Abstract
Background
Hematopoietic cell signal transducer (HCST) and tyrosine kinase-binding protein (TYROBP) are triggering receptors expressed on myeloid cells 2 (TREM2), which are pivotal in the immune response to disease. Despite growing evidence underscoring the significance of TREM2, HCST, and TYROBP in certain forms of tumorigenesis, a comprehensive pan-cancer analysis of these proteins is lacking.
Methods
Multiple databases were synthesized to investigate the relationship between TREM2, HCST, TYROBP, and various cancer types. These include prognosis, methylation, regulation by long non-coding RNAs and transcription factors, immune signatures, pathway activity, microsatellite instability (MSI), tumor mutational burden (TMB), single-cell transcriptome profiling, and drug sensitivity.
Results
TREM2, HCST, and TYROBP displayed extensive somatic changes across numerous tumors, and their mRNA expression and methylation levels influenced patient outcomes across multiple cancer types. long non-coding RNA (lncRNA) -messenger RNA (mRNA) and TF-mRNA regulatory networks involving TREM2, HCST, and TYROBP were identified, with lncRNA MEG3 and the transcription factor SIP1 emerging as potential key regulators. Further immune analyses indicated that TREM2, HCST, and TYROBP play critical roles in immune-related pathways and macrophage differentiation, and may be significantly associated with TGF-β and SMAD9. Furthermore, the expression of TREM2, HCST, and TYROBP correlated with the immunotherapy markers TMB and MSI, and influenced sensitivity to immune-targeted drugs, thereby indicating their potential as predictors of immunotherapy outcomes.
Conclusion
This study offers valuable insights into the roles of TREM2, HCST, and TYROBP in tumor immunotherapy, suggesting their potential as prognostic markers and therapeutic targets for various cancers.
Keywords: Triggering receptor expressed on myeloid cells 2 (TREM2), Hematopoietic cell signal transducer (HCST), TYRO protein tyrosine kinase-binding protein (TYROBP), Pan-cancer, Tumor immunity
1. Introduction
Drug development for tumor therapeutic targets is of paramount importance because cancer is the leading cause of death worldwide [1,2]. Recently, the focus of research has shifted from tumor mutations to the tumor microenvironment, making tumor immunotherapy an essential anti-tumor treatment [3,4]. Consequently, investigation of new and combined immunotherapy targets is vital.
Triggering receptor expressed on myeloid cell 2 (TREM2) is an extracellular receptor that is predominantly expressed in myeloid lineage cells and plays a significant role in immune and metabolic processes. Despite lacking a signaling domain, TREM2 relies on hematopoietic cell signal transducers (HCST/DAP10) and the TYRO protein tyrosine kinase-binding protein (TYROBP/DAP12) for intracellular signal transduction. The TREM2 signaling network has been implicated in neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases and has been extensively studied in microglia [[5], [6], [7]].
Emerging research on TREM2, HCST, and TYROBP has gradually shifted its focus towards tumors, revealing their close relationship with immunosuppression. TREM2 has been identified as a specific marker of tumor-associated macrophages (TAM) in the tumor microenvironment, which is consistent across various human cancers and tumor models, including hepatocellular carcinoma, lung cancer, and breast cancer [[8], [9], [10]]. HCST is highly expressed in clear renal cell carcinoma [11] and correlates with low survival rates and cancer immune responses. TYROBP is highly expressed in gliomas and is associated with immune cell infiltration [12]. Multiple studies have confirmed that TREM2, HCST, and TYROBP are promising therapeutic targets, with significant potential for immunotherapy. However, research has primarily focused on specific tumor types and single genes, with limited studies on HCST and TYROBP. To date, no pan-cancer studies have investigated the relationship between TREM2, HCST, and TYROBP and various cancers.
In this study, we integrated multiple databases to analyze the association between TREM2, HCST, and TYROBP expression levels and various aspects of different cancer types, including prognosis, methylation, long non-coding RNA (lncRNA) and transcription factor regulation, immune signatures, pathway activity, microsatellite instability (MSI), tumor mutational burden (TMB), single-cell transcriptome profiling, and drug sensitivity. Our findings demonstrate that TREM2, HCST, and TYROBP can serve as prognostic factors for various cancers, are closely related to the regulation of immune and inflammation-related pathways, and play a crucial role in tumor immunity through immune cells, such as TAM, TMB, and MSI. Furthermore, these genes affect the sensitivity to various immune-targeted drugs. Our research contributes to a deeper understanding of the roles and potential of TREM2, HCST, and TYROBP in tumor immunotherapy.
2. Methods
2.1. Copy number variation (CNV) analysis
CNV data for 33 cancer types from The Cancer Genome Atlas (TCGA) database were collected using the UCSC Xena tool. Based on previous research, CNV values of 2 indicate amplifications, whereas values of −2 indicate deep deletions [13]. CNV ratios were calculated for each cancer type. The ComplexHeatmap R package was used to create OncoPrint plots to visualize the overall copy number variation.
2.2. mRNA expression analysis
Gene expression data for normal and tumor tissues were obtained from the TCGA dataset. The data were normalized to account for batch effects and log2(x + 1) was transformed. Of the 33 tumor types, we included only 17 with more than five pairs of tumor and normal samples in our differential expression analysis. We used the ggplot2 and ggpur packages to create box plots and determined p-values using t-tests.
We then sought to identify the expression-associated clusters of TREM2, HCST, and TYROBP through unsupervised consensus clustering using the PAM algorithm. We performed 1000 bootstrap runs, with each run including 80% of the patients and the number of clusters set between two and ten. Uniform cumulative distribution functions and delta areas were used to determine the optimal cluster count [14].
2.3. Survival analysis
We used the "survival" package to perform survival analysis. Based on the Cox regression analysis, hazard ratios were calculated for overall survival (OS), disease-free interval (DFI), and progression-free interval (PFI) for each tumor type. This helped identify high or low risks. We divided each tumor into two groups using the median value as a cutoff and conducted a log-rank test to determine the p-values. Instead of using Kaplan–Meier curves, we visualized the results with heat maps, showing only statistically significant tumor types. Red squares represent poor prognosis with high gene expression and blue squares indicate poor prognosis with low gene expression or high scores. Asterisks denote the significance levels: *p < 0.05, **p < 0.01, and ***p < 0.001.
2.4. Methylation analysis
We obtained DNA methylation data from TCGA samples using the UCSC Xena tool. We used the same method as for differential expression analysis to generate box plots and calculate p-values. We determined the hazard ratios for regulator methylation based on Cox regression analysis. A hazard ratio greater than one signified that high methylation was linked to poor OS, indicating a high risk. We divided each tumor type into two groups according to the median methylation level of each regulator and performed a log-rank test to calculate p-values. The heatmap used for visualizing the survival analysis was similar to that described in the previous section on the mRNA data.
2.5. LncRNAs, and transcription factors (TFs) analyses
We acquired lncRNA and TF regulation data from an earlier pan-cancer study that screened potential lncRNA-mRNA pairs and TF-mRNA pairs. We calculated the ratio of each pair for each tumor type using Spearman's correlation analysis to determine the expression relevance. We only kept pairs with an absolute correlation coefficient value > 0.3 and p < 0.05. We constructed lncRNA and TF regulatory networks using Cytoscape v3.8.2.
2.6. Immune characteristic analysis
We derived the abundance of 22 immune cell types based on the CIBERSORT algorithm from previous studies [15]. We determined the correlation between immune cell abundance and TREM2, HCST, and TYROBP expression in each tumor type. Additionally, we obtained immune activation-related genes, immune checkpoint-related genes, and TGF-β/EMT pathway-related genes from previously published studies [16].
The tumor immune microenvironment (TIME) is important for cancer prognosis and therapy. Studies have identified six immune subtypes of tumor types based on five representative immune signatures: wound healing (C1), IFN-γ dominant (C2), inflammatory (C3), lymphocyte depleted (C4), immunologically quiet (C5), and TGF-β dominant (C6) [17]. We performed differential expression analysis using the Kruskal–Wallis test with the "ggpur" software package to examine the mRNA expression levels of TREM2, HCST, and TYROBP in the six different immune tumor subtypes.
2.7. Pathway activity analyses
Gene set enrichment analysis (GSEA) was performed to identify the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in TREM2, HCST, and TYROBP across various cancer types. The GSVA gene set was obtained from the MSigDB database (v7.2, updated September 2020) using the "clusterProfiler" package. For each cancer type, samples were divided into two groups based on the median expression of the gene of interest. We calculated the log-fold change (logFC) between the groups and performed GSEA using logFC values and KEGG pathway gene sets. We retained only significant pathways (p < 0.05) and plotted the top enriched pathways. We also calculated the number of enriched pathways for each gene concerning cancer types and visualized pathways with over 20 enriched tumor types using bar charts. Finally, a Venn diagram was used to visualize the intersection of the enriched pathways for each gene with more than 20 enriched tumor types.
2.8. Correlation with TMB and MSI
TMB is a measurable immune response biomarker that represents the number of mutations in tumor cells. MSI resulting from MMR deficiency is associated with patient outcomes. TMB scores were calculated using the Perl script and adjusted by dividing them by the total exon length. We determined MSI scores for all samples using somatic mutation data from TCGA and analyzed the relationship between TREM2 expression, TMB, and MSI using the Spearman's rank correlation coefficient. We presented the results as radar diagrams using the "ggplot2″ package.
2.9. Single-cell transcriptional analysis
To explore the roles of TREM2, HCST, and TYROBP in specific cancer types, we analyzed single-cell sequencing data from colon cancer samples. We used the GSE161277 dataset from the GEO database and processed it using the Seurat package in R. Following the criteria of a previous study, we filtered out certain genes, standardized the data, and clustered the cells using principal component analysis [18]. We identified stromal and immune cells based on the specific markers reported in previous studies.
Next, we calculated the activity of the three key genes (TREM2, HCST, and TYROBP) in the dataset using the AUCell package, and visualized the scores using UMAP plots and boxplots. Statistical tests were performed to compare groups (adenoma, blood, carcinoma, normal, and paracarcinoma) using the "ggpur" package, considering the scores as different if the p-value was below 0.05. We conducted gene set variation analysis to investigate the differentially activated metabolic pathways in various cell types. To identify the intersection between the two sets of pathways, we compared the results of prior GSEA enrichment analysis and differential pathway analysis of normal and adenoma tissue macrophages. The intersections were visualized using a Venn diagram.
We also examined the subpopulation of cells with the highest AUC values for all seven genes and analyzed them in a pseudo-time sequence. The cells were sorted based on subpopulation markers from an earlier analysis. We evaluated the relationship between overall cellular expression changes and generated a single-cell pseudo-time trajectory using the monocle package in R. Finally, we visualized TREM2, HCST, and TYROBP expression at each developmental stage. The ggplot2 package was used to create violin plots to visualize the distribution of gene expression levels across various clusters of macrophage developmental trajectories. This methodology included statistical comparisons using the Wilcoxon test to assess differences between normal and other pathological tissues, with significant findings annotated directly in the plots.
2.10. Drug sensitivity analysis
CellMiner is an online tool that provides genomic and pharmacological information to researchers, enabling them to use transcript and drug response data from the NCI-60 cell line collected by the National Cancer Institute. We obtained RNA-seq spectral data for TREM2, HCST, and TYROBP, along with their pharmacological activities, from the CellMiner database (https://discover.nci.nih.gov/cellminer/). We used the "Impute" software package to pre-process the raw data. We examined the correlation between the transcriptional expression of TREM2, HCST, TYROBP, and compound sensitivity using Pearson correlation analysis. We considered relevant genes sensitive to the corresponding chemotherapeutic drugs if the p-value was less than 0.05, and the correlation coefficient was greater than 0.3.
3. Results
3.1. Genomic alterations of TREM2, HCST and TYROBP
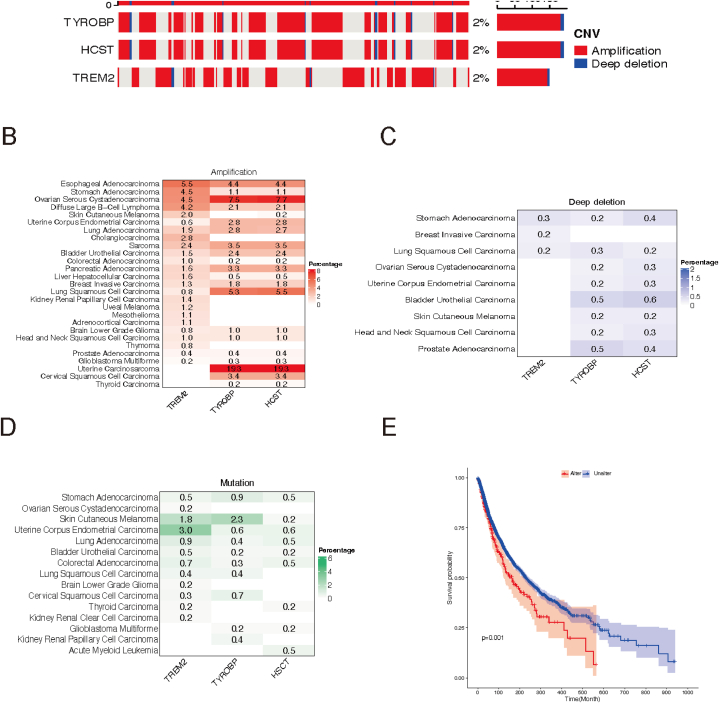
By analyzing CNV data from 10,680 pan-cancer samples, we identified genomic alterations in TREM2, HCST, and TYROBP in various tumors. These genes showed low CNV frequencies (2%), in 33 tumor types, mainly as amplifications (Fig. 1A). Specifically, TREM2, HCST, and TYROBP were amplified in 19 tumor types, including esophageal adenocarcinoma, stomach adenocarcinoma (STAD), ovarian serous cystadenocarcinoma (OV), diffuse large B-cell lymphoma (DLBC), skin cutaneous melanoma (SKCM), uterine corpus endometrial carcinoma (UCEC), lung adenocarcinoma (LUAD), cholangiocarcinoma (CHOL), sarcoma (SARC), bladder urothelial carcinoma (BLCA), colorectal adenocarcinoma, pancreatic adenocarcinoma (PAAD), liver hepatocellular carcinoma (LIHC), breast invasive carcinoma (BRCA), lung squamous cell carcinoma (LUSC), brain lower grade glioma, head and neck squamous cell carcinoma (HNSC), prostate adenocarcinoma, and glioblastoma multiforme (GBM). Notably, two gynecologic tumor types, uterine carcinosarcoma (UCS) (19.3%) and OV (7.5%, 7.7%), exhibited the highest amplification rates for HCST and TYROBP (Fig. 1B).
Fig. 1.
Genomic alterations in TREM2, HCST and TYROBP. (A) CNV landscape of TREM2, HCST and TYROBP in tumors. Each row represents a gene, and each column represents a patient. Only patients with TREM2, HCST and TYROBP CNV alterations are shown. The mutation rate for each gene is shown in the right label. (B–D) Frequency distributions of amplifications (B), deep deletions (C), and mutations (D) in different cancer types. The numbers in the figure represent the specific mutation rate, and the intensity of the color is proportional to the frequency. (E) Kaplan–Meier curve showing the overall survival difference between wild-type and mutated samples. Wild-type samples (Unalter) is represented by a blue line and mutated samples (Alter) is represented by a red line. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Deep deletions of these genes were infrequent and were found in less than 1% of stomach adenocarcinoma (STAD) and LUSC cases (Fig. 1C). Mutation analysis across tumor types revealed that all three genes were mutated in six tumor types (STAD, SKCM, UCEC, LUAD, BLCA and colorectal adenocarcinoma), with the highest TREM2 mutation frequency in UCEC (3.0%) and SKCM (1.8%). TYROBP mutations were most frequent in SKCM (2.3%) (Fig. 1D). Other tumor types had mutation frequencies of <1%. This comprehensive study highlights heterogeneous somatic cell changes in TREM2, HCST, and TYROBP across cancer types. Importantly, patients with these gene mutations had a poorer prognosis(p = 0.001) than those with the wild-type genes (Fig. 1E).
3.2. Gene expression patterns of TREM2, HCST and TYROBP
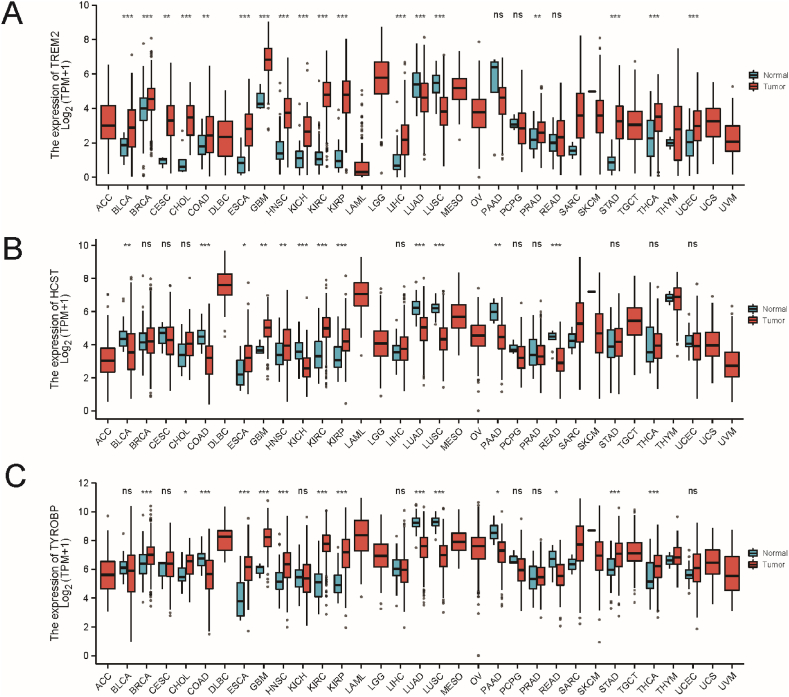
To characterize the gene expression patterns of TREM2, HCST, and TYROBP, paired differential expression analysis was performed between tumor and normal samples, which were found to be aberrantly expressed in multiple tumor types. TREM2 expression was upregulated in 16 tumors (BLAC, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LIHC, PRAD, STAD, THCA, and UCEC) and downregulated in two tumors (LUAD and LUSC); HCST was significantly upregulated in five tumors (ESCA, GBM, HNSC, KIRC, and KIRP) and downregulated in seven tumors (BLCA, COAD, KICH, LUAD, LUSC, PAAD, and READ), TYROBP was upregulated in nine tumors (BRCA, CHOL,ESCA,GBM, HNSC, KIRC, KIRP, STAD, and THCA) and downregulated in five tumors (COAD, LUAD, LUSC, PAAD, and READ) (Fig. 2A–C).
Fig. 2.
Differences in mRNA expression between tumor samples and normal samples (***p< 0.001, **p< 0.01, *p< 0.05). (A) TREM2, (B) HCST, (C) TYROBP. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; EMT, epithelial-mesenchymal transition; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, low-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, thymoma and uveal melanoma.
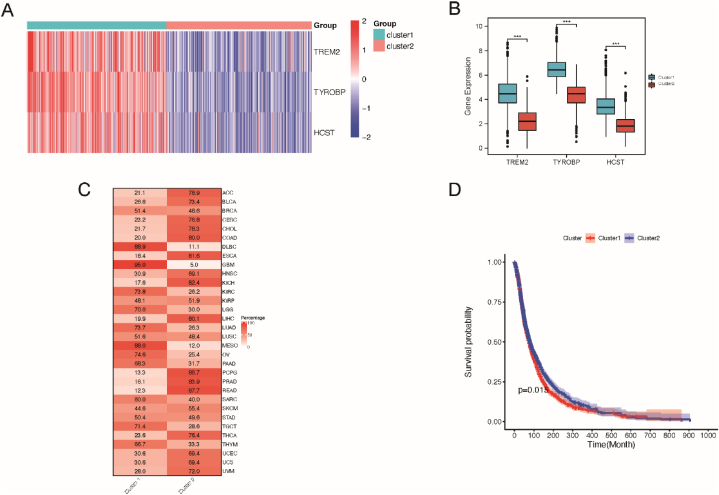
Furthermore, unsupervised consensus clustering based on the TREM2, HCST, and TYROBP mRNA expression was performed for all 33 tumor types to better understand their expression in tumors. All the tumors were clearly divided into two distinct clusters (Fig. 3A). The expression levels of TREM2, HCST, and TYROBP were significantly higher in cluster 1 than in cluster 2 (Fig. 3B). In terms of tumor type distribution, DLBC, GBM, KIRC, LGG, LUAD, LUSC, MESO, OV, PAD, SARCTGCT, and THYM were distributed in cluster 1. ACC, BLAC, CESC, CHOL, COAD, ESCA, HNSC, KICH, LIHC, PCPG, PRAD, READ, THCA, UCEC, UCS, and UVM were mostly distributed in cluster 2 (Fig. 3C). From the perspective of survival analysis, the overall prognosis of cluster 2 was significantly better than that of cluster 1 (Fig. 3D), indicating that the mRNA expression levels of TREM2, HCST, and TYROBP may affect the survival of patients with tumors.
Fig. 3.
Unsupervised consensus clustering based on mRNA expression of TREM2, HCST and TYROBP. (A) Two distinct clusters of TREM2, HCST and TYROBP. Each row represents a cuprotosis regulator and each column represents a patient. Red indicates high expression; blue indicates low expression. Expression data were normalized by z-score. (B) Boxplots showing differences in TREM2, HCST and TYROBP gene expression in the two clusters. Differences were tested by Student's t-test. ***p < 0.001, (C) Sample distributions in two clusters. Each row represents a tumor type, and each column represents a cluster. The numbers and red intensity in each box indicate the percentage of samples classified in the corresponding cluster. (D) Kaplan–Meier curve showing the overall survival difference between cluster 1 and cluster 2. Cluster 1 is represented by a blue line and cluster 2 is represented by a red line. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Relationship between prognosis of cancer patients and TREM2, HCST and TYROBP
Based on the impact of TREM2, HCST, and TYROBP on the survival of patients with tumors, overall survival (OS), disease-free survival (DFI), and progression-free survival (PFI) were examined to further elucidate their predictive roles in prognosis. Fig. 4A shows the relationship between TREM2, HCST, and TYROBP expression levels and OS, depending on the tumor type. For TREM2, poor OS was associated with low TREM2 expression in CESC, DLBC, SKCM, and THCA, and high TREM2 expression in LGG. For HCST, low expression of BRCA, CHOL, and SKCM indicated poor OS, whereas high expression of KIRC and LGG indicated poor OS. Low expression of TYROBP in SESC and SKCM led to poor OS, while high expression in LGG, LIHC, TGCT, THYM, and UVM led to poor OS.
Fig. 4.
Prognostic analysis of TREM2, HCST and TYROBP. (A) OS, (B) PFI, (C) DFI. *p < 0.05, **p < 0.01, ***p < 0.001. Only statistically significant tumor types are shown. Red squares indicate that high gene expression is associated with worse prognosis, and blue dotted squares indicate that low gene expression or high score is associated with worse prognosis. (D) Survival analysis of the TREM2, HCST and TYROBP across cancers. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In addition to overall survival, the expression of TREM2, HCST, and TYROBP was analyzed in relation to DFI and PFI(Fig. 4B and C). TREM2, HCST, and TYROBP expression levels and activity scores were closely correlated with the DFI of 11 tumors and the PFI of nine tumors, and the correlation depended on the tumor type. Cox regression analysis showed that TREM2 was a protective factor against CESC, DLBC, KIRP, THCA, and SKCM (p < 0.05, HR < 1) as also a protective factor for LGG and LIHC (p < 0.05, HR > 1). HCST was a protective factor for BRCA, CESC, HNSC, SARC, and SKCM (p < 0.05, HR < 1), and a risk factor for KIRC, LGG, and UVM (p < 0.05, HR > 1). TYROBP was a risk factor for THYM, UVM, LGG, and LUSC (p < 0.05, HR > 1) (Fig. 4D). These results revealed that TREM2, HCST, and TYROBP have different prognostic roles in different tumor types.
3.4. Methylation analysis of TREM2, HCST and TYROBP
The degree of methylation of the gene promoter region is closely related to the level of gene expression. Therefore, we explored methylation changes in TREM2, HCST, and TYROBP. First, methylation differences in the regulators were compared between paired normal and tumor tissues. The methylation levels of TREM2, HCST, and TYROBP were altered in all 16 tumors, except BLAC, SARC, and STAD, and the methylation changes of the regulators were heterogeneous in different tumor types (Fig. 5A–C). Subsequently, analysis of the correlation between TREM2, HCST, and TYROBP methylation levels and mRNA expression levels in 33 tumor types revealed that in most tumor types, HCST methylation levels were negatively correlated with mRNA expression levels, whereas in specific tumor types, the methylation levels of TREM2 and TYROBP were negatively or positively correlated with mRNA expression levels (Fig. 5D). Survival analysis revealed that the methylation levels of TREM2, HCST, and TYROBP were associated with OS in eight tumor types, and this association depended on the tumor type. Among them, hypermethylation of HCST was associated with poor OS in LIHC, SARC, SKCM, and UCE, but was associated with benign OS in KIRC, LGG, and UVM (Fig. 5E).
Fig. 5.
Methylation analysis of TREM2, HCST and TYROBP. (A–C) Differences in mRNA methylation between tumor samples and normal samples. (A) TREM2, (B) TYROBP, (C) HCST. (D) Correlation between methylation and mRNA expression. Blue squares represent negative correlation, red squares represent positive correlation, and the darker the color, the higher the correlation. *p < 0.05, **p < 0.01, ***p < 0.001. (E) Relationship between TREM2, HCST, TYROBP methylation and survival. Red squares indicate that high methylation is associated with decreased survival, and blue squares indicate that high methylation is associated with increased survival. *p < 0.05, **p < 0.01, ***p < 0.001. Only statistically significant tumor types are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
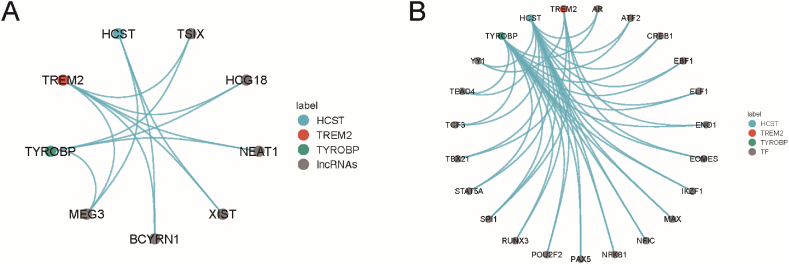
3.5. LncRNAs and TFs regulate the expression levels of TREM2, HCST and TYROBP
LncRNAs play a major role in transcription, translation, and post-translational modification and are essential regulatory factors for gene expression. TF are key regulators of gene transcription and expression, and dysregulated TF mediate abnormal gene expression, thereby representing a unique class of drug targets. Based on this, by combining the pan-cancer analysis of lncRNAs with the data from gene expression correlation analysis, potential lncRNA-mRNA pairs that regulate TREM2, HCST, and TYROBP in different tumor types were screened. A total of 62 lncRNA-mRNA pairs were identified, including 47 lncRNAs, 13 of which targeted two mRNAs in TREM2, HCST, and TYROBP, constituting the lncRNA regulatory network (Fig. 6A). A series of TFs listed in previous pan-cancer studies were analyzed, and 213 TF-mRNA pairs were found, including 171 different TFs, of which 42 targeted at least two: TREM2, HCST, and TYROBP (Fig. 6B). In particular, the nuclear factor SPI1 targets TREM2, HCST, and TYROBP, suggesting that it may be a key TF that mediates the transcriptional regulation of TREM2, HCST, and TYROBP in tumors.
Fig. 6.
LncRNA, and TF regulatory networks of TREM2, HCST and TYROBP. (A–C) Differences in mRNA methylation between (A) The lncRNA–mRNA regulatory network showing lncRNAs target at least two of TREM2, HCST and TYROBP. (B) The TF–mRNA regulatory network showing TFs target at least two of TREM2, HCST and TYROBP.
3.6. Immunological properties of TREM2, HCST and TYROBP
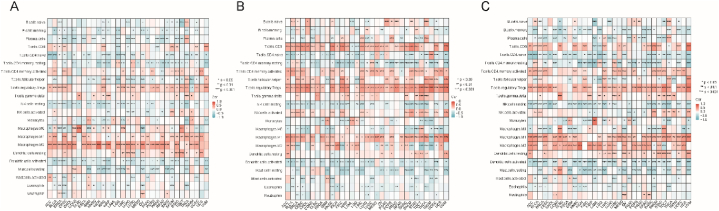
Considering that TREM2, HCST, and TYROBP are closely related to immune and inflammatory responses, we calculated the correlation between them and the level of immune cell infiltration in each tumor type, which better revealed their intrinsic connection with tumor immunity. The results (Fig. 7A–C) revealed a heterogeneous correlation between these factors and tumor-infiltrating immune cells. For example, in 27 tumor types, excluding DLBC, LAML, SKCM, TGCT, UCS, and UVM, TREM2 was negatively correlated with the abundance of M2 macrophages, whereas in 22 tumor types, TREM2 was negatively correlated with the abundance of resting memory CD4 T-cells. TREM2, HCST, and TYROBP are associated with an abundance of numerous immune cells, and this association may vary according to tumor type. Notably, all three genes strongly correlated with macrophages.
Fig. 7.
Heatmap of correlations with the abundance of 22 immune cells in each cancer type. Each column is a cancer type, and each row is a type of immune cell. Red represents positive correlation; blue represents negative correlation. (A) TREM2, (B) HCST, (C) TYROBP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Immunomodulators are critical to the response to immunotherapy, and the correlations between TREM2, HCST, and TYROBP and these immune modulators were calculated using pathway-related genes collected in previous studies. TREM2, HCST, and TYROBP were positively correlated with most immune regulators in most tumor types. It is worth noting that SMAD9 was inversely correlated with TREM2, HCST and TYROBP in most tumor types (Fig. 8A–C).
Fig. 8.
Heatmap of correlations with the immunomodulators in each cancer type. Each column is a cancer type, and each row is a type of immunomodulator. Red represents positive correlation; blue represents negative correlation. (A) TREM2, (B) HCST, (C) TYROBP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In our study, we categorized six immune subtypes (C1–C6) across samples from 33 tumor types, each displaying distinct immune characteristics within a pan-cancer context. We compared the expression levels of TREM2, HCST, and TYROBP among subtypes. According to our findings presented in Fig. 9A, subtype C6, characterized by TGF-β dominance, showed the highest expression of TREM2, HCST, and TYROBP. In contrast, subtype C5, known as the 'immunologically quiet' subtype, exhibited the lowest expression levels of these genes. This suggests a potential connection between the subdued immune response in tumors and higher expression of TREM2, HCST, and TYROBP, possibly linked to TGF-β signaling.
Fig. 9.
Differences in TREM2, HCST and TYROBP activity between immune subtypes (C1–C6). (A) Box plot of differences in TREM2, HCST and TYROBP activity between immune subtypes. The p-values between each of the two immune subtypes were less than 0.001. (B)Stacked bar graph showing the proportion of 33 tumor types in each immune subtype.
Further analysis revealed notable differences in the distribution of tumor types among these subtypes. Specifically, LUAD, MESO, LGG, and LIHC formed the majority of the samples in TGF-β-dominant immunosubtype C6. Conversely, in the immunologically quiet subtype C5, PRAD and LGG were predominantly represented (Fig. 9B). These observations underscore a broader relationship between the expression of TREM2, HCST, TYROBP, and tumor immune subtypes, enriching our understanding of tumor immunology in a pan-cancer scenario.
3.7. Pathway activity analyses of TREM2, HCST and TYROBP
For tumor-associated pathways involved in TREM2, HCST and TYROBP, we inferred pathway enrichment levels in more than 20 tumor types. Gene enrichment analysis of TREM2, HCST, and TYROBP enriched ten, 13, and 16 genes, respectively, that participate in many biological processes. There were a total of ten intersection pathways, most of which were enriched in cytokine receptor interaction, chemokine signaling pathway, T-cell receptor signaling pathway, intestinal immune network for IgA, and NOD-like receptor signaling pathway, among which TYROBP was involved in natural killer cell-mediated cytotoxicity and the B-cell receptor signaling pathway (Fig. 10A and B). These results were consistent with previous immune signature analysis results, confirming the important regulatory roles of TREM2, HCST, and TYROBP in tumor immunity.
Fig. 10.
Pathway analyses of TREM2, HCST and TYROBP. (A)Pathways enriched in more than 20 cancers in single-gene GSEA (TREM2, HCST, TYROBP). (B) Venn Diagram visualizing the interaction of enriched pathways in more than 20 cancers between TREM2, HCST and TYROBP.
3.8. Tumor mutation burden (TMB) and microsatellite instability (MSI) correlation with TREM2, HCST and TYROBP
TMB and MSI reflect the frequency of genomic mutations and are biomarkers for tumor immunotherapy [19]. Based on the immune characteristics of TREM2, HCST, and TYROBP, their association with TMB and MSI was studied. TREM2, HCST, and TYROBP are associated with TMB in some types of tumors. For TMB, all three were moderately positively correlated in CHOL, whereas TREM2 and HCST were negatively correlated in OV. TYROBP and HCST were positively correlated in UVM, TREM2 was negatively correlated in TGCT, and TYROBP was positively correlated with STAD (Fig. 11 A–C). TREM2 was positively correlated with MSI in UCS and PAAD, TYROBP and HCST were negatively correlated in STAD, and HCST was positively correlated with LUAD (Fig. 11D–F).
Fig. 11.
Tmb and MSI correlation of TREM2, HCST and TYROBP. (A) Correlation between TREM2 and TBM, (B) correlation between HCST and TBM, (C) correlation between TYROBP and TBM, (D) correlation between TREM2 and MSI (E) correlation between HCST and MSI, (F) correlation between TYROBP and MSI.
3.9. Single-cell transcriptome analysis of TREM2, HCST and TYROBP
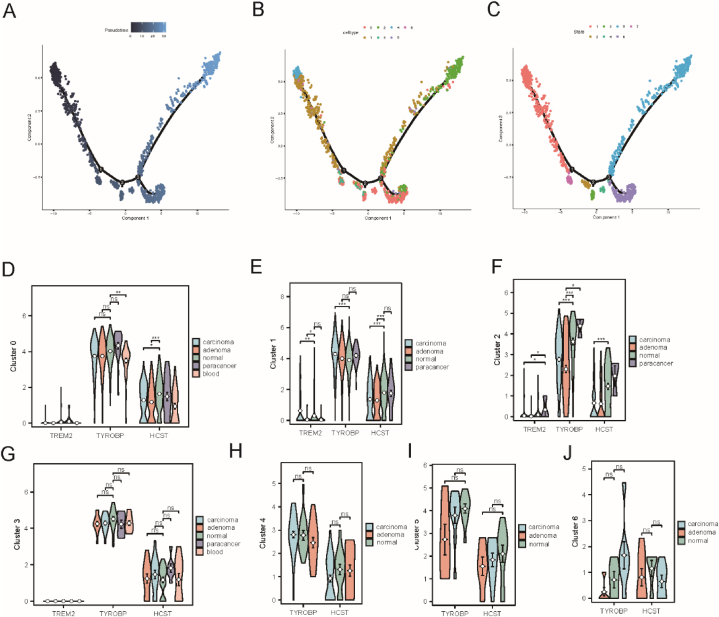
After identifying the associations of TREM2, HCST, and TYROBP with immune cells and immune-related pathways, we analyzed 13 single-cell samples (including three samples each from adenoma, blood, cancer, and normal tissues) using scRNA-seq to evaluate the activities of TREM2, HCST, and TYROBP in COAD single cells. Six cell types were identified, and the most important 5 marker genes for each cell type are displayed as dot plots (Fig. 12 A–B). The AUC score results showed that the AUC values of TREM2, HCST, and TYROBP were highest in macrophages and NK cells and lowest in the endothelium. In all cells, the AUC values of TREM2, HCST, and TYROBP were different among the adenoma, blood, cancer, normal tissue, and paracarcinoma groups (p < 0.05) (Fig. 12 C–D).
Fig. 12.
Single-cell analysis of TREM2, HCST and TYROBP. (A) UMAP map of GSE161277 cell clusters. (B) Most important 5 marker genes of 8 cell types. (C) AUC scores of TREM2, HCST and TYROBP across cell types. (D) UMAP plot of AUC score for each cell. (E) Venn diagram visualizing the intersection between GSVA results of differential pathways of macrophages between normal tissue and adenoma tissue and the enriched pathways of TREM2, HCST and TYROBP in single-gene gene set enrichment pathways (F) Bar graph visualizing the t value of GSVA of intersecting pathways.
To further explore the TREM2, HCST, and TYROBP pathways in single cells, the macrophages with the highest activity in adenoma and cancer tissues were selected for gene set variation analysis. There were differences between normal and adenoma tissues, and 90 pathways were obtained that intersected with the 10 pathways obtained in the previous GSEA enrichment analysis. Finally, eight pathways that are possibly related to TREM2, HCST, and TYROBP were identified (Fig. 12 E–F).
Pseudo-time analysis was performed to chart the developmental stages of the macrophage subpopulations, revealing that seven clusters corresponded to distinct differentiation states (Fig. 13). Early developmental stages were represented by clusters 4, 1, and 6, whereas cluster 2 encompassed cells in later stages of development (Fig. 13A–C). TYROBP expression was notably higher in clusters 0–6 (Fig. 13D–J). This suggests that the increased expression of TYROBP correlates with both the early and late stages of macrophage development and may play a role in their differentiation. HCST displays a developmental expression pattern similar to that of TYROBP, albeit at lower levels. In contrast, TREM2 expression was either low or absent in all tissues at all developmental stages. Within the single developmental trajectories, clusters 0, 1, and 2 showed marked differences in the expression of TREM2, TYROBP, and HCST among various macrophage subgroups, with the Wilcoxon test highlighting these disparities when compared with the normal group. Cluster 0 indicated significant differences in the HCST across the carcinoma, adenoma, and paracarcinoma groups. Cluster 1 revealed pronounced overexpression of TYROBP and HCST in carcinomas and a similar significant increase in TREM2 expression. Cluster 2 exhibited significant upregulation of all three genes in carcinoma tissues compared to normal tissues, and TREM2 expression differed significantly between normal and adenoma samples. The findings from these clusters revealed specific gene expression changes linked to disease states in the macrophage subsets.
Fig. 13.
Analysis of macrophage differentiation time of TREM2, HCST and TYROBP. (A) Cell differentiation time axis, (B) Distribution of macrophages in different clusters on the cell trajectory curve (C) Distribution of macrophages in different states on the cell trajectory curve (D–J) The expression of TREM2, HCST and TYROBP in cluster 0–6, and Wilcoxon test was performed between normal tissues and other pathological tissues(*p < 0.05, **p < 0.01, ***p < 0.001).
3.10. Drug Sensitivity Analysis of TREM2, HCST and TYROBP
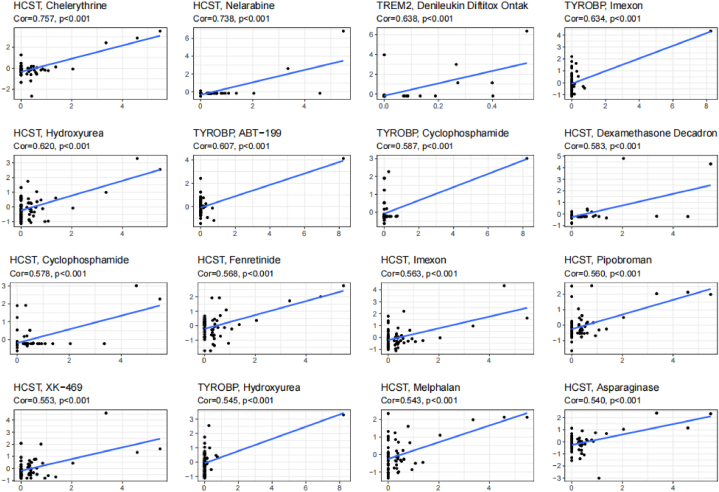
Finally, gene expression and drug sensitivity data were integrated to understand whether TREM2, HCST, and TYROBP affected patient response to anti-tumor therapy. The scatter plot shows the most significant sensitivity results to 16 drugs (Fig. 14), with TREM2 associated with sensitivity to one drug, HSCT associated with sensitivity to 11 drugs, and TYROBP associated with sensitivity to four drugs. There are a total of three drugs related to both HSCT and TYROBP (cyclophosphamide, imexon, and hydroxyurea). The mechanisms of action of these 13 drugs were focused on targeting inflammatory factors (chelerythrine and denileukin diftitox), cell cycle, apoptosis (hydroxyurea, venetoclax, fenretinide, XK-469, asparaginase, pipobroman, melphalan, and nelarabine), immunosuppression (cyclophosphamide and cecadron), and oxidative stress (imexon). This trend is consistent with previous findings, although little research has been conducted on the relationship of TREM2, HCST, and TYROBP with these drugs. These results provide preliminary evidence for the roles of TREM2, HCST, and TYROBP in tumor immunity and therapy.
Fig. 14.
Drug Sensitivity Analysis of TREM2, HCST and TYROBP. Scatter plot of drug sensitivity results in the top 16 of significance.
4. Discussion
Our study revealed that TREM2, HCST, and TYROBP undergo extensive changes in somatic cells in various tumors. The CVN landscape of TREM2 is amplified, leading to high mRNA expression levels in most cancers. This supports the notion that oncogene amplification fosters genetic heterogeneity within tumors, accelerates tumor evolution, and potentially causes RNA or protein overexpression [20,21].
Consistent with previous research, TREM2, HCST, and TYROBP were highly expressed in ESCA, GBM, HNSC, KIRC, KIRP [[22], [23], [24], [25], [26], [27]], and expressed at low levels in LUAD and LUSC. Studies have shown that their activation can mediate NK cell killing activity in lung cancer, which typically has reduced NK cell levels, possibly contributing to their low expression [28,29]; In BLCA and COAD, TREM2 is highly expressed [30,31], while HCST and TYROBP display opposite trends, potentially due to tumor heterogeneity, competitive binding of HCST and TYROBP to other receptors, or other factors that warrant further investigation.
Unsupervised consensus clustering of mRNA expression suggested that TREM2, HCST, and TYROBP mRNA expression levels may affect tumor patient survival, and further survival analysis corroborated this conclusion. High TREM2, HCST, and TYROBP expression is associated with poor prognosis in patients with LGG [12,23]. Conversely, low expression was associated with a poor prognosis in SKCM. Recent studies support our findings, suggesting that high TREM2 expression in SKCM may lead to longer survival periods and improved prognosis and that low DAP10 expression is observed in metastatic SKCM [32,33]. The expression of these three genes in LGG and SKCM exhibited a high degree of consistency, suggesting that the impact of TREM2, HCST, and TYROBP on prognosis is strongly correlated, and serves as a key predictive factor for both LGG and SKCM outcomes. However, the differential expression trends of these three genes in the context of poor prognosis in LGG and SKCM may be associated with differences in tumor types. Further studies are required to elucidate the underlying mechanisms. For LIHC, our study revealed that high TREM2 and TYROBP expression levels are associated with poor prognosis. However, some studies have suggested that TREM2 protects the liver from hepatocellular carcinoma [34], whereas others have indicated that TREM2 acts as a prognostic marker for HCC with immunosuppressive activity and promotes immune escape, correlating with disease progression [35]. These discrepancies may be related to the different stages of LIHC, where TREM2 limits inflammatory damage and tumor development in the early stages, but inhibits anti-tumor immune responses and promotes cancer progression in later stages.
We also found that the methylation levels of TREM2, HCST, and TYROBP in the promoter region of our gene could serve as biomarkers for cancer patient prognosis and are closely related to the expression levels of methylation regulator genes. Considering the prognostic impact of TREM2, HCST, and TYROBP methylation levels in multiple tumors, the simultaneous targeting of these genes may produce synergistic effects, requiring extensive basic and clinical research for validation.
Further investigation of lncRNA and TF regulation of gene expression led to the construction of lncRNA and TF mRNA regulatory networks. Notably, lncRNA MEG3 and TF SIP1 targeted TREM2, HCST, and TYROBP mRNAs in the network, suggesting that MEG3 and SIP1 may be the key lncRNAs and TF for TREM2, HCST, and TYROBP. MEG3 and SIP1 exert regulatory effects on various tumors via mechanisms involving tumor macrophage aggregation and polarization, immune infiltration, chemotherapy sensitivity, PTEN, CCL7 transcription, and p53 signaling pathways [[36], [37], [38], [39], [40]]. One study on the role of the SPI1-TYROBP-FCER1G network in osteosarcoma occurrence and prognosis and its relationship with immune infiltration integrated the GEO database [41]. However, the associations between MEG3 and TREM2, HCST, TYROBP, and between SIP1 and TREM2 and HCST have not yet been characterized. Thus, we hypothesized that MEG3 and SIP1 may also affect tumor development by regulating TREM2, HCST, and TYROBP, which warrants further investigation.
Our results highlight the strong correlations between TREM2, HCST, and TYROBP in immune-related pathways, cells, and signatures, suggesting that they play critical roles in cancer immunity. Immune signature analysis revealed that TREM2, TYROBP, and HCST are associated with numerous immune cells, varying by tumor type, and are strongly correlated with macrophages. Previous studies have reported a relationship between TREM2, HCST, and TYROBP and tumor immune infiltration. In HCC, TREM2 is predominantly expressed by macrophage subsets enriched in the tumor tissue, where TREM2+ macrophages recruit suppressive Treg cells and promote microenvironment remodeling [35]. HCST is associated with T-cell exhaustion in pediatric B-cell acute lymphoblastic leukemia [42], and TYROBP regulates macrophage polarization and the expression of immune checkpoints in osteosarcoma [43].
Further results showed that, in most tumor types, SMAD9 was negatively correlated with TREM2, HCST, and TYROBP. Studies have confirmed that SMAD9 and TGF-β are significantly downregulated in ESCA tissues [44], whereas TREM2, HCST, and TYROBP expression is significantly upregulated. Immune subtype analysis suggested that tumor immune responses with high TREM2, HCST, and TYROBP expression may be related to TGF-β. Pathway enrichment analysis also indicated that TREM2, HCST, and TYROBP mechanisms affecting tumors are related to immune-related pathways, such as cytokines, chemokines, and T-cell receptors. TGF-β, a well-known tumor immunomodulator, regulates immune cell infiltration, inflammatory cell recruitment, and chemokine-chemokine receptor interactions in the tumor microenvironment, affecting both the tumor microenvironment and tumor parenchyma. The dual role of TGF-β as both a repressor and a promoter in tumor development may be linked to differential regulation of signaling pathways by tumor-autonomous TGF-β signaling. TGF-β signaling primarily involves the TGF-β/SMAD pathway, in which SMAD9 is a receptor-regulated SMAD. The TGF-β/SMAD-dependent pathway mediates growth inhibition in early tumor stages but has also been reported to participate in tumor-promoting activity [45,46]. Combined with our results, the role of TREM2, HCST, and TYROBP in tumor immunity is accomplished by regulating tumor macrophages, and the mechanism may be closely related to TGF-β, SMAD9, and cytokines and chemokines in tumor immunity. Further research is required to elucidate their specific roles and mechanisms of action.
TMB and MSI are crucial predictive factors for immune suppression [19]. Our study suggests that TREM2, HCST, and TYROBP2 expression correlates with TMB and MSI in different tumor types, indicating that their expression levels can affect TMB and MSI in tumors, thereby influencing patient response to immune checkpoint suppression therapy. These findings provide a new reference for prognosis after immunotherapy.
Single-cell and drug sensitivity analyses confirmed our hypothesis. Single-cell results showed that TREM2, HCST, and TYROBP2 expression was highest in macrophages and NK cells, with differences across tissues. HCST and TYROBP are involved in macrophage differentiation. Pathways related to TREM2, HCST, and TYROBP2 expression still focus on cytokine receptor interactions, chemokines, T-cell receptors, and B-cell receptors. Drug sensitivity results showed that drugs highly sensitive to TREM2, HCST, and TYROBP2 were related to inflammatory factors, cell cycle, apoptosis, immunosuppression, and oxidative stress. These findings were also supported by the immunoassay results, providing a direction for drug research related to TREM2, HCST, and TYROBP2.
5. Conclusion
In conclusion, our study revealed that TREM2, HCST, and TYROBP play essential roles in tumor immunity, and that their interactions with various immune-related pathways, cells, and signatures are crucial for understanding cancer development. The expression levels of these genes affect the survival of patients with tumors, and their methylation levels can be used as biomarkers for cancer prognosis. The regulatory effects of MEG3 and SIP1 on TREM2, HCST, and TYROBP require further investigation, as these factors may influence tumor development. Moreover, the role of TGF-β, SMAD9, cytokines, and chemokines in tumor immunity, in conjunction with TREM2, HCST, and TYROBP, warrants additional research to elucidate the specific mechanisms at play.
Our findings also emphasize the importance of TMB and MSI as predictive factors for immune suppression and suggest that targeting TREM2, HCST, and TYROBP simultaneously may have synergistic effects. Single-cell and drug sensitivity analyses confirmed the involvement of these genes in macrophage differentiation and their relationship with various immune-related pathways. This study provides valuable insights for future drug research and development, potentially leading to novel therapeutic strategies targeting TREM2, HCST, and TYROBP for cancer treatment.
Data availability statement
This study conducted analyses on publicly available datasets. The sources of these datasets are described and provided at the respective locations in the Methods section. All datasets utilized in this study are openly accessible and available for public use.
Funding
This work was supported by Hunan Cancer Hospital Climb Plan (No. IIT2021001), Science and Technology Program of Henan, China (No.192102310161 and NO.182102310291), Natural Science Foundation of Hunan Province (No. 2023JJ40877) and Hunan Provincial Education Commission Foundation (No.19B068, No.20A056).
CRediT authorship contribution statement
Piao Zheng: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Funding acquisition, Formal analysis. Yejun Tan: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Formal analysis, Data curation. Qing Liu: Writing – review & editing, Conceptualization. Changwu Wu: Writing – review & editing. Jing Kang: Writing – review & editing. Shuzhi Liang: Writing – review & editing. Lemei Zhu: Writing – review & editing, Funding acquisition. Kuipo Yan: Writing – review & editing, Funding acquisition. Lingfeng Zeng: Writing – review & editing, Funding acquisition. Bolin Chen: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Lancet T. Global cancer: overcoming the narrative of despondency. Lancet. 2023;401(10374):319. doi: 10.1016/S0140-6736(23)00234-9. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., et al. Global cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Maiorino L., et al. Innate immunity and cancer Pathophysiology. Annu. Rev. Pathol. 2022;17:425–457. doi: 10.1146/annurev-pathmechdis-032221-115501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu T., et al. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021;14(1):98. doi: 10.1186/s13045-021-01103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond T.R., Marsh S.E., Stevens B. Immune signaling in neurodegeneration. Immunity. 2019;50(4):955–974. doi: 10.1016/j.immuni.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konishi H., Kiyama H. Microglial TREM2/DAP12 signaling: a Double-Edged Sword in neural diseases. Front. Cell. Neurosci. 2018;12:206. doi: 10.3389/fncel.2018.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basha S.C., Ramaiah M.J., Kosagisharaf J.R. Untangling the role of TREM2 in Conjugation with microglia in Neuronal Dysfunction: a hypothesis on a novel pathway in the Pathophysiology of Alzheimer's disease. J Alzheimers Dis. 2023 doi: 10.3233/JAD-221070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binnewies M., et al. Targeting TREM2 on tumor-associated macrophages enhances immunotherapy. Cell Rep. 2021;37(3) doi: 10.1016/j.celrep.2021.109844. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., et al. Immunosuppressive TREM2(+) macrophages are associated with undesirable prognosis and responses to anti-PD-1 immunotherapy in non-small cell lung cancer. Cancer Immunol. Immunother. 2022;71(10):2511–2522. doi: 10.1007/s00262-022-03173-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassez A., et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27(5):820–832. doi: 10.1038/s41591-021-01323-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y., et al. The immune-related gene HCST as a novel biomarker for the Diagnosis and prognosis of clear cell renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.630706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J., et al. Elevated TYROBP expression predicts poor prognosis and high tumor immune infiltration in patients with low-grade glioma. BMC Cancer. 2021;21(1):723. doi: 10.1186/s12885-021-08456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C., et al. Pan-cancer analyses reveal molecular and clinical characteristics of cuproptosis regulators. iMeta. 2023;2(1):e68. doi: 10.1002/imt2.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830 e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng D., et al. Tumor microenvironment Characterization in Gastric cancer Identifies prognostic and Immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 17.Pinero J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., et al. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis. Signal Transduct Target Ther. 2022;7(1):60. doi: 10.1038/s41392-022-00881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z., et al. Correlation between TMB and MSI in patients with solid tumors. J. Clin. Oncol. 2020;38(15_suppl) [Google Scholar]
- 20.Joshi R.S., et al. High tumor amplification burden is associated with TP53 mutations in the pan-cancer setting. Cancer Biol. Ther. 2022;23(1):1–6. doi: 10.1080/15384047.2022.2128608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero Llobet S., et al. An mRNA expression-based signature for oncogene-induced replication-stress. Oncogene. 2022;41(8):1216–1224. doi: 10.1038/s41388-021-02162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duggan S.P., et al. siRNA Library screening Identifies a Druggable immune-signature Driving esophageal adenocarcinoma cell growth. Cell Mol Gastroenterol Hepatol. 2018;5(4):569–590. doi: 10.1016/j.jcmgh.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu M., et al. TREM2 is associated with tumor immunity and implies poor prognosis in glioma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1089266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang b., et al. 2021. TREM2 Is a Prognostic Biomarker and Correlates in Immune Infiltrates in Kidney Renal Papillary Cell Carcinoma and Liver Hepatocellular Carcinoma. [Google Scholar]
- 25.Ford J.W., et al. Tumor-infiltrating myeloid cells Co-express TREM1 and TREM2 and Elevated TREM-1 Associates with disease progression in renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.662723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., et al. The roles and potential mechanisms of HCST in the prognosis and immunity of KIRC via comprehensive analysis. Am J Transl Res. 2022;14(2):752–771. [PMC free article] [PubMed] [Google Scholar]
- 27.Lv X.Q., et al. Higher TYROBP and lower SOX6 as predictive biomarkers for poor prognosis of clear cell renal cell carcinoma: a pilot study. Medicine (Baltim.) 2022;101(51) doi: 10.1097/MD.0000000000030658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donatelli S.S., et al. TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014;111(11):4203–4208. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhi L., et al. A chimeric switch-receptor PD1-DAP10-41BB augments NK92-cell activation and killing for human lung Cancer H1299 Cell. Biochem. Biophys. Res. Commun. 2022;600:94–100. doi: 10.1016/j.bbrc.2022.02.052. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., et al. Combined single-cell RNA-seq and bulk RNA-seq to analyze the expression and role of TREM2 in bladder cancer. Med. Oncol. 2022;40(1):23. doi: 10.1007/s12032-022-01885-8. [DOI] [PubMed] [Google Scholar]
- 31.Yang T., et al. Platinum-based TREM2 inhibitor Suppresses tumors by remodeling the immunosuppressive microenvironment. Angew Chem. Int. Ed. Engl. 2023;62(2) doi: 10.1002/anie.202213337. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X., et al. TREM2 as a potential immune-related biomarker of prognosis in patients with skin cutaneous melanoma microenvironment. Dis. Markers. 2023;2023 doi: 10.1155/2023/8101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirjacic Martinovic K., et al. Decreased expression of pSTAT, IRF-1 and DAP10 signalling molecules in peripheral blood lymphocytes of patients with metastatic melanoma. J. Clin. Pathol. 2016;69(4):300–306. doi: 10.1136/jclinpath-2015-203107. [DOI] [PubMed] [Google Scholar]
- 34.Esparza-Baquer A., et al. TREM-2 defends the liver against hepatocellular carcinoma through multifactorial protective mechanisms. Gut. 2021;70(7):1345–1361. doi: 10.1136/gutjnl-2019-319227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L., et al. Integrated analysis highlights the immunosuppressive role of TREM2(+) macrophages in hepatocellular carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.848367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du Y., et al. LncRNA MEG3 promotes cisplatin sensitivity of cervical cancer cells by regulating the miR-21/PTEN axis. BMC Cancer. 2022;22(1):1145. doi: 10.1186/s12885-022-10188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Z.F., et al. UXT, a novel DNMT3b-binding protein, promotes breast cancer progression via negatively modulating lncRNA MEG3/p53 axis. Mol Ther Oncolytics. 2022;24:497–506. doi: 10.1016/j.omto.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z., et al. LINC01094/SPI1/CCL7 Axis promotes macrophage Accumulation in lung adenocarcinoma and tumor cell Dissemination. J Immunol Res. 2022;2022 doi: 10.1155/2022/6450721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng H., et al. SPI1 is a prognostic biomarker of immune infiltration and immunotherapy efficacy in clear cell renal cell carcinoma. Discov Oncol. 2022;13(1):134. doi: 10.1007/s12672-022-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J., et al. Comprehensive analysis of immune Implications and prognostic value of SPI1 in Gastric cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.820568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., et al. The role of SPI1-TYROBP-FCER1G network in oncogenesis and prognosis of osteosarcoma, and its association with immune infiltration. BMC Cancer. 2022;22(1):108. doi: 10.1186/s12885-022-09216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi N., et al. DAP10 predicted the outcome of pediatric B-cell acute lymphoblastic leukemia and was associated with the T-cell exhaustion. J Oncol. 2021;2021 doi: 10.1155/2021/4824868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang T., et al. TYROBP, TLR4 and ITGAM regulated macrophages polarization and immune checkpoints expression in osteosarcoma. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-98637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song W., et al. Comprehensive analysis of the expression of TGF-beta signaling regulators and prognosis in human esophageal cancer. Comput. Math. Methods Med. 2021;2021 doi: 10.1155/2021/1812227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Pang Y., Moses H.L. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31(6):220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson C., et al. TGF-beta: a master immune regulator. Expert Opin. Ther. Targets. 2020;24(5):427–438. doi: 10.1080/14728222.2020.1744568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study conducted analyses on publicly available datasets. The sources of these datasets are described and provided at the respective locations in the Methods section. All datasets utilized in this study are openly accessible and available for public use.