Abstract
In the past decade, the field of LC-MS based metabolomics has transformed from an obscure specialty into a major “-omics” platform for studying metabolic processes and biomolecular characterization. However, as a whole the field is still very fractured, as the nature of the instrumentation and of the information produced by the platform essentially creates incompatible “islands” of datasets. This lack of data coherency results in the inability to accumulate a critical mass of metabolomics data that has enabled other –omics platforms to make impactful discoveries and meaningful advances. As such, we have developed a novel algorithm, called Disparate Metabolomics Data Reassembler (DIMEDR), which attempts to bridge the inconsistencies between incongruent LC-MS metabolomics datasets of the same biological sample type. A single “primary” dataset is postprocessed via traditional means of peak identification, alignment, and grouping. DIMEDR utilizes this primary dataset as a progenitor template by which data from subsequent disparate datasets are reassembled and integrated into a unified framework that maximizes spectral feature similarity across all samples. This is accomplished by a novel procedure for universal retention time correction and comparison via identification of ubiquitous features in the initial primary dataset, which are subsequently utilized as endogenous internal standards during integration. For demonstration purposes, two human and two mouse urine metabolomics datasets from four unrelated studies acquired over 4 years were unified via DIMEDR, which enabled meaningful analysis across otherwise incomparable and unrelated datasets.
Introduction:
In the past decade, metabolomics has risen to become a crucial platform for in-depth analysis of metabolic processes and small molecule characterization in biological systems. The US National Institutes of Health’s establishment of six Regional Comprehensive Metabolomics Resource Cores, the UK Medical Research Council’s National Phenome Center, as well as investments in metabolomics core facilities by countries all over the world 1 attests to the immense potential of the field for making transformative discoveries in biological research. However, little progress has been made towards building the critical mass of harmonized data that has enabled other “-omics” platforms, namely genomics and the GenBank database, to make truly meaningful discoveries and impactful advances. This is largely due to the nature of the instrumentation and of the information produced by the platform resulting in “islands” of datasets that are often incomparable to one another. Bridging these islands to create more harmonized data frameworks is an essential step towards the field’s maturation.
Much of the field’s growth can be attributed to advances in high performance liquid chromatography (LC) coupled with high resolution mass spectrometry (MS). The proliferation of these new technologies has enabled the development of high-throughput analytical workflows that offer an unprecedented level of comprehensive quantitative insight into the metabolome. However, these new analytical techniques, and the rapid pace of technological progress itself, has drawbacks with respect to data coherence. While liquid chromatography has vastly simplified sample preparation procedures (to the point where “dilute-and-shoot” methods have been advocated) 2, especially in comparison to established separation technologies such as gas chromatography, fundamental properties of LC make retention times highly variable. This variability is so great, even within the same laboratory, that retention time values are effectively irreproducible when considered as a means for aiding compound identification 3. This, coupled with a lack of best practice standards for LC method development, makes quantitatively meaningful intra- and inter-laboratory comparisons of retention times nearly impossible. While inter-instrument data coherence for mass spectrometers is less of an issue, the plethora of technologies promulgated by mass spectrometry manufacturers in recent years (MSALL, SWATH, MMDF, MSn) 4 further muddies the water in regard to data standardization and compatibility. Thus, the rapid pace of development in both the instrumentation and methodologies creates a moving target for standardization and impedes data coherency.
The rapid pace of development in LC-MS based metabolomics has also resulted in a dearth of informatics tools and workflows for data standardization and coherence. Efforts thus far have focused on analysis of single batches of experimental data for the purposes of identifying statistically significant analytes that may serve as biomarkers or elucidating biologically relevant outcomes via metabolic pathway analysis. The vast majority of existing bioinformatics tools and workflows for metabolomics, which include XCMS 5, MZmine 6, MetaboAnalyst 7, Workflow4metabolomics 8, and MetaboLyzer 9, are geared towards this single batch oriented analysis. Several data repositories have also been developed for storing, organizing, and curating metabolomics datasets such as the EBI MetaboLights 10 and NIH Metabolomics Workbench 11 resources. However, none of these efforts attempt to tackle the problem of integrating multiple batches of data from numerous experiments to form a single coherent dataset. While such an endeavor may initially seem to have limited use cases, it has profound implications when considering the “big picture” of metabolomics and its ultimate goal of studying the totality of information contained within the metabolome, which necessitates an integrated database consisting of mutually coherent datasets. Such an endeavor exceeds the scope and resources of any single laboratory or institution, requiring a concerted and collaborative effort by the metabolomics community as a whole, which includes developing tools for increased data standardization and coherency.
With these goals in mind, Disparate Metabolomics Data Reassembler (DIMEDR) was developed. DIMEDR reassembles incongruent datasets that have been acquired across multiple unrelated experiments into a single coherent data matrix. To do so, DIMEDR prioritizes the identification of mutual spectral features across all datasets that are being reassembled, and accomplishes this by inspecting features at the individual chromatogram level within each dataset. In doing so, both intra- and inter-dataset biases and irregularities can be taken into account, which can include intra-set retention time drifts and inter-set shifts, and systematic inter-set m/z value biases. Initially, a user-defined primary sample set is selected from the sets that are being reassembled. This primary set undergoes a standard feature selection workflow, which includes peak picking and retention time correction. This is the basis for the unified data matrix template that data from all subsequent sets will be integrated into. Ultimately, the path towards universal harmonized metabolomics databases is a challenge that can only be solved through concerted community driven efforts that involve both logistical and informatics solutions. DIMEDR is an initial step towards bridging these “islands” of incompatible datasets through novel informatics methodologies, which will hopefully spur the field to further these goals.
Methods and Tools:
DIMEDR was written in Python utilizing a variety of open source libraries and tools. These include Matplotlib 12, NumPy 13, and the R statistical computing environment 14 via Rpy2 15. DIMEDR relies on the XCMS CentWave algorithm 16 to conduct peak picking and peak integration via R. All code was developed in a Unix environment via Ubuntu 18.04 LTS, and is freely available at https://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:dimedr along with detailed installation instructions.
General Workflow Overview:
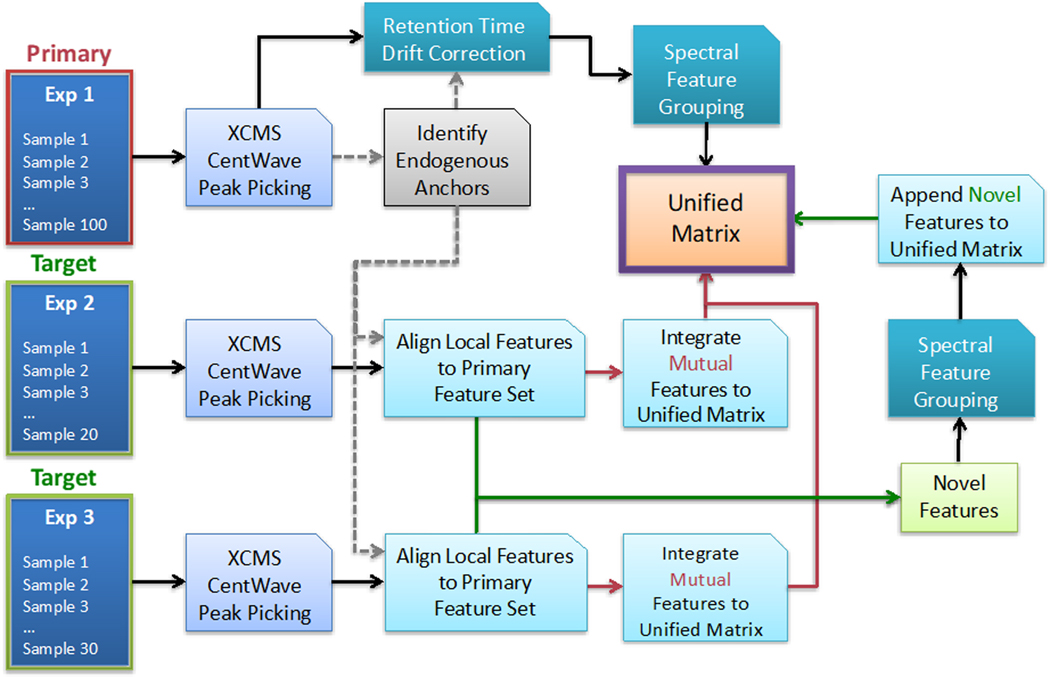
DIMEDR’s strategy for bridging the inconsistencies between two or more incongruent LC-MS metabolomics datasets relies on the use of persistent spectral features that are utilized as reference points, called endogenous anchors, for data correction and adjustment procedures. Initially, the user chooses a “primary” dataset, which may possess the largest sample size or is determined to be of the highest quality. All other datasets are designated as “target” sets whose data will be integrated into the unified matrix template, creating a unified matrix. This integration is facilitated by endogenous anchors that are initially derived from the primary dataset, and subsequently identified in each target set, which act as reference points that enable the target spectral features to be assimilated into the primary set. Figure 1 presents an overview of this workflow.
Figure 1.
An overview of the Disparate Metabolomics Data Reassembler (DIMEDR) workflow for integrating potentially incongruent datasets originating from multiple experiments. The algorithm relies on an initial selection of a “primary” dataset, with all other datasets designated as “target” sets. The primary dataset can be the largest or, what is determined to be the highest quality, and serves as the template for creating the unified matrix that data from all other target sets will be integrated into. This is facilitated by extracting persistent spectral features from the primary dataset, called endogenous anchors, that are utilized as reference points for universal data correction. Endogenous anchors are initially identified in each target dataset, and subsequently used to align, identify, and integrate mutual spectral features that are shared with the primary dataset into the unified matrix. Novel features, for which retention times have also been corrected via this process, are collected across all target datasets, grouped, and appended to the unified matrix as well.
Data extraction procedures on the primary dataset are initially conducted, which involves peak extraction, retention time correction, grouping, and endogenous anchor elucidation. Peak picking and integration is independently conducted for each sample in the set via the XCMS CentWave algorithm, a widely used and well documented peak extraction algorithm in metabolomics. The set of peaks extracted for each sample is then sequentially analyzed by run order, examining for the most commonly recurrent matching peaks. This matching is conducted via user-defined ppm based m/z error window (e.g. 20 ppm), and a percentage error per unit time based retention time window (detailed in the next section). The sequential nature of this procedure enables retention time drift and sudden shifts (that may be the result of external factors that interrupt the run) to be detected and corrected for. The subset of matched peaks that are found to be present in a high percentage of the samples in the primary set (e.g. 90%) are utilized as endogenous anchors for retention time correction of all other peaks. These corrected peaks subsequently undergo peak grouping via a bottom-up consensus clustering method wherein all potential peak groups are calculated utilizing the same peak matching parameters for endogenous anchor elucidation, with final groups selected based on a voting schema (detailed in Supporting Information). The results are outputted into a data matrix consisting of grouped spectral features, defined by its averaged m/z value coupled with a corrected retention time, and their abundance values for each sample in the set. This matrix serves as the primary data template by which data extracted from every subsequent target dataset is integrated into.
Data extraction of subsequent target datasets is conducted independently for each set, and is reliant on the endogenous anchors elucidated from the primary dataset extraction. Similar to the previous workflow, XCMS CentWave based peak picking and integration is first conducted for each sample in a target set. The extracted peak set for each sample is analyzed for the presence of endogenous anchors (via m/z value and retention time matching procedures identical to the previous workflow), and retention time corrections are made utilizing the subset of endogenous anchors found in each sample. All corrected peaks are then matched to the primary dataset spectral feature list, and its data incorporated into the unified matrix template. As with the primary extraction workflow, this procedure is conducted sequentially for the purposes of correcting for retention time drifts and shifts. Once all samples in the target set have been analyzed, the subset of peaks that were matched to a primary spectral feature are re-analyzed for systemic data errors such as m/z value biasing and post-correction retention time shifts that are endemic to the target set as a whole. These results are utilized to make additional corrections to the target data to maximize the identification of matching features.
The final step of analysis is the incorporation of novel spectral features not found in the primary dataset into the unified data matrix. The unmatched corrected peaks that remain after analysis of each target set are pooled, and undergo peak grouping identical to the initial formation of the primary spectral feature groups. These novel features are then appended to the unified matrix. The final result, illustrated in Figure 2, is a unified matrix that incorporates data from multiple disjoint metabolomics datasets, enabling analysis across biological samples from independent experiments.
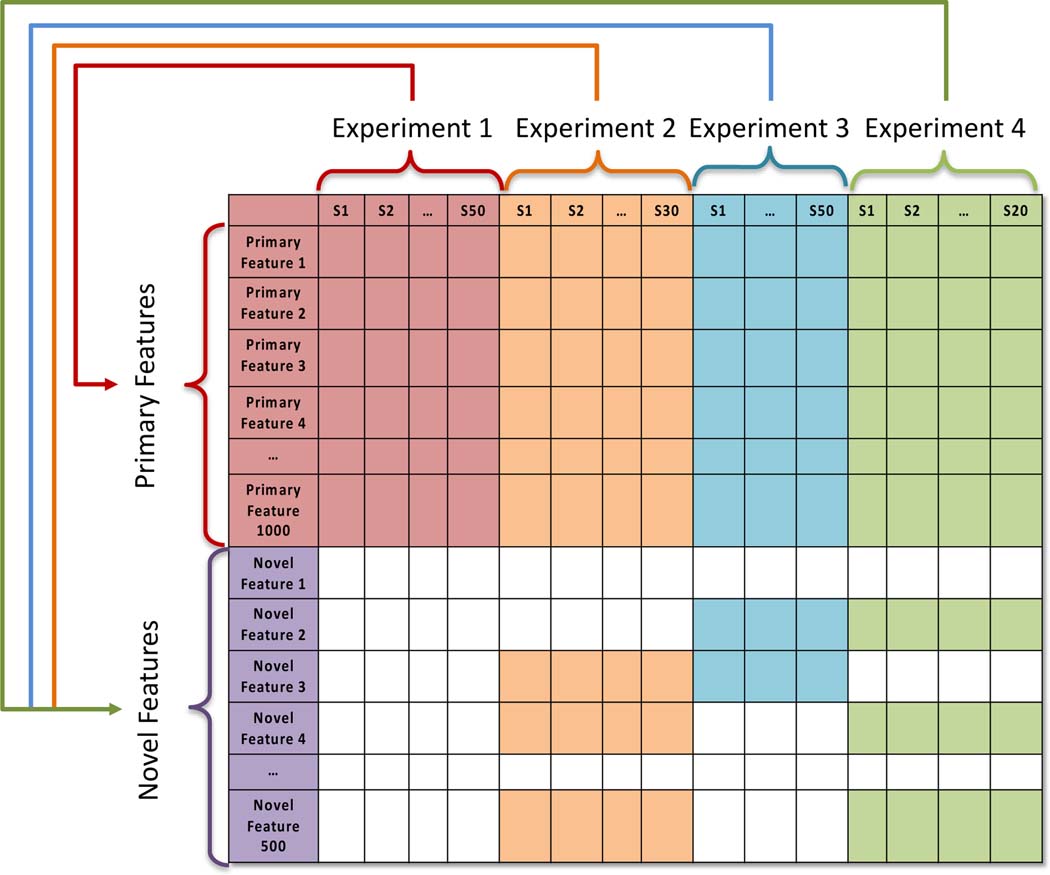
Figure 2.
The topology of a unified matrix that has been created by DIMEDR from the integration of disjoint datasets from multiple experiments. The template for the unified matrix is initially constructed from the primary dataset (Experiment 1), from which all primary spectral features originate. Data from all target datasets (Experiments 2–4) are integrated into this template, with emphasis placed on maximizing the identification of primary features in each sample of a target dataset. Features that do not match any of the primary spectral features are considered novel. These novel spectral features are pooled across all target datasets, analyzed, and integrated into the unified matrix as well.
Endogenous Anchor Elucidation and Retention Time Correction:
The most difficult hurdle in disparate dataset integration is overcoming the high degree of variability and fluctuation exhibited by retention time measurements. 17 DIMEDR attempts to address this challenge by identifying persistent spectral features in the primary dataset that are utilized as endogenous anchors. These endogenous anchors facilitate universal retention time correction both within the primary set as well as in all target sets and enables the datasets to be unified in a coherent manner. Initial endogenous anchor elucidation in the primary dataset involves robust methodologies for calculating retention time error windows and identifying and correcting for retention time drifts and shifts. Subsequent retention time correction procedures utilizing relative retention times based on these endogenous anchors is conducted on both primary and target datasets.
Endogenous anchor elucidation begins with identifying persistent spectral features in the primary dataset. Extracted peaks for each sample are analyzed sequentially according to their run order. In the first sample, all extracted peaks are initially treated as progenitor endogenous anchors. An attempt is then made to find a matching peak in the second sample for each progenitor anchor, forming endogenous anchors that consist of matched peak sets. Peak matching relies on user defined thresholds for determining similar m/z values (e.g. < 20 ppm), and retention times as determined by the time differential as a function of the current chromatographic run time. For two given retention times, a maximum error per unit run time threshold ((), and a maximum allowable retention time differential (), the error per unit run time calculated between two retention times (, ) must fulfill:
The correction factor (CF) for the calculation includes an adjustment term at the very beginning of the chromatographic run time, but also prevents the absolute retention time difference from exceeding a user defined threshold (). Nonmatching peaks in the second sample are also treated as progenitor endogenous anchors. This procedure is sequentially repeated for the extracted peaks in each subsequent sample, resulting in the expansion of existing endogenous anchor matched peak sets and the formation of new ones. However, a matched peak set is discarded if the maximum threshold for missingness is exceeded. For instance, if the minimum threshold for a matched peak set to be considered as an endogenous anchor is 90%, and the primary dataset consists of 100 samples, then the missingness threshold is 10 samples, i.e. the spectral feature can be missing from no more than 10 samples.
The sequential nature of the endogenous anchor elucidation procedure allows for systemic errors to be detected and corrected for. These errors may result from retention time drift which is endemic to liquid chromatography. An attempt is made to account for drift by utilizing the most recent retention time in the matched peak set according to the run order when matching an extracted peak in the current sample. Errors may also be due to non-ideal runtime conditions such as mid-run column cleaning or switching, which can cause sudden retention time shifts. Shifts are accounted for by simultaneously utilizing the retention time from the earliest extracted peak added to the spectral group to elucidate a potential match for the current sample. While drift based matching takes precedence, if a shift based match occurs when a drift based match fails, all subsequent drift based matching will reset to the original retention time from the earliest peak added to the group. This non-parametric methodology, illustrated in Figure 3, is utilized during initial endogenous anchor discovery in the primary dataset, as well as for anchor searching in the target datasets.
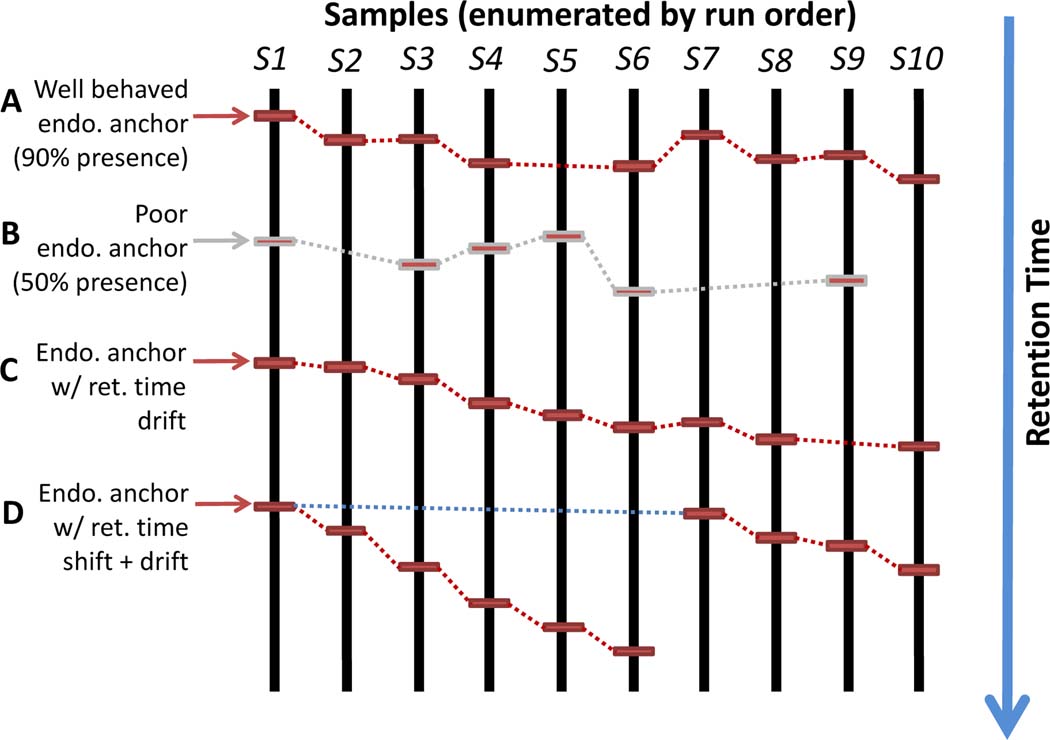
Figure 3.
An illustrative example of DIMEDR’s endogenous anchor elucidation. Spectral features that have been identified to be present in a high percentage (e.g. 90%) of the samples in the primary dataset are utilized as endogenous anchors (A). Features that are missing in too many samples are not considered (B). Retention time drift is compensated for by comparing feature retention times sequentially according to sample run order, even if the feature is missing in a sample (C). Even retention time shifts, which may be caused by mid-run interruptions, can be accounted for by comparing to the retention time from the earliest sample that the feature was detected in (D).
Once endogenous anchors have been acquired from the primary dataset, retention time correction can be conducted on all extracted peaks. The endogenous anchors identified in each sample act as internal standards by which retention times are calibrated to. While each member peak in an endogenous anchor group will have a localized retention time for a given sample (), the endogenous anchor will be represented by the mean retention time () in the final unified matrix. For a raw extracted peak in sample , its retention time () is reinterpreted as the signed difference of a nearby endogenous anchor (via its localized retention time), and then recalculated using the endogenous anchor’s mean retention time:
Corrected retention times are calculated for the closest endogenous anchors for peak , with calculated as half the total number of available endogenous anchors available for sample . The final corrected retention time for peak is the mean of these calculated retention times after excluding outliers via 1.5 interquartile range based filtering. This procedure is conducted for each extracted peak in every sample in the primary set, as well as in each sample for target sets once endogenous anchors have been identified.
Inter-Set Systemic Error Correction Procedures:
Inherent to any experimental metabolomics dataset are systemic errors that result from real-world factors that cannot be accounted, much less controlled for. These errors, which may arise from a combination of factors such as instrument miscalibration and human error, can vary in intensity and prevalence from experiment to experiment, even when conducted by the same laboratory. Though the result of these errors can be profound and immediately noticeable, its effects to the data may be subtle. However, even these subtleties can have a major impact when considering the high sensitivity nature of the metabolomics platform, contributing to the inter-experimental incompatibility of the resulting data. When integrating data from target datasets into the unified matrix, DIMEDR attempts to identify set specific systemic errors in the m/z values and retention times and correct for them to maximize the number of shared inter-set spectral features in the final unified data. Figure 4 illustrates this integration procedure.
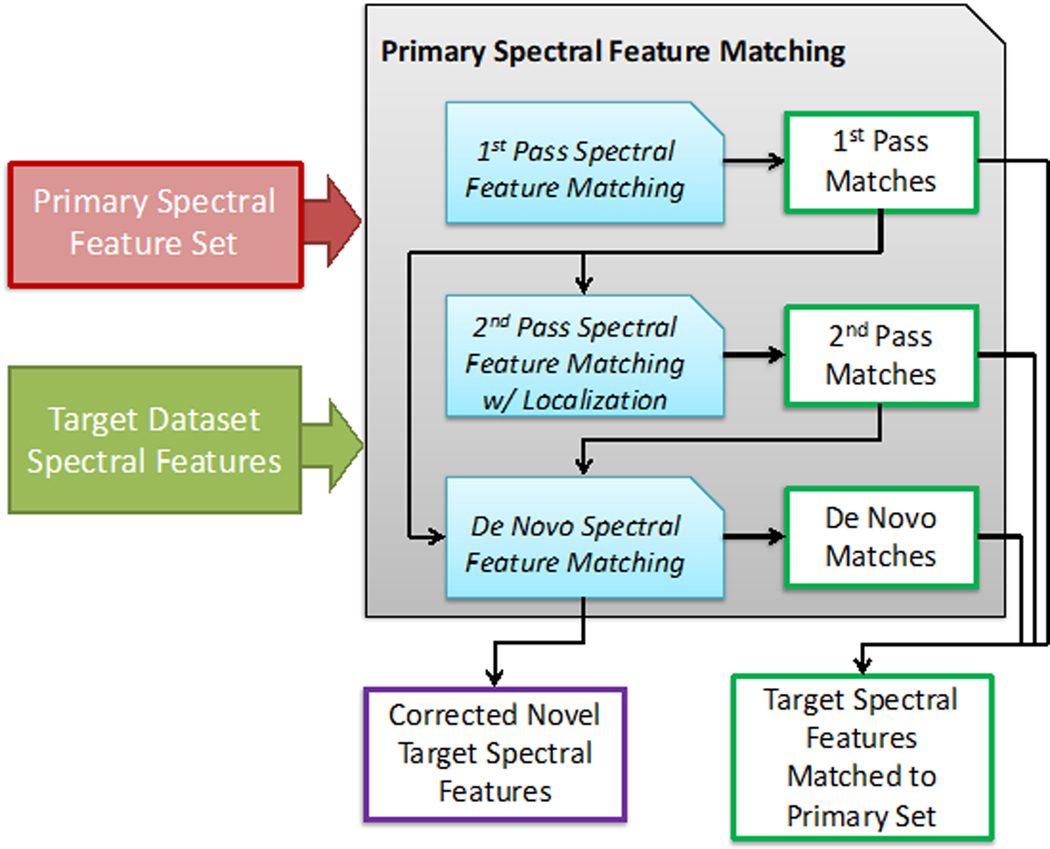
Figure 4.
The workflow for primary spectral feature matching that is conducted for all target datasets. A first-pass matching of target spectral features is conducted based on the original m/z and retention time values from the primary feature set. These initial matches are then used to derive localized values for first-pass matched primary features that better reflect biases in the current target dataset, and are used to find second-pass matches. These first and second-pass matches are both used to make further localizations to unmatched primary spectral features to find de novo matches. These localization procedures are also utilized to correct novel target spectral features for improved coherency with the primary spectral feature set and eventual integration into the unified matrix.
Systemic errors are characterized for a target dataset after an initial analysis of its extracted peaks for matches to primary spectral feature groups. These first-pass matches, which are conducted on extracted peaks that have already undergone endogenous anchor based retention time correction, are matched based on primary dataset derived m/z and retention time parameters. For a given primary feature (), its matches for the target set () are re-examined to derive an averaged m/z value () and retention time () that is more representative for the set being analyzed:
These localized parameters, which are derived for each primary dataset spectral feature group, are used to find second-pass matches in the remaining unmatched peaks in the target set. In doing so, set-specific biases in the m/z values and retention times can be accounted for.
These aggregate matches undergo additional analysis to extrapolate localized m/z biases for unmatched primary spectral feature groups. Initially, the m/z bias offset for each primary spectral feature () with target matches is calculated (in ppm) as a function of the primary averaged m/z value () and the target averaged m/z value () which now includes the second-pass matches:
In considering m/z bias as a function of the m/z value itself, the localization procedure involves calculating an inferenced bias offset for an unmatched primary spectral feature () by averaging the m/z biases for the subset of existing matched primary spectral features () within a small range (e.g. +/−10 m/z) of the unmatched primary spectral feature:
This extrapolated bias () is then utilized to find de novo first-pass matches for the primary spectral feature. Second-pass matches are subsequently found via averaging of the first-pass match m/z values and retention times as described previously.
As a final measure of maximizing coherency between the target and primary datasets, all remaining unmatched peaks undergo m/z bias correction utilizing the totality of target peaks that have been matched to primary spectral features. This correction procedure is identical to the de novo matching procedure as previously described, wherein existing matches are utilized to extrapolate potential bias in the m/z values of unmatched peaks, and is calculated utilizing matched features that are within a 10 m/z range of the peak being corrected. This m/z bias detection and adjustment procedure is crucial for the final stage of the DIMEDR workflow, wherein unmatched peaks across all target sets undergo peak grouping to form novel spectral features that were not found in the primary dataset.
Analysis of Experimental Data:
For demonstration purposes, DIMEDR was used to integrate data from four unrelated studies consisting of two human and two mouse urine metabolomics datasets acquired over 4 years. The human datasets originate from a radiobiology study consisting of 304 human urine samples collected from 95 patients undergoing total body irradiation (TBI) at the Memorial Sloan Kettering Cancer Center, NYC18 and a colorectal cancer (CRC) recurrence study consisting of 40 human urine samples collected from 40 patients at the Georgetown Lombardi Cancer Center, DC19. The mouse datasets originate from a radiobiology study consisting of 21 urine samples collected from C57BL/6N 8–10 week old male mice, and a lipopolysaccharide (LPS) exposure study consisting of 24 urine samples from C57BL/6N 8–10 week old male mice, both conducted at the Georgetown University Medical Center20. All 389 samples were stored, prepared, and analyzed at the Georgetown Lombardi Cancer Center Proteomics and Metabolomics Shared Resource between 2010 to 2014. All urine samples were stored at −80°C and analyzed utilizing Ultra Performance Liquid Chromatography coupled to time-of-flight mass spectrometry utilizing a Waters Corporation QTOF Premier. Samples were run in both positive and negative ionization modes, however only the positive mode data was analyzed.
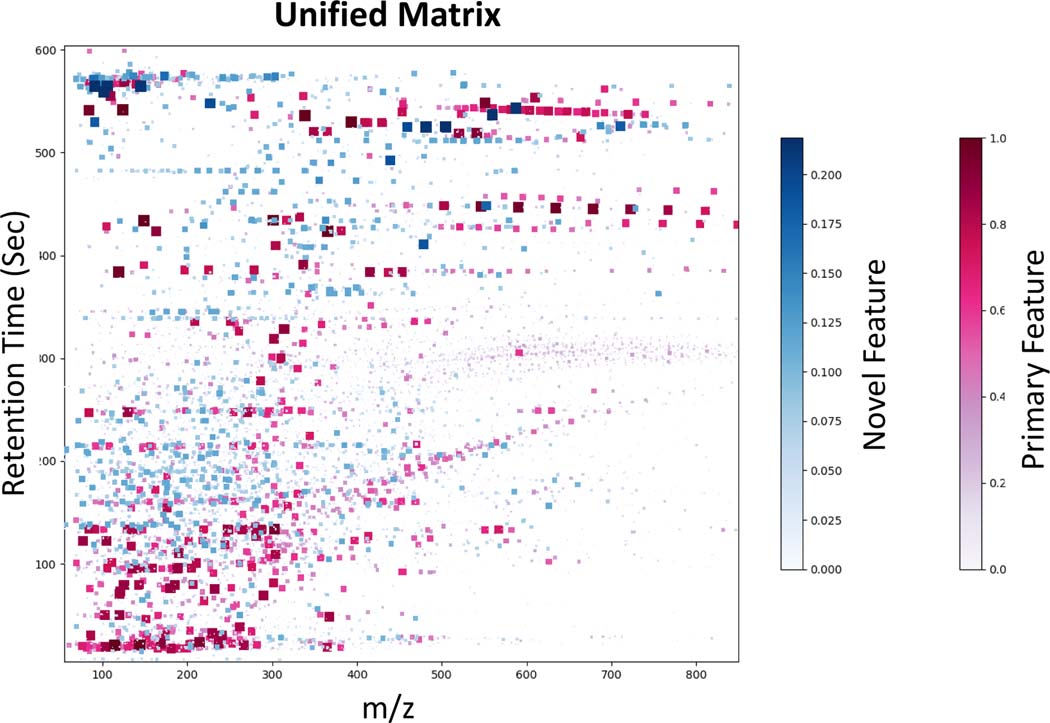
DIMEDR was able to integrate data from all datasets into a single unified matrix consisting of 35091 spectral features across 389 biological samples. The TBI dataset was designated as primary due to its high sample count, and an endogenous anchor threshold of 90% (i.e. a spectral feature found in at least 274 of the 304 TBI samples) was used, resulting in 108 elucidated anchors. An average of 62 anchors per sample were found in the human CRC dataset, but only 20 anchors per sample in the mouse LPS and radiation datasets. A total of 23,066 spectral features were extracted from the initial primary dataset analysis and unified matrix template construction, with an additional 12,025 novel features found in the 3 target datasets. Figure 5 is a visual representation of the unified matrix, with each feature represented as either a red (primary) or a blue (novel) marker, and plotted according to its m/z value and retention time. The intensity and size of each marker is a function of the fraction of all samples in which it was found to be present. The scale for novel feature presence for this analysis is limited to a maximum of 0.2185 as this represents the fraction of samples that are from target sets.
Figure 5.
A visual representation of the unified matrix created from the integration of 4 datasets by DIMEDR. Each marker represents a spectral feature, plotted by its m/z value (X-axis) and retention time (Y-axis). Markers shaded in red represent primary features that were originally found in the primary TBI dataset. Blue shaded markers represent novel features that were found to be present only in the 3 target sets. The size and hue of each marker is a function of the fraction of the total sample count the spectral feature was found to be present in. As such, the upper limit of the novel feature fraction is 85 out of 389 (0.2185), which is reflected in the novel feature color bar.
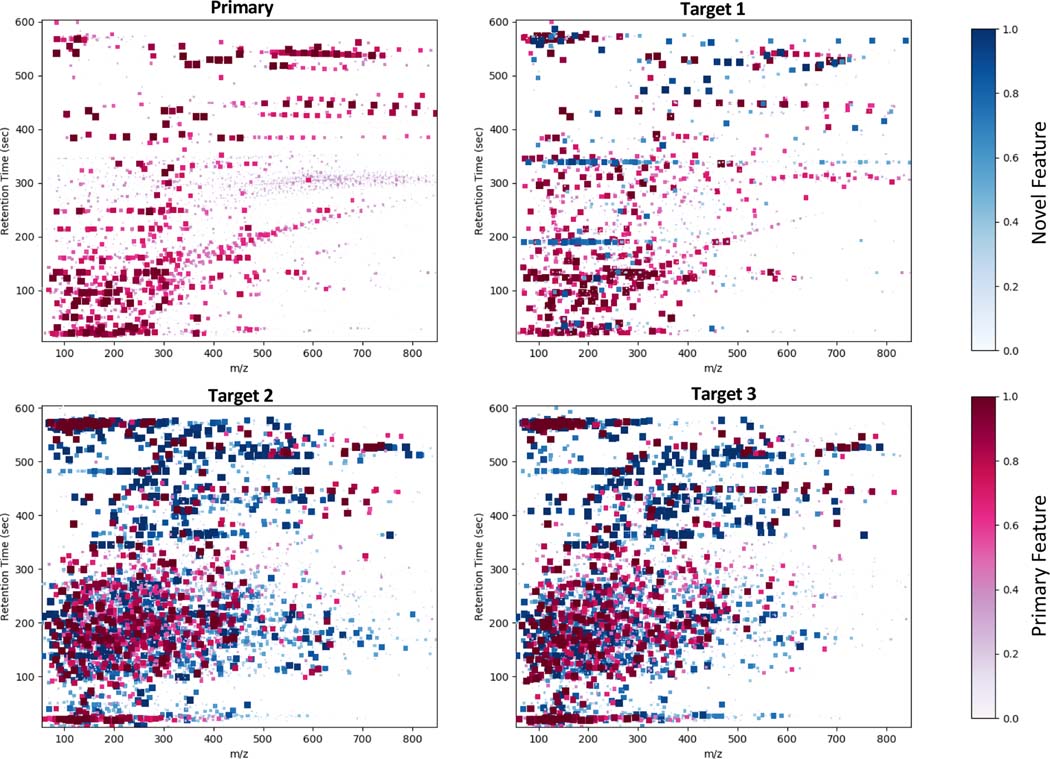
Approximately 63% of spectral features in the CRC dataset were matched to the primary feature set, while roughly 30% of the features in either of the mouse datasets matched. Table 1 provides summary statistics of the analysis. DIMEDR provides a visual representation of the breakdown of the unified matrix by each of its constituent datasets, as shown in Figure 6. As in Figure 5, each marker represents a spectral feature, however in this representation the color intensity is normalized to the sample size for each dataset. This breakdown representation enables quick visual comparisons to be made between all constituent datasets. From this it is easy to identify the largest contributor of novel spectral features (blue markers) as being from the two mouse datasets. Furthermore, by examining the distribution of the features it is apparent that there are qualitative similarities between the two human datasets, and the two mouse datasets. Overall, these results indicate a greater degree of concordance between the two human datasets in comparison to the two mouse sets, with the greatest differentiator being the high number of novel features originating from the mouse sets. This unified matrix can be used to explore a wide range of research topics that extends far beyond the scope of the original experiments that generated the constituent datasets.
Table 1.
Summary statistics for the integration of 2 human and 2 mouse datasets via DIMEDR. The human TBI dataset was designated as primary due to its large sample size, with all other sets designated as targets. Based on the number of endogenous anchors found in the samples of the target sets coupled with the percentage of matched primary features, the human CRC dataset unsurprisingly bore the greatest similarity with the primary set, while the 2 mouse datasets yielded the largest number of unmatched novel features.
| Primary | Target 1 | Target 2 | Target 3 | |
|---|---|---|---|---|
| Description | TBI | CRC | Radiation | LPS |
| Sample count | 304 | 40 | 21 | 24 |
| Sample type | Human Urine | Human Urine | Mouse Urine | Mouse Urine |
| Endogenous anchors | 108 total anchors | 62.4 anchors/sample (avg) | 20.4 anchors/sample (avg) | 20.8 anchors/sample (avg) |
| % matched to primary features | - | 62.7% | 31.8% | 29.5% |
| Unmatched novel features (avg. features/sample) | - | 502.25 | 1918.95 | 2335.83 |
| Total extracted features (avg. features/sample) | 1358.82 | 1348.30 | 2815.24 | 3309.92 |
Figure 6.
A visual representation breaking down the unified matrix into its 4 constituent datasets. Each graph consists of spectral features, represented by square markers plotted by their m/z (X-axis) and retention times (Y-axis), that are present for the specified dataset. As with Figure 5 the markers are colorized as either red (primary features) or blue (novel features), however the size and hue are determined by the fraction of samples contained in each constituent dataset, rather than the total sample count. Visual inspection reveals obvious similarities in both the distribution and presence of spectral features between the two human sets (Primary and Target 1) and also the two mouse sets (Target 2 and 3). Furthermore, it is apparent the vast majority of novel features originate from the mouse datasets.
Discussion:
By incorporating pragmatic approaches into a logical framework for data integration, DIMEDR can unify otherwise incomparable metabolomics datasets from multiple experiments into a single coherent data matrix. In doing so, DIMEDR extends the utility of metabolomics datasets beyond their original experimental design, enabling new avenues of research to be pursued with existing resources. This is not the first attempt at metabolomics data harmonization, with MetMatch 21 having many of the same m/z and retention time correction capabilities and even incorporating adduct deconvolution capabilities that DIMEDR lacks. However, DIMEDR’s scope is far broader, emphasizing the harmonization of large numbers of potentially disparate experimental datasets, accounting for bias at multiple levels of granularity (e.g. individual sample versus dataset specific).
More importantly, taking such an expansive approach can eventually lead to greater applications than merely the improved utilization of existing datasets. Data harmonization is a critical evolution of the metabolomics platform that will enable large-scale, multi-institutional studies with heterogenous data acquisition platforms yielding fully unified datasets, which is currently not feasible. DIMEDR is a purely informatics driven approach to data integration, focusing on the reduction of confounding factors originating primarily from the instrumentation and intrinsic limitations of the technology. But these goals cannot ultimately be achieved through technical solutions alone, and they necessitate coordinated efforts by the metabolomics community to develop standard protocols, methodologies, and shared resources that work in tandem with informatics tools.
As DIMEDR represents only an initial step towards metabolomics data harmonization, there are indeed significant limitations to its capabilities. The most substantial shortcoming stems from the lack of standardization in the field, especially regarding sample preparation and LC separation methods. As such, DIMEDR cannot accommodate datasets that have been acquired using different LC methods and/or sample preparation procedures. For example, a dataset acquired with 30-minute LC runs cannot be compared to data with 10-minute runs due to potential differences in operating pressure, nor can DIMEDR handle datasets with different gradient elution methods, or different LC techniques such as reverse phase versus hydrophilic interaction chromatography (HILIC). Furthermore, reliance on endogenous spectral feature anchors for universal data correction requires at least some degree of baseline similarity between the constituent datasets, and thus the best results are achieved when all sets originate from the same biofluid type, e.g. all urine or all blood serum samples, though it is not a critical restriction of the algorithm. Differences in mass accuracy and sensitivity between QTOF and Orbitrap instruments also restricts DIMEDR to processing datasets from the same instrument type, preferably the same make and model. In its current version, DIMEDR cannot take advantage of biological/technical replicates, or diagnostic samples (e.g. QC, pooled, blank) to enhance harmonization, though internal standards would necessarily be incorporated as endogenous anchors if present in the primary dataset. However, these enhancements can be incorporated in future releases.
A significant aspect of data harmonization that DIMEDR ignores are any batch effects that are exhibited in the abundance values of the spectral features when comparing different experimental sample sets. For a given sample and spectral feature, DIMEDR outputs the raw abundance as extracted by the XCMS CentWave algorithm. No attempt is made to normalize or fill in missing values. It was a deliberate design choice during DIMEDR’s development to focus solely on maximizing m/z and retention time coherency. Any attempt to “normalize” abundance was deemed to have too many pitfalls, as the inherent nature of the metabolomics platform and its extreme sensitivity blurs the line between what can be safely deemed “batch effects” and biological significance. As such, abundance normalization must be conducted during statistical analysis, which many informatics workflows are already capable of.
Despite these shortcomings, DIMEDR is nonetheless an enormously capable tool for data integration and harmonization. DIMEDR has immediate applications in metabolomics core facilities and other shared resource environments where standardized procedures for sample preparation and instrument operation are in place. Integrating the high volume of data that is produced by a core facility into unified frameworks enables critical insight to be provided on a customer’s individual dataset that would otherwise be impossible to glean from isolated analysis. DIMEDR expedites this evaluation by providing summary statistics on the average number of novel versus incorporated primary features per sample for each target dataset, as well as graphs that visualize the entire dataset either as a unified matrix or broken down into its constituent sets. Furthermore, meta-data from this unified framework can be utilized by the core for quality control purposes, protocol improvement, and even expediting method development for recurrent spectral feature identification. This integrated approach can potentially accelerate intra- and inter-institutional collaborations as well by identifying correlations between unrelated experimental datasets from different labs.
Thus far, data harmonization has been an overlooked issue in metabolomics, with far more attention given to the pursuit of biologically meaningful results in individual datasets. Indeed, the rich datasets that are produced by the platform from even modest experiments provide enough “low hanging fruit” to satiate most investigators, and thus the vast majority of informatics tools have been designed to analyze data only at the level of a single experiment. While the diversity of these solutions indicates that many of the difficulties in analyzing individual datasets are by no means completely solved, it is nonetheless vital to look at the bigger picture. The ability to bridge these “islands” of datasets was the impetus behind DIMEDR’s development, and thereby advances a critical but often unnoticed aspect of the field.
Conclusion:
One of the greatest shortcomings of metabolomics is its inability to harmonize metabolomics datasets into coherent unified frameworks. While not a comprehensive solution, DIMEDR nonetheless makes significant strides in the pursuit of this goal. DIMEDR can incorporate multiple experimental datasets, while taking into account the biases and idiosyncrasies of each set, to create a single coherent data matrix that maximizes the number of shared spectral features. In doing so, DIMEDR permits the exploration of data originating from multiple experiments at a far deeper level than traditional meta-analysis techniques and lays the groundwork for more ambitious goals of large-scale unified metabolomics data frameworks.
Supplementary Material
Acknowledgements:
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Footnotes
Supporting Information Available:
Expanded methods on spectral feature grouping
Competing Financial Interests:
The authors declare no competing financial interests.
References
- (1).Sumner LW; Hall RD Metabolomics across the globe. 2013,
- (2).Deventer K; Pozo OJ; Verstraete AG; Van Eenoo P Dilute-and-shoot-liquid chromatography-mass spectrometry for urine analysis in doping control and analytical toxicology. TrAC Trends in Analytical Chemistry. 2014, 55, 1–13. [Google Scholar]
- (3).Scalbert A; Brennan L; Fiehn O; Hankemeier T; Kristal BS; van Ommen B; Pujos-Guillot E; Verheij E; Wishart D; Wopereis S Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009, 5, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhu X; Chen Y; Subramanian R Comparison of information-dependent acquisition, SWATH, and MS(All) techniques in metabolite identification study employing ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal Chem. 2014, 86, 1202–1209. [DOI] [PubMed] [Google Scholar]
- (5).Smith CA; Want EJ; O’Maille G; Abagyan R; Siuzdak G XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006, 78, 779–787. [DOI] [PubMed] [Google Scholar]
- (6).Olivon F; Grelier G; Roussi F; Litaudon M; Touboul D MZmine 2 Data-Preprocessing To Enhance Molecular Networking Reliability. Anal Chem. 2017, 89, 7836–7840. [DOI] [PubMed] [Google Scholar]
- (7).Chong J; Soufan O; Li C; Caraus I; Li S; Bourque G; Wishart DS; Xia J MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Giacomoni F; Le Corguillé G; Monsoor M; Landi M; Pericard P; Pétéra M; Duperier C; Tremblay-Franco M; Martin JF; Jacob D; Goulitquer S; Thévenot EA; Caron C Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015, 31, 1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mak TD; Laiakis EC; Goudarzi M; Fornace AJ MetaboLyzer: a novel statistical workflow for analyzing Postprocessed LC-MS metabolomics data. Anal Chem. 2014, 86, 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Haug K; Salek RM; Conesa P; Hastings J; de Matos P; Rijnbeek M; Mahendraker T; Williams M; Neumann S; Rocca-Serra P MetaboLights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic acids research. 2012, 41, D781–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sud M; Fahy E; Cotter D; Azam K; Vadivelu I; Burant C; Edison A; Fiehn O; Higashi R; Nair KS Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic acids research. 2015, 44, D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hunter JD Matplotlib: A 2D graphics environment. Computing in science & engineering. 2007, 9, 90. [Google Scholar]
- (13).Van Der Walt S; Colbert SC; Varoquaux G The NumPy array: a structure for efficient numerical computation. Computing in Science & Engineering. 2011, 13, 22. [Google Scholar]
- (14).Team, R. C. R language definition. Vienna, Austria: R foundation for statistical computing. 2000, [Google Scholar]
- (15).Gautier L rpy2: A Simple and Efficient Access to R from Python. URL http://rpy.sourceforge.net/rpy2.html. 2008,
- (16).Smith CA; Want EJ; O’Maille G; Abagyan R; Siuzdak G XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical chemistry. 2006, 78, 779–787. [DOI] [PubMed] [Google Scholar]
- (17).Barwick VJ Sources of uncertainty in gas chromatography and high-performance liquid chromatography. Journal of Chromatography A. 1999, 849, 13–33. [Google Scholar]
- (18).Laiakis EC; Mak TD; Anizan S; Amundson SA; Barker CA; Wolden SL; Brenner DJ; Fornace AJ Jr Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiation research. 2014, 181, 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Madhavan S; Gusev Y; Natarajan TG; Song L; Bhuvaneshwar K; Gauba R; Pandey A; Haddad BR; Goerlitz D; Cheema AK Genome-wide multi-omics profiling of colorectal cancer identifies immune determinants strongly associated with relapse. Frontiers in genetics. 2013, 4, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Laiakis EC; Hyduke DR; Fornace AJ Jr Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors γ radiation and lipopolysaccharide. Radiation research. 2012, 177, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Koch S; Bueschl C; Doppler M; Simader A; Meng-Reiterer J; Lemmens M; Schuhmacher R MetMatch: A Semi-Automated Software Tool for the Comparison and Alignment of LC-HRMS Data from Different Metabolomics Experiments. Metabolites. 2016, 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.