Abstract
Insulin and insulin-like growth factor 1 (IGF-1) evoke diverse biological effects through receptor-mediated tyrosine phosphorylation of insulin receptor substrate (IRS) proteins. We investigated the elements of IRS-1 signaling that inhibit apoptosis of interleukin 3 (IL-3)-deprived 32D myeloid progenitor cells. 32D cells have few insulin receptors and no IRS proteins; therefore, insulin failed to inhibit apoptosis during IL-3 withdrawal. Insulin stimulated mitogen-activated protein kinase in 32D cells expressing insulin receptors (32DIR) but failed to activate the phosphatidylinositol 3 (PI 3)-kinase cascade or to inhibit apoptosis. By contrast, insulin stimulated the PI 3-kinase cascade, inhibited apoptosis, and promoted replication of 32DIR cells expressing IRS-1. As expected, insulin did not stimulate PI 3-kinase in 32DIR cells, which expressed a truncated IRS-1 protein lacking the tail of tyrosine phosphorylation sites. However, this truncated IRS-1 protein, which retained the NH2-terminal pleckstrin homology (PH) and phosphotyrosine binding (PTB) domains, mediated phosphorylation of PKB/akt, inhibition of apoptosis, and replication of 32DIR cells during insulin stimulation. These results suggest that a phosphotyrosine-independent mechanism mediated by the PH and PTB domains promoted antiapoptotic and growth actions of insulin. Although PI 3-kinase was not activated, its phospholipid products were required, since LY294002 inhibited these responses. Without IRS-1, a chimeric insulin receptor containing a tail of tyrosine phosphorylation sites derived from IRS-1 activated the PI 3-kinase cascade but failed to inhibit apoptosis. Thus, phosphotyrosine-independent IRS-1-linked pathways may be critical for survival and growth of IL-3-deprived 32D cells during insulin stimulation.
Members of the insulin receptor (IR) family play important roles throughout evolution, since they are involved in the development of flies, worms, and mammals (10, 11, 24, 27). In mammals, IRs are essential for carbohydrate metabolism, insulin growth factor 1 (IGF-1) receptors promote cell growth, and both may contribute to cell survival and cellular transformation (32, 35, 47, 59, 60). Disruption of IGF-1 receptors causes severe developmental defects, and without insulin receptors neonates cannot survive (1, 25, 36). Moreover, partial inhibition of the IR kinase is associated with type 2 diabetes (61).
IR substrate (IRS) proteins coordinate and amplify signals from IR family members (44, 72). IRS-1 and IRS-2 are widely expressed members of the IRS protein family which are tyrosine phosphorylated during insulin and IGF-1 stimulation (57, 58). IRS-3 and IRS-4 have similar structures, but their expression is largely restricted to adipocytes and pituitary-thyroid, respectively (33, 34). Various cytokine receptors, including those for interleukin 4 (IL-4), growth hormone, interferon, and others, also stimulate tyrosine phosphorylation of IRS proteins, usually through the activation of Janus (JAK) kinases (13, 14). Physiologically, IRS proteins are crucial for normal development and metabolic control. Mice lacking IRS-1 grow poorly in utero and remain small throughout life (5); partial disruption of both the IR and IRS-1 causes hyperinsulinemia and diabetes (12). Without IRS-2, mice grow to normal size but develop type 2 diabetes, owing to insulin resistance and a progressive reduction of β-cell mass (68). Disruption of both IRS proteins is embryonic lethal, suggesting that together they are essential for normal development and growth (67).
Tyrosine phosphorylated IRS proteins bind to the Src homology 2 domains in a variety of signaling proteins, including phosphatidylinositol 3 (PI 3)-kinase, Grb-2, SHP2, crk, and fyn (72). PI 3-kinase consists of a catalytic domain and a regulatory domain; the kinase is activated when the regulatory domain binds to tyrosine-phosphorylated motifs in activated growth factor receptors or docking proteins such as IRS-1 and IRS-2 (72). The binding of p55/p85 regulatory subunits to the IRS proteins is the principal mechanism that activates PI 3-kinase during insulin and IGF-1 stimulation (7, 43, 49). In certain systems, the catalytic domain directly binds to activated ras, which contributes to the activation of PI 3-kinase (39, 51).
The PI 3-kinase cascade promotes metabolic and growth responses during growth factor, cytokine, or insulin/IGF-1 stimulation (18, 26, 50). Products of PI 3-kinase are coupled to many biological responses through various serine kinases, including PDK1, PKB/akt, p70s6k, PKCγ, and others (3, 40, 43, 64, 66). PKB/akt is a serine/threonine kinase which contains a pleckstrin homology (PH) domain that binds to PtdIns-3,4,5-P3 (2). Activation of PI 3-kinase recruits PKB/akt to the plasma membrane, where it is activated by phosphorylation of Thr308 and Ser473 (2, 3, 19, 31). Thr308 is phosphorylated by phosphoinositide-dependent kinase 1 (PDK1), and Ser473 is phosphorylated by a distinct membrane-associated kinase, which has tentatively been called PDK2 (17). Activated PKB/akt mediates various metabolic effects of insulin (19, 30, 28a). Mutation of the phosphorylation sites in PKB/akt blocks activation and prevents insulin-stimulated protein synthesis (28a). Moreover, PKB/akt is particularly important for IGF-1-dependent neuronal survival, since it phosphorylates the bcl2 family member BAD and inhibits apoptosis (22, 23).
Although considerable evidence suggests that activation of PI 3-kinase is critical for survival and growth, this hypothesis fails to explain the inability of the Drosophila IR to promote survival and growth of IL-3-deprived 32D cells during insulin stimulation (71). The Drosophila IR is similar to the mammalian IR. Both receptors catalyze tyrosine phosphorylation of Shc, which recruits Grb2/mSOS to activate mitogen-activated protein (MAP) kinase (53). However, the Drosophila IR has an extended COOH-terminal tail with homology to a region of IRS-1 that directly binds p85 and activates the PI 3-kinase cascade. Despite the ability to stimulate PI 3-kinase in 32D cells without IRS-1, the Drosophila IR fails to promote growth and survival of these 32D cells during insulin stimulation (71). Apparently, the Drosophila IR lacks crucial signaling elements that are provided by IRS proteins.
We designed two classes of recombinant proteins to identify the essential elements provided by IRS-1 that mediate survival and growth of IL-3-deprived 32D cells. First, a region of IRS-1 that contains four PI 3-kinase-binding YMXM motifs and one Grb2-binding YVNI motif (5Y region) was attached to the end of the human IR β-subunit; this chimera was called IR5Y. The 5Y region resembles the tail of the Drosophila IR; thus, IR5Y is expected to activate PI 3-kinase without IRS-1 (71). For comparison, truncated IRS-1 proteins composed of the PH and phosphotyrosine binding (PTB) domains alone (PP domain) or including the 5Y region (PP5Y) were coexpressed with the IR in 32D cells. Our results reveal that stimulation of the PI 3-kinase cascade by IR5Y did not inhibit apoptosis or promote long-term replication of 32D cells during insulin stimulation. Additional pathways mediated by the NH2 terminus of IRS-1, possibly including transient phosphorylation-activation of PKB/akt or other kinases, may inhibit apoptosis and promoted replication of 32DIR cells during insulin.
MATERIALS AND METHODS
Construction of chimeric molecules.
An IR chimera (IR5Y) was constructed by adding a cDNA fragment encoding residues 563 to 898 from rat IRS-1 (5Y region) onto the COOH terminus of the human IR. A 3.7-kb HindIII and AflIII fragment of IR and an AflIII/XbaI PCR fragment of IR corresponding to amino acids Asp1178 to Ser1342 were subcloned into pBluescript to produce the vector called IR end. A PCR fragment encoding the 5Y region was blunt-end subcloned into the BstXI and XbaI site of the pCMVhis expression vector to create pCMVhis5Y (63). The IR end and pCMVhis5Y constructs were then digested with XbaI and ligated to generate the chimeric IR containing the IRS-1-derived extension in the pCMVhis expression vector (IR5Y). With an IRS-1 template containing five tyrosine-to-phenylalanine substitutions (amino acids 608, 628, 658, 727, and 895), a similar subcloning procedure was used to generate the IR chimera with a tyrosine-deficient IRS-1-derived extension (IR5F) (46). Both constructs were confirmed by DNA sequencing in the Joslin Diabetes Center DNA Core facility (ABI sequencing).
The IRS-1-derived substrates were prepared by attaching the 5Y region directly to the COOH terminus of the PP domain (residues 1 to 309 of IRS-1). For PP5Y and PP5F, an IRS-1/pBluescript template, either rat IRS-1 or IRS1F18 (rat IRS-1 with 18 tyrosine-to-phenylalanine point mutations), was digested with EcoRI and AatII and religated in the presence of an oligonucleotide linker containing EcoRI- and AatII-compatible overhangs and an intervening SalI site for confirmation of linker insertion (5′-AATTAGTCGACATTCATGACGT-3′ and 5′-CATGAATGTCGACT-3′). The resulting constructs and PCR fragments of IRS-1 corresponding to amino acids 1 to 309 were digested with BspEI and PflmI and ligated together. The subsequent inserts were subcloned into the pCMVhis expression vector by using SacI and HindIII. The expression vector containing the PP domain of IRS-1 was generated by introducing a stop codon at amino acid 310 of IRS-1. All constructs were confirmed by DNA sequencing.
Generation of 32D cell lines.
All 32D cells lines are grown and maintained in RPMI 1640 medium (GIBCO) containing 10% fetal bovine serum (FBS) and 5% WEHI-conditioned medium to provide IL-3; 32DIR and 32DIR/IRS-1 cell lines have been previously described (63). 32D cell lines expressing the IR chimeras and IRS-1-derived substrates were generated by similar techniques. Briefly, cesium chloride-purified DNA (10 μg) of each construct was used to transfect 5 × 106 32D cells by electroporation. The cells were then plated in 15 ml of nonselective medium for 18 h and then transferred to selective medium and diluted into two 24-well plates. Antibiotic-resistant colonies were then expanded and screened for protein expression. Five or more lines of each cell type were selected for further analysis.
p85 and Grb-2 association.
32D cell lines were starved for 4 h, stimulated with insulin (100 nM), and lysed as described elsewhere (45). Proteins were immunoprecipitated with specific antibodies against the IR or IRS-1 for 1 h at 4°C (71). Immune complexes were collected with protein A-Sepharose. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunoblotted with antiserum against p85 (49) or Grb-2 (C-23; Santa Cruz).
ERK, PKB/akt, and p70s6k phosphorylation.
32D cells were incubated without IL-3 for 4 h, stimulated with insulin (100 nM) for 5 min (ERK experiments) or 30 min (p70s6k) or the indicated times (PKB/akt), and lysed as described elsewhere (45). Lysates were resolved on 10% polyacrylamide gels, transferred to nitrocellulose, and immunoblotted with phosphospecific antibodies against ERK1/ERK2, PKB/akt (New England Biolabs), or a p70s6k-specific antibody prepared in our laboratory as previously described (71).
MTT assay.
32D cells were washed and counted, and 10,000 cells/well were seeded into 96-well tissue culture dishes with the growth factor treatments indicated. When indicated, various concentrations of drug LY294002 were added at the beginning of the incubation time. 3-(4,5-Dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) assays were performed as described elsewhere (42). Cells were incubated for the indicated times; MTT (50 μg/well) was added, and the cells were incubated for an additional 3 h. The cells were lysed, and the formazan dye was dissolved in 150 μl of HCl-isopropanol (2 ml of 1 N HCl/48 ml of isopropanol). Absorbance at 570 nM was measured on a BIO-TEK EL311-SX microplate reader.
[3H]thymidine incorporation.
Insulin-stimulated thymidine incorporation was assayed as previously described (71). Briefly, cells in log-phase growth were washed, and 2 × 105 cells were seeded into 1 ml of medium in each of 24 wells containing RPMI with 10% FBS alone or 100 nM insulin or IL-3-containing conditioned medium (WEHI). Cells were incubated for 48 h at 37°C. [3H]thymidine (ICN) was added to a final concentration of 0.5 mCi/ml, and incubation was continued for 2 h. The cells were collected onto glass microfiber filters, lysed, and washed repeatedly with water to remove unincorporated nucleotide. The filters were dried, and retained radioactivity was measured for 1 min in scintillation fluid.
Gel fragmentation assay.
Cells (5 × 106) were incubated for 18 h in RPMI 1640 containing 10% FBS alone, 10% FBS with 100 nM insulin, or 10% FBS with 5% IL-3-conditioned medium as indicated. The cells were then lysed in 400 μl of lysis buffer (10 mM Tris-HCl, 10 mM EDTA, 0.2% Triton X-100 [pH 7.5]), and cell debris was removed by centrifugation. Lysates were then extracted once with an equal volume of phenol and once with phenol-chloroform (1:1), precipitated in the presence of glycogen carrier, and treated with RNase (1 h at 37°C). DNA was then separated on a 1.5% agarose gel and stained with ethidium bromide; staining intensity was determined by scanning the entire lane with Multi-Analyst (BioRad).
PI 3-kinase assays.
32D cell lines were grown, stimulated, lysed, and immunoprecipitated as for immunoprecipitations as described above. Immune complexes were precipitated from the supernatant with protein A-Sepharose (Pharmacia) and washed successively in phosphate-buffered saline containing 1% Nonidet P-40 (NP-40) and 2 mM Na3VO4 (three times), 100 mM Tris-HCl (pH 7.5) containing 500 mM LiCl and 2 mM Na3VO4 (three times), and 10 mM Tris-HCl (pH 7.5) containing 100 mM NaCl, 1 mM EDTA, and 2 mM Na3VO4 (twice). The pellets were resuspended in 50 μl of 10 mM Tris-HCl (pH 7.5) containing 100 mM NaCl and 1 mM EDTA and were combined with 10 μl of 100 mM MnCl2 and 10 μl of 2 μg of PI (Avanti) per μl sonicated in 10 mM Tris-HCl (pH 7.5) containing 1 mM EGTA. The phosphorylation reaction was started by adding 10 μl of 440 μM ATP containing 30 μCi of [γ-32P]ATP (NEN DuPont). After 10 min at 22°C, the reaction was stopped with 20 μl of 8 N HCl and 160 μl of CHCl3-methanol (1:1). The samples were centrifuged, and the lower organic phase was removed and applied to a silica gel thin-layer chromatography (TLC) plate (Merck) which had been coated with 1% potassium oxalate. TLC plates were developed in CHCl3-CH3OH-H2O-NH4OH (60:47:11.3:2), dried, and visualized and quantified on a Molecular Dynamics PhosphorImager.
PKB/akt kinase activity assay.
32D cell lines were starved for 4 h in serum-free medium followed by stimulation with insulin (100 nM) for 30 min. Cells were lysed (20 mM Tris [pH 7.4], 5 mM EDTA, 1% NP-40, Na4P2O7, 100 mM NaF, 2 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 μM microcystin, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml), and cellular debris was removed by centrifugation at 14,000 rpm. Following normalization for protein levels, between 1.5 and 2.5 mg of total cellular protein was added to 20 μl of Gamma Bind G Sepharose (Pharmacia Biotech, Piscataway, N.J.), which was incubated with 3 μl of anti-rat akt-1 sheep polyclonal antibody (Upstate Biotech, Inc., Lake Placid, N.Y.). Samples were incubated on a rocker at 4°C overnight, and the immune complexes were washed three times in lysis buffer and twice in kinase buffer (20 mM Tris [pH 7.4] 10 mM MgCl2, 1 mM dithiothreitol [DTT]). The kinase reaction was prepared by adding 50 μl of 50 mM Tris [pH 7.4] containing 10 mM MgCl2, 1 mM DTT, 5 μM ATP, and 1 μM protein kinase inhibitor (Sigma) to each immunoprecipitate; the phosphorylation was initiated by adding 10 μCi of [32P]ATP (NEN DuPont) containing 30 μM the PKB/akt-specific substrate Crosstide (GRPRTSSFAEG; Upstate Biotech, Inc.). Samples were incubated for 30 min at room temperature with gentle agitation. Reactions were stopped by the addition of 10 μl of stop buffer (1% bovine serum albumin [BSA], 1 mM ATP, 0.6% HCl), and the supernatant (20 μl) was spotted on phosphocellulose paper (P81; Whatman, Hillsboro, Oreg.). The paper was dried, washed four times in 75 mM phosphoric acid, once in acetone, dried, and analyzed by Cerenkov counting (Beckman).
p70s6k activity assay.
Quiescent cells were stimulated for 30 min with 100 nM insulin and collected as described above. The cells were lysed in ice-cold 10 mM potassium phosphate–1 mM EDTA (pH 7.05) containing 0.5% NP-40, 5 mM EGTA, 10 mM MgCl2, 50 mM glycerophosphate, 1 mM Na3VO4, 2 mM DTT, 0.1 mM PMSF, and 10 mg each of aprotinin and leupeptin per ml. Insoluble material was removed by centrifugation at 10,000 × g for 10 min. Anti-p70s6k antibodies were added for 2 h and collected on protein A-Sepharose beads for 1 h at 4°C. The immunoprecipitates were washed and incubated with [32P]ATP (final concentration, 50 mM; 20 mCi per reaction) containing 20 mg per reaction as described (16, 43, 69). The reactions were stopped by the addition of 10 μl of stop buffer (1% bovine serum albumin, 1 mM ATP, 0.6% HCl), and the supernatant (20 μl) was spotted on phosphocellulose paper (P81; Whatman). The paper was dried, washed four times in 75 mM phosphoric acid and once in acetone, dried, and analyzed by Cerenkov counting (Beckman).
Cell replication.
Insulin-stimulated cell replication was assayed in a Coulter counter. 32D cell clones in log-phase growth were washed in calcium-magnesium-free PBS, collected by centrifugation, and resuspended in RPMI 1640 medium containing 10% FBS. Cells were distributed into each well of a 24-well tray at 50,000 cells per well. The cells were treated in duplicate with IL-3 (5% WEHI medium as the source of IL-3) or 100 nM insulin or with no addition. After 24, 48, and 72 h, the entire contents of each well was diluted in saline and measured in a Coulter counter, with the upper and lower limits set at 15 and 5 μ, respectively. The increase in cell number between 24 and 72 h was taken as the insulin- or IL-3-stimulated cell growth. The results are reported as a ratio of insulin-stimulated growth to IL-3-stimulated growth.
RESULTS
The function of chimeric IRs and truncated IRS-1 proteins.
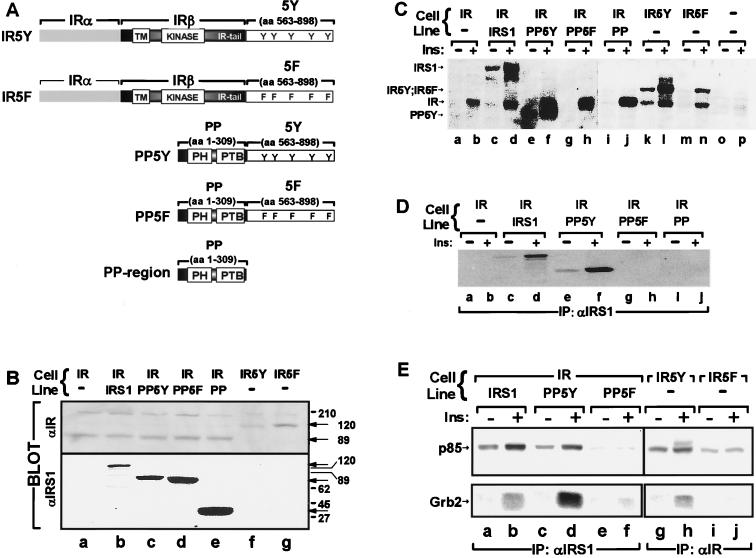
We attached a region of IRS-1 (5Y region) that contains four YMXM motifs and one YVNI motif to the COOH terminus of the human IR β-subunit (Fig. 1A). This chimeric IR, which was called IR5Y, and a mutant receptor containing five Tyr→Phe substitutions, which was called IR5F, were expressed to equal levels in 32D cells (Fig. 1B). Insulin stimulated tyrosine phosphorylation of the wild-type IR and both chimeric receptors (Fig. 1C).
FIG. 1.
(A) Schematic representation of the chimeric molecules used in this study showing the various domains and phosphorylation sites. TM, transmembrane domain; 5Y region, amino acids (aa) 555 to 898 of wild-type IRS-1; 5F region, amino acids 555 to 898 of IRS-1 containing five Tyr→Phe substitutions. (B) Cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with antibodies against the COOH terminus of the IR (αIR) or the NH2 terminus of IRS-1 (αIRS1) as described in Materials and Methods. (C, D, and E) The indicated cell lines were starved for 4 h, stimulated with 100 nM insulin (Ins) for 5 min, and lysed as described in Materials and Methods. (C) Cell lysates directly resolved by SDS-PAGE and immunoblotted with αPY; (D) cell lysates immunoprecipitated (IP) with αIRS1, separated by SDS-PAGE, and immunoblotted with αPY; (E) cell lysates immunoprecipitated (IP) with αIRS1 or αIR, separated by SDS-PAGE, and immunoblotted with antiserum against p85 (top panel) or Grb-2 (bottom panel) as indicated. All results presented are representative of at least two separate experiments.
The 5Y region resembles the COOH tail of the Drosophila IR, which directly binds p85 and activates PI 3-kinase (71). As expected, insulin recruited p85 to IR5Y, whereas IR5F and the wild-type human IR did not bind p85 (Fig. 1E). Unlike the Drosophila IR tail, the 5Y region of IRS-1 also contains a YVNI motif that binds Grb2 during insulin stimulation. As expected, IR5Y recruited Grb-2 during insulin stimulation, whereas IR5F did not bind Grb2 (Fig. 1E). Thus, the 5Y region and the 5F region displayed the expected function when attached to the COOH terminus of the IR β-subunit.
The COOH-terminal tail was removed from IRS-1 to construct the PP domain, which contains the first 309 amino acid residues of IRS-1, including the PH and PTB domains (Fig. 1A). In addition, the 5Y region or the 5F region was attached to the PP domain to create truncated IRS-1 proteins, called PP5Y or PP5F, respectively (Fig. 1A). 32D cells expressing the human IR (32DIR) were transfected, and cell lines that expressed equal amounts of IRS-1, PP5Y, PP5F, or PP domain were selected by immunoblotting with an antibody against the PH domain (Fig. 1B). During insulin stimulation, IRS-1 or PP5Y was tyrosine phosphorylated, whereas PP5F and PP domain were not (Fig. 1C and D). Consistent with these results, PP5Y bound p85 and Grb-2, whereas PP5F and the PP domain did not recruit these signaling proteins (Fig. 1E). Thus, the 5Y region behaved as predicted when it was attached to the PP domain of IRS-1 (Table 1).
TABLE 1.
Summary of insulin-stimulated signaling, viability, and growth of 32D cell linesa
| Transfectantc | Relative insulin stimulation by the following assaysb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ERK phosphorylation (100 nM, 5 min, [Fig. 2A]) | PI-3 K activation (100 nM, 5 min [Fig. 2B]) | p70s6k activation (100 nM, 30 min [Fig. 2F]) | PKB/akt phosphorylation (100 nM, 0 to 48 h [Fig. 2C]) | PKB/Akt activation (100 nM, 30 min [Fig. 2E]) | DNA synthesis (100 nM, 48 h [Fig. 3C]) | Antiapoptosis (100 nM, 18 h [Fig. 4]) | Viability (MTT) (0 to 100 nM, 0 to 48 h [Fig. 3]) | Cell replication (100 nM, 24 to 72 h [Fig. 4]) | |
| −/− | −* | −* | −* | − | −* | −* | − | − | − |
| −/IRS1 | −* | −* | −* | NT | NT | −* | −* | −* | NT |
| IR/− | + | − | − | − | − | − | −/+ | −/+ | − |
| IR/IRS1 | + | +++ | +++ | +++d | +++ | +++ | +++ | +++ | +++ |
| IR/PP5Y | + | +++ | +++ | +++d | +++ | +++ | +++ | ++ | ++ |
| IR/PP5F | + | − | − | ++e | − | −* | +++ | + | + |
| IR/PP | + | − | − | ++e | − | − | +++ | + | + |
| −/PP | −* | −* | −* | NT | −* | − | − | − | − |
| IR5Y | + | +++ | +++ | +++d | +++ | ++ | −¶ | −/+ | − |
| IR5F | + | − | −/+ | − | − | − | −/+ | −/+ | − |
| IR5Y/PP | NT | NT | NT | NT | NT | NT | − | −* | NT |
| IR5F/PP | NT | NT | NT | NT | NT | NT | +++ | +* | NT |
Summary of the biochemical, growth, and survival assays for 32D cells expressing the IRS-1-derived, truncated substrates and chimeric IRs indicated (see Fig. 1 for details).
Concentrations of insulin and times of stimulation are indicated in parentheses, with the figures used to compile these data indicated in brackets. The relative response in each cell type is compared qualitatively to the response in 32DIR cells. +, Magnitude of stimulation in 32DIR cells; −, no response as observed in 32D cells; ++ and +++, stimulation greater than that observed in 32DIR cells; −¶, negative response; NT, not tested; *, data not shown in this paper.
Absence (−) or presence of indicated isoforms. −/−, parental 32D cells; −/IRS1, wild-type IRS-1; IR/IRS1, wild-type IR/IRS-1; IR/PP5Y, wild-type IR/truncated IRS-1; IR/PP5F, wild-type IR/truncated IRS-1 with Tyr-to-Phe point mutations in PI 3-K binding region; IR/PP, wild-type IR/PH-PTB domains of IRS-1; −/PP, PH-PTB domains of IRS-1 only; IR5Y, chimeric IR containing PI 3-K and Grb2 binding regions of IRS-1; IR5F, chimeric IR containing PI 3-K and Grb2 binding regions of IRS-1 with Tyr-to-Phe point mutations; IR5Y/PP, chimeric IR containing PI 3-K and Grb2 binding regions of IRS-1/PH-PTB domains of IRS-1; IR5F/PP, chimeric IR containing PI 3-K and Grb2 binding regions of IRS-1 with Tyr-to-Phe point mutations/PH-PTB domains of IRS-1. See text for complete description of isoforms.
Phosphorylation was maintained for longer than 60 min.
Phosphorylation at 2 to 60 min.
MAP kinase phosphorylation by the chimeric IRs and truncated IRS-1 proteins.
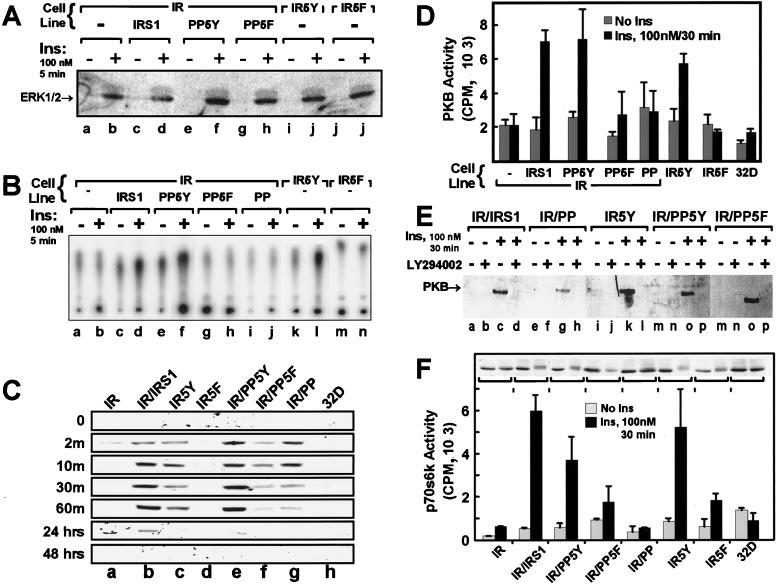
Previous reports show that insulin stimulates the ERK-MAP kinase cascade in 32DIR cells through tyrosine phosphorylation of Shc and its association with Grb2, which stimulates ERK1 and ERK2 (45). Shc phosphorylation occurs independently of IRS proteins, so the inclusion of the 5Y region in the COOH terminus of the IR or expression of the PP domain with the IR should be unnecessary for ERK1 or ERK2 activation in 32D cells. The activation of ERK1 and ERK2 was detected with a specific antibody that recognizes a pair of phosphorylated residues, Thr202 and Tyr204, in the active enzyme (see Materials and Methods). Insulin-stimulated phosphorylation of ERK1 and ERK2 in all of the 32D cell lines expressing the wild-type or chimeric IRs (Fig. 2A), which was consistent with our previous results (45).
FIG. 2.
The indicated 32D cell lines were incubated without IL-3-containing WEHI medium for 4 h, stimulated with 100 nM insulin for the indicated time intervals, and lysed as described in Materials and Methods. (A) Cells were stimulated with insulin (Ins) for 5 min, lysed, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with phosphospecific ERK antibodies (New England Biolabs, model no. 9105S). Similar results were observed in two separate experiments. (B) Cells were stimulated with insulin (Ins) for 5 min, and lysates were subjected to immunoprecipitations with p85-specific antisera. Each immune complex was assayed for PI 3-kinase activity as described in Materials and Methods. The results are representative of three separate experiments. (C) Cells were starved for 4 h and stimulated with 100 nM insulin for the indicated time intervals. Lysates were immunoblotted directly with phosphospecific PKB/akt antibody and detected by enhanced chemiluminescence. (D) PKB/akt kinase activity in immunoprecipitates prepared with a general PKB/akt antibody (UBI) was measured as described in Materials and Methods. Data are the averages ± standard deviations of triplicate determinations obtained in three separate experiments. (E) PKB/akt phosphorylation was measured as described for panel C, but the cells were treated with 10 μM LY294002 for 30 min before 100 nM insulin (Ins) stimulation. (F) The activity of p70s6k and its migration during SDS-PAGE in the indicated cell lines were measured following 30 min of insulin stimulation (100 nM). Lysates were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-p70s6k. Kinase activities are the averages ± standard deviations of triplicate determinations. Identical results were obtained in two separate experiments.
Regulation of the PI 3-kinase cascade by the chimeric IRs and truncated IRS-1 proteins.
The activation of PI 3-kinase was measured in p85-specific immunoprecipitates as previously described (46). Without IRS-1, the IR failed to mediate insulin-stimulated PI 3-kinase activity in 32DIR cells. By contrast, cells expressing IRS-1 displayed strong activation of PI 3-kinase (Fig. 2B). Consistent with the ability of insulin to stimulate the association of p85 with IR5Y in 32D cells, insulin also stimulated PI 3-kinase activity in p85 immunoprecipitates from these cells (Fig. 2B). However, IR5F was ineffective, since it did not engage p85 (Fig. 1E and 2B). Similarly, the PP domain or PP5F expressed in 32DIR cells did not mediate activation of PI 3-kinase during insulin stimulation (Fig. 2B). These results support the hypothesis that association of p85 with phosphorylated YMXM motifs, which are ordinarily located in the tail of IRS-1 or alternatively in the tail of IR5Y, is a principal mechanism to activate PI 3-kinase during insulin stimulation (8, 52).
Several studies indicate that PI 3-kinase is an upstream mediator of several serine kinases, including PDK1, PKB/akt, and p70s6k (2, 3, 17, 20, 43). PKB/akt is activated in part by phosphorylation of Thr308 by PDK1 and Ser473 by an unknown kinase; both phosphorylation events are sensitive to wortmanin treatment, suggesting that products of the PI 3-kinase are involved (2). Immunoblotting with an antibody that recognizes phosphorylated Ser473 revealed that insulin did not stimulate phosphorylation of Ser473 in 32D cells or in 32D cells expressing the IR (Fig. 2C). However, expression of IRS-1 or PP5Y in 32DIR cells promoted phosphorylation of Ser473, which persisted strongly during the first hour of insulin stimulation and which in the case of IRS-1 was still detected 24 h later (Fig. 2C). IR5Y also mediated Ser473 phosphorylation during insulin stimulation, but the intensity was slightly reduced compared to IRS-1, and phosphorylation was not observed at 24 h (Fig. 2C); IR5F was ineffective. Unexpectedly, PKB/akt was phosphorylated in 32DIR cells expressing PP domain or PP5F. This phosphorylation reached maximal levels after 10 min of insulin stimulation but decreased by nearly 80% after 30 min and was close to the basal level at 60 min; it was not detected after 24 h of insulin stimulation (Fig. 2C).
To investigate whether phosphorylation of PKB/akt on Ser473 correlates with its activity, the phosphorylation of Crosstide (GRPRTSSFAEG) was measured in PKB/akt immunoprecipitates following 30 min of insulin stimulation. Consistent with the detection by immunoblotting of Ser473 phosphorylation, PKB/akt was strongly stimulated by insulin in 32DIR cells expressing IRS-1 or PP5Y or in 32D cells expressing IR5Y (Fig. 2D). By contrast, expression of the wild-type IR or IR5F alone did not mediate insulin-stimulated PKB/akt activation. These results are consistent with their inability to bind and activate PI 3-kinase (Fig. 2D). Similarly, activation of PKB/akt was not detected 30 min after insulin stimulation of 32DIR cells expressing PP domain and was not significantly activated in 32DIR cells expressing PP5F (Fig. 2E). Interestingly, variable activation of PKB/akt was detected 10 min after insulin stimulation in 32DIR cells expressing PP domain or PP5F, which corresponded to the time of maximal Ser473 phosphorylation (data not shown).
Considerable evidence suggests that products of PI 3-kinase mediate activation of PKB/akt (2a). Thus, activation of PKB/akt by insulin in 32DIR cells expressing PP domain was unexpected, because insulin-stimulated PI 3-kinase activity was not detected (Fig. 2B and C). However, basal activity of PI 3-kinase may be sufficient to facilitate transient phosphorylation of Ser473. This possibility was tested by a 30-min pretreatment of the cells with 5 μM LY294002, which inhibits PI 3-kinase activity (15). LY294002 inhibited insulin-stimulated phosphorylation of PKB/akt (Ser473) in 32DIR cells expressing IRS-1, PP5Y, or IR5Y (Fig. 2E), consistent with previous data from other systems with another PI 3-kinase inhibitor, wortmanin (2). However, LY294002 also inhibited phosphorylation of the PKB/akt in 32DIR cells expressing the PP domain or PP5F (Fig. 2E). These results suggest that basal products of PI 3-kinase may be required for insulin-stimulated PKB/akt phosphorylation even though PI 3-kinase was not activated in these cells. Alternatively, another kinase regulated through the PP-domain may be inhibited by LY294002.
Activation of p70s6k by the chimeric IRs and truncated IRS-1 proteins.
Many reports demonstrate that the activation of p70s6k is controlled by multistep phosphorylation mediated by both wortmanin- and rapamycin-sensitive pathways (6). Previous experiments with 32DIR cells expressing IRS-1 suggest that activation of PI 3-kinase is required for activation of p70s6k kinase (43). Moreover, both PDK1 and PKB/akt have been implicated in the phosphorylation of some of the regulatory sites of p70s6k (4, 29, 54). Recent experiments demonstrate that the enzymatic activity of p70s6k requires phosphorylation of both Thr252 and Thr412 and that significant in vivo activation correlates with the phosphorylation of Thr412 (66). Moreover, activation of p70s6k correlates with retarded migration during SDS-PAGE (66). Insulin-stimulated activation of p70s6k in the various 32D cell lines was assessed in specific immunoprecipitates with a synthetic peptide substrate (RRRLSSLRA) (48). Insulin did not stimulate p70s6k activity or retard the migration of p70s6k during SDS-PAGE in 32D or 32DIR cells; however, expression of IRS-1 or PP5Y in 32DIR cells promoted p70s6k activation during insulin stimulation. In these cells, the migration of p70s6k during SDS-PAGE was retarded, which was consistent with multisite phosphorylation (Fig. 2F). Moreover, p70s6k was strongly activated in 32D cells expressing IR5Y, which was consistent with the strong activation of PI 3-kinase and PKB/akt. Consistent with a lack of detectable activation of PI 3-kinase by insulin, the activity and migration of p70s6k in 32DIR cells expressing the PP domain or PP5F or in 32D cells expressing IR5F were barely changed (Fig. 2F).
Viability of IL-3-deprived 32D cells during insulin stimulation.
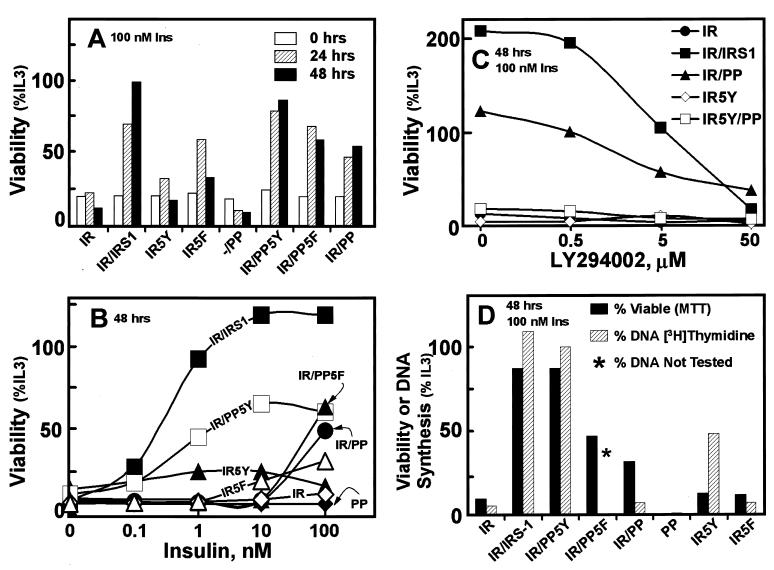
32D cells require IL-3 for survival and replication, and removal of IL-3 causes rapid death by apoptosis (9, 37). Previous work demonstrates that the expression of both the IR and IRS-1 promotes long-term growth of IL-3-deprived 32D cells during insulin stimulation (43, 63). We used the MTT assay to determine the viability of transfected 32D cells during IL-3-deprivation. This assay measures the activity of a mitochondrial reductase, which is fully dependent on cell viability (42). Consistent with previous results, insulin-stimulated 32DIR cells were barely viable 48 h after removal of IL-3 (Fig. 3A and B). By contrast, insulin-stimulated 32DIR cells expressing either IRS-1 or PP5Y were viable during this interval, with IRS-1 mediating the strongest and most sensitive response to insulin (Fig. 3B). During the first 24 h without IL-3, insulin increased viability of 32D cells expressing IR5Y or IR5F to a slightly greater extent than that observed with the wild-type IR; however, this response was lost after 48 h. Unexpectedly, IL-3-deprived 32DIR cells expressing the PP domain or PP5F were remarkably viable after 48 h with 100 nM insulin (Fig. 3A and B). As expected, 32DIR cells expressing PP5Y were more sensitive to insulin, suggesting that activation of PI 3-kinase by IRS proteins enhanced the sensitivity of the response (Fig. 3B).
FIG. 3.
Viability and DNA synthesis in 32D cell lines were measured. Insulin-stimulated cell viability was measured by the MTT assay and compared with the viability of the cells incubated with IL-3-containing WEHI medium (%IL3). The standard deviation for each point was less than 5% in all cases, and the bars were omitted for clarity. (A) Time course of cell viability during 100 nM insulin stimulation in the absence of IL-3. (B) Dose response of insulin-stimulated viability 48 h after removal of IL-3. The results represent at least five independent experiments. (C) The effect of LY294002 on 100 nM insulin-stimulated viability 48 h after IL-3 withdrawal. The results represent two independent experiments, and data are expressed relative to the viability of cells incubated without LY294002 and with IL-3. (D) The indicated cell lines were analyzed for insulin-stimulated DNA synthesis by [3H]thymidine incorporation. The values represent a 2-h pulse or [3H]thymidine incorporation accumulated after 48 h of insulin stimulation of IL-3-deprived cells. The results are compared with the viability of the cells determined in parallel by the MTT assay. The results are expressed as percentages of incorporation obtained with cells incubated continuously with IL-3.
Although the PP domain did not mediate the stimulation of PI 3-kinase during insulin stimulation, basal PI 3-kinase activity may be required for the viability of IL-3-deprived 32D cells. To investigate this possibility, 32D cells were treated with LY294002 during insulin stimulation. LY294002 (5 μM) inhibited the insulin-stimulated viability of 32DIR cells expressing IRS-1 by 50% (Fig. 3C). Interestingly, it also decreased by 50% the viability of 32DIR cells expressing PP domain. By contrast, it had no effect on 32D cells expressing either the IR or IR5Y, since these cells display little viability. These results suggest that PI 3-kinase may contribute to the insulin-stimulated viability of IL-3-deprived 32DIR cells expressing the PP domain, even though PI 3-kinase was not activated. This result may also reflect the inhibition of PKB/akt phosphorylation or the inhibition of an unknown kinase (Fig. 2E).
DNA synthesis in IL-3-deprived 32D cells during insulin stimulation.
Our previous results suggest that 32D cells display robust insulin-stimulated DNA synthesis, which is mediated at least partially by stimulation of PI 3-kinase (41, 46). Consistent with these results, IRS-1 or PP5Y mediated DNA synthesis and increased the number of viable 32DIR cells during insulin stimulation (Fig. 3D). However, in 32DIR cells expressing the PP domain, insulin-stimulated (100 nM, 48 h) DNA synthesis was not detected during the 2-h [3H]thymidine pulse, even though these cells were viable under these conditions (Fig. 3D). The discordance between DNA synthesis and cell viability was further emphasized by the finding that IR5Y mediated detectable insulin-stimulated DNA synthesis, although these cells had low viability; IR5F did not promote insulin-stimulated DNA synthesis in 32D cells (Fig. 3D). Together, these results suggest that insulin-stimulated DNA synthesis required activation of PI 3-kinase, through association either with PP5Y or IRS-1 or with IR5Y. By contrast, long-term viability during insulin stimulation required expression of the PP domain and was most sensitive to insulin when PI 3-kinase was recruited directly to the PP domain by the 5Y region in PP5Y or intact IRS-1.
Cell replication and apoptosis.
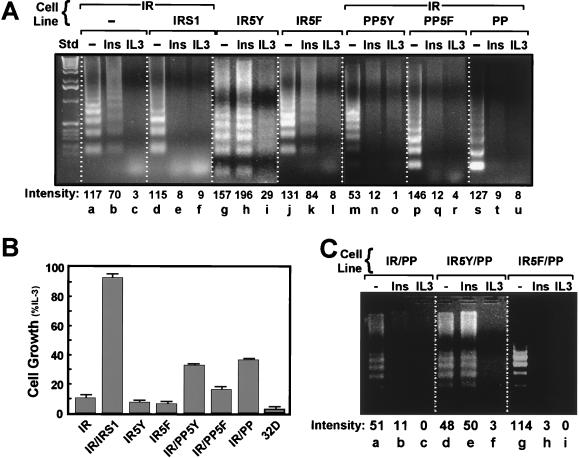
Since the regulation of cell number is determined by the relative rates of cell division and cell death, we examined the ability of the chimeric receptors (IR5Y and IR5F) and truncated IRS-1 proteins (PP, PP5Y, and PP5F) to inhibit apoptosis and to stimulate the replication of IL-3-deprived 32D cells. Apoptosis was assessed by detecting DNA fragmentation, a diagnostic characteristic of apoptotic cells (70). 32D cells begin to undergo spontaneous apoptosis within 8 to 12 h after IL-3-withdrawal (9, 37). After 18 h without IL-3, the DNA extracted from 32D cells was fragmented in a pattern typical of apoptosis, whereas fragmentation was completely absent when cells were incubated in IL-3-containing medium (Fig. 4A, lanes a to c). The addition of insulin weakly but reproducibly inhibited apoptosis in IL-3-deprived 32DIR cells, whereas insulin was as effective as IL-3 in 32DIR cells expressing IRS-1 (Fig. 4A, lanes d to f). The replication of 32DIR cells expressing IRS-1 was increased equally during insulin or IL-3 stimulation, which was consistent with the strong inhibition of apoptosis and stimulation of DNA synthesis (Fig. 4B).
FIG. 4.
Apoptosis and cell replication were determined as described in Materials and Methods. (A) Cells were assayed for apoptosis by gel fragmentation. All cells were incubated with 10% FBS in the absence or presence of 100 nM insulin (Ins) or IL-3 for 18 h. Cells were then collected, and the fragmented DNA was isolated, separated on a 1.5% agarose gel, and visualized by ethidium bromide staining. The relative intensity of DNA bands in each lane was determined with Molecular Analyst (BioRad). Similar results were observed in three separate experiments. (B) Cell replication was measured in a Coulter Counter. Cells (50,000/well) were incubated with 10% FBS, in the absence or presence of 100 nM insulin or IL-3 for 24, 48, and 72 h. Each bar shows the increase in insulin-stimulated cell numbers (± standard deviations) between 72 and 24 h relative to the IL-3-stimulated cell numbers during the same interval. (C) The indicated cell lines were assayed for apoptosis as described for panel A, and similar results were observed in three experiments.
In the absence of IL-3, 32DIR cells expressing PP domain, PP5Y, or PP5F were viable during insulin stimulation (100 nM), suggesting that apoptosis was suppressed. This prediction was confirmed by the DNA fragmentation assay (Fig. 4A). These truncated IRS-1 proteins also promoted an increase in the number of 32DIR cells, although not as rapidly as wild-type IRS-1 (compare Fig. 3C and 4B). Insulin did not suppress apoptosis in IL-3-deprived 32D cells expressing IR5Y, although PI 3-kinase cascade was activated (Fig. 4A, lanes g to i). Moreover, IR5Y lacked the weak inhibition of apoptosis ordinarily observed with the wild-type IR, and IL-3 did not completely block apoptosis in these cells (Fig. 4A, compare lanes b and h). By contrast, insulin weakly inhibited apoptosis of 32D cells expressing IR5F, and, as expected, IL-3 completely inhibited apoptosis (Fig. 4A, lanes i and l). These results suggest that recruitment of PI 3-kinase directly to the IR activated an apoptotic pathway that was not inhibited by activation of PI 3-kinase-dependent pathways, including PKB/akt.
Since the PP domain inhibited apoptosis and IR5Y-mediated DNA synthesis, we tested whether together they promote survival and proliferation of IL-3-deprived 32D cells during insulin stimulation. However, IR5Y promoted apoptosis of 32D cells expressing the PP domain, supporting the idea that activation of PI 3-kinase by association with IR5Y activates an apoptotic pathway (Fig. 4C). Consistent with these results, 32D cells expressing IR5Y and the PP domain were not viable in the MTT assay (data not shown). Thus, survival and growth by insulin required expression of the PP domain and were most sensitive to insulin when PI 3-kinase was recruited to PP5Y or IRS-1. By contrast, recruitment of PI 3-kinase to IR5Y promoted apoptosis.
DISCUSSION
A large body of work supports the hypothesis that activation of PI 3-kinase is a critical first step to inhibit apoptosis during insulin and IGF-1 stimulation and stimulation by a variety of other growth factors and cytokines (26, 38, 65). PI 3-kinase mediates the activation of many cellular processes, in part through the serine/threonine kinases including PDK1 and PKB/akt, atypical PKC isoforms, and p70s6k (2, 3, 17, 20, 41, 43). One well-defined antiapoptotic pathway in neuronal systems proceeds through PI 3-kinase to PDK1 and PKB/akt and ultimately to the phosphorylation of the bcl2 family member BAD (3, 21, 22). IRS proteins are important because their association with p85/p55 is the principal mechanism through which insulin and IGF-1 activate PI 3-kinase (41, 43, 49). Our results are important because they reveal a unique aspect of IRS1-mediated PI 3-kinase activity that may not be shared by other mechanisms. Activation of PI 3-kinase without IRS-1 through the chimeric mammalian IRs or the Drosophila IR (71) does not inhibit apoptosis in IL-3-deprived 32DIR cells (Table 1). Moreover, the PH and PTB domains of IRS-1 appear to regulate signaling pathways that inhibit apoptosis; however, the intensity of these signals may be increased by IRS-1-mediated PI.3-kinase but not by IR5Y-mediated PI 3-kinase activity (Table 1). These novel IRS protein signaling pathways may be essential for development, since disruption of both IRS-1 and IRS-2 in mice is embryonic lethal (67).
To explore the role of IRS-1 in the survival and growth responses of insulin, we constructed a series of truncated IRS-1 proteins or chimeric IRs and analyzed them in 32DIR cells. 32D cells are ideal for these experiments, since they undergo apoptosis shortly after IL-3 withdrawal and they do not contain detectable levels of IR or IRS proteins (63, 73). The chimeric IRs (IR5Y and IR5F) are similar to the Drosophila IR, since it contains an ∼400-residue IRS-1-derived tail with four tyrosine phosphorylation sites that bind p85 and one that binds Grb-2 (71). Our results with 32D cells expressing the chimeric IR5Y indicate that insulin stimulation of the PI 3-kinase cascade without IRS-1, including p70s6k and PKB/akt, is not sufficient to prevent apoptosis during IL-3 withdrawal. Apparently, the rate of cell death is more rapid than the rate of cell growth, even though DNA synthesis is stimulated. These results suggest that IRS-1-independent activation of the MAP kinase and PI 3-kinase cascades is not sufficient to inhibit apoptosis of IL-3-deprived 32D cells during insulin stimulation.
Interestingly, IR5Y is less antiapoptotic than the wild-type IR expressed without IRS-1 in 32D cells. Moreover, 32D cells expressing IR5Y display a low level of apoptosis even in the presence of IL-3. This phenotype apparently arises from phosphorylation of the p85-binding sites in the IRS-derived tail. Accordingly, mutation of the COOH-terminal tyrosines to phenylalanine in IR5F eliminates apoptosis, which persists during IL-3 stimulation, and restores the moderate insulin-stimulated protection from apoptosis observed in cells expressing the wild type IR. This result may be related to recent findings in Caenorhabditis elegans. The C. elegans IR (DAF-2) is similar to the Drosophila IR and IR5Y, since it contains a COOH-terminal extension that has PI 3-kinase binding sites (28, 62). Decreases in DAF-2 signaling induce metabolic and developmental changes, which increase the life span of C. elegans. Thus, by analogy with our results with 32D cells, recruitment of PI 3-kinase to DAF-2 may reduce the life span of C. elegans by promoting apoptosis.
Activation of PI 3-kinase may not be essential to inhibit apoptosis of IL-3-deprived 32DIR cells during insulin stimulation. This unexpected conclusion arises from observations with 32DIR cells expressing the PP domain. The PP domain is composed of the first 309 amino acid residues of IRS-1, which lacks all of the COOH-terminal tyrosine phosphorylation sites of IRS-1. The PP domain is not tyrosine phosphorylated during insulin stimulation and does not activate PI 3-kinase assayed in p85 immunoprecipitates (Table 1). However, the PP domain inhibits apoptosis of IL-3-deprived 32DIR cells during insulin stimulation, suggesting that activation of the PI 3-kinase is not required. Similar results are observed in 32DIR cells expressing PP5F and a tyrosine phosphorylation-deficient (IRS1F18) version of full-length IRS-1 (data not shown). The sensitivity of antiapoptosis to insulin in these cells is low, so activation of PI 3-kinase through PP5Y or IRS-1 clearly facilitates the insulin response by increasing insulin sensitivity; the sensitization to insulin may arise through the recruitment of kinases, including PDK1, PDK2, PKB/akt, and others essential to the plasma membrane by products of PI 3-kinase.
IR5Y may promote apoptosis because it activates the PI 3-kinase cascade without engaging signaling pathways regulated by the PP domain of IRS-1. Coexpression of both the chimeric IR5Y receptor and the PP domain also fails to inhibit apoptosis or to mediate cell replication, indicating that together they cannot reconstitute the full function of IRS-1 or PP5Y. In contrast, cells coexpressing the PP domain and the chimeric IR5F display the same viability observed with the PP domain and the wild-type IR. Thus, the apoptotic stimulus observed with IR5Y is due to the IRS-1-derived tyrosine phosphorylation sites and blocks the protective effects of the PP domain. By contrast, activation of PI 3-kinase by IRS-1 or PP5Y promotes survival and replication. This difference may be due to distinct compartmentalization; however, an exact explanation is unclear, since similar kinases are activated in each case.
Our results suggest that novel phosphotyrosine-independent signaling pathways may be regulated by the PP domain. The nature of the signaling pathways remains unknown but may include serine/threonine kinases that associate with the PH domain or the PTB domains. Interestingly, LY294002, a specific inhibitor of PI 3-kinase, reduces the insulin-stimulated viability of IL-3-deprived 32DIR cells expressing either IRS-1 or the PP domain. The 50% effective dose (ED50) for this effect of LY294002 is approximately 5 μM, which is comparable to the Ki for inhibition of PI 3-kinase activity (15). Thus, activation of PI 3-kinase may increase the sensitivity of the IR, whereas the inhibition of apoptosis requires other pathways mediated by the PP domain that depend on low levels of PI 3-kinase products.
The hypothesis that the PP domain regulates signaling pathways is supported by the finding that insulin transiently stimulates PKB/akt phosphorylation in 32DIR cells expressing the PP domain or PP5F. PKB/akt contains two regulatory phosphorylation sites, Thr308 and Ser473, and phosphorylation of both sites is required for activity (2). Thr308 is phosphorylated by PDK1, whereas Ser473 is phosphorylated by an unidentified kinase. Both kinases are thought to also require PI 3-kinase for activity, since phosphorylation of both sites is sensitive to the PI 3-kinase inhibitor wortmanin (3). Consistent with these results, Ser473 of PKB/akt is strongly phosphorylated in all 32D cell lines in which insulin stimulates PI 3-kinase, including those expressing IR5Y, or the IR and IRS-1; as expected, the wild-type IR alone or IR5F fails to mediate this response. Surprisingly, expression of the PP domain or PP5F with the IR mediates a strong but transient phosphorylation of PKB/akt. In all cases, this insulin-stimulated phosphorylation is inhibited by 5 μM LY294002. Thus, low levels of PI 3-kinase products may mediate transient insulin-stimulated PKB/akt phosphorylation. However, this mechanism predicts the existence of unique pathways that are regulated by the PP domain. Undoubtedly, activation of PI 3-kinase contributes to the sustained activation of PKB/akt by ensuring that essential components are recruited to the plasma membrane. Although prolonged phosphorylation of Ser473 correlates with sustained activation of PKB/akt, it is not required for antiapoptosis mediated by the PP domain and does not inhibit apoptosis of cells expressing chimeric IR5Y.
p70s6k is another important downstream effector in the PI 3-kinase pathway. This serine/threonine kinase is activated by insulin and other cytokines and is critical for many cellular responses, including protein synthesis and cell cycle progression (6). It is regulated by multiple phosphorylation events mediated by several distinct kinases through wortmanin and rapamycin-sensitive pathways. Recent work demonstrates that significant activation of this enzyme requires the phosphorylation of Ser252 and Thr412 and that these phosphorylation events are sensitive to the PI 3-kinase inhibitor wortmanin (65). Ser252 is phosphorylated in vitro by PDK1 (3). In vivo, activation of p70s6k correlates most closely with phosphorylation of Thr412, which is catalyzed by an unknown kinase. Our results are consistent with the hypothesis that activation of PI 3-kinase is required to mediate significant and sustained insulin-stimulated p70s6k in 32D cells; however, weak activation of p70s6k is consistently detected in 32DIR cells expressing the PP domain or PP5F. Since strong sustained phosphorylation of p70s6k does not occur in 32DIR cells expressing the PP domain, it may not play an important role in the inhibition of apoptosis or replication of these cells during long-term insulin treatment.
Experiments with 3T3-L1 cells indicate that insulin mediates novel signaling pathways that are independent of IRS-1 tyrosine phosphorylation and MAP kinase activation (55). This hypothesis is based on hyperexpression of the PTB domain of IRS-1 in 3T3-L1 adipocytes, which inhibits tyrosine phosphorylation of endogenous IRS proteins (55). During this experiment, insulin-stimulated activation of PI 3-kinase and p70s6k was completely blocked, whereas PKB/akt activation and 2-deoxyglucose uptake persisted. These results suggest that alternate IRS-1-independent pathways regulate a subset of insulin’s bioeffects (55, 56). Our results are consistent with these earlier findings but suggest that the tyrosine phosphorylation-independent signals may be dependent on the PH and PTB domains of IRS-1 but continue to occur through IRS-1.
In summary, our results indicate that activation of the PI 3-kinase pathway, including PKB/akt and p70s6k, by IRS-1-derived phosphorylation sites at the COOH terminus of the insulin receptor (IR5Y) does not inhibit apoptosis during insulin stimulation. Moreover, association of PI 3-kinase with the IR may promote apoptosis. In contrast, 32DIR cells expressing the PP domain are fully protected by insulin from apoptosis during IL-3 withdrawal and display a moderate level of replication. Thus, activation of PI 3-kinase and p70s6k is not required for this response. It is not clear how the PP domain mediates the antiapoptotic signal, but the transient activation of a serine/threonine kinase which phosphorylates Ser473 of PKB/akt may provide a clue.
ACKNOWLEDGMENTS
This work was supported by DK38712 and DK43808 (M.F.W.). L.Y. was supported by DK07260, and D.B. was supported by an FPI fellowship from the Spanish Government. C.Z. is a research associate of the Howard Hughes Medical Institute.
Footnotes
Corresponding author. Mailing address: Joslin Diabetes Center, Howard Hughes Medical Institute, 1 Joslin Place, Boston, MA 02215. Phone: (617) 732-2578. Fax: (617) 732-2593. E-mail: whitemor@joslab.harvard.edu.
REFERENCES
- 1.Accili D, Drago J, Lee E J, Johnson M D, Cool M H, Salvatore P, Asico L D, Jose P A, Taylor S I, Westphal H D. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 2a.Alessi D R, Cohen P. Mechanism of activation and function of protein kinase B. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R J, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 4.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 5.Araki E, Lipes M A, Patti M E, Brüning J C, Haag III B L, Johnson R S, Kahn C R. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 6.Avruch J. Insulin signal transduction through protein kinase cascades. Mol Cell Biochem. 1998;182:31–48. [PubMed] [Google Scholar]
- 7.Backer J M, Myers M G, Jr, Shoelson S E, Chin D J, Sun X J, Miralpeix M, Hu P, Margolis B, Skolnik E Y, Schlessinger J, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backer J M, Myers M G, Jr, Shoelson S E, Chin D J, Sun X J, Miralpeix M, Hu P, Margolis B, Skolnik E Y, Schlessinger J, White M F. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baffy G, Miyashita T, Williamson J R, Reed J C. Apoptosis induced by withdrawal of IL-3 from an IL-3 dependent hematopoietic cell line is associated with repartitioning of intracellular calcium and is blocked by enforced Bcl-2 oncoprotein production. J Biol Chem. 1993;268:6511–6519. [PubMed] [Google Scholar]
- 10.Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 11.Baserga R. Oncogenes and the strategy of growth factors. Cell. 1994;79:927–930. doi: 10.1016/0092-8674(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 12.Bruning J C, Winnay J, Bonner-Weir S, Taylor S I, Accili D, Kahn C R. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;86:561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 13.Burfoot M S, Rogers N C, Watling D, Smith J M, Pons S, Paonessaw G, Pellegrini S, White M F, Kerr I M. Janus kinase-dependent activation of insulin receptor substrate 1 in response to interleukin-4, oncostatin M and the interferons. J Biol Chem. 1997;272:24183–24190. doi: 10.1074/jbc.272.39.24183. [DOI] [PubMed] [Google Scholar]
- 14.Carter-Su C, Schwartz J, Smit L S. Molecular mechanism of growth hormone action. Annu Rev Physiol. 1996;58:187–207. doi: 10.1146/annurev.ph.58.030196.001155. [DOI] [PubMed] [Google Scholar]
- 15.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung J, Kuo C J, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinase. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 17.Cohen P, Alessi D R, Cross D A. PDK1, one of the missing links in insulin signal transduction? Growth Reg. 1997;410:3–10. doi: 10.1016/s0014-5793(97)00490-0. [DOI] [PubMed] [Google Scholar]
- 18.Collins M K, Perkins G R, Rodriguez Tarduchy G, Nieto M A, Lopez Rivas A. Growth factors as survival factors: regulation of apoptosis. BioEssays. 1994;16:133–138. doi: 10.1002/bies.950160210. [DOI] [PubMed] [Google Scholar]
- 19.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 20.Cross D A E, Watt P W, Shaw M, van der Kaay J, Downes C P, Holder J C, Cohen P. Insulin activates protein kinase B, inhibits glycogen synthase kinase-3 and activates glycogen synthase by rapamycin-insensitive pathways in skeletal muscle and adipose tissue. Growth Reg. 1997;406:211–215. doi: 10.1016/s0014-5793(97)00240-8. [DOI] [PubMed] [Google Scholar]
- 21.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 22.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 23.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez, R., D. Tabarini, N. Azpiazu, M. Frasch, and J. Schlessinger. The Drosophila insulin receptor homologue: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 25.Hone J, Accili D, Psiachou H, Alghband-Zadeh J, Mitton S, Wertheimer E, Sinclair L, Taylor S I. Homozygosity for a null allele of the insulin receptor gene in a patient with leprechaunism. Hum Mutat. 1995;6:17–22. doi: 10.1002/humu.1380060105. [DOI] [PubMed] [Google Scholar]
- 26.Kapeller R, Cantley L C., Jr Phosphatidylinositol 3-kinase. BioEssays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 27.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 28.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. . (See comments.) [DOI] [PubMed] [Google Scholar]
- 28a.Kitamura T, Ogawa W, Sakaue H, Hino Y, Kuroda S, Takata M, Matsumoto M, Maeda T, Konishi H, Kikkawa U, Kasuga M. Requirement for activation of the serine-threonine kinase Akt (protein kinase B) in insulin stimulation of protein synthesis but not of glucose transport. Mol Cell Biol. 1998;18:3708–3717. doi: 10.1128/mcb.18.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohn A D, Barthel A, Kovacina K S, Boge A, Wallach B, Summers S A, Birnbaum M J, Scott P H, Lawrence J C J, Roth R A. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J Biol Chem. 1998;273:11937–11943. doi: 10.1074/jbc.273.19.11937. [DOI] [PubMed] [Google Scholar]
- 30.Kohn A D, Summers S A, Birnbaum M J, Roth R A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 31.Kohn A D, Takeuchi F, Roth R A. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 32.Lammers R, Gray A, Schlessinger J, Ullrich A. Differential signalling potential of insulin- and IGF-1-receptor cytoplasmic domains. EMBO J. 1989;8:1369–1375. doi: 10.1002/j.1460-2075.1989.tb03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavan B E, Fantin V R, Chang E T, Lane W S, Keller S R, Lienhard G E. A novel 160 kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:21403–21407. doi: 10.1074/jbc.272.34.21403. [DOI] [PubMed] [Google Scholar]
- 34.Lavan B E, Lane W S, Lienhard G E. The 60-kDa phosphotyrosine protein in insulin-treated adipocytes is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:11439–11443. doi: 10.1074/jbc.272.17.11439. [DOI] [PubMed] [Google Scholar]
- 35.Lee A D, Hilsenbeck S G, Yee D. IGF system components as prognostic markers in breast cancer. Breast Cancer Res Treat. 1998;47:295–302. doi: 10.1023/a:1005915420341. [DOI] [PubMed] [Google Scholar]
- 36.Liu J P, Baker J, Perkins J A, Robertson E J, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (lgf-1) and type 1 IGF receptor (lgf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 37.Magnelli L, Cinelli M, Turchetti A, Chiarugi V P. Apoptosis induction in 32D cells by IL-3 withdrawal is preceded by a drop in the intracellular calcium level. Biochem Biophys Res Commun. 1993;194:1394–1397. doi: 10.1006/bbrc.1993.1979. [DOI] [PubMed] [Google Scholar]
- 38.Marte B M, Downward J. PKB/Akt: connecting phosphoinositide 3-kinase to cell survival and beyond. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/s0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 39.Marte B M, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr Biol. 1996;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- 40.Mendez R, Kollmorgen G, White M F, Rhoads R E. Requirement of protein kinase C zeta for stimulation of protein synthesis by insulin. Mol Cell Biol. 1997;17:5184–5192. doi: 10.1128/mcb.17.9.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendez R, Myers M G, Jr, White M F, Rhoads R E. Stimulation of protein synthesis, eukaryotic translation initiation factor 4E phosphorylation, and PHAS-1 phosphorylation by insulin requires insulin receptor substrate-1 and phosphotidylinositol-3-kinase. Mol Cell Biol. 1996;16:2857–2864. doi: 10.1128/mcb.16.6.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 43.Myers M G, Jr, Grammer T C, Wang L M, Sun X J, Pierce J H, Blenis J, White M F. IRS-1 mediates PI 3′-kinase and p70s6k signaling during insulin, IGF-1 and IL-4 stimulation. J Biol Chem. 1994;269:28783–28789. [PubMed] [Google Scholar]
- 44.Myers M G, Jr, Sun X J, White M F. The IRS-1 signaling system. Trends Biochem Sci. 1994;19:289–294. doi: 10.1016/0968-0004(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 45.Myers M G, Jr, Wang L M, Sun X J, Zhang Y, Yenush L, Schlessinger J, Pierce J H, White M F. The role of IRS-1/GRB2 complexes in insulin signaling. Mol Cell Biol. 1994;14:3577–3587. doi: 10.1128/mcb.14.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers M G, Jr, Zhang Y, Aldaz G A I, Grammer T C, Glasheen E M, Yenush L, Wang L M, Sun X J, Blenis J, Pierce J H, White M F. YMXM motifs and signaling by an insulin receptor substrate 1 molecule without tyrosine phosphorylation sites. Mol Cell Biol. 1996;16:4147–4155. doi: 10.1128/mcb.16.8.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne C K, Bolan G, Monaco M E, Lippman M E. Hormone responsive human breast cancer in long-term tissue culture: effect on insulin. Proc Natl Acad Sci USA. 1976;73:4536–4540. doi: 10.1073/pnas.73.12.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelech S L, Krebs E G. Mitogen-activated S6 kinase is stimulated via protein kinase C-dependent and independent pathways in Swiss 3T3 cells. J Biol Chem. 1987;262:11598–11606. [PubMed] [Google Scholar]
- 49.Pons S, Asano T, Glasheen E M, Miralpeix M, Zhang Y, Fisher T L, Myers M G, Jr, Sun X J, White M F. The structure and function of p55PIK reveals a new regulatory subunit for the phosphatidylinositol-3 kinase. Mol Cell Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiss K, Porcu P, Sell C, Pietrzkowski Z, Baserga R. The insulin-like growth factor 1 receptor is required for the proliferation of hemopoietic cells. Oncogene. 1992;7:2243–2248. [PubMed] [Google Scholar]
- 51.Rodriguez-Viciana P, Warne P H, Vanhaesebroeck B, Waterfield M D, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 52.Rordorf-Nikolic T, Van Horn D J, Chen D, White M F, Backer J M. Regulation of phosphatidylinositol 3-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85 kDa regulatory subunit. J Biol Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 53.Ruan Y, Chen C, Cao Y, Garofalo R S. The Drosophila insulin receptor contains a novel carboxyl-terminal extension likely to play an important role in signal transduction. J Biol Chem. 1995;270:4236–4243. doi: 10.1074/jbc.270.9.4236. [DOI] [PubMed] [Google Scholar]
- 54.Scott P H, Brunn G J, Kohn A D, Roth R A, Lawrence J C J. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma P M, Egawa K, Gustafson T A, Martin J L, Olefsky J M. Adenovirus-mediated overexpression of IRS-1 interacting domains abolishes insulin-stimulated mitogenesis without affecting glucose transport in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:7386–7397. doi: 10.1128/mcb.17.12.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma P M, Egawa K, Huang Y, Martin J L, Huvar I, Boss G R, Olefsky J M. Inhibition of phosphatidylinositol 3-kinase activity by adenovirus-mediated gene transfer and its effect on insulin action. J Biol Chem. 1998;273:18528–18537. doi: 10.1074/jbc.273.29.18528. [DOI] [PubMed] [Google Scholar]
- 57.Sun X J, Pons S, Wang L M, Zhang Y, Yenush L, Burks D, Myers M G, Jr, Glasheen E, Copeland N G, Jenkins N A, Pierce J H, White M F. The IRS-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol Endocrinol. 1997;11:251–262. doi: 10.1210/mend.11.2.9885. [DOI] [PubMed] [Google Scholar]
- 58.Sun X J, Wang L M, Zhang Y, Yenush L, Myers M G, Jr, Glasheen E M, Lane W S, Pierce J H, White M F. Role of IRS-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka S, Ito T, Wands J R. Neoplastic transformation induced by insulin receptor substrate-1 overexpression requires an interaction with both Grb2 and Syp signaling molecules. J Biol Chem. 1996;271:14610–14616. doi: 10.1074/jbc.271.24.14610. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka S, Wands J R. Insulin receptor substrate 1 overexpression in human hepatocellular carcinoma cells prevents transforming growth factor β1-induced apoptosis. Cancer Res. 1996;56:3391–3394. [PubMed] [Google Scholar]
- 61.Taylor S I, Accili D. Mutations in the genes encoding the insulin receptor and insulin receptor substrate-1. In: LeRoith D, Taylor S I, Olefsky J M, editors. Diabetes mellitus: a fundamental and clinical text. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 575–583. [Google Scholar]
- 62.Tissenbaum H A, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L M, Myers M G, Jr, Sun X J, Aaronson S A, White M F, Pierce J H. IRS-1: essential for insulin and IL-4-stimulated mitogenesis in hematopoietic cells. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 64.Weng Q P, Andrabi K, Klippel A, Kozlowski M T, Williams L T, Avruch J. Phosphatidylinositiol 3-kinase signals activation of p70 S6 kinase in situ through site-specific p70 phosphorylation. Proc Natl Acad Sci USA. 1995;92:5744–5748. doi: 10.1073/pnas.92.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weng Q P, Kozlowski M, Belham C, Zhang A, Comb M J, Avruch J. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]
- 66.Weng Q P, Kozlowski M, Belham C, Zhang A, Comb M J, Avruch J. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]
- 67.Withers, D., M. F. White, et al. Unpublished details.
- 68.Withers D J, Sanchez-Gutierrez J C, Towery H, Ren J M, Burks D J, Previs S, Zhang Y, Bernal D, Pons S, Shulman G I, Bonner-Weir S, White M F. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–903. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 69.Wood K W, Sarnecki C, Roberts T M, Blenis J. ras mediates nerve growth factor receptor modulation of three signal-transducing protein kinases: MAP kinase, Raf-1, and RSK. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 70.Wyllie A H, Kerr J F R C A R. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 71.Yenush L, Fernandez R, Myers M G, Jr, Grammer T C, Sun X J, Blenis J, Pierce J H, Schlessinger J, White M F. The Drosophila insulin receptor activates multiple signaling pathways but requires insulin receptor substrate proteins for DNA synthesis. Mol Cell Biol. 1996;16:2509–2517. doi: 10.1128/mcb.16.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yenush L, White M F. The IRS-signaling system during insulin and cytokine action. BioEssays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 73.Zamorano J, Wang H Y, Wang L M, Pierce J H, Keegan A D. IL-4 protects cells from apoptosis via the insulin receptor substrate pathway and a second independent signaling pathway. J Immunol. 1996;157:4926–4934. [PubMed] [Google Scholar]