Abstract
Background
Skin cutaneous melanoma (SKCM) is one of the most lethal skin malignancies worldwide. Sphingosine 1-phosphate (S1P) regulates tumor cells through S1P receptors (S1PRs). Unlike S1PR1/2/3/5, whose anti-apoptotic effects have been widely studied, the regulatory effect of S1PR4 on tumors has not been studied extensively. In this study, we aimed to investigate the correlation between S1PR4 expression and survival, clinical manifestations, tumor microenvironment, and immune infiltration in patients with SKCM.
Results
Low S1PR4 expression was associated with poor prognosis in patients with SKCM. Patients in the high-expression group had significantly longer disease survival and progression-free survival than those in the low-expression group.
Conclusion
High S1PR4 expression was highly associated with better prognosis and milder clinical manifestations; thus, S1PR4 may be used as a prognostic marker to help physicians monitor patients with SKCM.
Keywords: S1PR4, Skin cutaneous melanoma, Prognosis, Immunotherapy
1. Introduction
Skin cutaneous melanoma (SKCM) is one of the most common malignant dermatological tumors. Over 50,000 people die of SKCM annually [1], accounting for 0.7% of all cancer-related deaths and 72% of all skin malignancies [2]. The incidence and mortality rates of cutaneous melanoma vary among countries. The incidence of melanoma is higher in fair-skinned individuals, with an annual increasing trend. A systematic review of information from 20 countries found that personal and family histories of melanoma were associated with sunburn, tanning, melanocytic nevi, dysplastic nevi, and cutaneous melanoma and were positively correlated with socioeconomic status [3]. The clinical diagnosis of melanoma is not challenging. Owing to its prominent morphological features, 90% of SKCM cases can be accurately diagnosed by experienced physicians in the clinic via visual diagnosis or dermoscopy. Local surgery is the first-choice treatment for SKCM in the early stages, whereas adjuvant drugs or immunotherapy are needed for patients with stage 2 or higher. Most patients with clinical diagnoses miss the opportunity for early local treatment.
Sphingolipids, including ceramide and sphingosine-1-phosphate (S1P), play opposing roles in regulating tumor apoptosis and survival. Ceramide is hydrolyzed by ceramidase to produce sphingosine, which is phosphorylated by sphingosine kinase to produce S1P. S1P binds to five specific G protein-coupled receptors, S1PR1–5, to play various biological roles. One study found that sphingolipid metabolism is strongly altered in cutaneous melanoma [4]. The expression and activity of several metabolic enzymes are disordered. , which limits the accumulation of ceramide and anti-tumor metabolites, and promoting the production of S1P, a tumor metabolite. S1P regulates a series of cellular processes by activating S1PRs and their downstream pathways, such as cell proliferation, survival and migration, immune cell recruitment, angiogenesis, and lymph angiogenesis pathways.
Abnormal S1P metabolism, receptor expression, and signal transduction are associated with the initiation, progression, and prognosis of various tumors [[5], [6], [7], [8]]. Targeting the S1P receptor potentially affects the development of skin cancer [9]. Since S1PR4 is mainly expressed in immune cells, S1PR4 may play an important role in the immune process of tumors. Studies have shown that S1PR4 can affect tumor growth rate and CD8+ T cell abundance and play a role in a variety of tumors [[10], [11], [12]]. Therefore, studying the molecular mechanisms underlying the occurrence and development of SKCM and exploring new molecular diagnostic markers and immunosuppression sites for early diagnosis, disease progression, and new inhibition targets are of great significance.
To investigate the expression of S1PR4 in melanoma and its potential clinical value, this study aimed to analyze the expression and functional enrichment of S1PR4 and tumor-infiltrating immune cells and their prognostic role in 472 melanoma patients using The Cancer Genome Atlas (TCGA) database. Moreover, the relationship between the S1PR4 expression level and drug sensitivity was assessed. Our study is critical for understanding the therapeutic potential of targeting S1PR4 in cancer therapy and developing personalized cancer drugs.
2. Results
2.1. S1PR4 expression in melanoma and effect on survival and prognosis
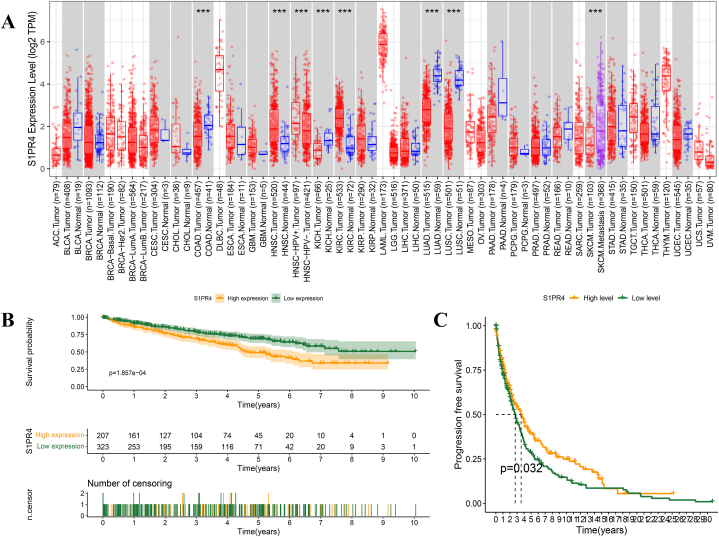
The Timer 2.0 public database showed that the S1PR4 mRNA expression in eight tumor types differed from that in normal tissues adjacent to the tumor, and the high or low expression of S1PR4 mRNA was heterogeneous across different tumor types (Sup Table. 1). The number of normal melanoma samples was too small to demonstrate significance. S1PR4 expression was significantly reduced in primary tumor tissues compared with that in metastatic tumor tissues (Fig. 1A, Sup Fig. 1). The expression level of S1PR4 in tumor tissues was lower than that in normal tissues.
Fig. 1.
Sphingosine 1-phosphate receptor 4 (S1PR4) expression in melanoma and its effect on survival and prognosis. (A) Comparison of S1PR4 expression in 33 tumors and their adjacent tissues. Relationship between high and low expression of S1PR4 and survival (B) and progression-free survival (C) in patients with skin cutaneous melanoma (SKCM). Statistical significance was calculated using Wilcoxon test and annotated as *p < 0.05, **p < 0.01, ***p < 0.001. Red, tumor; blue, normal; purple, metastasis of SKCM.
We mined TCGA database to explore the impact of S1PR4 on the prognosis of patients with melanoma. Kaplan–Meier curves showed that low S1PR4 expression was associated with a poor prognosis in patients with SKCM (p = 0.007; Fig. 1B). Progression-free survival (PFS) analysis showed that, compared with those in the S1PR4 low-expression group, the patients in the high-expression group had significantly prolonged PFS and a reduced risk of disease progression or death (p = 0.032; Fig. 1C).
2.2. Relationship between S1PR4 expression and clinical manifestations of SKCM
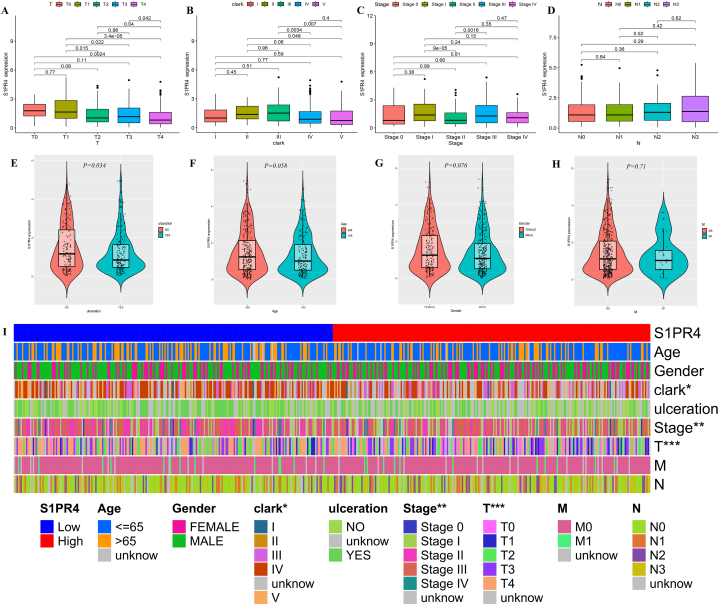
We investigated the correlation between different clinical traits and S1PR4 expression to elucidate the cause of the poor prognosis in patients with SKCM. S1PR4 expression decreased with increasing T stage (p < 0.001; Fig. 2A). There was no significant difference in S1PR4 expression between Clark grades I and III, with a significant downward trend for grades higher than III (p < 0.01; Fig. 2B). The expression of S1PR4 in Stage II was significantly lower than that in Stages I and III, and there was no significant difference from that in Stage IV (p < 0.01; Fig. 2C). Ulcerated lesions also resulted in reduced S1PR4 expression (p = 0.034; Fig. 2E). These results differ slightly from previous results. This may be because of the comprehensive scoring of multiple factors at the clinical stage, which is affected by multiple factors unrelated to S1PR4 expression. N score (p > 0.05; Fig. 2D) at Tumor Node Metastasis (TNM) staging, age (p = 0.058; Fig. 2F), sex (p = 0.076; Fig. 2G), and M score (p = 0.071; Fig. 2H) were not significantly correlated with S1PR4 expression. We then compared the clinical parameters of the groups with high and low S1PR4 expression and found significant differences in the Clark grades, stages, and T-scores between the two groups (Fig. 2I).
Fig. 2.
Relationship between sphingosine 1-phosphate receptor 4 (S1PR4) expression and clinical manifestations of skin cutaneous melanoma (SKCM). Effects of T (A) stages; Clark grade (B); clinical stage (C); N (D) stages; ulceration (E); age (F); sex (G); M (H) stages on the expression of S1PR4. (I) Distribution of each clinical trait in S1PR4 high- and low-expression groups is shown using thermography. Statistical significance is indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.
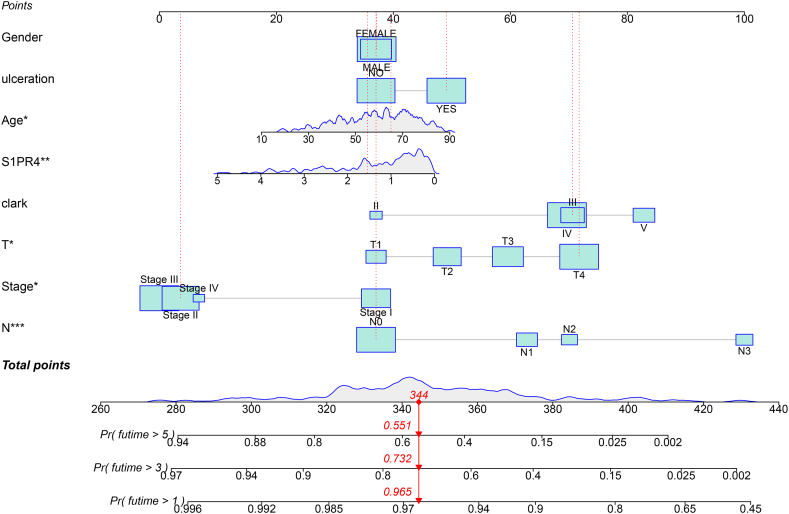
Logistic regression analysis was used to construct the Nomo plot (Fig. 3). To estimate the risk of SKCM for different S1PR4 expression levels and clinical traits, the observed values of each variable were assigned corresponding scores, and the sum of the scores for each variable is shown in the Nomo plot. An increase in age and T and N scores was positively correlated with the risk score, and the expression of S1PR4 was negatively correlated with the risk score.
Fig. 3.
Visualization of impact of each clinical trait on survival prognosis using Nomo plots, with corresponding risk coefficient predicted by sum of scores for each clinical trait. Statistical significance is indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.
2.3. Co-expression gene and functional enrichment analysis of S1PR4
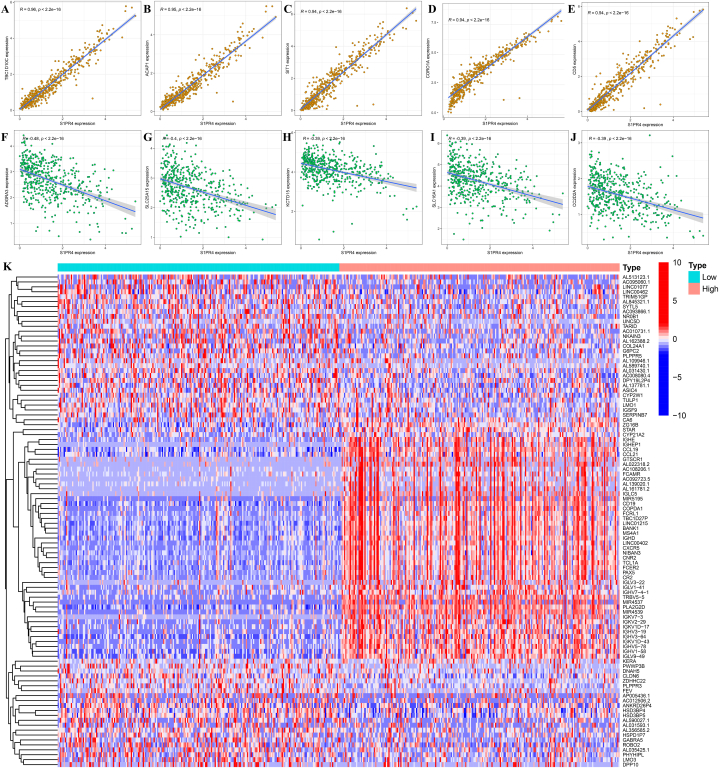
The results of the correlation coefficient analysis were filtered based on the absolute value of the correlation coefficient |log2(fold change)| > 0.3 (p < 0.001), and five genes with the strongest positive and negative correlations were selected for visualization. The five genes with the highest positive correlation were TBC1D10C (Cor = 0.96; Fig. 4A), ACAP1 (Cor = 0.95; Fig. 4B), Sit1 (Cor = 0.94; Fig. 4C), CORO1A (Cor = 0.94; Fig. 4D), and CD5 (Cor = 0.94; Fig. 4E), whereas the genes with the highest negative correlations were ADGRA3 (Cor = −0.48; Fig. 4F), SLC23A15 (Cor = −0.4; Fig. 4G), KCTD15 (Cor = −0.39; Fig. 4H), SLC16A1 (Cor = −0.39; Fig. 4I), and CC2D2A (Cor = −0.39; Fig. 4J). We then divided the samples into high and low S1PR4 expression groups and visualized the significantly different co-expressed genes using a heat map (Fig. 4K).
Fig. 4.
Analysis of co-expressed genes of sphingosine 1-phosphate receptor 4 (S1PR4). The five genes most positively associated with S1PR4 (A–E) and those most negatively associated with S1PR4 (F–J). Correlation is expressed as an R-value (positive correlation: R > 0; negative correlation: R < 0). p-value <0.05 represents significant difference. (K) Expression of all co-expressed genes at low and high levels of S1PR4 expression is depicted using a heat map with red indicating high expression and blue indicating low expression.
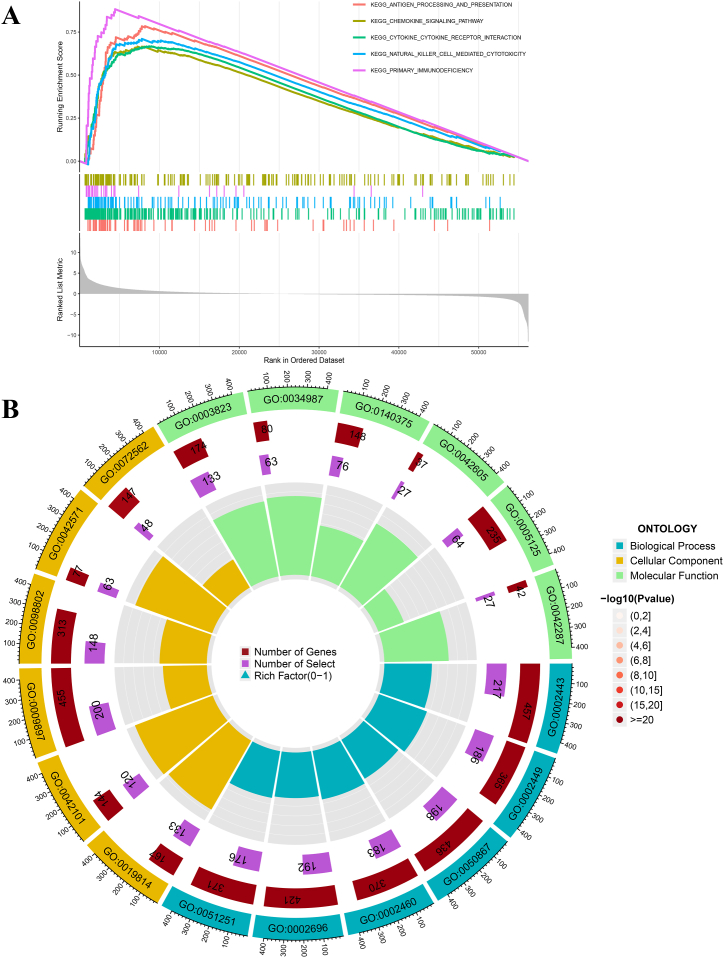
To determine the potential functions and related pathways of S1PR4 and its co-expressed genes, we divided the samples into high- and low-expression groups and performed a gene set enrichment analysis (GSEA) on the gene sets for the KEGG pathway. The first five pathways of enrichment: antigen processing and presentation, chemokine signaling, cytokine receptor interaction, natural killer cell-mediated cytotoxicity, and primary immunodeficiency, were significantly active when S1PR4 was highly expressed (Fig. 5A). Gene ontology (GO) enrichment analysis was performed for the previously acquired genes whose expression differed between the high and low S1PR4 expression groups, visualizing the functions of the first six dimensions of BP (Biological Process), CC (Cellular Component), and MF (Molecular Function) as circles (Fig. 5B). The cycle graphs show the function encoding, number of genes, number of selections, and rich factor (0–1) from the outside to the inside. The results showed a significant enrichment of immune receptors and immune cells.
Fig. 5.
Functional enrichment of sphingosine 1-phosphate receptor 4 (S1PR4) co-expressed genes. (A) Gene set enrichment analysis (GSEA) of S1PR4 co-expressed genes in The Cancer Genome Atlas (TCGA) dataset revealed the five most prominent signaling pathways. (B) Gene ontology (GO) enrichment analysis of differentially expressed genes between high and low levels of S1PR4 expression.
2.4. Correlation of S1PR4 expression level with proportion of tumor-initiating cells
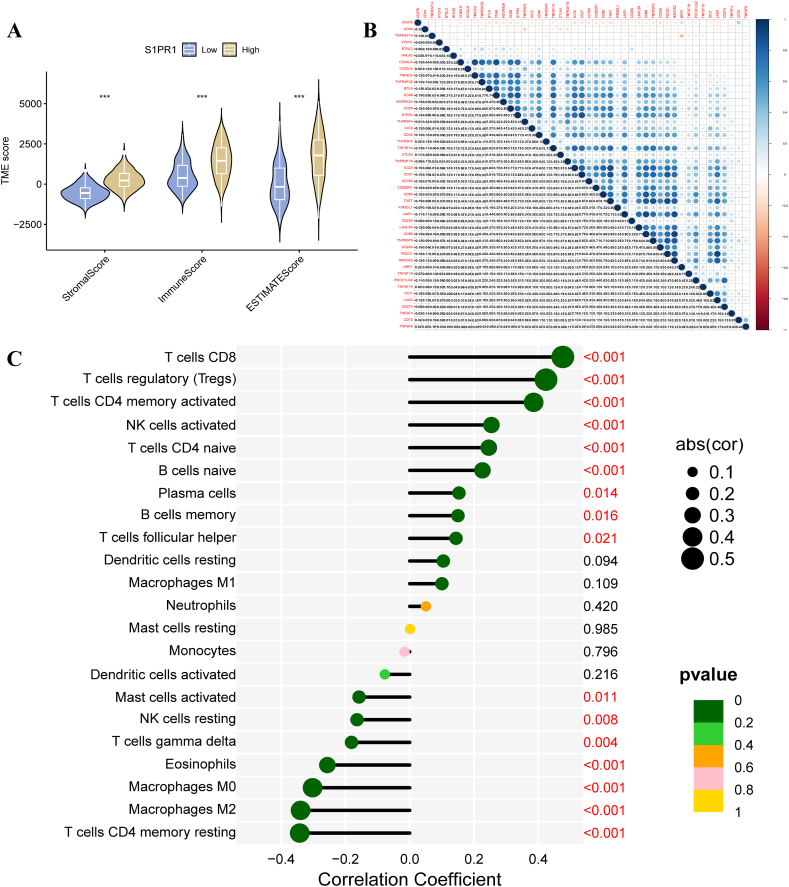
ESTIMATE calculations showed that the stromal, immune cell, and ESTIMATE scores were significantly higher in the tumor tissues of patients in the S1PR4-HIGH-EXPRESSION group than in those in the low-expression group (p < 0.001; Fig. 6A). We performed correlation tests on 47 common immune checkpoint genes and S1PR4. S1PR4 was positively correlated with most immune checkpoint genes (Fig. 6B). The results showed that S1PR4 was only negatively correlated with CD276 (R = −0.237, p = 2.04E−07) and positively correlated with other checkpoint genes. CD27 (R = 0.913, p = 1.28E−183), TIGIT (R = 0.865, p = 4.52E−142), PDCD1 (R = 0.852, p = 1.57E−133), CD48 (R = 0.835237, p = 2.26E−123) were the most relevant genes. Among them, higher expression of CD3, CD4, and CD8 suggested longer survival time and longer progression-free survival (Sup Fig. 2, Fig. 3). The CIBERSORT algorithm was used to analyze the proportion of infiltrating immune subsets, correlation between high and low expression of S1PR4, and content of infiltrating immune cells. T CD8 (R = 0.48, p = 5.8E−16), T regulatory (R = 0.42, p = 1.2E−12), and T CD4 memory activated cell (R = 0.39, p = 1.5E−10) levels were positively correlated with S1PR4 expression, while CD4 memory resting (R = −0.34, p = 1.8E−08), M2 macrophage (R = −0.34, p = 2.3E−08), and M0 macrophage (R = −0.3, p = 7.8E−07) levels were significantly negatively correlated with S1PR4 expression (Fig. 6C, Sup Fig. 4).
Fig. 6.
Effect of sphingosine 1-phosphate receptor 4 (S1PR4) expression on tumor microecology and immune infiltration in melanoma samples. (A) Effect of high and low S1PR4 expression on stromal, immune, and mixed ESTIMATE scores; (B) correlation between S1PR4 and common immune checkpoint genes; and (C) correlation between 22 tumor-initiating cells (TICs) and S1PR4 expression in skin cutaneous melanoma (SKCM) tumor samples (*p < 0.05; **p < 0.01; ***p < 0.001).
2.5. Co-expression of S1PR4 and immune-checkpoint genes and therapeutic sensitivity
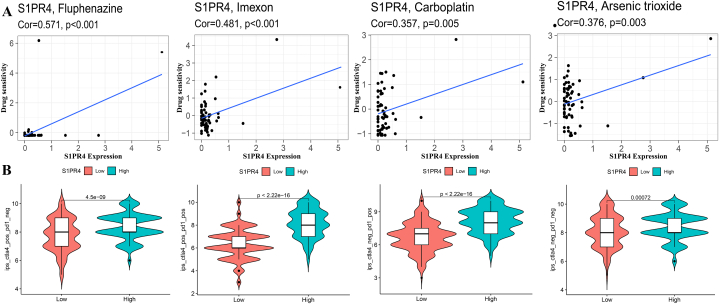
The correlation of S1PR4 expression level with drug sensitivity was similarly analyzed, and the four drugs showing a high correlation are shown in Fig. 7A. Higher S1PR4 expression corresponded with a higher sensitivity of tumor cells to chemotherapeutic drugs. The results showed that the most sensitive drugs to S1PR4 were Fluphenazine (R = 0.571, p < 0.001), Imexon (R = 0.481, p < 0.001), Carboplatin (R = 0.357, p = 0.005) and Arsenic trioxide (R = 0.376, p = 0.003).
Fig. 7.
Relationship of sphingosine 1-phosphate receptor 4 (S1PR4) expression with drug and immunotherapy sensitivity. (A) Four chemotherapeutics the most strongly correlated with S1PR4 expression all had positive correlations. (B) Effect of S1PR4 expression on CTLA4 and/or PD-1 immunotherapy. Correlation was expressed by R-value (positive correlation: R > 0; negative correlation: R < 0), and statistical significance was expressed via p-value, with p < 0.05 representing a significant difference.
We explored the effect of S1PR4 on the sensitivity of both CTLA-4 and PD-1 ICIS (Fig. 7B). Patients with a high S1PR4 expression level were more sensitive to either single CTLA-4 (p = 4.5e-09) or either single PD-1 (p < 2.2e-16) or composite immunotherapy (p < 2.2e-16).
3. Discussion
Sphingolipids regulate various biological processes by controlling signal transduction networks in cancer cells. In contrast to ceramide, which mediates cancer cell apoptosis, S1P produced by ceramide metabolism has anti-apoptotic and pro-tumorigenic effects. S1P acts through five S1P receptors (S1PR1–5), which affect the growth and drug resistance of tumor cells. However, each S1PR can direct different biological signals, thereby performing different functions; hence, therapies that neutralize S1P or target sphingosine kinases have limited effectivity.
BRAF inhibitors are currently used as immunotherapy for malignant melanoma and can significantly improve the immune response and overall survival rate of patients with malignant melanoma. Sandra et al. [13] reported S1P accumulation in melanomas. Garandeau et al. [9] found that patients with melanoma sensitive to BRAF inhibitors tend to have a significantly higher sphingolipid profile and ceramide ratio than healthy people, with a significantly lower expression of S1PR1 and S1PR3. Therefore, the S1P/S1PR axis may be related to the sensitivity of melanoma cells to BRAF inhibitors or may have independent therapeutic implications. However, in our study, we focused on the changes in S1PR4 expression instead of other S1P receptors. In contrast to the general distribution of S1PR1–3, S1PR4 is mainly expressed in immune cells but is rarely involved in lymphocyte transport. S1PR4-deficient mice showed no defect in lymphocyte composition and no obvious change in CD8+ T-cell migration [14,15]. Moreover, inhibition of S1PR4 did not interfere with lymphocyte accumulation in tumor tissues by affecting lymphocyte transport, leading to immune paralysis. Therefore, compared with other S1PR S1PR4 has potential immunotherapeutic significance.
Olesch et al. [10] reported that S1PR4 contributes to the survival and progression of breast cancer cells and negatively affects chemotherapy sensitivity. Cheng et al. [11] found that S1PR4 promoted prostate cancer invasion and metastasis. Moreover, S1PR4 has been shown to play a role in the anti-tumor immune response by regulating dendritic cells and macrophages, and the co-inhibition of SPHK1/PD-1 improved melanoma control in mice [12]. On this basis, Olesch et al. [10] found that S1PR4 ablation did not enhance the efficacy of PD-1 blockade. In our study, S1PR4 showed low or high expression in different tumor types; however, in melanoma, there was significant S1PR4 low expression in the primary tissue. The low expression of S1PR4 was associated with disease progression and poor prognosis in patients with SKCM; this, result was also supported by the positive correlation between levels of S1PR4 and CD4 and CD8 (Sup Fig. 2, Fig. 3).These findings seem to contradict those of previous studies [16,17] that the S1PR pan-antagonist (FYT-p) can slow melanoma growth and metastasis. We attribute this discrepancy to the fact that previous studies focused on the roles of S1PR1 and S1PR3 in anti-apoptosis and the promotion of neovascularization while ignoring the potential significance of S1PR4. Thus, the expression of S1PR4 may be used as a diagnostic criterion and an auxiliary factor for prognosing disease, and developing S1PR4 agonists may improve the prognosis of patients with SKCM.
The association between the expression of S1P and its receptor and the clinical traits of patients with SKCM was previously unreported. Our study found that age and sex did not influence S1PR4 expression, which is consistent with the previous risk factors for melanoma [18]. S1PR4 expression also did not differ between patients with M and N stages in terms of tumor stage, which may be related to the fact that most patients with SKCM are diagnosed and treated before the appearance of lymph node and distant metastases (i.e., stages N0 and M0). T stage (tumor thickness) plays a key role in assessing disease severity and prognosis in patients with SKCM. In our study, the T stage was inversely associated with S1PR4 expression, and ulcer formation resulted in reduced S1PR4 expression. Accordingly, there was a significant decrease in S1PR4 expression when the clinical stage reached Stage II. In contrast, there was no significant change in Stages III and IV, which was associated with how melanoma was graded, regardless of T stage. Stage II was assigned when lymph node (N) and distal metastasis (M) were absent. This broad grading approach may have led to significant bias in the statistical analysis. Clark staging showed a significant decrease in S1PR4 expression after tumor invasion into the dermis reticulum (Clark grade IV), suggesting a correlation between the tumor thickness, extent of invasion, and prognosis.
Co-expression studies have identified numerous genes with positive and negative correlations with S1PR4; the two genes with the strongest positive and negative correlations are TBC1D10C and ADGRA3, respectively. TBC1D10C belongs to the EPI64 family and promotes tumor development by inhibiting T cell-mediated anti-tumor immunity [19]. TBC1D10C deficiency can increase the number and cytotoxic activity of CD8+ T cells without affecting CD4+ T cells. Meanwhile, TBC1D10C is involved in lymphocyte activation and immune cell proliferation and can enhance the phagocytosis of macrophages [20]. However, we found that S1PR4 expression was positively correlated with the number of CD8+ T, CD4+ T, and Treg cells and negatively correlated with macrophage expression. At the same time, higher expression of CD4 and CD8 suggested longer survival time and longer progression-free survival. This contradiction suggests that S1PR4 and TBC1D10C may balance the immune environment in vivo through antagonism, whereas patients with melanoma present with S1PR4 expression inhibition, resulting in reduced numbers of CD8+ T cells and a decreased anti-tumor capacity. Therefore, S1PR4 may improve the prognosis of melanoma patients by enhancing the transformation and activation of CD8+ T cells; however, this requires further experimental confirmation.
ADGRA3, a subtype of adhesion G-protein-coupled receptors, plays a role in tumor proliferation, invasion, and metastasis [21]. Its upregulation promotes the formation of medullary sarcomas [22] and is associated with worse PFS in endometrial cancer [23]; however, its suppressed expression in patients with colorectal cancer [24] is associated with poor prognosis. S1PR4, a member of the G protein-coupled receptor family, also exhibits heterogeneity depending on the tumor type. In our study, S1PR4 expression was associated with a good prognosis in melanoma.
To investigate the possible molecular mechanisms of S1PR4 and its co-expressed genes in SKCM, we performed a GO analysis and GSEA using data from public databases. The upregulation of S1PR4 and its co-expressed genes was associated with NK cell-mediated cytotoxicity and primary immunodeficiency. GO enrichment analysis showed that co-expressed genes were associated with the positive regulation of white blood cells and lymphocyte activation, regulation of immunoglobulin receptor binding, enhancement of immune receptor activity, and aggregation in the T cell receptor complex. Therefore, S1PR4 may alter disease prognosis mainly through the immune microenvironment. In this study, we found that the abundances of both immune and stromal cells were significantly increased in S1PR4-overexpressing samples. The tumor microenvironment (TME) plays a critical role in the progression and prognosis of malignancies [25], and immune and stromal cells have a major impact on tumor invasion and prognosis. High S1PR4 expression may inhibit disease progression and improve prognosis by increasing the abundance of S1PR4.
Immune checkpoint inhibitors play an important role in anti-tumor immunity by correcting immune disorders, activating CD8+ T cells, and exerting cytotoxic effects. Immune checkpoint inhibitors, such as anti-CTLA4 and anti-PD-1 antibodies, have revolutionized the treatment of many cancers, particularly advanced melanoma [26]. We found that S1PR4 was positively correlated with the most common immune checkpoint genes, including CTLA4; however, when S1PR4 was highly expressed, the immunotherapeutic efficacy of anti-CTLA4 and anti-PD-1 antibodies, alone or in combination, was superior to that of the low-expression group. This result partly supports the hypothesis that S1PR4 antagonizes pro-tumor signals and maintains the immune balance. In addition to immunotherapy, an increase in S1PR4 expression can improve the sensitivity to chemotherapeutic agents, therapeutic effect, and prognosis of patients.
Specific chemotherapeutic agents, such as Carboplatin, induce metastatic behavior through stromal and tumor-derived cytokine and angiogenic factor signaling, and researchers have identified sphingosine kinase and S1P lyase as potential targets for improving the efficacy of cisplatin in human tumors [27]. S1PR1 antibody can enhance the cytotoxicity of Carboplatin on breast cancer cells and also enhance the anti-proliferation effect of S1P [28]. Arsenic trioxide has been found to have a wide range of anti-tumor activities, and studies have found that the anti-cancer activity of Arsenic trioxide may be achieved by altering the sphingolipid pathway in human multiple myeloma cell lines and gastric cancer cell lines [29]. Several drugs most related to S1PR4 unearthed in this study still lack relevant studies in melanoma, and further studies are needed to confirm.
This study had some limitations. First, the cancer data retrieved from TCGA database were almost exclusively from white populations, and the effect of skin color on melanoma was significant. For example, individuals with darker skin tones were more likely to develop mucosal melanoma, a type of cancer not included in the database. Therefore, the results may have been biased. Second, we focused on the total cellular content, pathway expression, disease presentation, and prognosis; thus, the interactions between upstream regulators, downstream effector molecules, and pathway opening were not investigated. Finally, no validation trials have been conducted to verify the effects of S1PR4 on melanoma-related clinical features, prognosis, and immune infiltration; mature preparations that specifically antagonize S1PR4 (rather than the pan-antagonists of the S1PR family) remain lacking.
In future studies, we hope to clinically apply our findings to SKCM treatment by establishing cell or animal models based on S1PR4 deletion or overexpression and developing specific agonists and antagonists,.
4. Conclusions
Our study found a low expression of S1PR4 in SKCM tumor samples, with a high expression indicating better prognosis, PFS, and milder clinical manifestations. Our study revealed the role of S1PR4 in SKCM progression and provided new ideas for finding novel immunosuppressive genes involved in SKCM from the perspectives of immune infiltration and drug resistance. Moreover, our findings contribute to developing personalized therapeutic cancer drugs. In conclusion, S1PR4 can potentially be a prognostic marker for patients with SKCM, helping clinicians formulate diagnosis and treatment plans and serving as a therapeutic target. However, further studies are needed to confirm this.
5. Materials and methods
5.1. Data source
In 2005, the use of TCGA was proposed to map the genome and variation of all human cancers via genomics and bioinformatic analyses to better understand the mechanisms underlying cancer occurrence and development. TCGA hosts sequencing data corresponding to 33 of the most common cancers and more than 11,000 tumor samples. Data from patients with SKCM, including RNA sequencing, clinical, and survival data, were obtained from TCGA database (portal.gdc.cancer.gov/), which hosts data on 472 SKCM tissues and one normal non-tumor tissue as of November 26, 2022. All data were collated using Perl v5.30.2.
5.2. Differential expression of S1PR4 and establishment of survival-risk curve for SKCM
First, we used the DE module of the Timer 2.0 open software (http://timer.comp-genomics.org/timer/) to perform pan-cancerous genetic differential analysis of S1PR4 to visualize the differential expression between tumors and attached normal tissues. Notably, because the number of normal melanoma samples was less than five, the database statistically visualized the differential expression between primary and metastatic melanomas.
Additionally, we divided the acquired patient data into high- and low-expression groups according to the median value of S1PR4 expression. According to the survival time and status data, the survival difference between the two groups was compared using R 4.2.1 software via the Kolmogorov–Smirnov test, producing a curve. The differences in PFS between the patients in the high- and low-expression groups were analyzed, and the PFS curve was drawn.
5.3. Correlation analysis between S1PR4 and clinical traits
To explore the relationship between S1PR4 expression and the clinical characteristics of melanoma, we used the patient's age, sex, clinical stage, Clark grade, ulceration, and TMN stage as variables. Box plots of each clinical characteristic were drawn. Next, we divided the patients into high- and low-expression groups according to the median value of S1PR4 expression, analyzed the correlation between clinical traits and high and low expression of S1PR4 using the chi-square test, and plotted a correlation heat map.
To make the results of the prediction model more readable and facilitate the evaluation of patients, we assigned the influence of each clinical trait and S1PR4 expression level on survival (i.e., regression coefficient) through multivariate regression analysis. We plotted nomograms to visualize the obtained data to clearly show the relationship between each clinical trait, gene expression level, and patient survival.
5.4. Functional enrichment of S1PR4 co-expressed genes in melanoma patients
To explore the correlation between the expression of S1PR4 and other genes further, the Pearson correlation coefficient was used to analyze the relationship between the expression of each gene and the expression of S1PR4, and the correlation coefficient and p-values were obtained. A correlation coefficient greater than and less than 0 was considered positive and negative, respectively. The top five genes that were positively and negatively correlated with S1PR4 expression were identified.
Next, we divided patients into high- and low-expression groups based on their expression level of S1PR4. The Wilcoxon test was used to analyze whether there was a difference between high and low S1PR4 expression for each gene. The average expression level (lowMean and highMean), logFC, and p-value of each gene in the two groups were calculated. Results with significant differences in p-values were corrected to obtain the fdr value (adjusted p-value). Genes with significantly different fdr values were visualized by heatmap analysis using the pheatmap package.
5.5. Pathway enrichment analysis
GO enrichment analysis was performed for genes that showed differences between high and low S1PR4 expression groups, and significantly enriched functions were obtained. The results of the significant top six genes in BP, CC, and MF were visualized. GSEA is a computational method that determines whether an a priori-defined set of genes shows statistically significant concomitant differences between two biological states. We used the median value of S1PR4 expression to classify patients into high- and low-expression groups and performed GSEA to identify and visualize the five pathways most associated with S1PR4.
5.6. Correlation analysis of tumor microenvironment and immune infiltration
The estimation of stromal and immune cells in malignant tumors using the expression data (ESTIMATE) algorithm uses gene expression information to infer the proportion of stromal and immune cells in a tumor sample. We scored the level of the infiltration of immune and stromal cells in each sample using the ESTIMATE algorithm and calculated a composite ESTIMATE score. Then, the samples were divided into high- and low-expression groups according to the expression of S1PR4. The correlation between high and low S1PR4 expression in tumor samples was analyzed using the Wilcoxon test, and a box plot was drawn for visualization.
To explore the correlation between S1PR4 expression and immune cell infiltration, we analyzed the proportion of infiltrating immune subsets in each sample using the CIBERSORT calculation, filtering the results based on p-value to retain immune cell types with p < 0.05. Similarly, the samples were grouped according to the expression level of S1PR4, and the correlation between high and low expression and immune cell content was analyzed and visualized using the Spearman test.
An immune checkpoint is an immunosuppressive molecule that can prevent damage and destruction of normal tissues by regulating the immune response. Immune checkpoints are one of the main causes of immune tolerance. To explore the co-expression relationship between S1PR4 and immune checkpoint genes in melanoma, we performed correlation tests between common immune checkpoint genes and S1PR4 and obtained correlation coefficients and p-values for visualization.
5.7. Drug sensitivity and immunotherapy sensitivity
Pearson's correlation test was used to explore the relationship between S1PR4 expression and drug sensitivity. Only 263 FDA-approved and clinically tested drugs were included in the correlation analysis.
Immune checkpoint inhibitors (ICIs) target dysfunctional immune systems and induce CD8+ T-cells to kill cancer cells. The targeting of CTLA4 and PD-1 by ICIs has revolutionized the management of advanced melanoma and many other malignancies. To investigate the effect of S1PR4 expression on the sensitivity and efficacy of immune checkpoint therapy, we performed a differential analysis between the expression of S1PR4 and immunotherapy efficacy. The samples were divided into high- and low-expression groups according to the expression level of S1PR4. The immune score data of SKCM tumor samples were downloaded from The Cancer Imaging Archive (TCIA) database to compare the effect of high and low expression of S1PR4 on the sensitivity of CTLA4 and PD-1 cells. Violin charts were constructed for visualization.
Statistical analysis
All statistical analyses were performed using SPSS (version 22.0, Chicago, IL, USA) and R 4.2.1 (https://www.r-project.org/). The Wilcoxon rank-sum test was used to analyze the differences in gene expression between the tumor and normal tissues. Patients with SKCM in different datasets were divided into high- and low-expression groups according to their median S1PR4 expression, and the OS of the patients was analyzed using the Kaplan–Meier method. Co-expression relationships were explored using the “ggplot2” package in R. The correlation between gene expression and drug susceptibility was determined by calculating Pearson correlation coefficients. Statistical significance was defined as p < 0.05.
Ethics declarations
Review and/or approval by an ethics committee was not needed for this study because the research was based on the human sample data in the database.
Consent for publication
Not applicable.
Data availability statement
The datasets generated and/or analyzed during the current study are available in TCGA database (portal.gdc.cancer.gov/).
Funding
NA
Authors’ contributions
Zi Wang drafted and revised the manuscript. Fei Pan and Guangzhong Zhang contributed equally to the study, including data collection and analysis. All authors reviewed and approved the final version of the manuscript.
CRediT authorship contribution statement
Zi Wang: Data curation, Methodology, Writing – original draft. Fei Pan: Visualization, Writing – review & editing. Guangzhong Zhang: Supervision, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study is based on a public database and does not require financial support. Due to the guarantee of scientific integrity, we honestly do not name any project or funding source.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27505.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Arnold M., Singh D., Laversanne M., Vignat J., Vaccarella S., MeheusA F., Cust E., de Vries E., Whiteman D.C., Bray F. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022;158:495–503. doi: 10.1001/jamadermatol.2022.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenlee R.T., Hill-Harmon M.B., Murray T., Thun M. Cancer statistics, 2001. CA A Cancer J. Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Lopes F.C.P.S., Sleiman M.G., Sebastian K., Bogucka R., Jacobs E.A., Adamson A.S. UV exposure and the risk of cutaneous melanoma in skin of color: a systematic review. JAMA Dermatol. 2021;157:213–219. doi: 10.1001/jamadermatol.2020.4616. [DOI] [PubMed] [Google Scholar]
- 4.Carpinteiro A., Becker K.A., Japtok L., Hessler G., Keitsch S., Požgajovà M., Schmid K.W., Adams C., Müller S., Kleuser B., Edwards M.J., Grassmé H., Helfrich I., Gulbins E. Regulation of hematogenous tumor metastasis by acid sphingomyelinase. EMBO Mol. Med. 2015;7:714–734. doi: 10.15252/emmm.201404571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagahashi M., Abe M., Sakimura K., Takabe K., Wakai T. The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci. 2018;109:3671–3678. doi: 10.1111/cas.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyne N.J., El Buri A., Adams D.R., Pyne S. Sphingosine 1-phosphate and cancer. Adv. Biol. Regul. 2018;68:97–106. doi: 10.1016/j.jbior.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang P., Yuan Y., Lin W., Zhong H., Xu K., Qi X. Roles of sphingosine-1-phosphate signaling in cancer. Cancer Cell Int. 2019;19:295. doi: 10.1186/s12935-019-1014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garandeau D., Noujarède J., Leclerc J., Imbert C., Garcia V., Bats M.L. Targeting the sphingosine 1-phosphate axis exerts potent antitumor activity in BRAFi-resistant melanomas. Mol. Cancer Therapeut. 2019;18:289–300. doi: 10.1158/1535-7163.MCT-17-1141. [DOI] [PubMed] [Google Scholar]
- 10.Olesch C., Sirait-Fischer E., Berkefeld M., Fink A.F., Susen R.M., Ritter B. S1PR4 ablation reduces tumor growth and improves chemotherapy via CD8+ T cell expansion. J. Clin. Invest. 2020;130:5461–5476. doi: 10.1172/JCI136928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.F., Dang A., Hernandez E., Pong R.C., Chen B., Sonavane R. Activation of sphingosine kinase by lipopolysaccharide promotes prostate cancer cell invasion and metastasis via SphK1/S1PR4/matriptase. Oncogene. 2019;38:5580–5598. doi: 10.1038/s41388-019-0833-3. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty P., Vaena S.G., Thyagarajan K., Chatterjee S., Al-Khami A., Selvam S.P. Pro-survival lipid sphingosine-1-phosphate metabolically programs T cells to limit anti-tumor activity. Cell Rep. 2019;28:1879–1893. doi: 10.1016/j.celrep.2019.07.044. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colié S., Van Veldhoven P.P., Kedjouar B., Bedia C., Albinet V., Sorli S.C. Disruption of sphingosine 1-phosphate lyase confers resistance to chemotherapy and promotes oncogenesis through Bcl-2/Bcl-xL upregulation. Cancer Res. 2009;69:9346–9353. doi: 10.1158/0008-5472.CAN-09-2198. [DOI] [PubMed] [Google Scholar]
- 14.Dillmann C., Mora J., Olesch C., Brüne B., Weigert A. S1PR4 is required for plasmacytoid dendritic cell differentiation. Biol. Chem. 2015;396:775–782. doi: 10.1515/hsz-2014-0271. [DOI] [PubMed] [Google Scholar]
- 15.Schulze T., Golfier S., Tabeling C., Räbel K., Gräler M.H., Witzenrath M., Lipp M. Sphingosine-1-phosphate receptor 4 (S1P₄) deficiency profoundly affects dendritic cell function and TH17-cell differentiation in a murine model. Faseb. J. 2011;25:4024–4036. doi: 10.1096/fj.10-179028. [DOI] [PubMed] [Google Scholar]
- 16.LaMontagne K., Littlewood-Evans A., Schnell C., O'Reilly T., Wyder L., Sanchez T. Antagonism of sphingosine-1-phosphate receptors by FTY720 inhibits angiogenesis and tumor vascularization. Cancer Res. 2006;66:221–231. doi: 10.1158/0008-5472.CAN-05-2001. [DOI] [PubMed] [Google Scholar]
- 17.Tay K.H., Liu X., Chi M., Jin L., Jiang C.C., Guo S.T. Involvement of vacuolar H(+)-ATPase in killing of human melanoma cells by the sphingosine kinase analogue FTY720. Pigment Cell Melanoma Res. 2015;28:171–183. doi: 10.1111/pcmr.12326. [DOI] [PubMed] [Google Scholar]
- 18.Schadendorf D., van Akkooi A.C.J., Berking C., Griewank K.G., Gutzmer R., Hauschild A. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9.Erratumin. Lancet. 393 (2019) 746. [DOI] [PubMed] [Google Scholar]
- 19.Cohen A.O., Woo S.H., Zhang J., Cho J., Ruiz M.E., Gong J. Tbc1d10c is a selective, constitutive suppressor of the CD8 T-cell anti-tumor response. OncoImmunology. 2022;11 doi: 10.1080/2162402X.2022.2141011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villagomez F.R., Diaz-Valencia J.D., Ovalle-García E., Antillón A., Ortega-Blake I., Romero-Ramírez H. TBC1D10C is a cytoskeletal functional linker that modulates cell spreading and phagocytosis in macrophages. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., An S., Ward R., Yang Y., Guo X.X., Li W., Xu T.R. G protein-coupled receptors as promising cancer targets. Cancer Lett. 2016;376:226–239. doi: 10.1016/j.canlet.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Fu J.F., Yen T.H., Chen Y., Huang Y.J., Hsu C.L., Liang D.C., Shih L.Y. Involvement of Gpr125 in the myeloid sarcoma formation induced by cooperating MLL/AF10(OM-LZ) and oncogenic KRAS in a mouse bone marrow transplantation model. Int. J. Cancer. 2013;133:1792–1802. doi: 10.1002/ijc.28195. [DOI] [PubMed] [Google Scholar]
- 23.Lei P., Wang H., Yu L., Xu C., Sun H., Lyu Y. A correlation study of adhesion G protein-coupled receptors as potential therapeutic targets in uterine corpus endometrial cancer. Int. Immunopharm. 2022;108 doi: 10.1016/j.intimp.2022.108743. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Chen W., Gong L., Ke C., Wang H., Cai Y. Elevated G-protein receptor 125 (GPR125) expression predicts good outcomes in colorectal cancer and inhibits Wnt/β-catenin signaling pathway. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018;24:6608–6616. doi: 10.12659/MSM.910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swartz M.A., Iida N., Roberts E.W., Sangaletti S., Wong M.H., Yull F.E. Tumor microenvironment complexity: Emerging roles in cancer therapy. Cancer Res. 2012;72:2473–2480. doi: 10.1158/0008-5472.CAN-12-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlino M.S., Larkin J., Long G.V. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002–1014. doi: 10.1016/S0140-6736(21)01206-X. [DOI] [PubMed] [Google Scholar]
- 27.Min J., Van Veldhoven P.P., Zhang L., Hanigan M.H., Alexander H., Alexander S. Sphingosine-1-phosphate lyase regulates sensitivity of human cells to select chemotherapy drugs in a p38-dependent manner. Mol. Cancer Res. 2005;3:287–296. doi: 10.1158/1541-7786.MCR-04-0197. [DOI] [PubMed] [Google Scholar]
- 28.Xiao S., Yang J. Preclinical study of the antitumor effect of sphingosine-1-phosphate receptor 1 antibody (S1PR1-antibody) against human breast cancer cells, Investig. N. Drugs. 2019;37:57–64. doi: 10.1007/s10637-018-0618-5. [DOI] [PubMed] [Google Scholar]
- 29.Zou J., Ma X., Zhang G., Shen L., Zhou L., Yu Y., Zhu F., Chen Z. Evaluation of the change in sphingolipids in the human multiple myeloma cell line U266 and gastric cancer cell line MGC-803 treated with arsenic trioxide. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2015;1004:98–107. doi: 10.1016/j.jchromb.2015.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in TCGA database (portal.gdc.cancer.gov/).