Abstract
Coronavirus disease (COVID-2019) is emerging in Wuhan, China in 2019. It has spread throughout the world since the year 2020. Millions of people were affected and caused death to them till now. To avoid the spreading of COVID-2019, various precautions and restrictions have been taken by all nations. At the same time, infected persons are needed to identify and isolate, and medical treatment should be provided to them. Due to a deficient number of Reverse Transcription Polymerase Chain Reaction (RT-PCR) tests, a Chest X-ray image is becoming an effective technique for diagnosing COVID-19. In this work, the Hybrid Deep Learning CNN model is proposed for the diagnosis COVID-19 using chest X-rays. The proposed model consists of a heading model and a base model. The base model utilizes two pre-trained deep learning structures such as VGG16 and VGG19. The feature dimensions from these pre-trained models are reduced by incorporating different pooling layers, such as max and average. In the heading part, dense layers of size three with different activation functions are also added. A dropout layer is supplemented to avoid overfitting. The experimental analyses are conducted to identify the efficacy of the proposed hybrid deep learning with existing transfer learning architectures such as VGG16, VGG19, EfficientNetB0 and ResNet50 using a COVID-19 radiology database. Various classification techniques, such as K-Nearest Neighbor (KNN), Naive Bayes, Random Forest, Support Vector Machine (SVM), and Neural Network, were also used for the performance comparison of the proposed model. The hybrid deep learning model with average pooling layers, along with SVM-linear and neural networks, both achieved an accuracy of 92%.These proposed models can be employed to assist radiologists and physicians in avoiding misdiagnosis rates and to validate the positive COVID-19 infected cases.
Keywords: Visual Geometry Group-16 (VGG16), Visual Geometry Group-19 (VGG-19), COVID- coronavirus disease 2019 (COVID-19), X-rays, Hybrid Deep Learning CNN model, Neural network (NN)
Highlights
-
•
The feature dimensions of the vgg-16 and vgg-19 pre-trained models are reduced by max and average pooling layers.
-
•
The dropout layer is supplemented to avoid overfitting.
-
•
The hybrid deep learning model (average pooling layer 2 × 2) outperforms the state-of-the art CNN models.
-
•
The hybrid deep learning model assists radiologists and physicians in diagnosing COVID-19-infected cases.
1. Introduction
COVID-19 began as an unknown cause of Pneumonia in Wuhan, Hubei state of China, in 2019. Later, it became a pandemic in 2020 [[1], [2], [3]]. This virus is named COVID-19, and is also phrased as SARS-CoV-2. This disease starts in Wuhan and spreads throughout china within 30 days [4]. As of March 5, 2023, around 675 million COVID-19 cases and 6.87 million deaths were reported throughout the world [5]. In Ethiopia, 500000 people have been affected, and 7572 people were dead. In January 2023, 5,35,14,115 people were vaccinated as per the World Health Organization (WHO) report [6]. COVID-19 can cause stern respiratory problems and the death of a human being. The warning sign of COVID-19 includes sore throat, fever, headache, cough, fatigue, muscle pain and shortness of breath [7].
Currently, Real-time reverse transcription-polymerase chain reaction (RT-PCR) is used to diagnose COVID-19. The sensitivity of RT-PCR ranges from 60% to 70% even if false negative results are obtained. The existing PCR-based testing is time-consuming and produces more false negatives. On the other hand, radiographic imaging like X-rays is used for COVID detection [8]. The COVID-affected patients are scanned through X-ray or CT machines to assess the disease's severity and spread in the lungs. Due to the manual analysis of chest X-rays and the increase in the spread of COVID diseases, the radiologists have a significant burden. As a result, it is necessary to develop an automatic system to diagnose COVID infections quickly.
CT is a sensitive technique to detect COVID-19, and is used as a screening tool along with the RT-PRC [8]. The lung disease is observed on the CT scan only after ten days from the onset of symptoms [9]. The radiologic images taken from COVID-19 patients contain important information for diagnostics.
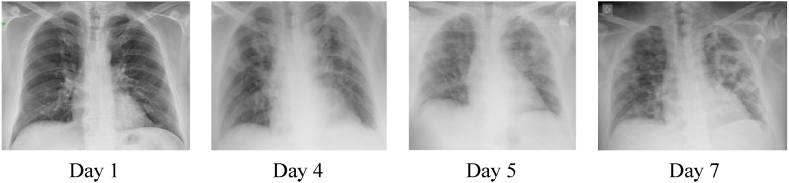
Kong et al. stated that right infra hilar airspace opacities are present in the COVID-19 cases [10]. Yoon et al. observed that one in every three cases had a single nodular opacity present in the left lower lung area [11]. The remaining two cases had five irregular opacities there in both lungs. Zheo et al. found ground-glass opacities (GGO) in almost all persons and also noted the consolidation and vascular dilation in the lesion [12]. Li and Xia stated that interlobular septal thickening with or without vascular development, GGO and air bronchogram signs were present as general features of COVID cases [13]. In another study, Multifocal GGO or peripheral focal affects both the lungs up to 50%–70% [14]. Chung et al. stated that 33% of chest CT images had rounded lung opacities. In Fig. 1, COVID-19 patient chest X-ray images were taken on days 1, 4, 5 and 7 for a 50-year-old person [15].
Fig. 1.
COVID-19 patient chest X-ray images [15].
Deep learning is one of the popular research areas of Artificial intelligence (AI). It creates end-to-end models to obtain promising results without manual, feature extraction [16,17]. It is applied successfully to many problems such as skin cancer classification [18,19], breast cancer detection [20,21], Pneumonia detection from chest X-rays images [22], Lung segmentation [23,24], brain disease classification, fundus image segmentation and arrhythmia detection. The COVID-19 pandemic necessitated the need for proficient doctors in this field. This problem shows the way for the automated detection system based on AI techniques. Due to the limited number of radiologists, it is diffficult to provide expert clinicians in every hospital. Accurate, simple, high-speed AI models with timely assistance to the patient may be provided in all hospitals to overcome this emergency. Thus, the AI model is useful in eliminating disadvantages such as test cost; delay in the medical test reports and an insufficient number of available RT-PCR. The objective of this research work is to design a new deep learning CNN model and examine its efficiency with state-of-the art deep learning. CNN models for automatic detection of COVID-19 using chest X-ray images.
1.1. Related works
Narin et al. Presented ResNet50 model to predict COVID-19 using chest x-ray images and accomplished 98% recognition [25]. Setty and Behera extract the features from the various convolution neural network, and their results show that the ResNet50 model, along with the SVM classifier, endows with the best performance [26]. Ardakani et al. used VGG-16, VGG-19, Alex Net, Google Net, SqueezeNet, Mobile Net-V2, Xception and ResNet-101 to diagnose the COVID-19 and compare their results [27]. Ferhat ucar et al. introduced a novel method named Deep Bays-SqueezeNet to diagnose COVID-19 [28]. Singh et al. used image augmentation, and preprocessing steps and fine-tuned the VGG-16 architecture to extract the features from CT images. Four different classifiers, such as Extreme Learning Machine (ELM), CNN, Online Sequential ELM and Bagging Ensemble with SVM were employed for classification proposes. Bagging Ensemble with SVM, achieved a maximum accuracy of 95.7% [29]. Dalvi et al. proposed DenseNet-1169 model along with Nearest-Neighbor interpolation technique for diagnosing the COVID-19 diseases from the X-ray images. The DenseNet-1169 model attains 96.37% accuracy and outperforms existing transfer learning techniques such as ResNet-50, VGG-16 and VGG-19 [30]. Kathamuthu et al. projected different deep-learning CNN models for diagnosis of COVID-19 diseases in CT images. VGG19, VGG16, InceptionV3, Densenet121, Xception and Resnet50 models were used in this work. The existing pre-trained VGG16 model outperforms other models in this experiment with an accuracy of 98.00 % [31]. Alhares et al. introduced a multi-source adversarial transfer learning model (AMTLDC), which was developed from the CNN-model. It was generalizable between multiple data sources. The accuracy of AMTLDC surpasses the other existing pre-trained models [32]. Kumar et al. design a proposed ensemble model which detects COVID-19 infection by integrating various transfer learning models such as GoogLeNet, EfficientNet, and XceptionNet. The proposed ensemble model improves the classifier's performance for multiclass and binary COVID-19 datasets [33]. Chow et al. has investigated 18 different CNN models which include AlexNet, DarkNet-53, DenseNet-201, DarkNet-19, Inception-ResNet-v2, GoogLeNet, MobileNet-v2, Inception-v3, NasNet-Large, NasNet-Mobile, ResNet-50, ResNet-18, ResNet-101, ShuffleNet, SqueezeNet, VGG-19, VGG-16, and Xception. Among them, only ResNet-101, VGG-16, SqueezeNet and VGG-19 models attain accuracy higher than 90%. Finally, the author recommended both SqueezeNet and VGG-16 as additional tools for the diagnosis of COVID-19 [34].
In related work, the use of transfer learning models has shown its ability to enhance the accuracy rate compared to other machine learning techniques. Still, the issues of accuracy, high dimensional features and execution time was not attained the optimal results in related works. In this work, we examine the efficiency of existing multi CNN, such as VGG 16, VGG 19, EfficientNet B0 and ResNet50, with our proposed hybrid deep learning model for automatic detection of COVID-19. From the related works, it is inferred that the performances of VGG16 and VGG19 are better compared to other pertained models. The discriminative features from the last layer of VGG16 and VGG19 models were reduced by different pooling layers. Finally, dimension-reduced features were passed into three fully connected dense neural network layers to predict of COVID-19. As an outcome, a fast and dependable intelligence device has been provided to detect of COVID-19. These models can be exploited to help radiologists and physicians in the decision-making process. The misdiagnosis rate of COVID-19 can be reduced by using our proposed model.
The remaining section of this study is organized as follows: materials and methods are presented in Section 2. In Section 3, the experimental results of hybrid deep learning models were evaluated with existing pre-trained CNN models. In Section 4, the discussion is presented. Finally, Section 5 presents the conclusions.
2. Materials and methods
2.1. COVID-19 databases
The total number of samples and distribution of the classes have an impact on proposed model. The morphological features like shape, color and texture-based features affect the performance of the proposed models. Pattern such as bilateral, peripheral, multifocal patchy consolidation, crazy-paving pattern and predominant ground-glass opacity (GGO) were observed in chest X-ray images.
The COVID-19 Radiography database (Winner of the COVID-19 Database award by the Kaggle community) was used in this research work [35]. This database is created by a team of researchers from Qatar University and the University of Dhaka, Bangladesh, in collaboration with doctors from Pakistan and Malaysia. In the second update, the database consists of 3616 COVID-19-positive cases along with 10,192 normal cases. From the database, 9220 images were taken from two classes for our research, as shown in Table 1. The normal images from the COVID-19 Radiography database were collected from RSNA [36] and Kaggle [37]. Similarly COVID-19 images from the COVID-19 Radiography database were collected from the padchest dataset [38], German medical school [39], SIRM, Github, Kaggle & Tweeter [[40], [41], [42]] from another Github source [43].
Table 1.
Sample distribution between two classes.
| Class | Number of samples for training | Number of samples for testing | Total |
|---|---|---|---|
| COVID-19 | 1689 | 724 | 2413 |
| Normal | 4765 | 2042 | 6807 |
| Total | 6454 | 2766 | 9220 |
2.2. Proposed hybrid deep learning model
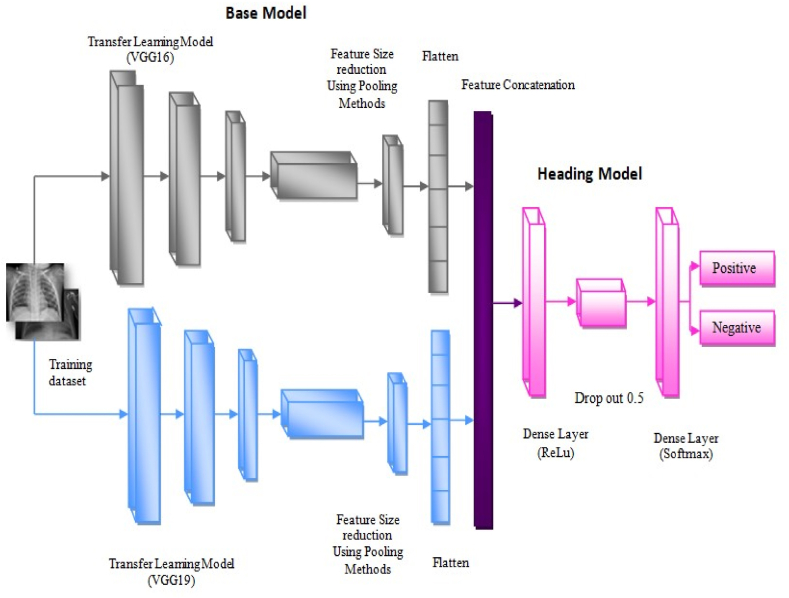
The proposed hybrid deep learning model is presented in Fig. 2. It includes two base models followed by a heading model to perform the classification process on COVID-19 Radiography. The deeper layers of pre-trained CNN models, such as VGG16 and VGG19, are concatenated to form a single feature vector. The deep neural network classifier is employed to investigate the output metrics. Two pre-trained models are preferred due to structural simplicity and ease of training time.
Fig. 2.
Illustration of proposed hybrid deep learning model.
The covid-19 detection process consists of three stages: feature extraction by a pre-trained transfer learning model, reducing the feature size by different pooling methods, and lastly to find the labels of the X-ray images using a deep neural network classifier. The final features were obtained by concatenating the pooling layer output from the VGG16 and VGG19 models. 4096 concatenated feature vector is obtained finally. Since, the pooling layers at the last part of each pre-trained CNN model aim to acquire the best features that classify the target class rather than unrelated features.
Therefore, the recital of classification was improved. In this work, max and average pooling layers are used to achieve higher accuracy and lower computational cost by minimizing the feature sizes. The total feature size is reduced from 16384 1 to 4096 1 after applying the pooling layer. Then feature set values were normalized with zero mean. The deep neural network is used for classification. 70%, 30% and 20% of the datasets were used for training, testing and validation. The Deep neural network provides a suitable plan which classifies the classes efficiently. The evaluation metrics and accuracy of the existing CNN models and the proposed model were compared. Google Colab environment was used to perform the experiments. CPU and GPU were used for computation. Tesla K80 accelerator, 12 GB GDDR5 VRAM Intel Xeon CPU @2.20 GHz, and 13 GB RAM were used. The proposed hybrid deep learning model steps are described below.
Step 1 The first step was image acquisition. Primarily, X-ray images of non-COVID-19 and the COVID-19 patients were taken from the public dataset such as COVID-19 Radiography database [35]. Step 2 The dataset was loaded, and all images were resized to 150 150. Step 3 Then, one-hot encoding was performed by label binaries. It takes categorical data and returns a numpy array [44]. Step 4 The entire dataset was divided into 70-30%. 70% is used for validation and training sets and 30% is used for the testing sets. In Step 5, the pre-trained model, such as VGG16 and VGG19, were initialized with weights generated from the ‘imagenet’ database separately. Nevertheless, neither the head nor the top of the pre-trained models was loaded. Step 6 Then, build a single head for the two pre-trained base models, such as VGG16 and VGG19. In this step, the head was constructed, and it is appended to the top of the two per-trained base models. The head model consists of (i) The discriminative features from last the layer of these two models were reduced by different pooling layers such as average and a max of size 2 2. (ii) Then, the head models were flattened. VGG16 and VGG19 features were concatenated to form a final feature vector. It was followed by a dense layer of size 1000 neurons with the activation of “ReLU”. The dropout is applied at the rate of 0.5 in the head model to avoid over fitting. At last, a dense layer has 2 neurons with a softmax activation function. The head model is placed at the top of the base model. The entire model is ready for training and testing the data. Step 7, the proposed model was supported by Adam optimizer. It was a combination of RMSProp and AdaGrad algorithms [45]. These algorithms perform well for noisy data. The initial learning rate is 3e-4. Step 8 The model is trained by a batch size of 64 and epochs of 25. Step 9 Finally, the model is tested with the remaining 30% dataset.
2.3. Statistical analysis
The results, such as recall, precision, accuracy, specificity, sensitivity and F1-score, were derived from the confusion matrix to calculate the quantitative performance of proposed hybrid deep learning models. The columns and rows of the confusion matrix show the predicted classes, and actual classes respectively. The samples classified correctly were distributed on the diagonal cells of the confusion matrix. The remaining parts of the cells contain incorrectly classified samples. The rightmost column of the confusion matrix is for calculating precision metrics. The bottom row of the confusion matrix is for calculating the recall metrics. The evaluation metrics were defined as follows in equations [[1], [2], [3], [4], [5], [6], [7], [8], [46]]:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
Here, (1-TPR) and (1-PPV) are correspond to the proportional of type II and type I errors.
3. Results
Table 2 shows the parameter setting for existing pre-trained models and proposed hybrid deep learning models. The first, second and third dense layer of the neural network consists of 1000, 500 and 150 neurons. Finally, the last layer consists of 2 neurons respectively. Various kernels, such as linear, Radial Basis Function (RBF), and sigmoid, are used in SVM classifiers. The random_state in Scikit-Learn is set to 0. It considers the same train and test sets across different executions. Gaussian Naive Bayes models are used in scikit-learn for Naive Bayes. n_estimators is set to 20 for Random Forest. It represents the number of trees required to be built before taking the averages of the predictions. The K value and distance measure of the KNN classifier are set to 2 and euclidean, respectively.
Table 2.
Model parameters setting for experimentation.
| Parameters [31] | vgg16 | vgg19 | EfficiebtnetB0 | ResNet50 | Hybrid Deep Learning Models |
|---|---|---|---|---|---|
| Image size | 150 × 150 × 3 | 150 × 150 × 3 | 150 × 150 × 3 | 150 × 150 × 3 | 150 × 150 × 3 |
| Batch size | 64 | 64 | 64 | 64 | 64 |
| Optimizer | Adam | Adam | Adam | Adam | Adam |
| Learning rate | 3e-4 | 3e-4 | 3e-4 | 3e-4 | 3e-4 |
| epochs | 25 | 25 | 25 | 25 | 25 |
| 1st Dense layer Activation Function(1000) | Relu activation | Relu activation | Relu activation | Relu activation | Relu activation |
| 2nd and 3rd Dense layer Activation Function(500,150) | Sigmoid activation | Sigmoid activation | Sigmoid activation | Sigmoid activation | Sigmoid activation |
| Dropout | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Output Layer(2) | Categorical/Softmax | Categorical/Softmax | Categorical/Softmax | Categorical/Softmax | Categorical/Softmax |
| Activation/Loss Function | Cross Entropy | Cross Entropy | Cross Entropy | Cross Entropy | Cross Entropy |
3.1. Performance of proposed hybrid deep learning model
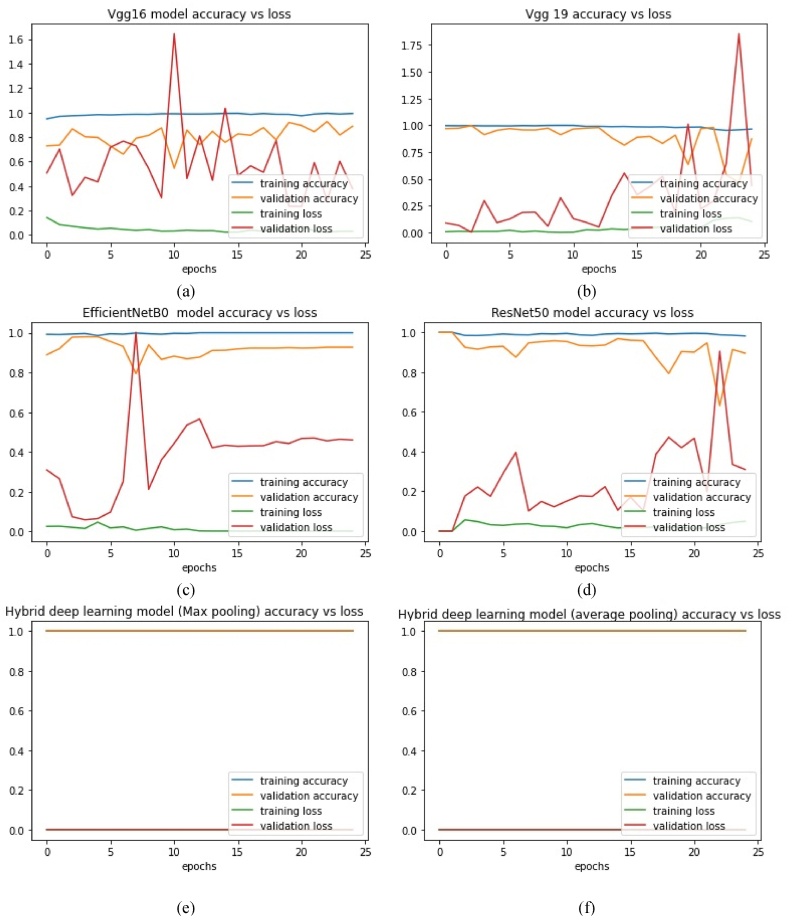
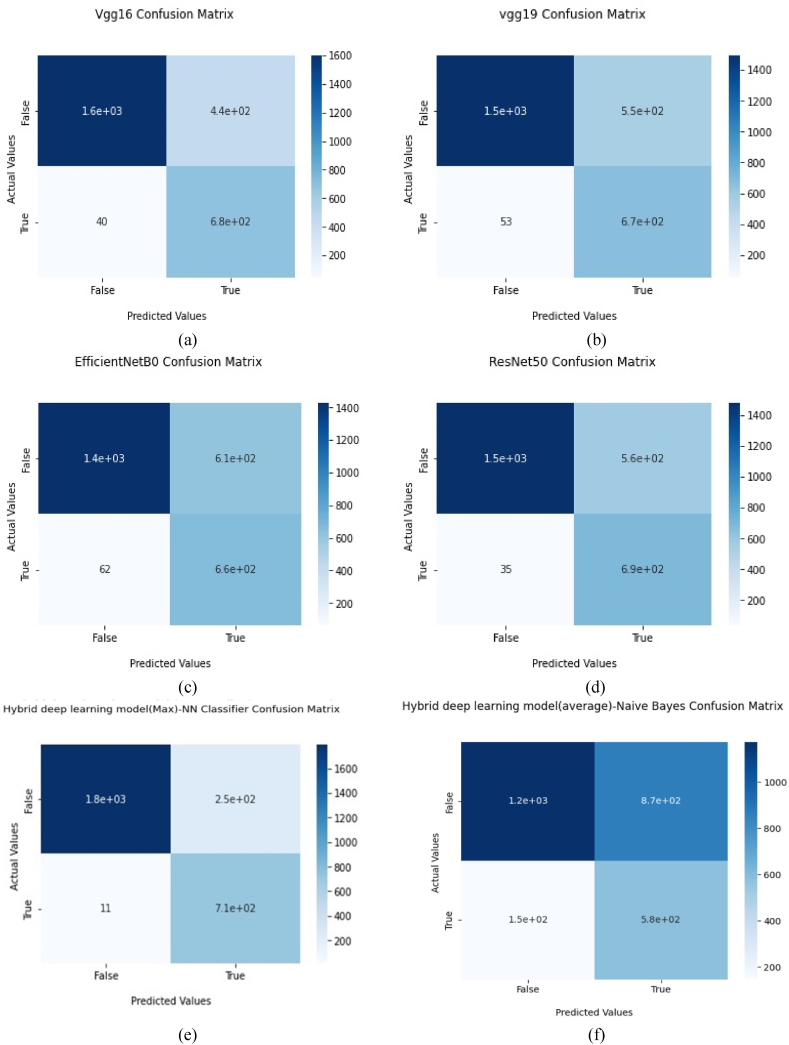
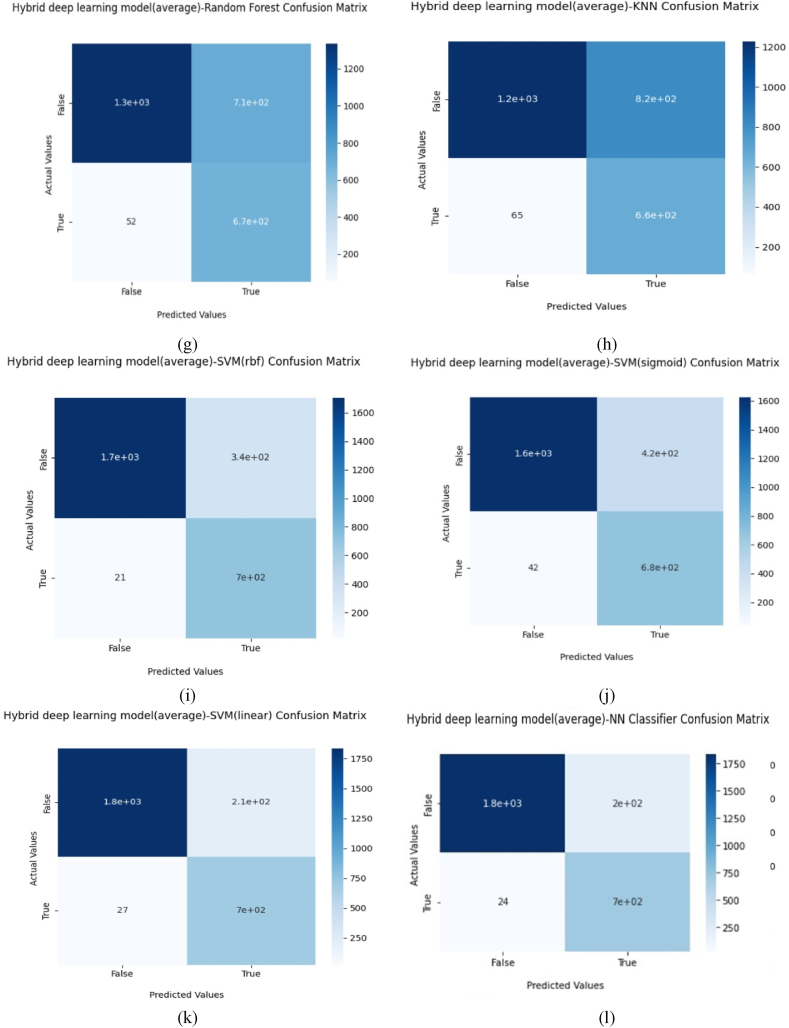
The performance of the proposed model and different existing pre-trained models was compared in this subsection. Fig. 3 (a)-(d) shows the accuracy and loss for both training and validation of existing methods. Fig. 3 (e) and (f) show that the proposed hybrid deep learning models attained 100% validation accuracy and zero validation loss. Fig. 4 (a)-(d) shows the binary-class confusion matrix of existing transfer learning model with deep neural network. Fig. 4 (e)–(l) shows the binary-class confusion matrix of hybrid deep learning models with various classifiers. Fig. 4 (l) demonstrates 224 misclassification samples among 2766 test samples. The covid-19 class was misclassified more compared to the normal class. Still, the misclassification sample of the proposed model was less compared to all other models. The comprehensive classification results for the proposed model and other pre-trained existing models are given in Table .3. The proposed models show a remarkable performance improvement in all classes. Both normal and COVID-19 classes attained the best f1-score results. The COVID-19 class of the proposed hybrid deep Learning Model (average pooling) had attained higher values of precision, recall and F1 score as 77%, 97% and 86% respectively. The overall classification scores of the deep neural network were given in Table 4.
Fig. 3.
Training and Validation evaluation of proposed hybrid deep learning models and existing pre-trained –CNN model for COVID-19 Radiography database [35]. (a) VGG16 model accuracy vs loss plot. (b) VGG19 model accuracy vs loss plot. (c) EfficientNet B0 model accuracy vs loss plot. (d) ResNet50 model accuracy vs loss plot. (e) Hybrid deep learning model (Max pooling) model accuracy vs loss plot. (f) Hybrid deep learning model (average pooling) model accuracy vs loss plot.
Fig. 4.
The confusion matrix of each transfer model (a) VGG16 model confusion matrix. (b) VGG19 model confusion matrix. (c) EfficientNet B0 model confusion matrix. (d) ResNet50 model confusion matrix. (e) Hybrid deep learning model (Max pooling) model confusion matrix. (f) Hybrid deep learning model (average pooling)-naive bayes confusion matrix. (g) Hybrid deep learning model (average pooling)-Random Forest confusion matrix. (h) Hybrid deep learning model (average pooling)-KNN confusion matrix. (i) Hybrid deep learning model (average pooling)-SVM (rbf) confusion matrix. (j) Hybrid deep learning model (average pooling)-SVM(sigmoid). (k) Hybrid deep learning model (average pooling)-SVM(linear). (l) Hybrid deep learning model (average pooling)-NN confusion matrix.
Table 3.
Performance comparison of the proposed hybrid deep learning model and the other existing pre-trained CNN model for each class.
| Deep learning models | Class | Precision | recall | f1-score |
|---|---|---|---|---|
| Vgg16 | Normal | 98 | 78 | 87 |
| Covid-19 | 61 | 94 | 74 | |
| Vgg19 | Normal | 97 | 73 | 83 |
| Covid-19 | 55 | 93 | 69 | |
| EfficientNetB0 | Normal | 96 | 70 | 81 |
| Covid-19 | 52 | 91 | 66 | |
| ResNet50 | Normal | 98 | 72 | 83 |
| Covid-19 | 55 | 95 | 70 | |
| Hybrid deep learning model (max pooling layer)-NN | Normal | 99 | 88 | 93 |
| Covid-19 | 74 | 98 | 85 | |
| Hybrid deep learning model (average pooling layer)-Naive Bayes | Normal | 89 | 57 | 70 |
| Covid-19 | 40 | 80 | 53 | |
| Hybrid deep learning model(average pooling layer)-Random Forest | Normal | 96 | 65 | 78 |
| Covid-19 | 49 | 93 | 64 | |
| Hybrid deep learning model(average pooling layer)-KNN | Normal | 95 | 60 | 74 |
| Covid-19 | 45 | 91 | 60 | |
| Hybrid deep learning model(average pooling layer)-SVM(RBF) | Normal | 99 | 83 | 90 |
| Covid-19 | 68 | 97 | 80 | |
| Hybrid deep learning model(average pooling layer)-SVM(sigmoid) | Normal | 97 | 80 | 88 |
| Covid-19 | 62 | 94 | 75 | |
| Hybrid deep learning model(average pooling layer)-SVM(linear) | Normal | 99 | 90 | 94 |
| Covid-19 | 77 | 96 | 86 | |
| Hybrid deep learning model (average pooling layer)-NN | Normal | 99 | 90 | 94 |
| Covid-19 | 77 | 97 | 86 |
Table 4.
The overall classification scores of proposed hybrid deep learning models and the other existing pre-trained CNN model for COVID-19 Radiography database [35].
| Deep learning models | Accuracy | Precision | recall | Specificity | Sensitivity | f1-score | MCC | JI | CSI |
|---|---|---|---|---|---|---|---|---|---|
| Vgg16 | 83% | 88% | 83% | 60.71% | 97.56% | 84% | 0.655 | 0.761 | 0.855 |
| Vgg19 | 78% | 86% | 78% | 54.92% | 96.59% | 80% | 0.582 | 0.713 | 0.782 |
| EfficientNetB0 | 76% | 84% | 76% | 51.97% | 95.76% | 77% | 0.540 | 0.676 | 0.754 |
| ResNet50 | 78% | 87% | 78% | 55.2% | 97.72% | 80% | 0.600 | 0.715 | 0.787 |
| Hybrid deep learning model (max pooling layer)-NN | 91% | 93% | 91% | 73.96% | 99.39% | 91% | 0.796 | 0.873 | 0.906 |
| Hybrid deep learning model(average pooling layer)- Naive Bayes | 63% | 76% | 63% | 79.45% | 57.97% | 66% | 0.328 | 0.541 | 0.636 |
| Hybrid deep learning model(average pooling layer)-Random Forest | 73% | 84% | 73% | 92.97% | 64.67% | 74% | 0.507 | 0.631 | 0.721 |
| Hybrid deep learning model(average pooling layer)-KNN | 68% | 82 % | 68% | 91.03% | 59.40% | 70% | 0.446 | 0.575 | 0.678 |
| Hybrid deep learning model(average pooling layer)-SVM (rbf) | 87% | 91% | 87% | 97.08% | 83.33% | 88% | 0.729 | 0.824 | 0.869 |
| Hybrid deep learning model(average pooling layer)-SVM (sigmoid) | 83% | 88% | 83% | 94.18% | 79.20% | 84% | 0.659 | 0.776 | 0.834 |
| Hybrid deep learning model(average pooling layer)-SVM (linear) | 92% | 93% | 92% | 96.29% | 89.55% | 92% | 0.805 | 0.883 | 0.913 |
| Hybrid deep learning model (average pooling layer)-NN | 92% | 93% | 92% | 77.77% | 98.68% | 92% | 0.814 | 0.889 | 0.917 |
The empirical results demonstrate that proposed hybrid deep Learning model (average pooling layer 2 2) with 4096 feature size attains the best total metric values for the COVID-19 Radiography database. The proposed hybrid deep learning model (average pooling layer) reached an accuracy of 92% for the COVID-19 Radiography database. The remaining performance metrics, such as precision, recall, specificity, sensitivity and F1 score, were 93%, 92%, 77.77%, 98.68, and 92%, respectively. By considering the data in Table 4, the radar chart was plotted in Fig. 5. It shows the performance evaluation of proposed and existing CNN models using different metrics. The proposed hybrid deep Learning model (average pooling layer 2 2) outperforms existing models in all metrics. It provides 9%, 14%, 16% and 14% accuracy improvement over VGG16, VGG19, EfficientNetB0 and ResNet50, respectively. Table 5 Shows the Computational Time
Fig. .5.
Radar chart of the performance in terms of all output metrics.
Table 5.
Computational time.
| Deep learning models | Convolution Layer output | Feature size | Testing time milliseconds(ms) |
|---|---|---|---|
| Vgg16 | 4, 4, 512 | 8192 | 101549.47 |
| Vgg19 | 4, 4, 512 | 8192 | 11425.91 |
| EfficiebtnetB0 | 5, 5, 1280 | 32000 | 406196.04 |
| ResNet50 | 5, 5, 2048 | 51200 | 609294.78 |
| Hybrid deep Learning model (max pooling layer 2 )-NN | Vgg16 (2, 2, 512) Vgg19 (2, 2, 512) |
4096 | 30217.18 |
| Hybrid deep learning model(average pooling layer)- Naive Bayes | 30069.80 | ||
| Hybrid deep learning model(average pooling layer)-Random Forest | 28290.65 | ||
| Hybrid deep learning model(average pooling layer)-KNN | 364.87 | ||
| Hybrid deep learning model(average pooling layer)-SVM (rbf) | 424.26 | ||
| Hybrid deep learning model(average pooling layer)-SVM (sigmoid) | 38316.83 | ||
| Hybrid deep learning model(average pooling layer)-SVM (linear) | 171267.67 | ||
| Hybrid deep learning model (average pooling layer 2 )-NN | 32185.88 |
4. Discussion
In this paper, a novel hybrid deep learning model is formed with optimized features from two pre-trained CNN models such as VGG16 and VGG19, respectively. The optimized features were obtained by different pooling layers. These pooling layers are employed to attain higher accuracy and lower computation costs by extracting the best features. The efficacy of the proposed model is evaluated by an independent test dataset. It consists of 2766 sample X-ray images, where 2042 were normal and the remaining 724 were COVID-19 images. The proposed Hybrid deep learning model (average pooling layer) attains a diagnostic f-score of 94% and 86 for both normal and COVID-19 classes in Table .2.
All over the world, the total number of COVID-19 cases and mortality rate are increased rapidly. Diseases like Pneumonia and lower respiratory tract contagion may be involved with COVID-19. Generally, the normal and COVID-19 images were clinically confused. It is more complex to distinguish normal and COVID-19 patients through X-ray images during the pandemic period. It is vital to diagnose the COVID-19 patient at an earlier stage to avoid mortality [47]. At present, RT-PCR is used as the reference standard to diagnose the COVID-19 patient. Similarly, it is seen that X-ray images are used as a fast and reliable approach to the diagnosing of COVID-19 patient. Features like peripheral, scattered ground, bilateral, glass opacities and consolidations are observed in the CT image of a COVID-19 patient [48]. Radiologists need to be familiar with the above X-ray image features for the new infection. At the same time, the shortage of radiologists to analyze X-ray images during the pandemic situation. It makes encumber for chest disease specialists who are proverbial with lung radiology also. To support chest specialist doctors, Artificial Intelligence (AI) applications are applied to the diagnosis of COVID-19 disease during pandemic situation.
The efficiency of AI algorithms in detecting COVID-19 images from chest X-rays has been examined in various studies. Ismael et al. proposed pre-trained deep CNN models such as ResNet50, ResNet101, ResNet18, VGG19, and VGG16 to diagnose COVID-19. The pre-trained CNN models were employed to extract deep features from the CT images. Various kernel functions such as Gaussian Linear, Cubic and Quadratic were used in Support Vector Machines (SVM) classifier to classify the COVID-19 images. The ResNet50 model, along with the SVM classifier, obtained a maximum accuracy score of 94.7% compared to other models. Here, COVID-19 Dataset has 200 images from normal patients and 180 images from COVID-19-infected patients. It is noticed that 45 COVID-19 CT images were used for the testing process [49]. Arora et al. apply pre-trained models such as InceptionV3, XceptionNet, DenseNet, ResNet50, MobileNet, and VGG 16 to classify the COVID-19 images. Super-resolution operation is used in CT image acquisition to improve performance metrics. The pre-trained CNN Mobilenet model attains 94.12% accuracy. Here, COVID-CT-Dataset has 463 CT images from normal patients and 349 COVID-19 CT images from 216 infected patients. 69 and 93 images from COVID-19 and normal datasets were employed for testing. It is noticed that diagnostic effectiveness was attained high by considering a very small dataset [50]. Shaik et al. proposed an ensemble approach that collectively improves the strength of deep learning architecture such as VGG16, ResNet50, ResNet50V2, Xception, InceptionV3, VGG19, and MobileNet. They used the COVID-CT dataset for training and testing. The data set contains 349 COVID-19 cases and 397 normal cases. The Proposed ensembling method attains an accuracy of 93.54%. 746 images were used in this research [51]. Singh et al. developed a Fine-tuning transfer learning-CoronaVirus 19 (Ftl-CoV19) for detecting COVID-19 X-ray images. The idea was derived from the VGG16 model, with includes various combinations of max-pooling, convolution and dense layers. The efficacy of the Ftl-CoV19 method is evaluated by publicly available datasets named “Curated dataset for COVID-19 posterior-anterior chest radiography images”. It contains 1481 normal posterior-anterior X-ray images and 1281 COVID-19 images. The proposed method attains training and validation accuracy of 98.82% and 99.27%. Still, the total images used for the evaluation were very less [52]. Umair et al. compared four pre-trained models such as MobileNet, VGG16, ResNet-50, and DenseNet-121 for diagnosis of COVID-19. In this research, 7232 images were utilized for evaluating the pre-trained network. The dataset consists of 3616 normal and 3616 COVID-19 images. Here, 2170 images were utilized for testing. The pre-trained MobileNet attained 96.48% accuracy [53]. In our study, more COVID-19 and normal X-ray images were employed to evaluate the efficacy of the proposed method. 9220 images were used for evaluate the robustness of the proposed method. 6454 and 2766 images were used for training and testing. Our testing dataset was quite large compared to the above research papers.
Incorporating large-scale images in the dataset is the significance of our research. A combined layer from VGG16 and VGG19 is applied to attain more detailed and complex features. The pooling layers are employed to remove the unrelated features from the X-ray images and to extract the best features for classification. The imbalanced dataset distribution between COVID-19 and normal images caused untruthful accuracy improvement. This dilemma has been resolved by minimizing the temporal problem in the feature set. Finally, the classification rate is improved. Accordingly, the proposed Hybrid deep learning model (average pooling layer)-NN attains sensitivity; specificity and accuracy in the finding of COVID-19 diseases were 98.68%, 77.77% and 92%, respectively. Similarly, the hybrid deep learning model (average pooling layer)-SVM (linear) also attain similar results as Neural Network. But sensitivity is an important metric for positive findings for patients with a disease. Here, the hybrid deep learning model (average pooling layer)-NN outperforms the SVM-linear classifier in sensitivity. Even though the proposed model yields laudable performance compared to other existing CNN models. The dataset was constructed by considering different X-ray images of the same class. These samples could be useful for the proposed model in the testing stage. The testing time of the proposed models shows better improvement compared to existing models Table .5. The feature size of proposed models was less compared to existing models. The performance of the proposed model is improved due to these factors.
The testing time of the proposed model is less compared to existing models. The features of VGG16 and VGG19 were reduced by pooling layers. This minimizes the computation time during testing (Table 5).
5. Conclusion
Early diagnosis of COVID-19 infection is very important to avoid the spreading of the disease to the public. Here, four pre-trained existing deep CNN models, such as VGG16, VGG19, ResNet, and EfficientNetB0, are employed to predict COVID-19 disease without human intervention. In this paper, hybrid deep learning CNN models are proposed to increase the accuracy rate and also condense the feature dimension. Various classification techniques, such as K-Nearest Neighbor (KNN), Naive Bayes, Random Forest, Support Vector Machine (SVM), and Neural Network, were used to ascertain the robustness of the proposed method. Empirical results show that the proposed hybrid deep learning model (average pooling layer 2 × 2)-NN yielded the highest MCC, JI, and CSI as 0.814, 0.889, and 0.917 compared with the existing pre-trained CNN models. Further, this will be carried out by considering the number of subjects in the dataset to evaluate the robustness of the proposed method. In the future, this work will intend to apply other optimization approaches to increase convergence speed. This investigation is helps motivate the researchers to classify COVID-19 patients from other possible diseases like tuberculosis and pneumonia.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Code availability
The codes used to support the findings of this study are available from the corresponding author upon request.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon request.
https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database.
CRediT authorship contribution statement
Mohan Abdullah: Writing – original draft, Validation, Software, Methodology. Ftsum berhe Abrha: Data curation, Conceptualization. Beshir Kedir: Supervision, Software, Resources, Methodology. Takore Tamirat Tagesse: Writing – original draft, Software, Resources, Methodology, Investigation, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to express my special thanks to Electrical and Computer Engineering Department of Wachemo University, Ethiopia for their support.
Contributor Information
Mohan Abdullah, Email: abdullahbinmohan@gmail.com.
Ftsum berhe Abrha, Email: abrhaf@wcu.edu.et.
Beshir Kedir, Email: Kedir.temem@wcu.edu.et.
Takore Tamirat Tagesse, Email: Tamirat.tagesse@wcu.edu.et.
List of Abbreviations
- ELM
Extreme Learning Machine
- CNN
Convolutional Neural Network
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- COVID
Coronavirus Disease 2019 (COVID-19)
- CT
Computer tomography
- RT-PCR
Real-time reverse transcription-polymerase chain reaction
- VGG-16
Visual Geometry Group-16
- VGG
Visual Geometry Group-19
- AI
Artificial Intelligence
- CPU
central processing unit
- GPU
graphics processing unit
- Ftl-CoV19
Fine tuning transfer learning-coronavirus 19
- SVM
Support Vector Machines
- KNN
K-Nearest Neighbor
- AMTLDC
A multi-source adversarial transfer learning model
- NN
Neural Network
- RBF
Radial basis function
- JI
Jaccard Index
- MCC
Matthews Correlation Coefficient
- CSI
Classification Succes Index
References
- 1.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Pneumonia of unknown cause–China. Emergencies preparedness, response, disease outbreak news. World Health Organization (WHO) 2020 [Google Scholar]
- 4.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 5.Batah S.S., Fabro A.T. Pulmonary pathology of ARDS in COVID-19: a pathological review for clinicians. Respir. Med. 2021;176 doi: 10.1016/j.rmed.2020.106239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.https://covid19.who.int/region/afro/country/et.
- 7.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee E.Y., Ng M.Y., Khong P.L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan F., Ye T., et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon S.H., Lee K.H., et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J. Radiol. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am. J. Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Xia L. Coronavirus Disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 14.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. 2020 doi: 10.1148/radiol.2020200527. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar Lorente, COVID-19 pneumonia - evolution over a week. https://radiopaedia.org/cases/COVID-19-pneumonia-evolution-over-a-week-1?lang¼us..
- 16.LeCun Y., Bengio Y., Hinton G. Deep learning, Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 18.Esteva A., Kuprel B., Novoa R.A., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codella N.C., Nguyen Q.B., Pankanti S., Gutman D.A., Helba B., Halpern A.C., Smith J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 2017;61(4/5) 5-1. [Google Scholar]
- 20.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recogn. Lett. 2020;133:232–239. [Google Scholar]
- 21.Cruz-Roa A., Basavanhally A., et al. March). Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. Medical Imaging 2014: Digital Pathology. 2014;9041 International Society for Optics and Photonics. [Google Scholar]
- 22.Rajpurkar P., Irvin J., et al. 2017. Chexnet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 23.Gaal G., Maga B., Lukacs A. 2020. Attention U-Net Based Adversarial Architectures for Chest X-Ray Lung Segmentation. arXiv preprint arXiv:2003.10304. [Google Scholar]
- 24.Souza J.C., Diniz J.O.B., Ferreira J.L., da Silva G.L.F., Silva A.C., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Progr. Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (COVID- 19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethy P.K., Behera S.K. 2020. Detection of Coronavirus Disease (COVID-19) Based on Deep Features. [Google Scholar]
- 27.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ucar F., Korkmaz D., Covidiagnosis-Net Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh M., Bansal S., Ahuja S., Dubey R.K., Panigrahi B.K., Dey N. Transfer learning–based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Med. Biol. Eng. Comput. 2021 Apr;59:825–839. doi: 10.1007/s11517-020-02299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalvi Pooja Pradeep, Reddy Edla Damodar, Purushothama B.R. Diagnosis of coronavirus disease from chest X-ray images using DenseNet-169 architecture. SN Computer Science. 2023;4(3):1–6. doi: 10.1007/s42979-022-01627-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kathamuthu Nirmala Devi, Subramaniam Shanthi, Quynh Hoang Le, Muthusamy Suresh, Panchal Hitesh, Christal Mary Sundararajan Suma, Jawad Alrubaie Ali, Maher Abdul Zahra Musaddak. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Adv. Eng. Software. 2023;175 doi: 10.1016/j.advengsoft.2022.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alhares Hadi, Tanha Jafar, Ali Balafar Mohammad. AMTLDC: a new adversarial multi-source transfer learning framework to diagnosis of COVID-19. Evolving Systems. 2023:1–15. doi: 10.1007/s12530-023-09484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar N., Gupta M., Gupta D., Tiwari S. Novel deep transfer learning model for COVID-19 patient detection using X-ray chest images. J. Ambient Intell. Hum. Comput. 2023;14(1):469–478. doi: 10.1007/s12652-021-03306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow Li Sze, Tang Goon Sheng, Iwan Solihin Mahmud, Muhammad Gowdh Nadia, Ramli Norlisah, Rahmat Kartini. Quantitative and qualitative analysis of 18 deep convolutional neural network (CNN) models with transfer learning to diagnose COVID-19 on chest X-ray (CXR) images. SN Computer Science. 2023;4(2):1–17. doi: 10.1007/s42979-022-01545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database [Accessed 27 November. 2022].
- 36.https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data.
- 37.https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 38.https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590858128006-9e640421-6711.
- 39.https://github.com/ml-workgroup/covid-19-image-repository/tree/master/png.
- 40.https://sirm.org/category/senza-categoria/covid-19/.
- 41.https://eurorad.org.
- 42.https://github.com/ieee8023/covid-chestxray-dataset.
- 43.https://github.com/armiro/COVID-CXNet.
- 44.Hossin M., Sulaiman M. A review on evaluation metrics for data classification evaluations. Data Mining & Knowledge Management Process. 2015;5(2):1–11. [Google Scholar]
- 45.Ghoshal B., Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (covid-19) detection. 2020. https://arxiv.org/abs/2003.10769
- 46.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ismael Aras M., Şengur Abdulkadir. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arora Vinay, Ng Eddie Yin-Kwee, Singh Leekha Rohan, Darshan Medhavi, Singh Arshdeep. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaik Nagur Shareef, Cherukuri Teja Krishna. Transfer learning based novel ensemble classifier for COVID-19 detection from chest CT-scans. Comput. Biol. Med. 2022;141 doi: 10.1016/j.compbiomed.2021.105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh Tarishi, Saurabh Praneet, Bisen Dhananjay, Kane Lalit, Pathak Mayank, Sinha G.R. Ftl-CoV19: a transfer learning approach to detect COVID-19. Comput. Intell. Neurosci. 2022:2022. doi: 10.1155/2022/1953992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umair Muhammad, Khan Muhammad Shahbaz, Ahmed Fawad, Baothman Fatmah, Alqahtani Fehaid, Alian Muhammad, Ahmad Jawad. Detection of COVID-19 using transfer learning and Grad-CAM visualization on indigenously collected X-ray dataset. Sensors. 2021;21(17):5813. doi: 10.3390/s21175813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database.