Z (segment C [Fig. 1]). One of the CpG’s in this area is HhaI site 3 (44), which is methylated in AG ES cells and unmethylated in PG ES cells (Fig. 7). Some CpG sites showed differential methylation; others were methylated on both alleles. The in vivo DNase I patterns were different from the in vitro patterns but identical between cell lines with the exception of a hypersensitive site mapping to a C/EBP recognition sequence. The same recognition sequence had increased DMS modification in PG ES cells. C/EBP is a liver- and adipocyte-specific transcription factor (11, 17). Differences in DMS and DNase I reactivity at two C/EBP consensus sites (Fig. 3A, 4, and 7) may indicate that these sites are somehow marked for liver-specific expression of H19 (15). We observed similar methylation differences on the lower strand with primer set Z but found no differences in DMS or DNase I patterns between cell lines (data not shown).
FIG. 7.
Footprinting of the differentially methylated region. The upper strand is shown. The TY primer set was used. The C/EBP site is indicated on the right. For other details, see the legend to Fig. 2.
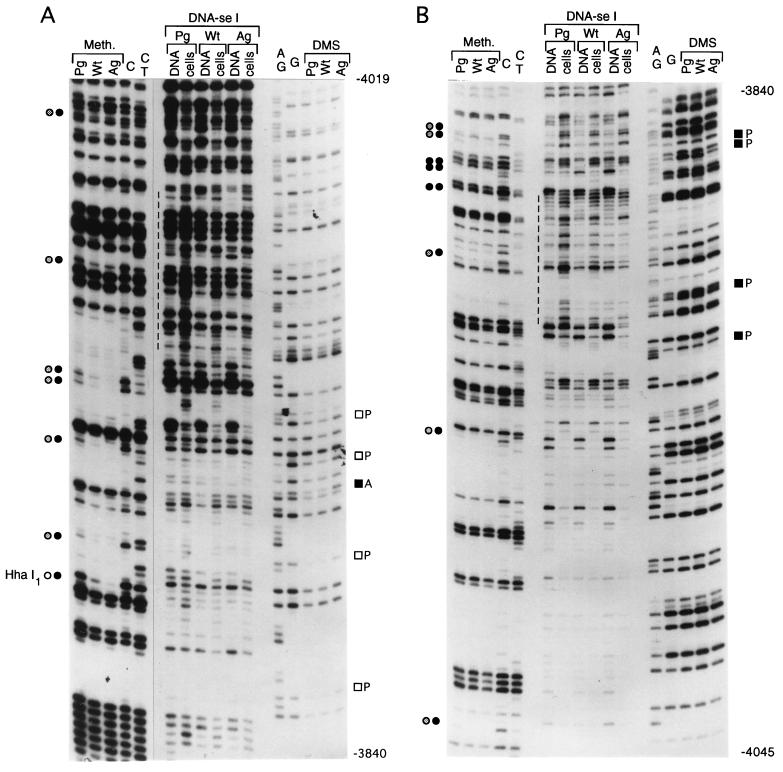
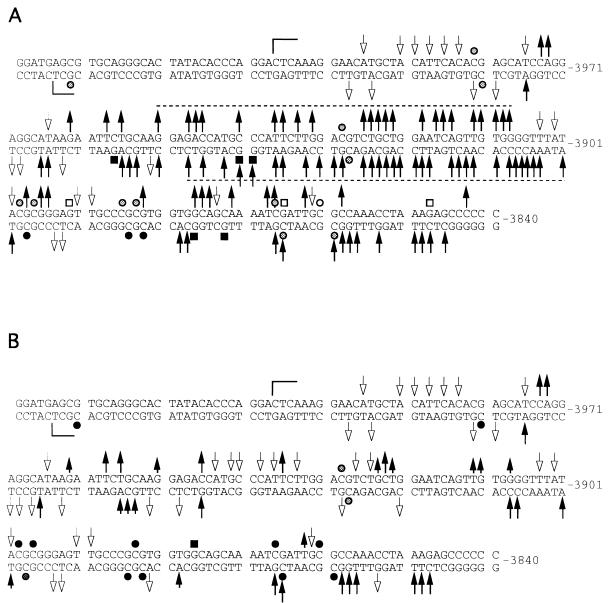
Analysis of the boundary of the methylated region.
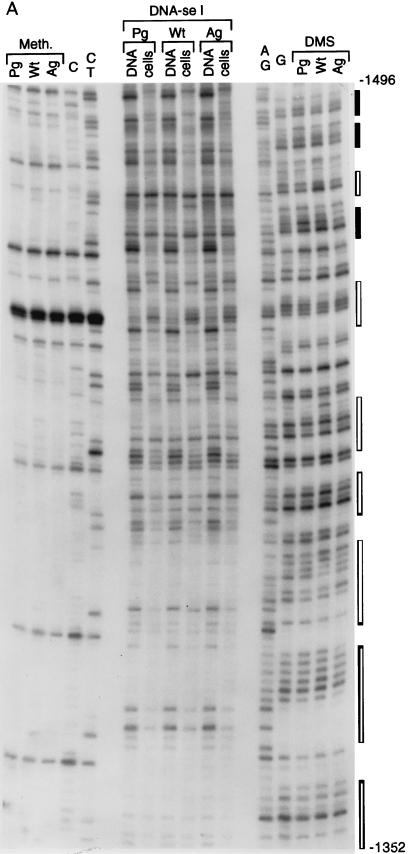
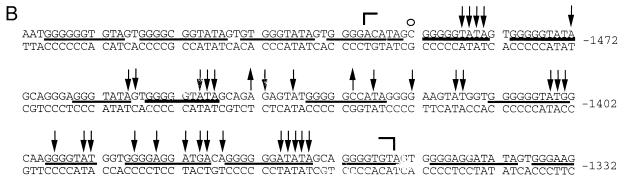
We analyzed a 200-bp sequence at the 5′ boundary of the 2-kb upstream paternally methylated region with primer sets I and J (segment D [Fig. 1]). Analysis of the upper strand is shown in Fig. 8A. At a CpG site designated HhaI-1 (45), there is complete methylation in AG ES cells, while a very strong band present in PG ES cells indicates that the site is fully unmethylated in these cells. All other CpG sites are completely methylated in AG ES cells and either unmethylated or only partially methylated in PG ES cells. The DNase I and DMS modification patterns for this region differ between PG ES cells and AG ES cells. The maternal chromosomes of the H19 gene have a series of DNase I-hypersensitive sites (from nt −3962 to −3843) that are much weaker or missing from the paternal chromosomes. Several bands have reduced intensity in the DMS ladder in PG ES cells, which could indicate binding of protein factors at these sites. On the opposite strand (Fig. 8B), an area of increased DNase I reactivity is seen between nt −3956 and −3848. The DMS modification patterns on the lower strand were similar between naked DNA and AG ES cells treated with DMS in vivo (Fig. 8B). However, some G’s on this strand were hyperreactive in PG ES cells compared to naked DNA or AG ES cells, which suggests protein binding at several positions in the area. However, DNase I hypersensitivity at the same sequences contradicts this possibility and instead suggests an altered DNA structure at these sites. WT DNA always exhibited intermediate intensities. A summary of DNase I and DMS footprinting experiments of the area between nt −4034 and −3840 is shown in Fig. 9. The DNase I hyperreactivity spanning 40 nt (between nt −3948 and −3908) on both strands suggested an unusual chromatin or DNA structure. To determine if this structure would consist of melted DNA, we analyzed the same region with the oxidative agent KMNO4. KMNO4 oxidizes pyrimidine bases, mostly T’s, preferentially in single-stranded or melted DNA (39). Reactivities of in vivo- and in vitro-treated samples were equal for all of the three cell lines (data not shown); therefore, we can exclude stable melting of these sequences.
FIG. 8.
Footprinting of the far upstream boundary of the differentially methylated sequences. (A) Upper strand, the J primer set used for LMPCR; (B) lower strand, the I primer set used. Filled and open squares indicate increases and decreases, respectively, in DMS reactivity in PG ES cells; dashed lines indicate the regions with the highest DNase I sensitivity. For other details, see the legend to Fig. 2.
FIG. 9.
Summary of the analysis of the far upstream sequences. (A) Maternal chromosome, PG ES cells; (B) paternal chromosome, AG ES cells. Arrows pointing up or down indicate increased or decreased DNase I sensitivity of the in vivo-treated versus the in vitro-treated samples; shorter arrows indicate lower DNase I sensitivity in AG cells compared to PG cells; filled or open squares indicate increases or decreases, respectively, in DMS reactivity; open or closed circles indicate hypomethylated or hypermethylated CpGs, respectively; grey circles stand for partially methylated sites; brackets indicate the sequences analyzed.
DISCUSSION
Previous observations of parent-specific methylation and experiments with transgenic mice have strongly suggested an important role for regions upstream of the H19 coding sequence in conferring monoallelic expression on this gene. Here we have examined some specific upstream sequences to determine if there exist additional parent-specific differences in chromatin structure that might also be involved in H19 imprinting.
Previous studies of H19 methylation were performed with methylation-sensitive restriction enzymes, which allow detection of the methylation status at a limited number of CpGs (3, 15, 16, 41). In addition, sodium bisulfite genomic sequencing revealed the methylation status at single-nucleotide resolution on individual maternal and paternal chromosomes (31, 44). Using a different sequencing method, we extend these previous analyses, showing the mean methylation level of the whole population of chromosomes at single-nucleotide resolution. This analysis has essentially confirmed the previous findings of hypo- and hypermethylation of maternally and paternally derived sequences, respectively, upstream of H19. It is significant that much of the paternal chromosome-specific methylation in AG ES cells must be laid down quickly during their derivation and early passage, as it has been shown that the CpGs in the H19 promoter are not paternally methylated in blastocysts (9, 44). This rapid de novo methylation occurs before the high-expression phase of H19, while its expression level is very low in all three types of ES cell. Thus, it is conceivable that this methylation is a response to the intrinsic state of paternal chromatin structure and may be prevented by transcription factor binding to the maternal chromosome.
Aside from differential methylation, we now describe the presence of a number of other differential epigenetic modifications at various regions upstream of the H19 coding sequence. The in vivo footprints were observed almost exclusively in PG ES cells or on maternally derived alleles of H19. When footprints were observed in AG ES cells, they were always weaker than the corresponding PG footprints. Partial footprints in AG ES cells at the CCAAT box and one Sp1 site perhaps may be explained by a lack of fully methylated CpGs near these sequences (Fig. 3B) and/or by the methylation independence of Sp1 binding (18).
The G-rich repetitive region at kb −1.3 (45) did not appear different in respect to the two parental chromosomes by DMS and DNase I footprinting. While these observations suggest that this repeat may not play a role in maintaining monoallelic expression of the H19 gene, they do not preclude the possibility that it has some role in initiation. Repeat elements are typically associated with imprinted genes (30), and it has been suggested that possible differences in their chromatin structure in the female and male germ lines may lead to different levels of methylation, which in turn leads to differential allelic expression (7).
In contrast to the negative observations for the repeat element, at the boundary of a far upstream region thought to harbor H19 imprints, we observed a localized region of DNase I hypersensitivity specific to maternally derived alleles and present on both strands of the DNA. This is most likely indicative of the presence of an unusual chromatin structure at this site. While further work is required to determine the precise nature of this structure, its presence raises the question of whether it might function after fertilization to inhibit methylation nearby, for example, in the putative imprinting region, and ultimately at more distant sites such as the promoter.
We provide the first evidence that transcription factors preferentially interact with the maternal allele of H19. The footprints within an approximately 100-bp region at the H19 promoter coincided with five transcription factor binding consensus sequences: two Sp1, one Sp1-like, one AP-2, and one CAATT box. These data suggest that binding at these sites is a requirement for maternal H19 expression and are consistent with the results of previous H19 promoter mapping studies in which a 127-bp fragment containing all five binding elements resulted in the highest level of reporter gene expression in hepatoma cells (46). Of interest are the similarities between these findings for the autosomal H19 gene and those for X-linked genes which exhibit monoallelic but random expression in XX somatic cells. The active allele of the human phosphoglycerate kinase-1 gene promoter is unmethylated and exhibits protein binding at a CCAAT sequence, an NF-1-like binding sequence, and at two GC boxes, each of which contains two Sp1 binding sequences. On the other hand, the inactive allele shows the opposite configuration: it is methylated and exhibits no protein binding at any of these sites (32, 34). Similar results were obtained for the human X-linked hypoxanthine guanine phosphoribosyltransferase gene (19) and the mouse inactive X-specific transcript gene (20).
While differences in parent-specific methylation and the additional differences in chromatin structure that we have described in this study might be involved in maintaining parent-specific expression of H19, challenges remaining are to determine if one of these epigenetic modifications determines the others and to establish their relationship to the primary determinant of monoallelic expression, the imprint, as imparted in the germ line. For example, it is possible that the unusual chromatin structure in the far upstream region of the H19 maternal allele is specific to oogenesis and somehow inhibits any potential for methylation in its vicinity before and after fertilization. Its absence in spermatogenesis and from the paternal allele after fertilization could then allow a gradual methylation at these sites by default.
The maternal allele-specific binding of transcription factors we have observed in upstream region of the H19 gene is consistent with the suggestion that protein factor-DNA binding could serve as an imprint (12) and could result from maternal germ line-specific expression of the relevant factors. Models such as this need not be considered unlikely on the basis that protein factor-DNA binding might be unstable throughout cell division (12). Female germ cells are arrested in meiosis I in the early fetus until just prior to ovulation in the adult, while male germ cells are arrested in mitosis in the early fetus until the early neonatal stage. This arrest could render differential transcription factor binding very stable in the germ line over long periods of developmental time and therefore able to inhibit the laying down of secondary or downstream epigenetic modifications which might ultimately be responsible for maintaining or stabilizing monoallelic expression in somatic cells. On this point, it has been suggested that transcription factors bound to promoter elements could outcompete nucleosomes and DNA methyltransferase and thereby inhibit methylation (2, 34). Indeed, Sp1 binding functions in a chromatin-dependent manner to augment human α-globin promoter activity (37) to protect the CpG island of the mouse adenine phosphoribosyltransferase gene and its human homolog in transgenes from methylation and to cause demethylation of artificially methylated sequences (8, 24). Also, it is worth pointing out that the differential methylation in gametes that is suggested to constitute imprints (40) would almost certainly be laid down during the time that at least the female germ line is arrested in cell division.
Elucidation of these various possibilities may become possible by extending the analyses we have performed here for PG and AG ES cells to the female and male germ lines of mice. This is a challenging prospect given the small amounts of material that can be recovered in respect to these cell lineages.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant RO1GM48103-04A2 from the National Institutes of Health.
REFERENCES
- 1.Allen N D, Barton S C, Hilton K, Norris M L, Surani M A. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 2.Antequera F, Bird A. CpG islands. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology and functional significance. Basel, Switzerland: Birkhäuser Verlag; 1993. pp. 169–185. [Google Scholar]
- 3.Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 4.Barton S C, Ferguson-Smith A C, Fundele R, Surani M A. Influence of paternally imprinted genes on development. Development. 1991;113:679–688. doi: 10.1242/dev.113.2.679. [DOI] [PubMed] [Google Scholar]
- 5.Beddington R S P, Robertson E J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 6.Beechey C V, Cattanach B M. Genetic and physical imprinting map of the mouse. Mouse Genome. 1997;95:100–105. [Google Scholar]
- 7.Bestor T H, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 8.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 9.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brook F A, Gardner R L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu A D, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 12.Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4:265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 13.Elson D A, Bartolomei M S. A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol Cell Biol. 1997;17:309–317. doi: 10.1128/mcb.17.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ephrussi A, Church G M, Tonegawa S, Gilbert W. B lineage specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 15.Feil R, Walter J, Allen N D, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 17.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 18.Höller M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 19.Hornstra I K, Yang T P. Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5′ region: implications for X chromosome inactivation. Mol Cell Biol. 1992;12:5345–5354. doi: 10.1128/mcb.12.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komura J, Sheardown S A, Brockdorff N, Singer-Sam J, Riggs A D. In vivo ultraviolet footprinting and dimethyl sulfate footprinting of the 5′ region of the expressed and silent Xist alleles. J Biol Chem. 1997;272:10975–10980. doi: 10.1074/jbc.272.16.10975. [DOI] [PubMed] [Google Scholar]
- 21.Latham K E, Doherty A S, Scott C D, Schultz R M. Igf2r and Igf2 gene expression in androgenetic, gynogenetic, and parthenogenetic preimplantation mouse embryos: absence of regulation by genomic imprinting. Genes Dev. 1994;8:290–299. doi: 10.1101/gad.8.3.290. [DOI] [PubMed] [Google Scholar]
- 22.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 23.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 24.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 25.Mann J R. Properties of androgenetic and parthenogenetic mouse embryonic stem cell lines; are genetic imprints conserved? Semin Dev Biol. 1992;3:77–85. [Google Scholar]
- 26.Mann J R, Gadi I, Harbison M L, Abbondanzo S J, Stewart C L. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: implications for genetic imprinting. Cell. 1990;62:251–260. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- 27.Mann J R, Stewart C L. Development to term of mouse androgenetic aggregation chimeras. Development. 1991;113:1325–1333. doi: 10.1242/dev.113.4.1325. [DOI] [PubMed] [Google Scholar]
- 28.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 29.Mueller P R, Wold B. In vivo footprinting of a muscle-specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 30.Neumann B, Kubicka P, Barlow D P. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- 31.Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer G P, Riggs A D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNaseI and ligation-mediated PCR. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer G P, Singer-Sam J, Riggs A D. Analysis of methylation and chromatin structure. Methods Enzymol. 1993;225:567–583. doi: 10.1016/0076-6879(93)25037-3. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeifer G P, Steigerwald S D, Mueller P R, Wold B, Riggs A D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- 36.Pfeifer K, Leighton P A, Tilghman S M. The structural H19 gene is required for transgene imprinting. Proc Natl Acad Sci USA. 1996;93:13876–13883. doi: 10.1073/pnas.93.24.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pondel M D, Murphy S, Pearson L, Craddock C, Proudfoot N J. Sp1 functions in a chromatin-dependent manner to augment human α-globin promoter activity. Proc Natl Acad Sci USA. 1995;92:7237–7241. doi: 10.1073/pnas.92.16.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripoche M-A, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 39.Rubin C M, Schmid C W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980;8:4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shemer R, Razin A. Establishment of imprinted methylation patterns during development. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 215–229. [Google Scholar]
- 41.Szabó P, Mann J R. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development. 1994;120:1651–1660. doi: 10.1242/dev.120.6.1651. [DOI] [PubMed] [Google Scholar]
- 42.Szabó P E, Mann J R. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 43.Törmänen V T, Swiderski P M, Kaplan B E, Pfeifer G P, Riggs A D. Extension product capture improves genomic sequencing and DNase I footprinting by ligation-mediated PCR. Nucleic Acids Res. 1992;20:5487–5488. doi: 10.1093/nar/20.20.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 46.Yoo-Warren H, Pachnis V, Ingram R S, Tilghman S M. Two regulatory domains flank the mouse H19 gene. Mol Cell Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]