Abstract
Graves' disease (GD) is an autoimmune disorder characterized by hyperthyroidism resulting from autoantibody-induced stimulation of the thyroid gland. Despite recent advancements in understanding GD's pathogenesis, the molecular processes driving disease progression and treatment response remain poorly understood. In this study, we aimed to identify crucial immunogenic factors associated with GD prognosis and immunotherapeutic response. To achieve this, we implemented a comprehensive screening strategy that combined computational immunogenicity-potential scoring with multi-parametric cluster analysis to assess the immunomodulatory genes in GD-related subtypes involving stromal and immune cells. Utilizing weighted gene co-expression network analysis (WGCNA), we identified co-expressed gene modules linked to cellular senescence and immune infiltration in CD4+ and CD8+ GD samples. Additionally, gene set enrichment analysis enabled the identification of hallmark pathways distinguishing high- and low-immune subtypes. Our WGCNA analysis revealed 21 gene co-expression modules comprising 1,541 genes associated with immune infiltration components in various stages of GD, including T cells, M1 and M2 macrophages, NK cells, and Tregs. These genes primarily participated in T cell proliferation through purinergic signaling pathways, particularly neuroactive ligand-receptor interactions, and DNA binding transcription factor activity. Three genes, namely PRSS1, HCRTR1, and P2RY4, exhibited robustness in GD patients across multiple stages and were involved in immune cell infiltration during the late stage of GD (p < 0.05). Importantly, HCRTR1 and P2RY4 emerged as potential prognostic signatures for predicting overall survival in high-immunocore GD patients (p < 0.05). Overall, our study provides novel insights into the molecular mechanisms driving GD progression and highlights potential key immunogens for further investigation. These findings underscore the significance of immune infiltration-related cellular senescence in GD therapy and present promising targets for the development of new immunotherapeutic strategies.
Keywords: Graves' disease, Immunotherapeutic biomarkers, Immune infiltration, WGCNA, Gene co-expression modules, Molecular mechanisms
Highlights
-
•
Identified key immunogenic factors for GD prognosis and therapy response
-
•
Utilized computational immunogenicity-potential scoring and multi-parametric cluster analysis
-
•
Co-expression modules related to immune infiltration and cellular senescence in CD4+ and CD8+ GD samples identified
-
•
Three genes (PRSS1, HCRTR1, P2RY4) were identified as potential key immunogens for GD therapy and overall survival prediction
-
•
Immune infiltration-related cellular senescence in GD underscores the importance of new immunotherapeutic strategies.
1. Introduction
Graves' disease (GD), a prevalent autoimmune disorder, affects the thyroid gland causing hyperthyroidism, often seen in individuals between the ages of 30 and 50 [1]. Women are more likely to be diagnosed with GD, with a female-to-male ratio of around 7:1 [2]. The incidence of GD is higher in industrialized countries such as the United States and Europe, with a prevalence of 1–2% and 0.5–2%, respectively [3,4]. However, the incidence of GD in Asia is relatively lower, with a reported rate of 0.3–0.7%. A study published in the Journal of Clinical Endocrinology & Metabolism has indicated an increasing trend of GD incidence in China, with a rate of 0.5–0.8% [5,6].
The standard treatment options for GD include antithyroid medications, radioactive iodine (RAI) therapy, and thyroidectomy [[7], [8], [9]]. Close monitoring and adjustments to the treatment plan may be necessary for some patients [10,11]. Although current treatments effectively manage hyperthyroidism, they do not address the underlying immune activation and autoantibody production [11]. Hence, there is a need for further research and development of treatments targeting the underlying immunological mechanisms in GD.
GD is characterized by immune pathology that involves the production of autoantibodies targeting the thyroid-stimulating hormone receptor (TSHR) in the thyroid gland, which results in hyperthyroidism due to excessive secretion of thyroid hormones [12,13]. The immune response in GD also involves the participation of T and B cells, which play critical roles in the disease progression [14]. T cells secrete cytokines that promote inflammation and stimulate the thyroid gland, while B cells produce autoantibodies against thyroid-specific antigens, which contribute to ongoing immune activation and excessive thyroid hormone secretion [14,15].
The exact cause of immune-mediated autoantibody production in autoimmune disorders is not yet fully understood, although genetic susceptibility, viral infections, and endocrine imbalances have been suggested as contributing factors. The initial triggers for autoantibody production and immune activation are still unknown, and the precise interactions between immune cells and autoantibodies in the thyroid gland require further investigation [12,16]. However, recent advances in computational tools, such as the immunogenicity-potential scoring (IPS) [17,18], microenvironment cell populations-counter (MCP) [[19], [20], [21]], X-cells algorithms [22,23], and weighted gene co-expression network analysis (WGCNA) [24,25], have enabled researchers to gain a deeper understanding of the molecular mechanisms underlying autoantibody production and the complex interactions between stromal and immune cells in the thyroid gland [26,27]. These cutting-edge tools allow for large-scale gene expression analysis to identify key biological pathways and processes involved in the path immunology of autoimmune disorders, providing valuable insights into the underlying mechanisms of these diseases [28].
In this study, we present a pioneering investigation into the immune landscape of GD utilizing cutting-edge computational tools. Our approach integrates advanced methodologies, including IPS, MCP, and X-cell algorithms, complemented by the well-established WGCNA [26,27]. This departure from conventional research strategies aims to fill crucial knowledge gaps regarding the molecular origins of immune-mediated autoantibody production in the GD [28]. By leveraging large-scale gene expression analysis, we uncover key biological pathways, unraveling the intricate immunological processes that characterize autoimmune disorders. This unique amalgamation of methodologies not only propels our comprehension of GD immunology but also lays the groundwork for revolutionary diagnostic and therapeutic avenues with profound implications. Specifically, we aim to determine the disrupted immune network associated with GD pathogenesis and evaluate the feasibility of utilizing a prognostic marker to predict the immune microenvironment status in GD patients. By combining the expression data of stromal and immune cells in GD-affected tissues, our study seeks to derive a novel understanding of the intricate interactions between immune cells and autoantibodies in GD and enhance our capacity to forecast and manage this debilitating autoimmune disorder. Our research has the potential to provide valuable insights into the pathophysiology of GD and inform the development of more effective diagnostic and therapeutic strategies.
2. Materials and methods
2.1. Study design
In this observational study, transcriptomic data of GD obtained from The Cancer Genome Atlas (TCGA) and The Cancer Genome Atlas Glioma (CGGA) collections were analyzed to identify cellular and molecular pathways associated with senescence in GD through an immune infiltration-guided strategy. The study employed IPS, MCP, and Xcell algorithms to comprehensively analyze the immune cell fractions and immune clustering of the immune infiltration components in GD datasets. The IPS algorithm was utilized to evaluate the overall abundance of immune cells within the transcriptomic data. This computational approach entails a thorough assessment of the expression profiles linked to various immune cell types. IPS offers a quantitative measure, enabling the determination of the global immune cell landscape in the context of GD [17,18]. Additionally, MCP was employed to scrutinize the specifics of immune cell populations present in the GD datasets. This algorithm utilizes advanced computational methods to deconvolute bulk transcriptomic data, providing insights into the quantitative distribution of distinct immune cell subsets. By leveraging gene expression signatures associated with various immune cell types, MCP enhances the resolution for characterizing specific immune cell populations contributing to the GD microenvironment [19,20]. Moreover, the distribution analysis of immune cell populations in the GD datasets was conducted using the Xcell algorithm. Xcell employs a sophisticated computational framework to evaluate the relative proportions of diverse immune cell types within the GD transcriptomic landscape [22,23]. By leveraging specific gene expression profiles associated with various immune cells, Xcell enhances our understanding of the spatial arrangement and interplay of immune cell populations in the context of GD. These algorithms collectively contribute to a comprehensive assessment of the immune landscape, providing valuable information on both the overall abundance and specific distribution patterns of immune cell populations associated with GD [26,27].
Furthermore, the WGCNA was used to identify genes related to the immune hub and involved in glioma immune invasion. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to uncover specific signaling pathways, and the reliability of the immune-hub genes was evaluated through clinical prognosis analysis. A protein-protein interaction analysis was also performed to predict potential mechanisms. The study aimed to provide a comprehensive understanding of the immune profile of GD and its potential implications for prognosis and therapy. The sample abundance scores for immune cells were separately calculated for patient samples from different clinical stages and ages [29]. The flowchart in Fig. 1 summarizes the preparation, processing, analysis, and validation of the data.
Fig. 1.
Flowchart describing the schematic overview of the study design for investigating immune infiltration characteristics in patients with GD.
2.2. Data acquisition
In this study, transcriptomic datasets were collected from the NCBI GEO database to investigate epigenetic profiling in CD4+ and CD8+ T cells of patients with GD. The datasets contained information on changes in genes related to T cell receptor signaling, providing valuable insights into the underlying mechanisms of GD. The study sample consisted of 38 GD patients and 31 healthy individuals from the GSE71956 cohort. The researchers extracted total RNA from sorted CD4+ and CD8+ T cells to analyze DNA methylation patterns of specific genes and examine their correlation with gene expression profiles in GD. This approach enabled the identification of potential targets for therapeutic intervention and a deeper understanding of the molecular pathways involved in GD pathogenesis. The study followed the PRISMA statement recommendations to ensure transparency and reproducibility of the research process.
2.3. Data pre-processing
Before the gene expression analysis, the raw data were preprocessed using the Limma package in R/Bioconductor, with version R 2.14.0. Quantile normalization was conducted on the data to ensure the accurate representation of gene expression levels, which was implemented using the Limma package. To match the microarray probe sets with their corresponding genes, an Affymetrix-released annotation file for each platform was used. To eliminate redundancy, probes that mapped to multiple genes were removed, and the average expression level of the probes that mapped to the same gene was used. Lastly, hierarchical clustering was carried out to eliminate any noisy data from the analysis.
2.4. Gene co-expression network reconstruction
The WGCNA approach was employed to identify co-expressed gene modules related to cellular senescence and immune infiltration in the GD [28,30]. To begin with, three biological co-expression networks of immune pathways were constructed using the WGCNA R package, and a weighted adjacency matrix and a topological overlap matrix were built to evaluate the correlation among the genes involved in the senescence [30,31]. The authors identified biologically relevant modules that had a minimum size of 30 and a split depth of 3. To understand the association of gene sets with the disease immunophenotype, the module-phenotype relationship was calculated, and the study highlighted that the glioma-immune microenvironment can impact cellular senescence's pro- or anti-immunopathology tendencies and therapy response. Additionally, the IPS, MCP, and Xcell algorithms were employed to evaluate the immune cell infiltration level, and the correlation between the immune score and hub genes was determined. Finally, the Chi-square test was carried out to assess the significance of immuno-module genes in each group, to comprehend the immune infiltration-related cellular senescence genes in glioma.
2.5. Gene and pathway enrichment analysis
To gain insights into the biological processes and molecular functions related to the targeted immune hubs in glioma, several analyses were conducted using multiple datasets such as DisGeNET [32], Clinical Variations [24], DisGeNET [25], BeFree [26], BioCyc [27], KEGG [28], and MSigDBBioCarta [30]. ToppGene and the clusterProfiler package were used to perform GO analysis and KEGG analysis to uncover enriched pathways across multiple datasets [[33], [34], [35]]. The gene set enrichment analysis (GSEA) method was used to conduct pathway enrichment analysis based on 50 hallmark pathways from the Molecular Signature Database [36]. The Enrichplot package was used to visualize the results, and Kaplan-Meier diagrams were created to determine the prognostic pattern of the top overlapping oncogenic pathways. These analyses aimed to reveal the underlying mechanisms of the targeted immune hubs and their impact on patient prognosis [37].
2.6. Protein-protein interaction network construction
To analyze the relationships between genes, a protein-protein interaction (PPI) network was constructed from the genes in the enriched modules. The PPI network was constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) and visualized using Cytoscape [38]. In the context of our analysis, STRING served as a valuable resource for comprehending the functional associations and potential interactions between genes. The constructed PPI network, visualized through Cytoscape, provided an insightful representation of gene connectivity and potential functional relationships. Hub genes were identified as those with node connectivity of ≥5, indicating that they play a crucial role in the network due to their many connections to other genes. The PPI network visualization allows for the identification of potential interactions and functional relationships between genes, providing valuable information for further analysis. The integration of STRING and Cytoscape in this analysis not only enhances the clarity of the PPI network but also contributes to the identification of crucial interactions, providing a foundation for further in-depth analysis of the functional relationships between genes.
2.7. Development of a comprehensive and predictive nomogram
To investigate the relationship between the expression of the key immune-related gene and clinical grade, the researchers utilized the survival package and survminer package in R for survival analysis. Clinicopathological grading and survival time records of the GD cases in the dataset were analyzed, and the correlation between the target immune-related gene and clinical grade was evaluated. The Kaplan-Meier method was employed to plot the survival curve, with the log-rank test used as a statistical significance test. A p-value of less than 0.05 was considered significant. Univariate and multivariate Cox regression analyses were conducted to identify independent prognostic factors. A comprehensive and predictive nomogram was developed using the "rms" R package, which incorporated the immune-score signature, age, gender, and stages. The performance and accuracy of the nomogram were evaluated using receiver operating characteristics (ROC) and calibration curves.
2.8. Data analysis
All data analysis in this study was carried out using the R software, version 4.0.3. Statistical tests such as the Wilcoxon signed-rank test were used to compare two groups, while the Kruskal-Wallis test was applied to compare three or more groups. A p-value less than 0.05 was considered statistically significant, and all tests were two-sided. Descriptive statistics such as mean ± standard deviation (SD) and medians (ranges) were used to present normally and non-normally distributed variables, respectively. Categorical variables were reported as numbers and percentages. T-tests or one-way ANOVA were used for data analysis, while Spearman's rank correlation coefficient test was used to identify the effect of the cut-off threshold and construct the ROC curve. The relationship between sensitivity and specificity was determined between groups. An adjusted p-value less than 0.05 was considered statistically significant.
3. Results
3.1. Identification of immune infiltration-related senescence genes
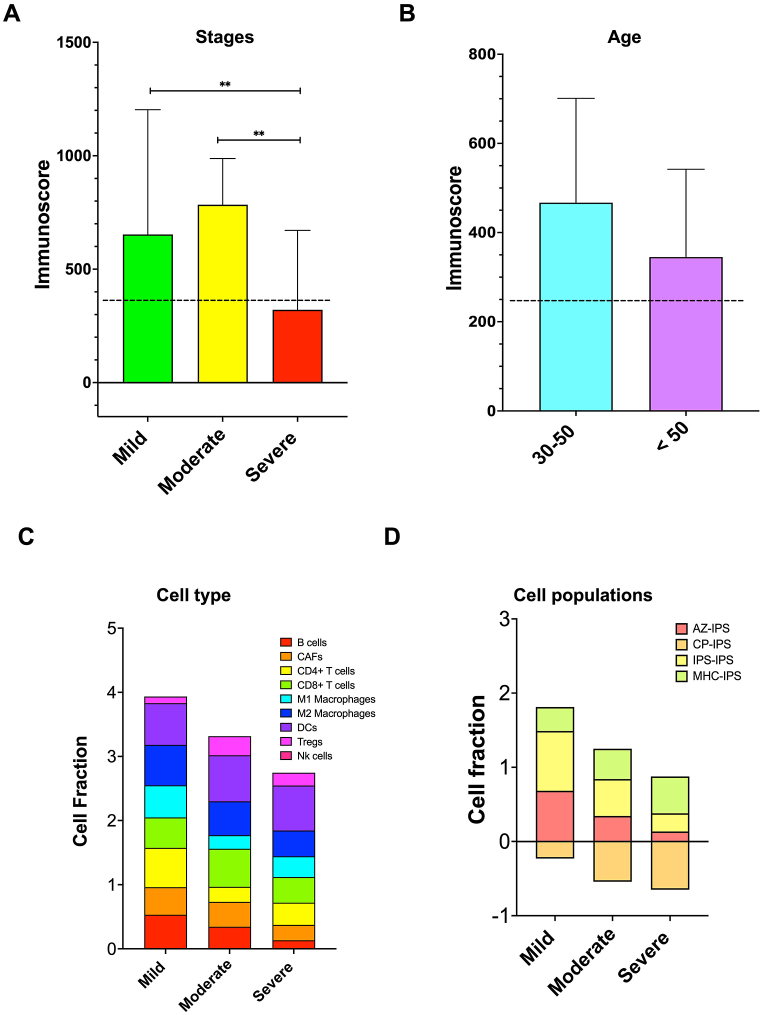
In this study, we aimed to identify senescence genes that have a role in the immune system and their connection with chronic diseases and GD. To achieve this, we utilized three algorithms - IPS, MCP, and Xcell - to measure different immune cell subsets in the TCGA cohort based on their phenotypic characteristics. S1 Figure shows the results of these analyses for each sample, and we examined the immune molecular characteristics at different stages of CD4+ and CD8+ GD samples. Our results indicated a significant decrease in immune scores as GD progressed, while purity decreased in higher grades (Fig. 2A). We also observed that GD was more prevalent in the 30–50 age group than in those above 50 years old (Fig. 2B). By using these algorithms, we evaluated the immune scores and infiltrations of various immune infiltration components such as B cells, M1 Macrophages, M2 Macrophages, NK cells, and Tregs at different stages of GD (Fig. 2C). Our analysis indicated that the infiltration of these immune cells was lower in the later stages of GD, except for DCs, which increased in severe stages. We also found a positive correlation between GA-related genes and immune scores. Additionally, we assessed the accuracy of the immunogenicity score of IPS, AZ-IPS, and CP-IPS and observed that different immunogenicity algorithms of GD decreased in the late stages of the disease. However, the MHC-I binding prediction component of the IPS algorithm (MHC 1-IPC) increased in the severe stage of GA, indicating an accurate prediction of MHC binding affinity for the development of effective cancer immunotherapies (Fig. 2D). Overall, our study provides insights into the role of senescence genes in the immune system and their association with chronic diseases and GD.
Fig. 2.
Differences in immune infiltration characteristics among patients with different stages of GD (A) and different ages (B). (C) The nine immune cell scores differed among the three stages based on IPS, MCP, and Xcell algorithms. (D) Immune cell fractions between different stages of GD. Values are mean ± SEM, *p < 0.05, **p < 0.001.
3.1.1. Immuno-module screening
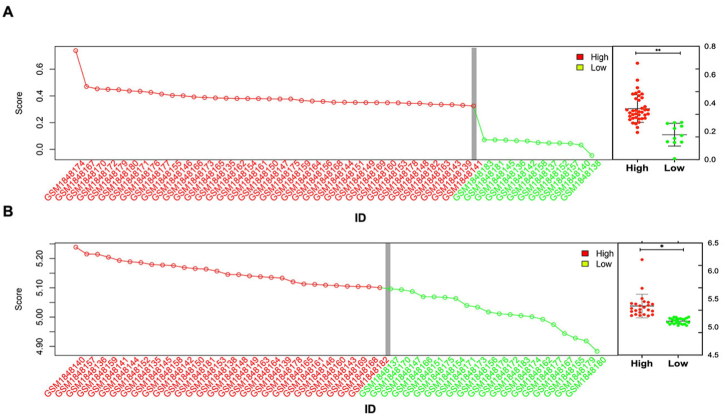
To analyze samples with high immune scores at the systems level, we utilized the WGCNA approach to construct a biological network. Samples were divided into high-expression outliers and low-expression groups based on the interquartile range, with values 1.5 times greater than the interquartile range being classified as high-expression outliers. Both IPC and MCP algorithms were used to analyze and select the high immune score samples (Fig. 3 A and 3B respectively). Out of the 38 samples, 11 samples with low immune scores were identified as outliers and removed from the study. The high immune expression group consisted of 68% (27/38) of the TCGA samples and 32% (12/38) of the samples (Fig. 3). For further investigation of immune-related genes and hubs, we performed additional analysis on the high immune score samples, given the focus of our study on GD, an autoimmune disorder. We sorted the sample results according to all immunoassay measurements, after screening them using both the IPC and MCP algorithms, as outlined in the S1 Table.
Fig. 3.
Immune screening of gene expression in CD4+ and CD8+ GD samples from the TCGA database was evaluated based on the IPS (A) and MCP (B) algorithms. The vertical gray line denotes a subset of high immune score expression outliers (right) determined by the 1.5 × interquartile range. Box plots (far right) display aggregate expression of the genes in high and low immune score groups. Statistical analysis was performed using the Mann-Whitney U test.
3.1.2. Immuno-module constructing
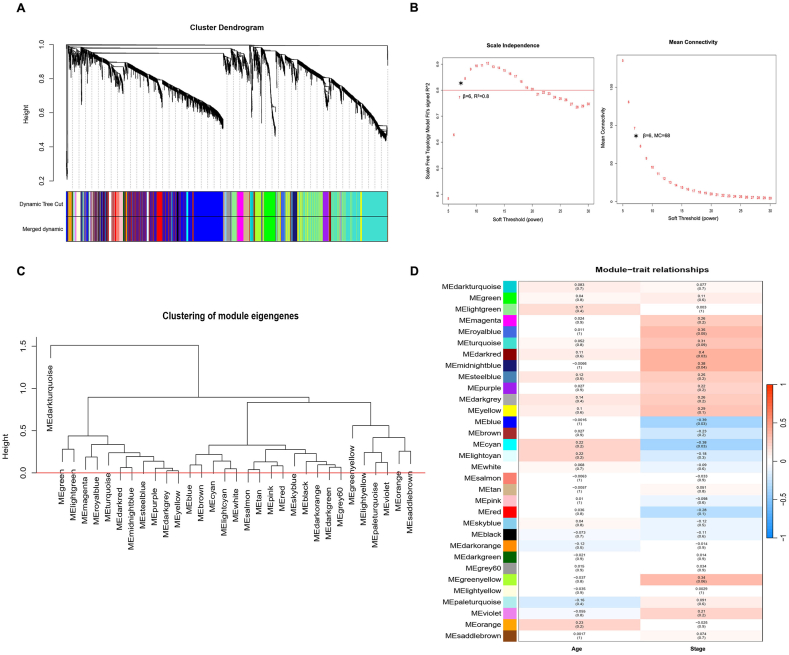
The study utilized the WGCNA method to construct a co-expression network of genes associated with the development of GD. Analysis of the microarray data identified 1,541 differentially expressed genes, and 21 co-expression modules were found to be linked with GD (Fig. 4A). We identified five modules through cluster dendrogram analysis, each labeled with a unique color: blue, cyan, dark red, midnight blue, and royal blue, which showed similar module integration (Fig. 4). Among the selected immuno-module, our analysis resulted in a scale-free network that displayed a relative balance between scale independence and means connectivity when we used a soft thresholding power of 6 (Fig. 4B). Interestingly, the blue module contained taxonomic genes that were different from the genes in the other modules. We employed eigengene co-expression analysis to merge the dynamic modules into three using a threshold of 0.5, and this process confirmed the reliability of the dark-red module, as illustrated in Fig. 4B. Our study's key finding was that 75% (52/69) of the genes in the dark-red module were significantly correlated with immune response in GD, making it the critical module between the normal mild GD and severe GD groups (p = 0.035). We have listed the genes of the selected immune module in the S2 Table. We also conducted dimensionless topological network analysis between different stages and age groups to identify potential candidate key immunogens linked to GD progression, which was used to scale the adjacency matrix (Fig. 4C). Our results indicated that the dark-red module was the most significant module linked to late-stage GD and contained 69 mRNAs that were potential candidates for further investigation (Fig. 4D; p = 0.004). Moreover, our module correlation analysis revealed statistically significant positive correlations between age and pathological stage characteristics with the dark red, midnight blue, and royal blue modules (all p < 0.05).
Fig. 4.
Weighted gene correlation network analysis (WGCNA) of CD4+ and CD8+ GD samples based on stage (mild, moderate, and severe) and age range. (A) Clustering dendrogram and module-trait analysis. Different colors of the column indicate different hub modules. (B) Analysis of the scale-free fit index and mean connectivity for various soft-threshold powers. (C) Clustering dendrogram of all genes with dissimilarity based on the topological overlap and assigned module colors. (D) Heatmap for the correlation between immune modules and traits. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Functional downstream analyses of immuno-module
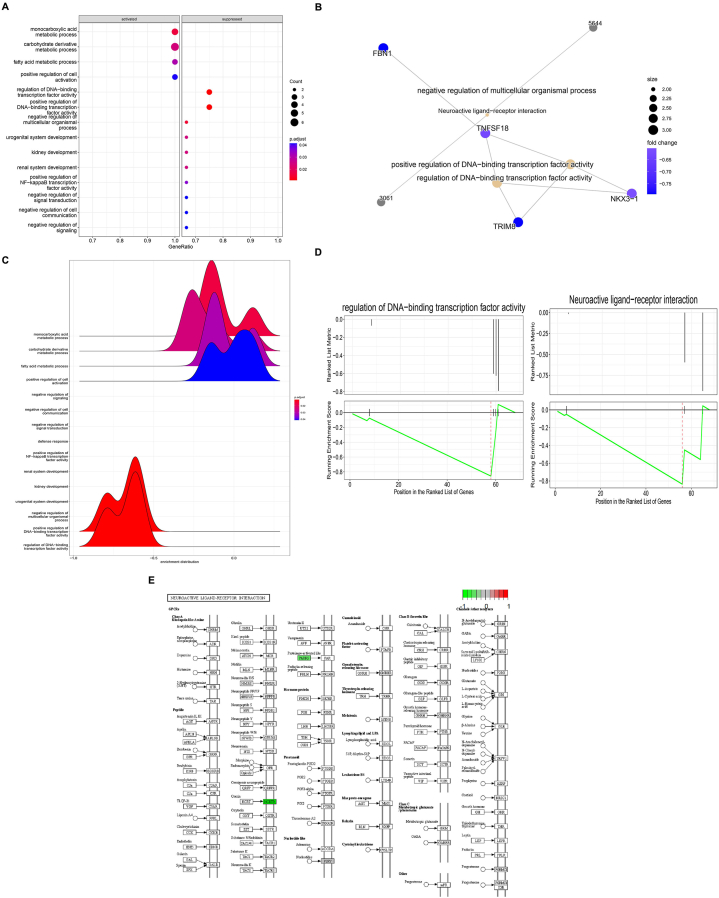
Our analysis revealed that genes within the dark-red module had stronger interactions with each other compared to genes in other modules. To gain insight into the biological processes and pathways associated with GD, we performed GO and KEGG pathway enrichment analyses on the genes in the dark-red module (Table 1 and Fig. 5). Our results identified several significant pathways and processes enriched in GD driver genes, including DNA binding transcription factor activity, Microcephaly, primary autosomal recessive cataracts, spastic paraparesis, speech delay, and peroxisomal fatty Acyl-Coa reductase disorder (Fig. 5A). These findings suggest that these biological processes and functions may play a critical role in the pathogenesis of GD. Additionally, our KEGG pathway analysis revealed that the most significant biological pathways related to neuroactive ligand-respecter interaction and DNA binding transcription factor activity of the purinergic signaling pathways, which have been linked to the regulation of immune response and inflammation in various autoimmune disorders, including GD (Fig. 5B and S2 Fig). Interestingly, we also identified two pathways in GD segregation that are associated with cognitive impairment, covering over 48% of the genes in the targeted immuno-module (Fig. 5C). The integration of the selected biological process of the purinergic signaling pathways, neuroactive ligand-respecter interaction, and DNA binding transcription factor activity is presented in Fig. 5D. This finding indicates that these pathways may contribute to the cognitive dysfunction observed in GD patients. The enriched pathways predominantly associated with the study are primarily involved in Autoimmune Thyroid Disease, Reproductive Health, Neurodevelopmental Disorders, Ocular and Neurological Disorders, Metabolic Disorders, and Hematological Disorders. Furthermore, our study identified the top enriched KEGG pathway related to neuroactive ligand-respecter interaction, which plays a crucial role in the production of thyroid-stimulating immunoglobulins in GD (Fig. 5E). These autoantibodies bind to and activate the TSHR on the surface of thyroid cells, leading to the overproduction of thyroid hormones and hyperthyroidism. Overall, our functional downstream analysis of the selected immuno-module revealed several significant biological processes and pathways associated with GD pathogenesis, including those related to immune response, inflammation, cognitive dysfunction, and hyperthyroidism. These findings may have implications for the development of targeted therapeutic approaches for the treatment of GD.
Table 1.
Pathways and processes enriched in GD driver genes within immune modules.
| Pathway | Sourcea | p-value | Hit Count in Genome | Mean Hit in Query List | |
|---|---|---|---|---|---|
| Pathway | Autoimmune Thyroid Disease | OMIM MedGen | 0.00165 | 20 | LRPPRC, |
| Immature Ovarian Teratoma | DisGeNET BeFree | 0.00247 | 24 | NDUFB9, PARRS | |
| DNA Bibding Trasciption factor Activilty | DisGeNET Curated | 0.00609 | 32 | FAR1, ATP13A2 | |
| Microcephaly, Primary Autosomal Recessive | DisGeNET Curated | 0.00109 | 19 | LRRC57, LRPPRC | |
| Cataracts, Spastic Paraparesis, And Speech Delay | Clinical Variations | 0.00129 | 23 | HCRT, RPS28 | |
| Peroxisomal Fatty Acyl-Coa Reductase Disorder | OMIM MedGen | 0.00129 | 15 | CDK5RAP2 | |
| Congenital Lactic Acidosis | Clinical Variations | 0.0049 | 21 | NDUFB9, RPS28, SLCO3A1 | |
| Diamond-Blackfan Anemia 15 | OMIM MedGen | 0.00179 | 5 | FAR1, GPAM | |
| Microcephaly 3, Primary, Autosomal Recessive | Clinical Variations | 0.00121 | 7 | LRPPRC, CDK5RAP2 | |
| Neuroactive ligand -respecter interaction | OMIM MedGen | 0.00106 | 43 | P2RY4, PCDHA1 | |
| Biological Process. | Monocarboxylic acid metabolic process | OMIM MedGen | 0.0024 | 46 | LRPPRC,P2RY4 |
| Carbohydrate derivative metabolic process | DisGeNET BeFree | 0.0027 | 46 | P2RY4, SLC38A8 | |
| Fatty acid metabolic process | Clinical Variations | 0.0072 | 29 | CDK5RAP2 | |
| Positive regulation of cell activation | OMIM MedGen | 0.0029 | 26 | USP8, SSPN | |
| Regulation of DNA−binding transcription factor activity | DisGeNET BeFree | 0.0039 | 25 | P2RY4, HCRTR | |
| Positive regulation of DNA binding transcription | DisGeNET Curated | 0.0037 | 42 | PARRS, P2RY4 | |
| Multicellular organismal process | DisGeNET Curated | 0.038 | 42 | RPS28 | |
| Urogenital system development | DisGeNET BeFree | 0.0451 | 55 | CDK5RAP2 | |
| Kidney development | DisGeNET Curated | 0.0451 | 124 | NDUFB9 | |
| Renal system development | DisGeNET BeFree | 0.0451 | 325 | SEC14L1 | |
| Positive regulation of NF−kappa B transcription factor activity | DisGeNET Curated | 0.0671 | 56 | LRRC57, LRPPRC | |
| Negative regulation of signal transduction | DisGeNET BeFree | 0.0951 | 56 | PFKFB2 | |
| Negative regulation of cell communication | DisGeNET BeFree | 0.0451 | 69 | FAR1, HCRTR | |
| Negative regulation of signaling | OMIM MedGen | 0.0521 | 126 | REPS1, P2RY4 |
Fig. 5.
Functional analyses of the selected immune module. (A) Pathway enrichment and main pathway networks of top-selected immune module dark-red module. (B) The PPI bipartite network analysis for the dark-red module. (C) Cognitive impairment analysis of the selected immune module. (D) Integration of the selected biological processes of purinergic signaling pathways, neuroactive ligand-receptor interaction, and DNA binding transcription factor activity. (E) Signaling enriched KEGG pathway analysis of purinergic signaling pathways. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Potential key immune-therapeutics mRNAs
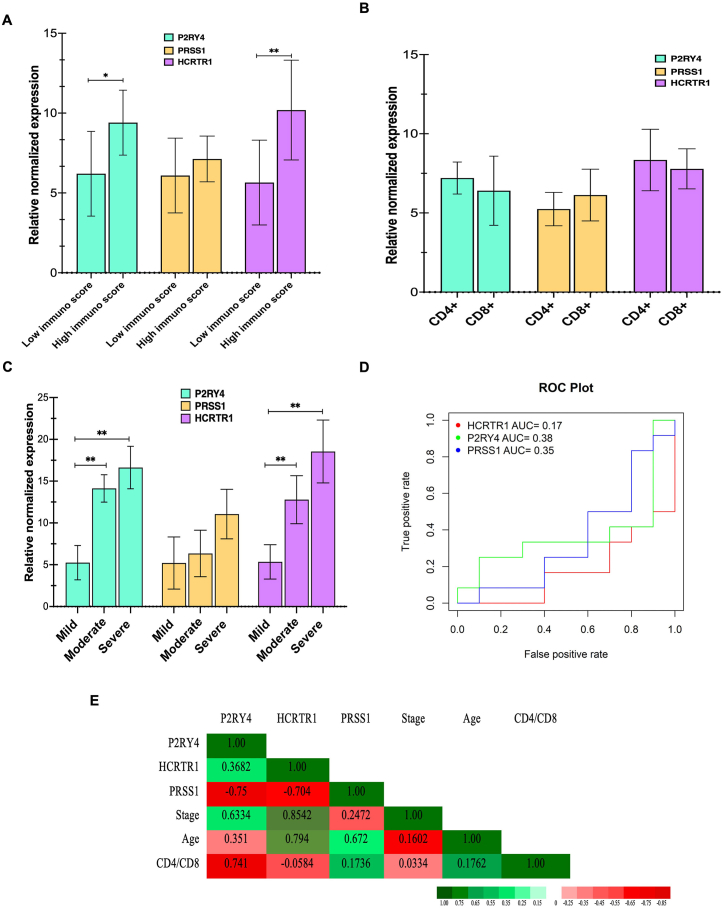
The co-expression and PPI networks analyzed in this research identified three hub genes that are associated with immune-related processes in GD (Table 2). These genes are protease serine 1 (PRSS1), hypocretin receptor 1 (HCRTR1), and purinergic receptor p2y, g protein-coupled, 4 (P2RY4), which were identified as hub genes based on co-expression network nodes ≥5. Gene ontology analysis showed that these genes are mainly involved in immune-related processes via the purinergic signaling pathways, specifically through the neuroactive ligand-receptor interaction and DNA binding transcription factor activity. The study found that the mean expression of HCRTR1 and P2RY4 was significantly higher in samples with high immune scores in GD compared to low-immune GD samples (Fig. 6A). However, when comparing the levels of these genes between CD4+ and CD8+ GD samples, no significant differences were found. Specifically, there was no significant difference in the mean expression of PRSS1, HCRTR1, and P2RY4 genes between the CD4+ (n = 25) and CD8+ (n = 24) DG samples (Fig. 6B; p > 0.05). Further analysis showed that the expression of HCRTR1 was significantly increased in severe and moderate stages of GD compared to the mild group (Fig. 6C; p ≤ 0.001). Additionally, P2RY4 levels were significantly higher in the severe-stage GD group (n = 18) compared to the mild-stage samples (n = 11; p = 0.003).
Table 2.
List of the top hub genes in the selected immune module.
| Substance Gene | Log 2 (FC) | P-value | Co-expression node | Nodes of PPI-networks notes |
|---|---|---|---|---|
| P2RY4 | 0.32342 | 0.0023 | 43 | 8 |
| PARRS | 0.3114 | 0.0032 | 24 | 5 |
| FAR1 | 0.1936 | 0.0451 | 34 | 4 |
| LRRC57 | 0.2298 | 0.0312 | 16 | 4 |
| HCRTR | 0.3481 | 0.0012 | 26 | 6 |
| CDK5RAP2 | 0.3006 | 0.0453 | 10 | 4 |
| LRPPRC | 0.2104 | 0.0239 | 18 | 3 |
| NDUFB9 | 0.2208 | 0.0353 | 11 | 4 |
Fig. 6.
Diagnostic accuracy analysis of targeted mRNA. (A) Boxplots of the targeted potential key mRNA drivers T cell proliferation, PRSS1, HCRTR1, and P2RY4 between high- and low-immune score samples (A), between CD4+ and CD8+ T cell samples (B), and different stages of GD samples (C) groups. These three genes were overexpressed in the metastatic groups. Horizontal bars represent mean ± SD. (D) Receiver operating characteristic (ROC) curve for PRSS1, HCRTR1, and P2RY4 expression. (E) Correlation of PRSS1, HCRTR1, and P2RY4 with other clinical trials of the samples.
3.4. Prognostic value of potential key mRNAs
We also analyzed the relative expression levels of targeted novel mRNAs associated with GD driver genes that have interacted with disease survival. In addition, we presented the ROC curves for the expressions of PRSS1, HCRTR1, and P2RY4. The area under the ROC curve (AUC) for PRSS1, HCRTR1, and P2RY4 were 0.35, 0.17, and 0.38, respectively (Fig. 6D). However, the study did not find any significant distinctive biomarkers among different stages and other groups of samples. To analyze the correlation of these genes with GD stages, patient age, and CD4+/CD8+ profiles, we used Spearman-rank correlation. The results showed that HCRTR1 and P2RY4 genes were significantly correlated with each other and with other risk factors (Fig. 6E). Specifically, higher levels of HCRTR1 were positively correlated with patient age and disease stage (r = 0.794 and r = 0.8542, respectively, both p ≤ 0.001). Meanwhile, P2RY4 expression was found to be correlated with the stage of the disease (r = 0.6334), but PRSS1 did not show any correlation with these clinical parameters. Overall, our findings indicate that higher levels of HCRTR1 and P2RY4 are associated with greater disease severity in GD patients. This suggests that these genes may serve as potential diagnostic biomarkers for distinguishing GD patients with different stages. However, it should be noted that further research is needed to confirm these findings, and a larger sample size may be required.
4. Discussion
In this study, we utilized sophisticated computational tools such as IPS, MCP, X-cell algorithms, and WGCNA to identify key biological pathways and processes involved in GD. Our findings indicate that genes involved in downstream pathways of the immune response, inflammation, cognitive dysfunction, and hyperthyroidism play important roles in GD pathogenesis. The positive correlation between GD-related genes and immune scores is intriguing and suggests a potential role for GD-related pathways in modulating immune cell infiltration and activation in GD. Our analysis identified five main modules - blue, cyan, dark red, midnight blue, and royal blue - that exhibited high immune scores. Notably, these modules showed significant positive correlations with age and pathological stage, distinguishing them from other module genes. Of the identified hub genes, we found that PRSS1, HCRTR1, and P2RY4 may function as novel genes that increase in severe and moderate stages of GD compared to the mild group. These genes may play a role in CD4+ and CD8+ T infiltration and activation during the progression of GD. To our knowledge, this is the first study to report that P2RY4 and HCRTR1 levels are significantly higher in the severe-stage GD group and are significantly correlated with the disease stage.
Despite extensive research, the exact triggers for autoantibody production and immune activation in GD remain unknown [13,39]. The complexity of the interactions between immune cells and autoantibodies in the thyroid gland further complicates the understanding of the disease pathogenesis [40]. In GD, immune dysregulation is primarily mediated by T and B lymphocytes. Among T cells, CD4+ T helper cells and CD8+ cytotoxic T cells play a crucial role in the autoimmune attack on the thyroid gland. CD4+ T cells differentiate into various subsets, including Th1, Th2, and Th17 cells, upon recognizing antigens presented by antigen-presenting cells [41]. In GD, Th1 cells produce pro-inflammatory cytokines such as interferon-gamma and tumor necrosis factor-alpha, which activate macrophages and stimulate B cells to produce autoantibodies against the thyroid gland. These autoantibodies bind to the TSHR on the surface of thyroid cells and stimulate the thyroid gland to produce excess thyroid hormones [42]. In some patients with GD, the autoantibodies can also bind to other receptors such as the insulin-like growth factor-1 receptor (IGF-1R), and contribute to the development of ophthalmopathy [43]. On the other hand, Th2 cells produce anti-inflammatory cytokines such as interleukin-4 (IL-4) and interleukin-10 (IL-10), which can inhibit Th1 cells and suppress autoantibody production [41,44]. However, in some cases of GD, Th2 cells can also contribute to the autoimmune attack by secreting cytokines that stimulate B cells to produce autoantibodies [43]. Understanding these interactions is essential for developing targeted therapies that can modulate the immune response and improve the clinical outcomes of patients with GD [45]. There is a significant gap in our understanding of immunology of GD and further research is necessary to fill these gaps and develop effective therapies for this debilitating autoimmune disorder.
Our investigation embarks on a groundbreaking quest into the nuanced immunological terrain of GD, harnessing cutting-edge computational tools like immune landscape algorithms IPS, MCP, and Xcell algorithms. These tools enable the analysis of large-scale gene expression data to identify key biological pathways and processes involved in the immunology of GA [46]. Diverging from traditional approaches, we integrate novel methodologies such as IPS, MCP, and X-cell algorithms to comprehensively decode the molecular intricacies governing autoantibody production in GD [[17], [18], [19], [20],[22], [23], [24], [25]]. Through the amalgamation of expression data from stromal and immune cells within GD-affected tissues, we unveil a disrupted immune network, offering profound insights into the intricate interactions steering GD pathogenesis. In this study, we first sorted and filtered our samples using three different immune component algorithms, namely IPS, MCP, and Xcell, before conducting WGCNA analysis. IPS is a prognostic score that is based on the expression levels of 18 immune-related genes and is a reliable predictor of survival outcomes in various immune diseases, including GD [47]. MCP is a widely used gene expression deconvolution algorithm that quantifies the relative proportions of different immune cell types in a mixed population of cells based on their gene expression profiles [[19], [20], [21]]. The Xcell algorithm is a computational method that estimates the levels of gene expression and cell type abundance in bulk tissue samples, which is a crucial tool in immunology research for understanding the underlying mechanisms of immunological diseases [22,23]. Our downstream analysis of the selected immuno-module revealed several significant biological processes and pathways associated with GD pathogenesis, including those related to immune response, purinergic signaling, inflammation, cognitive dysfunction, and hyperthyroidism. These findings may have important implications for the development of targeted therapeutic approaches for the treatment of GD. This pioneering framework not only advances our comprehension of GD immunology but also leads the way in developing prognostic markers for predicting the immune microenvironment status in GD patients. Our research stands as a beacon in deciphering and addressing the intricate complexities inherent in this autoimmune disorder.
Our investigation into the dark-red module highlighted robust interactions among genes, indicating shared functional significance. Conducting GO and KEGG pathway analyses on this module unveiled crucial pathways associated with GD pathogenesis. Noteworthy findings encompass heightened DNA binding transcription factor processes, links to conditions like Microcephaly, primary autosomal recessive cataracts, spastic paraparesis, speech delay, and perturbations related to peroxisomal fatty Acyl-Coa reductase disorder [48]. These insights underscore the potential involvement of these processes in the mechanisms driving GD [48,49].
Further analysis through KEGG pathways identified significant associations between dark-red module genes and pathways integral to neuroactive ligand-respecter interactions and DNA binding transcription factor activity within purinergic signaling [50]. Recognized for their roles in regulating immune responses and inflammation, these pathways have implications for various autoimmune disorders, including GD [51]. Notably, our study unveiled pathways associated with cognitive impairment, suggesting a potential link between the molecular mechanisms identified in GD and observed cognitive dysfunction. The integrated analysis of purinergic signaling pathways, neuroactive ligand-respecter interaction, and DNA binding transcription factor activity offered a comprehensive view of their interplay in contributing to GD pathophysiology [[51], [52], [53]]. Moreover, our investigation revealed the paramount role of the neuroactive ligand-respecter interaction pathway in producing thyroid-stimulating immunoglobulins in the GD [50]. These autoantibodies play a pivotal role by binding to and activating the TSHR on the surface of thyroid cells, leading to the overproduction of thyroid hormones and hyperthyroidism [54,55]. This critical finding provides a molecular understanding of the mechanisms driving excessive thyroid hormone release in GD patients. Overall, our study sheds light on the intricate interplay of pathways associated with immune response, inflammation, cognitive dysfunction, and hyperthyroidism, offering valuable insights for potential targeted therapeutic approaches in GD treatment.
Our research utilized computational algorithms to identify that higher expression levels of PRSS1, HCRTR1, and P2RY4 immune hub genes were linked to specific biological processes in the severe stage of GD. These genes may potentially be used as prognostic biomarkers to differentiate between GD patients of varying ages and disease stages. Previous studies have demonstrated the involvement of HCRTR1 and P2RY4 receptors in the development of other autoimmune diseases, including multiple sclerosis (MS), rheumatoid arthritis (RA), inflammatory bowel disease (IBD), systemic lupus erythematosus (SLE), and type 1 diabetes (T1D) [52,53,[56], [57], [58]].
In MS, the activation of HCRTR1 and HCRTR2 receptors on immune cells in the brain and spinal cord is linked to inflammation and demyelination. Targeting these receptors with specific antagonists has been shown to reduce inflammation and improve neurological symptoms in animal models of MS. The role of HCRTR1 in GD is not yet fully understood, and ongoing research is being conducted to elucidate its involvement in this autoimmune disorder. Several studies have suggested that hypocretin and its receptors, including HCRTR1, may play a role in the pathogenesis of autoimmune diseases by regulating the production and function of immune cells [58,59]. Hypocretin signaling may also affect the activity of the thyroid gland and play a role in regulating thyroid hormone levels. However, further research is needed to fully understand the involvement of HCRTR1 in the GD [58,60].
Similarly, P2RY4 receptors have been shown to play a key role in the activation of immune cells and the production of pro-inflammatory cytokines in RA and IBD [59]. P2RY4 is a gene that encodes for a purinergic receptor, which binds to extracellular nucleotides and mediates various physiological responses, including immune responses [61]. While there is limited research on the role of P2RY4 in the immunology of GD specifically, there is evidence to suggest that purinergic signaling pathways may be involved in the regulation of immune responses, and dysregulation of these pathways may contribute to autoimmune diseases such as GD [52]. Purinergic signaling involves the release of nucleotides, such as adenosine triphosphate (ATP), from cells in response to various stimuli, including inflammation. These nucleotides can then bind to purinergic receptors on immune cells, leading to the activation of immune responses [52]. P2RY4 is one such purinergic receptor that is expressed on various immune cells, including T cells, B cells, and DCs. In GD disease, autoantibodies are produced that bind to and stimulate the TSHR on thyroid cells, leading to increased production of thyroid hormones and hyperthyroidism [62]. The underlying genetic predisposition and environmental factors, such as inflammation, are believed to trigger the production of autoantibodies in GD. Purinergic signaling pathways are involved in regulating inflammation and immune responses, and P2RY4-mediated signaling is suggested to play a role in the development and progression of GD. When purinergic receptors on immune cells are activated, they can produce pro-inflammatory cytokines and chemokines that contribute to the autoimmune response in GD.
Targeting P2RY4 receptors with specific antagonists has been shown to reduce inflammation and improve symptoms in animal models of these diseases. HCRTR1 has also been implicated in regulating T cell function and promoting autoantibody production in SLE [60]. In animal models of SLE, HCRTR1 antagonists have been shown to reduce disease severity and prevent organ damage. Furthermore, P2RY414 has been shown to promote inflammation and cell death in T1D. In animal models of T1D, targeting P2RY414 with specific antagonists has been shown to protect against beta cell destruction and improve glucose control [63].
Our hub-immunogens express the PRSS1 gene, which encodes for the serine protease enzyme trypsinogen [50,64,65]. This enzyme is activated to trypsin by enterokinase and is primarily involved in the digestion of proteins in the small intestine. Although there is no direct evidence that PRSS1 plays a role in the immunology of GDe, studies suggest that dysregulated proteases and protease inhibitors are associated with autoimmunity and inflammation in GD [66]. Proteases, including serine proteases like PRSS1, play important roles in various physiological processes, such as inflammation and tissue remodeling [67,68]. In GD, autoantibodies that bind to and stimulate the TSHR on thyroid cells are produced, leading to increased production of thyroid hormones and hyperthyroidism [69,70]. The production of these autoantibodies is believed to be triggered by a genetic predisposition and environmental factors. Proteases, such as PRSS1, may play a role in producing and regulating these autoantibodies [50,71]. For instance, proteases can activate pro-inflammatory cytokines involved in the immune response, and they can break down proteins that act as inhibitors of autoantibody production, thereby increasing autoantibody production [68].
Understanding the elevated expression of immune hub genes, notably PRSS1, HCRTR1, and P2RY4, in severe stages of GD presents a transformative opportunity for personalized medicine. These genes, indicative of disease severity, could revolutionize diagnostics by offering more precise biomarkers. The discovery of heightened expression levels of immune hub genes, specifically PRSS1, HCRTR1, and P2RY4, in severe stages of GD carries significant clinical implications. These genes demonstrate potential as robust biomarkers, offering a distinct diagnostic advantage by serving as reliable indicators of GD severity. This molecular insight can greatly improve diagnostic precision, enabling clinicians to initiate timely and accurate interventions. Beyond diagnostics, the observed upregulation of PRSS1, HCRTR1, and P2RY4 provides valuable prognostic insights into the trajectory of GD. Monitoring the expression levels of these genes could establish a predictive framework, allowing healthcare professionals to anticipate the severity trajectory and tailor management strategies accordingly [74,75]. This prognostic dimension contributes to a more informed and proactive approach to patient care, potentially mitigating complications associated with severe stages of GD. Moreover, they provide valuable prognostic insights, aiding in the anticipation and management of complications. The potential for tailored treatment strategies targeting PRSS1, HCRTR1, and P2RY4 emerges as a novel therapeutic avenue, promising improved treatment efficacy. This shift towards personalized medicine recognizes the unique genetic profiles of individual patients, fostering innovative approaches and the prospect of more effective interventions for GD. The insights gained from these analyses provide a deeper understanding of the complex interactions between stromal and immune cells in the thyroid gland, as well as the molecular mechanisms underlying autoantibody production and the immune activation [72,73]. The use of these advanced computational tools holds great promise in furthering our knowledge of the immunology of GDP and in the development of novel and more effective treatments for this autoimmune disorder [72]. HCRTR1 and P2RY4 receptors may hold promise as potential targets for the development of new treatments [74,75]. However, more research is needed to fully understand the roles of these receptors in autoimmune diseases and to develop safe and effective drugs targeting these receptors.
While challenges and considerations exist, the promise of individualized care underscores the significance of incorporating personalized medicine into the clinical landscape, paving the way for enhanced outcomes in GD management. However, the limitations of our study should be acknowledged, including the small sample size and the lack of functional studies to elucidate the underlying mechanisms of HCRTR1 and P2RY4 in GD. In addition to the acknowledged limitations, our study prompts further avenues for exploration to deepen our understanding of the immunological intricacies of GD. Firstly, while our bioinformatic analysis identified potential gene interactions, it remains imperative to delve into the cellular and molecular aspects of these interactions. Future studies could employ advanced cellular and molecular techniques to unravel the precise mechanisms through which PRSS1, HCRTR1, and P2RY4 influence immune responses in GD and potentially in other autoimmune disorders [74]. Secondly, extending our findings to in vitro and in vivo models would facilitate a more comprehensive exploration of the overexpression and downregulation dynamics of these genes. Such modeling efforts can unravel their roles in immune dysregulation pathways, shedding light on novel therapeutic targets. Thirdly, investigating the mutation landscape of PRSS1, HCRTR1, and P2RY4 in GD patients may uncover genetic variations contributing to disease susceptibility and progression [76]. A focused examination of genetic mutations could provide valuable insights into individualized treatment approaches and enhance our understanding of GD heterogeneity. Finally, comprehensive pathway analyses elucidating the molecular functions of these genes in GD pathogenesis would be instrumental [77]. Unraveling their roles in key pathways could unveil potential druggable targets and inform the development of targeted therapies. Ultimately, while recognizing our current limitations, these suggested directions for future research aim to unravel the intricate immunological landscape of GD and other autoimmune diseases, paving the way for more personalized and effective therapeutic interventions.
5. Conclusions
In summary, our study has developed a model that predicts the prognosis and immunotherapy effectiveness of GD based on an immunogenic and immune infiltration-associated strategy guided by senescence genes. The model's performance was validated using external transcriptome data and immunotherapy data, and a three-gene signature (PRSS1, HCRTR1, and P2RY4) was identified as a prognostic and therapeutic biomarker for GD. By delving into the potential roles of HCRTR1 and P2RY4 genes in GD pathogenesis and their potential use as diagnostic biomarkers to differentiate between different disease stages, our study sheds light on the underlying mechanisms of GD.
Funding statement
This work will receive any research grant The funders had no role in the study design, data collection, analysis, the decision to publish, or the preparation of the manuscript. There was no additional external funding received for this study.
Data availability statement
All relevant data are within the manuscript and its Supporting Information files.
CRediT authorship contribution statement
Nianrong Mi: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Zhe Li: Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. Xueling Zhang: Formal analysis, Investigation, Methodology, Supervision, Validation. Yingjing Gao: Data curation, Formal analysis, Investigation, Methodology, Validation. Yanan Wang: Data curation, Software, Visualization. Siyan Liu: Investigation, Resources, Visualization. Shaolian Wang: Project administration, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27175.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
The immunoassay measurements of the IPC and MCP algorithms.
The genes list of the selected immune module.
Fig. S1.
Immune cell subsets distribution by three algorithms - IPS, MCP, and Xcell.
Fig. S2.
Significance of Biological Pathways in Purinergic Signaling in GD. This Figure shows the significance of the most prominent biological pathways related to neuroactive ligand-receptor interactions and DNA binding transcription factor activity in the purinergic signaling pathways, which have been linked to the regulation of immune response and inflammation in GD.
References
- 1.Antonelli A., Fallahi P., Elia G., Ragusa F., Paparo S.R., Ruffilli I., et al. Graves' disease: clinical manifestations, immune pathogenesis (cytokines and chemokines) and therapy. Best Pract. Res. Clin. Endocrinol. Metabol. 2020;34(1) doi: 10.1016/j.beem.2020.101388. Epub 2020/02/16. PubMed PMID: 32059832. [DOI] [PubMed] [Google Scholar]
- 2.Pouso-Diz J.M., Abalo-Lojo J.M., Gonzalez F. Thyroid eye disease: current and potential medical management. Int. Ophthalmol. 2020;40(4):1035–1048. doi: 10.1007/s10792-019-01258-7. Epub 2020/01/11. PubMed PMID: 31919775. [DOI] [PubMed] [Google Scholar]
- 3.Hussain Y.S., Hookham J.C., Allahabadia A., Balasubramanian S.P. Epidemiology, management and outcomes of Graves' disease-real life data. Endocrine. 2017;56(3):568–578. doi: 10.1007/s12020-017-1306-5. Epub 2017/05/10. PubMed PMID: 28478488; PubMed Central PMCID: PMCPMC5435772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor P.N., Albrecht D., Scholz A., Gutierrez-Buey G., Lazarus J.H., Dayan C.M., et al. Global epidemiology of hyperthyroidism and hypothyroidism. Nat. Rev. Endocrinol. 2018;14(5):301–316. doi: 10.1038/nrendo.2018.18. Epub 2018/03/24. PubMed PMID: 29569622. [DOI] [PubMed] [Google Scholar]
- 5.Wang C., Li Y., Teng D., Shi X., Ba J., Chen B., et al. Hyperthyroidism prevalence in China after Universal Salt Iodization. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.651534. Epub 2021/06/15. PubMed PMID: 34122333; PubMed Central PMCID: PMCPMC8194401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Han B., Yu J., Chen Y., Cheng J., Zhu C., et al. Influence of rapid urbanization on thyroid autoimmune disease in China. Internet J. Endocrinol. 2021;2021 doi: 10.1155/2021/9967712. Epub 2021/06/15. PubMed PMID: 34122544; PubMed Central PMCID: PMCPMC8189768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch H.B., Perros P., Bednarczuk T., Cooper D.S., Dolman P.J., Leung A.M., et al. Management of thyroid eye disease: a consensus statement by the American thyroid association and the European thyroid association. Thyroid. 2022;32(12):1439–1470. doi: 10.1089/thy.2022.0251. Epub 2022/12/09. PubMed PMID: 36480280; PubMed Central PMCID: PMCPMC9807259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mooij C.F., Cheetham T.D., Verburg F.A., Eckstein A., Pearce S.H., Leger J., et al. 2022 European thyroid association guideline for the management of pediatric Graves' disease. Eur. Thyroid J. 2022;11(1) doi: 10.1530/ETJ-21-0073. Epub 2022/01/05. PubMed PMID: 34981748; PubMed Central PMCID: PMCPMC9142815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutterman S.L., Zwaveling-Soonawala N., Verberne H.J., Verburg F.A., van Trotsenburg A.S.P., Mooij C.F. The efficacy and short- and long-term side effects of radioactive iodine treatment in pediatric Graves' disease: a systematic review. Eur. Thyroid J. 2021;10(5):353–363. doi: 10.1159/000517174. Epub 2021/09/21. PubMed PMID: 34540705; PubMed Central PMCID: PMCPMC8406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semeniene K., Dauksa A., Makstiene J., Sarauskas V., Velickiene D. Sporadic medullary thyroid carcinoma in Graves' disease. Acta Endocrinol. 2022;18(3):368–374. doi: 10.4183/aeb.2022.368. Epub 2023/01/27. PubMed PMID: 36699162; PubMed Central PMCID: PMCPMC9867812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese B.M., Noureldine S.I., Gleeson E.M., Tufano R.P., Kandil E. What is the best definitive treatment for Graves' disease? A systematic review of the existing literature. Ann. Surg Oncol. 2013;20(2):660–667. doi: 10.1245/s10434-012-2606-x. Epub 2012/09/08. PubMed PMID: 22956065. [DOI] [PubMed] [Google Scholar]
- 12.Dwivedi S.N., Kalaria T., Buch H. Thyroid autoantibodies. J. Clin. Pathol. 2023;76(1):19–28. doi: 10.1136/jcp-2022-208290. Epub 2022/10/22. PubMed PMID: 36270794. [DOI] [PubMed] [Google Scholar]
- 13.Pujol-Borrell R., Gimenez-Barcons M., Marin-Sanchez A., Colobran R. Genetics of Graves' disease: special focus on the role of TSHR gene. Horm. Metab. Res. 2015;47(10):753–766. doi: 10.1055/s-0035-1559646. Epub 2015/09/12. PubMed PMID: 26361261. [DOI] [PubMed] [Google Scholar]
- 14.Salvi M., Covelli D. B cells in Graves' Orbitopathy: more than just a source of antibodies? Eye (Lond) 2019;33(2):230–234. doi: 10.1038/s41433-018-0285-y. Epub 2018/12/06. PubMed PMID: 30514895; PubMed Central PMCID: PMCPMC6367428 declares that she has no conflict of interest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kustrimovic N., Gallo D., Piantanida E., Bartalena L., Lai A., Zerbinati N., et al. Regulatory T cells in the pathogenesis of Graves' disease. Int. J. Mol. Sci. 2023;24(22) doi: 10.3390/ijms242216432. Epub 2023/11/25. PubMed PMID: 38003622; PubMed Central PMCID: PMCPMC10671795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombardi A., Menconi F., Greenberg D., Concepcion E., Leo M., Rocchi R., et al. Dissecting the genetic susceptibility to Graves' disease in a cohort of patients of Italian origin. Front. Endocrinol. 2016;7:21. doi: 10.3389/fendo.2016.00021. Epub 2016/03/26. PubMed PMID: 27014188; PubMed Central PMCID: PMCPMC4781855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroenke M.A., Milton M.N., Kumar S., Bame E., White J.T. Immunogenicity risk assessment for multi-specific therapeutics. AAPS J. 2021;23(6):115. doi: 10.1208/s12248-021-00642-5. Epub 2021/11/07. PubMed PMID: 34741215; PubMed Central PMCID: PMCPMC8571146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattei A.E., Gutierrez A.H., Martin W.D., Terry F.E., Roberts B.J., Rosenberg A.S., et al. In silico immunogenicity assessment for sequences containing unnatural amino acids: a method using existing in silico algorithm infrastructure and a vision for future enhancements. Front Drug Discov (Lausanne) 2022;2 doi: 10.3389/fddsv.2022.952326. Epub 2022/01/01. PubMed PMID: 36945694; PubMed Central PMCID: PMCPMC10026553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. Epub 2016/10/22. PubMed PMID: 27765066; PubMed Central PMCID: PMCPMC5073889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petitprez F., Levy S., Sun C.M., Meylan M., Linhard C., Becht E., et al. The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 2020;12(1):86. doi: 10.1186/s13073-020-00783-w. Epub 2020/10/08. PubMed PMID: 33023656; PubMed Central PMCID: PMCPMC7541325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturm G., Finotello F., Petitprez F., Zhang J.D., Baumbach J., Fridman W.H., et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i445. doi: 10.1093/bioinformatics/btz363. Epub 2019/09/13. PubMed PMID: 31510660; PubMed Central PMCID: PMCPMC6612828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. Epub 2017/11/17. PubMed PMID: 29141660; PubMed Central PMCID: PMCPMC5688663 The authors declare that they have no competing interests. PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D. Cell-type enrichment analysis of bulk transcriptomes using xCell. Methods Mol. Biol. 2020;2120:263–276. doi: 10.1007/978-1-0716-0327-7_19. Epub 2020/03/04. PubMed PMID: 32124326. [DOI] [PubMed] [Google Scholar]
- 24.Yao Q., Song Z., Wang B., Jia X., Song R., Zhang J. Identification of lncRNA and mRNA expression profile in relapsed Graves' disease. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.756560. Epub 2021/12/21. PubMed PMID: 34926448; PubMed Central PMCID: PMCPMC8673561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Q., Song Z., Wang B., Qin Q., Zhang J.A. Identifying key genes and functionally enriched pathways in sjögren's syndrome by weighted gene Co-expression network analysis. Front. Genet. 2019;10:1142. doi: 10.3389/fgene.2019.01142. Epub 2019/12/05. PubMed PMID: 31798636; PubMed Central PMCID: PMCPMC6863930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu K., Wang L., Fu Y., Li G., Zhang X., Cao M. Bioinformatics analysis identifies immune-related gene signatures and subtypes in diabetic nephropathy. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.1048139. Epub 2022/12/27. PubMed PMID: 36568106; PubMed Central PMCID: PMCPMC9768367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Peng W., Li C., Qin R., Zhong Z., Sun C. Identification of an immune-related signature indicating the dedifferentiation of thyroid cells. Cancer Cell Int. 2021;21(1):231. doi: 10.1186/s12935-021-01939-3. Epub 2021/04/25. PubMed PMID: 33892730; PubMed Central PMCID: PMCPMC8067302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundararajan Z., Knoll R., Hombach P., Becker M., Schultze J.L., Ulas T. Shiny-Seq: advanced guided transcriptome analysis. BMC Res. Notes. 2019;12(1):432. doi: 10.1186/s13104-019-4471-1. Epub 2019/07/20. PubMed PMID: 31319888; PubMed Central PMCID: PMCPMC6637470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahaly G.J., Bartalena L., Hegedüs L., Leenhardt L., Poppe K., Pearce S.H. 2018 European thyroid association guideline for the management of Graves' hyperthyroidism. Eur. Thyroid J. 2018;7(4):167–186. doi: 10.1159/000490384. Epub 2018/10/05. PubMed PMID: 30283735; PubMed Central PMCID: PMCPMC6140607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serban M. Exploring modularity in biological networks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375(1796) doi: 10.1098/rstb.2019.0316. Epub 2020/02/25. PubMed PMID: 32089119; PubMed Central PMCID: PMCPMC7061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L., Langfelder P., Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinf. 2012;13:328. doi: 10.1186/1471-2105-13-328. Epub 2012/12/12. PubMed PMID: 23217028; PubMed Central PMCID: PMCPMC3586947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piñero J., À Bravo, Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1) doi: 10.1093/nar/gkw943. D833-d9. Epub 2016/12/08. PubMed PMID: 27924018; PubMed Central PMCID: PMCPMC5210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang G., Liu S., Maghsoudloo M., Dehghan Shasaltaneh M., Jabbarzadeh Kaboli P., Zhang C., et al. 2021. PLA1A Expression as a Diagnostic Marker of BRAF-Mutant Metastasis in Melanoma Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan M.A., Tania M., Wei C., Mei Z., Fu S., Cheng J., et al. Thymoquinone inhibits cancer metastasis by downregulating TWIST1 expression to reduce epithelial to mesenchymal transition. Oncotarget. 2015;6(23):19580–19591. doi: 10.18632/oncotarget.3973. PubMed PMID: 26023736; PubMed Central PMCID: PMC4637306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imani S., Wei C., Cheng J., Khan M.A., Fu S., Yang L., et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8(13):21362–21379. doi: 10.18632/oncotarget.15214. Epub 2017/04/21. PubMed PMID: 28423483; PubMed Central PMCID: PMCPMC5400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. Epub 2013/01/18. PubMed PMID: 23323831; PubMed Central PMCID: PMCPMC3618321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y., Liu J., Lu J., Peng J., Juan L., Zhu X., et al. Joint detection of copy number variations in parent-offspring trios. Bioinformatics. 2016;32(8):1130–1137. doi: 10.1093/bioinformatics/btv707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Lu Q., Shao X., Mo B., Nie X., Liu W., et al. Development of A three-gene prognostic signature for hepatitis B virus associated hepatocellular carcinoma based on integrated transcriptomic analysis. J. Cancer. 2018;9(11):1989–2002. doi: 10.7150/jca.23762. PubMed PMID: 29896284; PubMed Central PMCID: PMCPMC5995946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morshed S.A., Latif R., Davies T.F. Delineating the autoimmune mechanisms in Graves' disease. Immunol. Res. 2012;54(1–3):191–203. doi: 10.1007/s12026-012-8312-8. Epub 2012/03/22. PubMed PMID: 22434518; PubMed Central PMCID: PMCPMC4504182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gimenez-Barcons M., Colobran R., Gomez-Pau A., Marin-Sanchez A., Casteras A., Obiols G., et al. Graves' disease TSHR-stimulating antibodies (TSAbs) induce the activation of immature thymocytes: a clue to the riddle of TSAbs generation? J. Immunol. 2015;194(9):4199–4206. doi: 10.4049/jimmunol.1500183. Epub 2015/03/25. PubMed PMID: 25801430. [DOI] [PubMed] [Google Scholar]
- 41.Esfahanian F., Naimi E., Doroodgar F., Jadali Z. Th1/Th2 cytokines in patients with Graves' disease with or without ophthalmopathy. Iran. J. Allergy, Asthma Immunol. 2013;12(2):168–175. Epub 2013/06/12. PubMed PMID: 23754356. [PubMed] [Google Scholar]
- 42.Cheng C.W., Fang W.F., Tang K.T., Lin J.D. Serum interferon levels associated with the disease activity in women with overt Graves' disease. Cytokine. 2021;138 doi: 10.1016/j.cyto.2020.155353. Epub 2020/10/31. PubMed PMID: 33121876. [DOI] [PubMed] [Google Scholar]
- 43.Morshed S.A., Davies T.F. Graves' disease mechanisms: the role of stimulating, blocking, and cleavage region TSH receptor antibodies. Horm. Metab. Res. 2015;47(10):727–734. doi: 10.1055/s-0035-1559633. Epub 2015/09/12. PubMed PMID: 26361259; PubMed Central PMCID: PMCPMC5047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu P.W., Tang X.X., Wang Y.N., Lin S., Wang M.M., Yin Q.L., et al. Comparison of two regimens for patients with thyroid-associated ophthalmopathy receiving intravenous methyl prednisolone: a single center prospective randomized trial. Exp. Ther. Med. 2020;20(6):153. doi: 10.3892/etm.2020.9282. Epub 2020/10/24. PubMed PMID: 33093891; PubMed Central PMCID: PMCPMC7571371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang S., Lu Y., Huang Y., Zhou H., Fan X. Mechanisms that underly T cell immunity in Graves' orbitopathy. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.648732. Epub 2021/04/20. PubMed PMID: 33868176; PubMed Central PMCID: PMCPMC8049604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auslander N., Gussow A.B., Koonin E.V. Incorporating machine learning into established bioinformatics frameworks. Int. J. Mol. Sci. 2021;22(6) doi: 10.3390/ijms22062903. Epub 2021/04/04. PubMed PMID: 33809353; PubMed Central PMCID: PMCPMC8000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv W., Zhan Y., Tan Y., Wu Y., Chen H. A combined aging and immune prognostic signature predict prognosis and responsiveness to immunotherapy in melanoma. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.943944. Epub 2022/08/30. PubMed PMID: 36034849; PubMed Central PMCID: PMCPMC9402914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su X., Chen X., Peng H., Song J., Wang B., Wu X. Novel insights into the pathological development of dyslipidemia in patients with hypothyroidism. Bosn. J. Basic Med. Sci. 2022;22(3):326–339. doi: 10.17305/bjbms.2021.6606. Epub 2021/11/17. PubMed PMID: 34784265; PubMed Central PMCID: PMCPMC9162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bikeyeva V., Abdullah A., Radivojevic A., Abu Jad A.A., Ravanavena A., Ravindra C., et al. Nonalcoholic fatty liver disease and hypothyroidism: what you need to know. Cureus. 2022;14(8) doi: 10.7759/cureus.28052. Epub 2022/09/22. PubMed PMID: 36127957; PubMed Central PMCID: PMCPMC9477544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F., Wang X., Wang L., Sun X., Tan G., Wei W., et al. Genetics, epigenetics, cellular immunology, and gut microbiota: emerging links with Graves' disease. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.794912. Epub 2022/01/22. PubMed PMID: 35059400; PubMed Central PMCID: PMCPMC8765724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X., Zhao Q., Li B. Current and promising therapies based on the pathogenesis of Graves' ophthalmopathy. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1217253. Epub 2023/11/30. PubMed PMID: 38035032; PubMed Central PMCID: PMCPMC10687425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Y., Lu D., Xue M. Purinergic signaling in thyroid disease. Purinergic Signal. 2023;19(1):221–227. doi: 10.1007/s11302-022-09858-2. Epub 2022/03/30. PubMed PMID: 35347568; PubMed Central PMCID: PMCPMC9984614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silveira G.F., Buffon A., Bruno A.N. New approaches to thyroid hormones and purinergic signaling. J. Thyroid Res. 2013;2013 doi: 10.1155/2013/434727. Epub 2013/08/21. PubMed PMID: 23956925; PubMed Central PMCID: PMCPMC3730180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Struja T., Kutz A., Fischli S., Meier C., Mueller B., Recher M., et al. Is Graves' disease a primary immunodeficiency? New immunological perspectives on an endocrine disease. BMC Med. 2017;15(1):174. doi: 10.1186/s12916-017-0939-9. Epub 2017/09/26. PubMed PMID: 28942732; PubMed Central PMCID: PMCPMC5611589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prabhakar B.S., Bahn R.S., Smith T.J. Current perspective on the pathogenesis of Graves' disease and ophthalmopathy. Endocr. Rev. 2003;24(6):802–835. doi: 10.1210/er.2002-0020. Epub 2003/12/13. PubMed PMID: 14671007. [DOI] [PubMed] [Google Scholar]
- 56.Couvineau A., Voisin T., Nicole P., Gratio V., Abad C., Tan Y.V. Orexins as novel therapeutic targets in inflammatory and neurodegenerative diseases. Front. Endocrinol. 2019;10:709. doi: 10.3389/fendo.2019.00709. Epub 2019/11/07. PubMed PMID: 31695678; PubMed Central PMCID: PMCPMC6817618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Orozco F.J., Vicario J.L., Villalibre-Valderrey I., De Andrés C., Fernández-Arquero M., Peraita-Adrados R. Narcolepsy with cataplexy and comorbid immunopathological diseases. J. Sleep Res. 2014;23(4):414–419. doi: 10.1111/jsr.12143. Epub 2014/03/22. PubMed PMID: 24645699. [DOI] [PubMed] [Google Scholar]
- 58.Mohamed A.R., El-Hadidy W.F. Effect of orexin-A (hypocretin-1) on hyperalgesic and cachectic manifestations of experimentally induced rheumatoid arthritis in rats. Can. J. Physiol. Pharmacol. 2014;92(10):813–820. doi: 10.1139/cjpp-2014-0258. Epub 2014/09/12. PubMed PMID: 25211278. [DOI] [PubMed] [Google Scholar]
- 59.Heidari Z., Salimi S., Rokni M., Rezaei M., Khalafi N., Shahroudi M.J., et al. Association of IL-1β, NLRP3, and COX-2 gene polymorphisms with autoimmune thyroid disease risk and clinical features in the Iranian population. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/7729238. Epub 2021/11/19. PubMed PMID: 34790822; PubMed Central PMCID: PMCPMC8592725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapica-Topczewska K., Pogorzelski R., Tarasiuk J., Drozdowski W., Lewczuk P., Kułakowska A. Excessive daytime sleepiness in a patient with coexisting myotonic dystrophy type 1, myasthenia gravis and Graves' disease. Neurol. Neurochir. Pol. 2017;51(2):190–193. doi: 10.1016/j.pjnns.2017.01.007. Epub 2017/02/18. PubMed PMID: 28209438. [DOI] [PubMed] [Google Scholar]
- 61.Chen Z., Wang G., Xie X., Liu H., Liao J., Shi H., et al. Ginsenoside Rg5 allosterically interacts with P2RY(12) and ameliorates deep venous thrombosis by counteracting neutrophil NETosis and inflammatory response. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.918476. Epub 2022/08/30. PubMed PMID: 36032109; PubMed Central PMCID: PMCPMC9411522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reichert K.P., Castro M.F.V., Assmann C.E., Bottari N.B., Miron V.V., Cardoso A., et al. Diabetes and hypertension: pivotal involvement of purinergic signaling. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111273. Epub 2021/02/02. PubMed PMID: 33524787; PubMed Central PMCID: PMCPMC7846467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinod K., Witsch T., Farley K., Gallant M., Remold-O'Donnell E., Wagner D.D. Neutrophil elastase-deficient mice form neutrophil extracellular traps in an experimental model of deep vein thrombosis. J Thromb Haemost. 2016;14(3):551–558. doi: 10.1111/jth.13239. Epub 2015/12/30. PubMed PMID: 26712312; PubMed Central PMCID: PMCPMC4785059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szatmary P., Grammatikopoulos T., Cai W., Huang W., Mukherjee R., Halloran C., et al. vol. 82. All funds received have been paid to the University of Liverpool and/or Liverpool University Hospitals NHS Foundation Trust; 2022. Acute pancreatitis: diagnosis and treatment. Drugs. (PubMed Central PMCID: PMCPMC9454414 Cypralis, Farsight, and Corvidia, with All Funds Paid to West China Hospital of Sichuan University. RS Has Consulted for AbbVie CalciMedica, GlaxoSmithKline (GSK), Novartis and Takeda, and Has Received Research Funding from CalciMedica, EA Pharma, GSK, Lilly, Merck/MSD, Pfizer as Well as Multiple Public Sources in the Last Three Years. RS Is Collaborating in the IMI2 TransBioLine Consortium with Janssen, Lilly, Merck/MSD, Novartis, Pfizer, Roche, and Sanofi-Aventis). 12):1251-1276. Epub 2022/09/09. PubMed PMID: 36074322; [DOI] [Google Scholar]
- 65.Németh B.C., Sahin-Tóth M. Human cationic trypsinogen (PRSS1) variants and chronic pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;306(6):G466–G473. doi: 10.1152/ajpgi.00419.2013. Epub 2014/01/25. PubMed PMID: 24458023; PubMed Central PMCID: PMCPMC3949028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akhtari M., Zargar S.J., Vojdanian M., Ashraf-Ganjouei A., Javinani A., Hamzeh E., et al. P2 receptors mRNA expression profiles in macrophages from ankylosing spondylitis patients and healthy individuals. Int J Rheum Dis. 2020;23(3):350–357. doi: 10.1111/1756-185x.13783. Epub 2019/12/31. PubMed PMID: 31884692. [DOI] [PubMed] [Google Scholar]
- 67.Song M.Y., Park S. Association of polygenetic risk scores related to immunity and inflammation with hyperthyroidism risk and interactions between the polygenetic scores and dietary factors in a large cohort. J. Thyroid Res. 2021;2021 doi: 10.1155/2021/7664641. Epub 2021/09/28. PubMed PMID: 34567510; PubMed Central PMCID: PMCPMC8457978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glanville K.P., Coleman J.R.I., O'Reilly P.F., Galloway J., Lewis C.M. Investigating pleiotropy between depression and autoimmune diseases using the UK biobank. Biol Psychiatry Glob Open Sci. 2021;1(1):48–58. doi: 10.1016/j.bpsgos.2021.03.002. Epub 2021/07/20. PubMed PMID: 34278373; PubMed Central PMCID: PMCPMC8262258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anees A., Ayeni F.E., Eslick G.D., Edirimanne S. TSH receptor autoantibody levels post-total thyroidectomy in Graves' ophthalmopathy: a meta-analysis. Langenbeck's Arch. Surg. 2023;408(1):415. doi: 10.1007/s00423-023-03153-3. Epub 2023/10/23. PubMed PMID: 37870639; PubMed Central PMCID: PMCPMC10593610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chistyakov D.A., Savost'anov K.V., Turakulov R.I., Nosikov V.V. Genetic determinants of Graves disease. Mol. Genet. Metabol. 2000;71(1–2):66–69. doi: 10.1006/mgme.2000.3042. Epub 2000/09/26. PubMed PMID: 11001797. [DOI] [PubMed] [Google Scholar]
- 71.Rivas A.M., Lado-Abeal J. Thyroid hormone resistance and its management. SAVE Proc. 2016;29(2):209–211. doi: 10.1080/08998280.2016.11929421. Epub 2016/04/02. PubMed PMID: 27034574; PubMed Central PMCID: PMCPMC4790576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., Ma X.M., Wang X., Sun X., Wang L.J., Li X.Q., et al. Emerging insights into the role of epigenetics and gut microbiome in the pathogenesis of Graves' ophthalmopathy. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.788535. Epub 2022/01/25. PubMed PMID: 35069441; PubMed Central PMCID: PMCPMC8766297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallo D., Piantanida E., Gallazzi M., Bartalena L., Tanda M.L., Bruno A., et al. Immunological drivers in Graves' disease: NK cells as a master switcher. Front. Endocrinol. 2020;11:406. doi: 10.3389/fendo.2020.00406. Epub 2020/08/09. PubMed PMID: 32765422; PubMed Central PMCID: PMCPMC7379480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pulver A., Kiive E., Kanarik M., Harro J. Association of orexin/hypocretin receptor gene (HCRTR1) with reward sensitivity, and interaction with gender. Brain Res. 2020;1746 doi: 10.1016/j.brainres.2020.147013. Epub 2020/07/12. PubMed PMID: 32652147. [DOI] [PubMed] [Google Scholar]
- 75.Bufalo N.E., Dos Santos R.B., Marcello M.A., Piai R.P., Secolin R., Romaldini J.H., et al. TSHR intronic polymorphisms (rs179247 and rs12885526) and their role in the susceptibility of the Brazilian population to Graves' disease and Graves' ophthalmopathy. J. Endocrinol. Invest. 2015;38(5):555–561. doi: 10.1007/s40618-014-0228-9. Epub 2014/12/30. PubMed PMID: 25543543. [DOI] [PubMed] [Google Scholar]
- 76.Pujol-Borrell R., Giménez-Barcons M., Marín-Sánchez A., Colobran R. Genetics of Graves' disease: special focus on the role of TSHR gene. Horm. Metab. Res. 2015;47(10):753–766. doi: 10.1055/s-0035-1559646. Epub 2015/09/12. PubMed PMID: 26361261. [DOI] [PubMed] [Google Scholar]
- 77.Stefan M., Faustino L.C. Genetics of thyroid-stimulating hormone receptor-relevance for autoimmune thyroid disease. Front. Endocrinol. 2017;8:57. doi: 10.3389/fendo.2017.00057. Epub 2017/04/20. PubMed PMID: 28421036; PubMed Central PMCID: PMCPMC5376554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The immunoassay measurements of the IPC and MCP algorithms.
The genes list of the selected immune module.
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.