Abstract
Traditional toxicity testing reliant on animal models is costly and low throughput, posing a significant challenge with the increasing numbers of chemicals that humans are exposed to in the environment. The purpose of this investigation was to build optimal prediction models for various human in vivo/organ level toxicity endpoints (extracted from ChemIDPlus) using chemical structure and Tox21 in vitro quantitative high-throughput screening (qHTS) bioactivity assay data. Several supervised machine learning algorithms were applied to model 14 human toxicity endpoints pertaining to vascular, kidney, ureter and bladder, and liver organ systems. Three metrics were used to evaluate model performance: area under the receiver operating characteristic curve (AUC-ROC), balanced accuracy (BA), and Matthews correlation coefficient (MCC). The top four models, with AUC-ROC values >0.8, were derived for endocrine (0.90±0.00), musculoskeletal (0.88±0.02), peripheral nerve and sensation (0.85±0.01) and brain and coverings (0.83±0.02) toxicities, whereas the best model AUC-ROC values were >0.7 for the remaining 10 toxicities. Model performance was found to be dependent on the specific dataset, model type and feature selection method used. In addition, chemical structure and assay data showed different levels of contribution to the prediction of different toxicity endpoints. Although in vitro assay data, when combined with chemical structure, slightly improved the predictive accuracy for most endpoints (11 out of 14), a noteworthy finding was the near equal success of the structure-only models, which do not require Tox21 qHTS screening data, and the relatively poor performance of assay-only models. Thus, the top performing structure-only models from this study could be applied for hazard screening of large sets of chemicals for potential human toxicity, whereas the largest assay contributions to models (i.e., cellular targets) could be used, along with the top-contributing structural features, to provide insight into toxicity mechanisms.
Keywords: machine learning, in vitro assay, Tox21, chemical structure, human in vivo toxic, hazard assessment
Graphical Abstract

Introduction
Various chemicals are widely distributed in the environment in the form of, e.g., food products, drugs, cosmetics and household products, making human exposure to such chemicals virtually inevitable. Chemicals can enter the body through inhalation, ingestion or dermal exposure, and many of them can cause adverse health effects 1, 2. Assessment of chemical hazards is necessary to ensure public safety. Traditional toxicity testing relies largely on animal models, which are expensive and time-consuming, and it is difficult to extrapolate results obtained from animals to human health effects because of inter-species differences3. With an ever-growing number of environmental chemicals, most of which lack comprehensive toxicity data, novel and more efficient assessment methods are needed to evaluate potential harmful effects of environmental chemicals on human health.
Recent advances in high-throughput screening (HTS) technologies have enabled the in vitro profiling of biological activity of large chemical libraries in cell-based assays in a cost-efficient and timely manner4. Such an approach has already been leveraged by the Toxicology in the 21st Century (Tox21) partnership 5-8. During the production phase of Tox21, a library of nearly 10,000 environmental chemicals and drugs (termed the Tox21 10K library) has been screened against approximately 70 cell-based assays in a quantitative HTS (qHTS) format, generating nearly 100 million data points to date 9, 10. In addition, computational toxicology provides tools for toxicity assessment that can organize, analyze, model, simulate, visualize and predict the toxicity of chemicals 11, 12. When chemical structure features are combined with in vitro qHTS assay data, computational models have demonstrated utility in screening large sets of chemicals for potential in vivo toxicity to prioritize a smaller set for in-depth toxicological evaluation 13-16.
Machine learning, one example among a multitude of computational methods, has already been utilized in developing prediction models for chemical toxicity 15, 17. Machine learning methods can build classification models to describe the complex relationships between structure and biological activity, including toxicity, of chemicals based on the information obtained from experimental data 15, 17. Machine learning algorithms commonly used for toxicity prediction include Naïve Bayes (NB), Random Forests (RF), Support Vector Machines (SVM), Neural Networks (NNET) and extreme gradient boosting (XGboost). Multitask modeling has been shown to provide significant improvement over single-output models. Sosnin et al. revealed that multitask learning can be very useful in improving the quality of acute toxicity models 18. Due to its speed and high performance, XGBoost was often used for modeling high-dimensional data, such as quantitative structure - activity relationships (QSAR) and quantitative structure - property relationships (QSPR)19. Nevertheless, prediction of animal or human in vivo apical endpoints poses a significant challenge to the most sophisticated modeling approaches. Liu et al. demonstrated limited success in predicting 35 animal toxicity endpoints in repeat-dose studies based on a combination of bioactivity data from ToxCast in vitro HTS assays and chemical descriptors using NB, SVM, RF, etc. 17. The study reported that the combination of bioactivity and chemical structure descriptors was more predictive of in vivo toxicity outcomes than either chemical structure or bioactivity descriptors alone 17. However, the total number of chemicals in each model was limited (a minimum of 100 total) and the datasets imbalanced (with many more negatives than positives). In addition, although Shah et al. found that inclusion of bioactivities yielded the best-performing models, structure-only models were not far behind15.
Previous modeling efforts focused primarily on predicting animal toxicity. In this study, we modeled human in vivo toxicity data that had been collected from the literature and made publicly available, and built models for each toxicity endpoint using chemical structure and Tox21 assay data. We employed various machine learning methods and different data combinations to obtain the optimal model for each of 14 human in vivo toxicity endpoints. The models also identified assay targets and structure scaffolds that contributed the most to the prediction. The models were validated by internal 5-fold cross validation with performance evaluated by three different metrics: area under the receiver operating characteristic curve (AUC-ROC), balanced accuracy (BA), and Matthews correlation coefficient (MCC). The top performing models could be applied to filter large sets of chemicals for their potential to cause toxicity in humans. Furthermore, the targets and chemical scaffolds identified as contributing to the prediction of toxicity could provide insight into mechanisms of toxicity or provide structure alerts for screening chemicals to minimize toxicity.
Materials and Methods
In vitro assay and structure data
The in vitro assay data were generated by screening the Tox21 10K chemical library against a panel of qHTS assays; the data are publicly available on the National Center for Advancing Translational Sciences (NCATS) website (https://tripod.nih.gov/tox21/assays/) and PubChem 20. Most of the qHTS assays used human cell lines (80.88%), followed by murine embryo fibroblast (7.35%), Chinese hamster ovary cell lines (5.88%) and others (5.88%). The majority of these assays measure pathway activities, including nuclear receptor signaling (NR, 55.90%) and stress response (SR, 11.80%) pathway assays. Other assays measure cytotoxicity directly (8.80%) or other targets or pathways related to toxicity (23.50%). For this study, we used data from 47 assays with 147 readouts. For modeling purposes, curve rank was used as a measure of chemical activity 16, 21, 22. Curve rank is a number between 9 and −9, where a positive value denotes activation and a negative value denotes inhibition. Compounds with a good quality concentration-response curve corresponding to high potency and efficacy are assigned large absolute curve rank values (> 0.5) and labeled as active (1), whereas compounds with absolute curve rank values 0.5 and below are labeled as inactive (0). Two common structure-based fingerprint sets (ToxPrints or ECFP4) were used as input features to build classification models in this study. ToxPrints are based on the publicly available ToxPrint chemotypes (v2.0_r711, https://toxprint.org/) generated within the associated ChemoTyper application (https://chemotyper.org/) 23. There are 729 uniquely defined chemical features coded in XML-based Chemical Subgraphs and Reactions Markup Language (CSRML) in ToxPrint chemotypes. The 1024-bit ECFP4 fingerprints were generated using the CDK package 24 in the Konstanz Information Miner (KNIME) v. 4.0.2 25. The structure for each chemical was represented as a bit vector where the presence or absence of the feature was recorded in a binary system as 1 or 0, respectively 23.
In vivo human toxicity data
Human in vivo toxicity data in this study were collected by manually querying each chemical in the ChemIDPlus Advanced database of the United States National Library of Medicine (available at http://chem.sis.nlm.nih.gov/chemidplus/). This data resource contains a large collection toxicity values from multiple laboratories extracted from the published literature. The raw data are provided in Supplemental Table S2. The population sampled included infant, child, man, and woman. The test type included toxic dose low (TDLo), toxic concentration low (TCLo), lethal dose low (LDLo), lethal dose fifty (LD50) and lethal concentration low (LCLo). The route of administration included oral, inhalation, skin, intravenous, unreported, intramuscular, rectal, intraaural, parenteral, ocular, subcutaneous, multiple routes, intradermal, intraperitoneal, intraarterial, intraspinal and intravaginal. A summary of the combined in vivo toxicity data study totals is provided in Supplemental Table S1, whereas the complete table of extracted data for each of 843 queried chemicals (2474 total study records) is provided in Table S2. There are 14 unique in vivo toxicity endpoints represented, including behavioral, blood, brain and coverings, cardiac, endocrine, gastrointestinal, kidney ureter and bladder, liver, lungs thorax or respiration, musculoskeletal, peripheral nerve and sensation, sense organs and special senses, skin and appendages skin, and vascular. For the purposes of the present study, if a chemical exhibited toxic effect in any test type and route at any dose, then it was categorized as toxic (1), and nontoxic (0) otherwise. The full listing of binarized human in vivo toxicity results for the 843 compounds are provided in Supplemental Table S3, along with their corresponding desalted SMILES structure and indication of whether the result was used in the present modeling exercise (Supplemental Table S4).
Feature selection
To optimize model performance, feature selection as a pre-processing step prior to modeling was performed using three methods, including Fisher’s exact test with p value, importance scores from XGboost and the RF algorithm. For Fisher’s exact test, the features selected by 30 independent p values ranging from 0.01 to 0.3 were used to train the models and the feature sets that achieved the best performance were selected to build the final model. The “xgboost” 26 and “Random Forest” 27 packages were applied to retrieve feature importance scores, respectively. These scores were then ranked to obtain the top 50 features. The optimal set of assays or structure features corresponding to each endpoint was then combined to re-build the model to determine whether the merged feature set was superior to the structure or assay feature set alone.
Supervised machine learning
After feature selection, models for each toxicity endpoint were trained and tested using the selected feature sets, i.e., assay activity (activity-only models), chemical structure (structure-only models), and models based on combinations of structure and activity data via supervised machine learning. Five different classification algorithms were used, including NB, SVM, RF, NNET and XGboost. Models were built and tested using R version 3.4.2, with “e1071” package for NB and SVM classifiers 28, with “Random Forest” package for RF classifier 27, with “nnet” package for NNET 29, and with “xgboost” package for XGboost classifier. The implementation of NB classifier was adapted with the settings of Laplace smoothing, and the Gaussian Radial Basis Function kernel was used for the SVM classifier. In addition, the optimal parameters for SVM, RF and NNET classifiers were selected using the “e1071” package, in which tunable functions included radial basis kernel width (g, settings: 0.01,0.05,0.1,0.5,1) and penalty for violating the soft margin (C, settings: 0.01, 0.1, 1, 10, 100, 1000) for SVM, the number of trees (ntree, settings: 20, 50, 80, 100, 150, 200, 300, 500) and the number of randomly selected variables (mtry, settings: 1:10) for RF, as well as parameter for weight decay (decay, settings: 0.0001, 0.001, 0.01, 0.1) and maximum number of iterations (maxit, settings: 10, 100, 200, 500, 800, 1000) for NNET 28. The optimal constant parameters for XGboost classifier were as follows: control the learning rate (0.01), maximum depth of a tree (3), and subsample ratio of columns when constructing each tree (0.5). To evaluate model performance, 5-fold cross validation was applied with the dataset randomly divided into five parts, four parts for training and one part for testing. To ensure the robustness of our results, the random partitioning process was repeated 20 times for each model. Model performance was measured by AUC-ROC and BA using the “ROCR” and “pROC” packages 30, 31, and MCC using the “mltools” package. Plots were generated using the “ggplot2” package in R.
Results
Data sets of Tox21 assay results, chemical structure, and in vivo toxicity
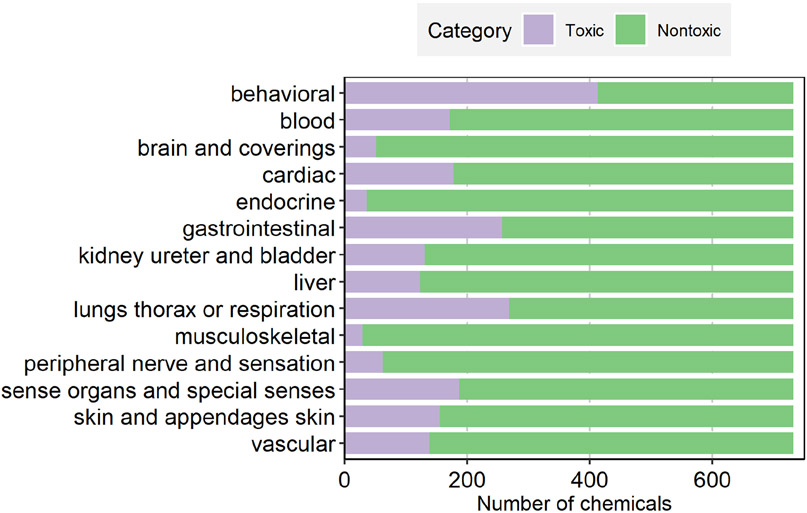
After merging the structure (ToxPrint or ECFP4) and assay data for the 843 chemicals for which toxicity endpoint data were available, a total of 732 chemicals (identified by CAS) remained. Mixtures and macro molecules without a defined structure were excluded from modeling. Structures were further salt stripped using the JChem package from ChemAxon. In cases of heavy metal salts and inorganics, the metal ion was retained after desalting. SMILES were converted to InChI keys in order to identify structure replicates. This resulted in 18 chemicals with duplicate desalted SMILES structures (i.e., collapsed to the same structure (InChI) after salts were removed). Data from replicated results for a single chemical as well as for duplicate InChI keys were averaged prior to modeling. The concordance rate of the in vivo toxicity data for the replicates is about 84%. The distributions of these selected chemicals across the 14 human toxicity endpoints are shown in Figure 1. The proportions of toxic chemicals corresponding to each endpoint, in ascending order, are musculoskeletal (3.96%), endocrine (4.92%), brain and coverings (6.97%), peripheral nerve and sensation (8.47%), liver (16.80%), kidney ureter and bladder (17.90%), vascular (18.85%), skin and appendages skin (21.17%), blood (23.36%), cardiac (24.32%), sense organs and special senses (25.55%), gastrointestinal (35.11%), lungs thorax or respiration (36.61%) and behavioral (56.42%). With the exception of “behavioral”, the number of non-toxic chemicals outweighed the toxic chemicals by a ~3.6 to 1 ratio across all toxicity endpoints. The various human toxicity endpoints were clustered based on the composition of toxic/nontoxic chemicals or significant ToxPrint features (Supplementary Figure S1 and S2), respectively. Some of the related toxicity endpoints are clustered together, such as “brain and coverings” and “peripheral nerve and sensation” by both methods, and “cardiac” and “vascular” by significant ToxPrint features. Overall, however, there appears to be no clear correlation between the organ/endpoint type and their co-clustering.
Figure 1.
Distribution of toxic and nontoxic chemicals in each human in vivo toxicity dataset.
The final structure dataset used for modeling was profiled based on the CPCat (Chemical and Product Categories) chemicals database use-category listing (downloaded from EPA’s CompTox Dashboard, https://comptox.epa.gov/dashboard/downloads) 32, as well as overlap with the DrugBank dataset (also accessed from the Dashboard) 33. Unsurprisingly, this human toxicity dataset contains a high proportion of drugs, with approximately half labeled as such in DrugBank, whereas nearly 85% are labeled as drugs (or are multi-purpose) in CPCat. Just the same, a cursory review of the list indicates quite a number of pesticides and industrial chemicals.
Predicting human in vivo toxicity
A total of five machine learning methods were applied to the Tox21 assay data and chemical structure data to build predictive models for the 14 organ level toxicity endpoints. Three different metrics, AUC-ROC, BA and MCC, were used to evaluate model performance. Subsets of features selected by three methods were used to build models and the optimal AUC-ROC value, together with BA and MCC obtained for each model, are provided in Supplemental Table S5. As one of the most widely used performance metrics, AUC-ROC was used as the primary reference metric for model performance in this study. AUC-ROC values varied for each endpoint depending on the specific machine learning method, type of data, and feature selection method used to build the model (Figure 2 and Supplemental Table S5). Models that included the ECFP4 feature set, either alone or in combination with assay data, showed optimal performance in the 14 in vivo toxicity predictions (Figure 2 and Supplemental Table S5). XGboost turned out to be the best feature selection method (13/14), followed by Fisher’s exact test (1/14) (Supplemental Table S5). To investigate whether adding assay data could improve model performance, we rebuilt models by combining the assays and ECFP4 features that each alone had produced optimal model performance. The performance metrics of the best performing models in each feature category (structure-alone, assay-alone, or combined structure and assay descriptors) for each target organ in terms of AUC-ROC are summarized in Table 1. According to the number of toxicity endpoints for which the method produced the optimal model with the best AUC-ROC, two classifiers, NB (8/14) and SVM (6/14), came out on top. Similarly, the top two data types that produced the best models were combined data (ECFP4 and assay,11/14) and ECFP4 data (3/14). The optimal models for four endpoints had AUC-ROC values greater than 0.8, including endocrine (0.90±0.00), musculoskeletal (0.88±0.02), peripheral nerve and sensation (0.85±0.01) and brain and coverings (0.83±0.02) toxicities. The remaining ten endpoints had models with AUC-ROC value greater than 0.7, including blood (0.79±0.01), sense organs and special senses (0.79±0.01), liver (0.78±0.01), vascular (0.78±0.01), kidney ureter and bladder (0.78±0.01), behavioral (0.77±0.01), skin and appendages skin (0.77±0.01), lungs thorax or respiration (0.75±0.01), cardiac (0.74±0.01) and gastrointestinal (0.72±0.01). Lastly, we applied the best model for each toxicity endpoint to predict the toxic potential of 7071 unique Tox21 compounds for which structures were available. The results, represented as a probability from 0 (not toxic) to 1 (toxic) for each chemical and endpoint, are provided in Supplementary Table S6.
Figure 2.
Model performance measured by AUC-ROC value. Five machine learning methods (NB, NNET, RF, SVM and XGboost) were applied to three types of data including qHTS assay data, ECPF4 and ToxPrint. Each row in the heat map is a toxicity endpoint and each column is a data type. The heat map is colored by the AUC-ROC value ranging from 0.5 to 1 such that a darker shade of red indicates better model performance (larger AUC-ROC).
Table 1.
The top-performing classification models for each toxicity endpoint according to AUC-ROC and model type: Structure + Assay (yellow), Structure-only (green), Assay-only (blue).
| In vivo toxicity | Method | Data set | Feature selection method |
AUC- ROC |
BA | MCC |
|---|---|---|---|---|---|---|
| behavioral | NB | ECFP4 | Fisher's exact test | 0.77±0.01 | 0.74±0.01 | 0.47±0.01 |
| NB | Assay + ECFP4 | all three methods | 0.77±0.01 | 0.74±0.01 | 0.47±0.02 | |
| SVM | Assay | XGboost | 0.69±0.01 | 0.67±0.01 | 0.34±0.02 | |
| blood | NB | Assay + ECFP4 | all three methods | 0.79±0.01 | 0.80±0.01 | 0.44±0.01 |
| NB | ECFP4 | XGboost | 0.78±0.01 | 0.75±0.01 | 0.42±0.01 | |
| NNET | Assay | Fisher's exact test | 0.67±0.01 | 0.64±0.01 | 0.30±0.02 | |
| brain and coverings | NB | ECFP4 | XGboost | 0.83±0.02 | 0.83±0.01 | 0.36±0.02 |
| NB | Assay + ECFP4 | all three methods | 0.83±0.01 | 0.81±0.01 | 0.36±0.03 | |
| NNET | Assay | XGboost | 0.64±0.05 | 0.57±0.02 | 0.21±0.06 | |
| cardiac | SVM | ECFP4 | XGboost | 0.74±0.01 | 0.70±0.01 | 0.37±0.02 |
| SVM | Assay + ECFP4 | all three methods | 0.74±0.01 | 0.67±0.02 | 0.37±0.03 | |
| XGboost | Assay | XGboost | 0.67±0.01 | 0.63±0.01 | 0.30±0.02 | |
| endocrine | NB | Assay + ECFP4 | all three methods | 0.90±0.01 | 0.91±0.01 | 0.41±0.02 |
| NB | ECFP4 | XGboost | 0.88±0.01 | 0.88±0.02 | 0.38±0.04 | |
| SVM | Assay | XGboost | 0.83±0.02 | 0.81±0.03 | 0.35±0.04 | |
| gastrointestinal | NNET | Assay + ECFP4 | all three methods | 0.75±0.01 | 0.70±0.01 | 0.41±0.02 |
| SVM | ECFP4 | XGboost | 0.73±0.01 | 0.69±0.01 | 0.39±0.02 | |
| XGboost | Assay | XGboost | 0.64±0.01 | 0.63±0.01 | 0.27±0.02 | |
| kidney ureter and bladder | NNET | ECFP4 | XGboost | 0.77±0.01 | 0.70±0.01 | 0.39±0.02 |
| NB | Assay + ECFP4 | all three methods | 0.77±0.01 | 0.77±0.01 | 0.40±0.02 | |
| SVM | Assay | XGboost | 0.72±0.02 | 0.73±0.01 | 0.31±0.02 | |
| liver | SVM | Assay + ECFP4 | all three methods | 0.78±0.01 | 0.75±0.01 | 0.39±0.02 |
| NB | ECFP4 | XGboost | 0.77±0.01 | 0.73±0.01 | 0.39±0.02 | |
| RF | Assay | XGboost | 0.68±0.02 | 0.61±0.01 | 0.27±0.02 | |
| lungs thorax or respiration | SVM | Assay + ECFP4 | all three methods | 0.75±0.01 | 0.71±0.02 | 0.41±0.02 |
| NB | ECFP4 | XGboost | 0.73±0.01 | 0.69±0.01 | 0.39±0.02 | |
| XGboost | Assay | XGboost | 0.65±0.01 | 0.64±0.01 | 0.28±0.01 | |
| musculoskeletal | NB | Assay + ECFP4 | all three methods | 0.88±0.01 | 0.86±0.04 | 0.39±0.06 |
| NB | ECFP4 | XGboost | 0.87±0.01 | 0.87±0.02 | 0.39±0.03 | |
| NNET | Assay | XGboost | 0.72±0.03 | 0.72±0.03 | 0.22±0.03 | |
| peripheral nerve and sensation | SVM | Assay + ECFP4 | all three methods | 0.84±0.02 | 0.83±0.01 | 0.40±0.02 |
| NNET | ECFP4 | XGboost | 0.83±0.02 | 0.75±0.01 | 0.37±0.02 | |
| SVM | Assay | XGboost | 0.79±0.02 | 0.78±0.02 | 0.34±0.02 | |
| sense organs and special senses | SVM | Assay + ECFP4 | all three methods | 0.79±0.01 | 0.71±0.02 | 0.45±0.03 |
| NB | ECFP4 | XGboost | 0.77±0.01 | 0.74±0.01 | 0.41±0.01 | |
| XGboost | Assay | XGboost | 0.68±0.01 | 0.64±0.01 | 0.30±0.02 | |
| skin and appendages skin | NB | ECFP4 | XGboost | 0.77±0.01 | 0.74±0.01 | 0.41±0.01 |
| NB | Assay + ECFP4 | all three methods | 0.77±0.01 | 0.77±0.01 | 0.40±0.01 | |
| SVM | Assay | XGboost | 0.69±0.02 | 0.64±0.01 | 0.32±0.03 | |
| vascular | NB | Assay + ECFP4 | all three methods | 0.78±0.01 | 0.66±0.01 | 0.39±0.02 |
| NB | ECFP4 | Fisher's exact test | 0.77±0.01 | 0.77±0.01 | 0.41±0.01 | |
| XGboost | Assay | XGboost | 0.69±0.01 | 0.62±0.01 | 0.30±0.02 |
Features for optimal AUC-ROC
In order to obtain the best model for each in vivo endpoint, feature selection was performed and the subset of features that achieved the best predictive performance measured by AUC-ROC is summarized in Figure 3 and listed in Table S7. The number of features selected for each in vivo endpoint, in ascending order, is brain and coverings (40), musculoskeletal (40), cardiac (50) , peripheral nerve and sensation (60) , sense organs and special senses (60) , blood (70) , lungs thorax or respiration (70) , vascular (70) , endocrine (80) , liver (80) , kidney ureter and bladder (80), skin and appendages skin (80), gastrointestinal (90), behavioral (94) (Figure 3 and Table S7). The toxicity endpoints with optimal models using only structural features include behavioral, brain and coverings and kidney ureter and bladder. The optimal models for other endpoints used both assays and structures, and the ratios of assay and structure features across different endpoints in ascending order are lungs thorax or respiration (40%), peripheral nerve and sensation (50%), sense organs and special senses (50%), endocrine (60%), skin and appendages skin (60%), blood (75%), vascular (75%), gastrointestinal (80%), kidney ureter and bladder (100%), musculoskeletal (100%), liver (167%) (Figure 3). To obtain features that contributed the most to the performance of the model, feature sets were selected using the Fisher's exact test. The top five assays or ToxPrint features for each in vivo endpoint are shown in Table 2 and Table 3. The details of these significant features are provided in Table S8 and S9.
Figure 3.
Distribution of feature (assay or structure) sets that generated the optimal model for each toxicity endpoint. Feature selection was performed as a pre-processing step prior to modeling. The histogram shows the number of assays and structure features in the optimal feature set that produced the best model.
Table 2.
The top five most significant assays for each toxicity endpoint.
| In vivo toxicity | Assay Readout | p value |
|---|---|---|
| behavioral | tox21_dt40_p1.aggregrated_100 | 3.47E-04 |
| tox21_car_antagonist_p1.aggregrated_activity | 6.82E-04 | |
| tox21_ar_bla_antagonist_p1.aggregrated_signal | 3.37E-03 | |
| tox21_rxr_bla_agonist_p1.aggregrated_viability | 5.04E-03 | |
| tox21_fxr_bla_antagonist_p1.aggregrated_viability | 7.19E-03 | |
| blood | tox21_rt_viability_hek293_p1.aggregrated_flor_16_hr | 2.78E-03 |
| tox21_h2ax_cho_p2.aggregrated_control | 5.84E-03 | |
| tox21_luc_biochem_p1.aggregrated_ratio | 8.14E-03 | |
| tox21_ap1_agonist_p1.aggregrated_control | 1.00E-02 | |
| tox21_dt40_p1.aggregrated_100 | 1.45E-02 | |
| brain and coverings | tox21_vdr_bla_antagonist_p1.aggregrated_viability | 4.54E-02 |
| tox21_vdr_bla_agonist_p1.aggregrated_control | 1.00E-01 | |
| tox21_er_bla_antagonist_p1.aggregrated_viability | 1.03E-01 | |
| tox21_h2ax_cho_p2.aggregrated_activity | 1.09E-01 | |
| tox21_ar_bla_antagonist_p1.aggregrated_control | 1.13E-01 | |
| cardiac | tox21_er_luc_bg1_4e2_antagonist_p1.aggregrated_viability | 2.99E-03 |
| tox21_ar_mda_kb2_luc_antagonist_p1.aggregrated_activity | 9.23E-03 | |
| tox21_are_bla_p1.aggregrated_signal | 1.26E-02 | |
| tox21_ppard_bla_agonist_p1.aggregrated_viability | 1.41E-02 | |
| tox21_car_agonist_p1.aggregrated_viability | 1.44E-02 | |
| endocrine | tox21_rar_antagonist_p2.aggregrated_viability | 1.22E-03 |
| tox21_rt_viability_hepg2_p1.aggregrated_glo_40_hr | 1.22E-03 | |
| tox21_rt_viability_hepg2_p1.aggregrated_glo_24_hr | 1.73E-03 | |
| tox21_fxr_bla_antagonist_p1.aggregrated_signal | 2.72E-03 | |
| tox21_rt_viability_hepg2_p1.aggregrated_flor_24_hr | 2.98E-03 | |
| gastrointestinal | tox21_luc_biochem_p1.aggregrated_ratio | 2.59E-03 |
| tox21_ar_bla_antagonist_p1.aggregrated_signal | 3.01E-03 | |
| tox21_pparg_bla_agonist_p1.aggregrated_signal | 3.07E-03 | |
| tox21_hre_bla_agonist_p1.aggregrated_control | 5.03E-03 | |
| tox21_ar_bla_agonist_p1.aggregrated_control | 4.00E-02 | |
| kidney ureter and bladde | tox21_luc_biochem_p1.aggregrated_ratio | 1.20E-02 |
| tox21_nfkb_bla_agonist_p1.aggregrated_signal | 1.42E-02 | |
| tox21_vdr_bla_agonist_p1.aggregrated_signal | 4.61E-02 | |
| tox21_hre_bla_agonist_p1.aggregrated_signal | 4.95E-02 | |
| tox21_fxr_bla_agonist_p2.aggregrated_signal | 5.28E-02 | |
| liver | tox21_pparg_bla_agonist_p1.aggregrated_signal | 7.36E-04 |
| tox21_ar_bla_agonist_p1.aggregrated_control | 2.46E-03 | |
| tox21_er_bla_agonist_p2.aggregrated_control | 6.82E-03 | |
| tox21_ar_bla_antagonist_p1.aggregrated_control | 9.03E-03 | |
| tox21_ar_bla_antagonist_p1.aggregrated_signal | 9.74E-03 | |
| lungs thorax or respiration | tox21_luc_biochem_p1.aggregrated_ratio | 8.02E-03 |
| tox21_ror_cho_antagonist_p1.aggregrated_viability | 1.54E-02 | |
| tox21_rar_agonist_p1.aggregrated_activity | 1.71E-02 | |
| tox21_rar_antagonist_p2.aggregrated_activity | 1.76E-02 | |
| tox21_gr_hela_bla_antagonist_p1.aggregrated_signal | 2.60E-02 | |
| musculoskeletal | tox21_aromatase_p1.aggregrated_viability | 5.76E-03 |
| tox21_elg1_luc_agonist_p1.aggregrated.aggregrated_viability | 7.43E-03 | |
| tox21_rt_viability_hek293_p1.aggregrated_flor_40_hr | 3.87E-02 | |
| tox21_vdr_bla_agonist_p1.aggregrated_activity | 6.57E-02 | |
| tox21_hse_bla_p1.aggregrated_control | 7.95E-02 | |
| peripheral nerve and sensation | tox21_er_luc_bg1_4e2_agonist_p2.aggregrated_activity | 6.54E-03 |
| tox21_ar_mda_kb2_luc_agonist_p1.aggregrated_activity | 7.74E-03 | |
| tox21_are_bla_p1.aggregrated_signal | 1.67E-02 | |
| tox21_esre_bla_p1.aggregrated_signal | 2.35E-02 | |
| tox21_gh3_tre_antagonist_p1.aggregrated_viability | 3.24E-02 | |
| sense organs and special senses | tox21_are_bla_p1.aggregrated_activity | 2.02E-04 |
| tox21_p53_bla_p1.aggregrated_signal | 1.21E-03 | |
| tox21_nfkb_bla_agonist_p1.aggregrated_viability | 1.72E-03 | |
| tox21_p53_bla_p1.aggregrated_activity | 5.08E-03 | |
| tox21_aromatase_p1.aggregrated_activity | 5.51E-03 | |
| skin and appendages skin | tox21_ar_bla_agonist_p1.aggregrated_control | 4.29E-04 |
| tox21_vdr_bla_agonist_p1.aggregrated_signal | 4.67E-04 | |
| tox21_pparg_bla_agonist_p1.aggregrated_activity | 7.18E-04 | |
| tox21_rt_viability_hek293_p1.aggregrated_flor_24_hr | 1.11E-03 | |
| tox21_ppard_bla_antagonist_p1.aggregrated_signal | 1.66E-03 | |
| vascular | tox21_hre_bla_agonist_p1.aggregrated_signal | 1.84E-02 |
| tox21_ahr_p1.aggregrated_viability | 2.31E-02 | |
| tox21_er_luc_bg1_4e2_antagonist_p1.aggregrated_viability | 2.53E-02 | |
| tox21_gr_hela_bla_agonist_p1.aggregrated_activity | 3.45E-02 | |
| tox21_hse_bla_p1.aggregrated_activity | 3.57E-02 |
Table 3.
The top five most significant chemical structure features for each toxicity endpoint.
|
In vivo toxicity |
p value | Feature Name | Feature Structure |
|---|---|---|---|
| behavioral | 6.94E-07 | bond:CC(=O)C_ketone_alkane_cyclic |
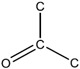
|
| 3.95E-06 | bond:CC(=O)C_ketone_alkene_cyclic_2-en-1-one_generic |

|
|
| 4.20E-06 | bond:CC(=O)C_ketone_aliphatic_generic |
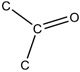
|
|
| 1.16E-05 | ring:aromatic_benzene |

|
|
| 1.65E-05 | bond:CC(=O)C_ketone_alkene_generic |

|
|
| blood | 7.12E-04 | group:ligand_path_4_bidentate_aminoethanol |
|
| 7.35E-04 | chain:alkeneCyclic_ethene_generic |
|
|
| 8.00E-04 | group:ligand_path_5-7_bidentate |
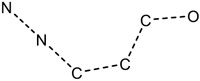
|
|
| 1.21E-03 | bond:CN_amine_pri-NH2_aromatic |
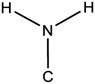
|
|
| 1.50E-03 | bond:CN_amine_pri-NH2_generic |
|
|
| brain and coverings | 5.10E-03 | ring:hetero_[6]_N_diazine_(1_3-)_generic |

|
| 5.77E-03 | bond:C=N_carboxamidine_generic |

|
|
| 2.19E-02 | ring:hetero_[6]_Z_1_3- |
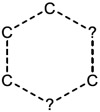
|
|
| 2.61E-02 | group:nucleobase_cytosine |

|
|
| 4.14E-02 | ring:hetero_[5]_N_pyrrole_generic |
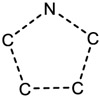
|
|
| cardiac | 5.76E-05 | bond:CN_amine_aliphatic_generic |
|
| 4.65E-04 | group:ligand_path_4_bidentate_aminoethanol |
|
|
| 1.30E-03 | bond:CN_amine_ter-N_aliphatic |

|
|
| 1.30E-03 | bond:CN_amine_ter-N_generic |

|
|
| 1.57E-03 | ring:hetero_[7]_generic_1-Z |
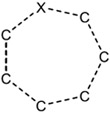
|
|
| endocrine | 6.53E-04 | bond:S(=O)N_sulfonamide |

|
| 8.29E-04 | bond:S(=O)N_sulfonylamide |

|
|
| 1.04E-03 | bond:S~N_generic |
|
|
| 2.35E-03 | ring:hetero_[5_6]_O_benzofuran |

|
|
| 5.17E-03 | bond:CX_halide_aromatic-X_ether_aromatic_(Ph-O-Ph) |
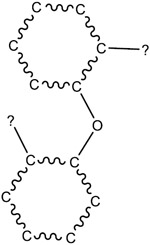
|
|
| gastrointestinal | 6.23E-04 | ring:hetero_[6]_Z_1_3_5- |

|
| 7.34E-04 | bond:CC(=O)C_ketone_alkene_generic |

|
|
| 1.44E-03 | bond:CC(=O)C_ketone_alkane_cyclic |
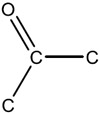
|
|
| 1.91E-03 | bond:C(=O)O_carboxylicEster_alkyl |

|
|
| 2.75E-03 | bond:C(=O)O_carboxylicEster_acyclic |

|
|
| kidney ureter and bladder | 1.20E-04 | ring:hetero_[6_6]_Z_generic |

|
| 7.01E-03 | bond:C(=O)O_carboxylicAcid_alkyl |

|
|
| 9.79E-03 | bond:C(=O)O_carboxylicAcid_generic |

|
|
| 1.95E-02 | bond:metal_metalloid_oxy |
|
|
| 4.25E-02 | atom:element_metal_metalloid |
|
|
| liver | 1.97E-03 | ring:hetero_[5]_S_thiophene |

|
| 3.02E-03 | ring:hetero_[5]_Z_1_3-Z |

|
|
| 3.33E-03 | bond:NN_hydrazine_acyclic_(connect_no Z) |
|
|
| 3.33E-03 | bond:S=O_sulfoxide |
|
|
| 4.65E-03 | chain:aromaticAlkane_Ar-C_ortho |
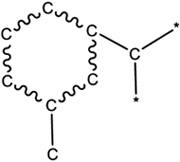
|
|
| lungs thorax or respiration | 1.72E-03 | ring:hetero_[6]_Z_generic |

|
| 2.51E-03 | bond:CN_amine_alicyclic_generic |
|
|
| 3.00E-03 | ring:hetero_[4]_Z_generic |
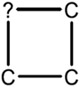
|
|
| 3.12E-03 | chain:aromaticAlkene_Ph-C2 |

|
|
| 5.16E-03 | ring:hetero_[4]_N_azetidine |

|
|
| musculoskeletal | 5.57E-03 | bond:COH_alcohol_allyl |

|
| 8.66E-03 | chain:alkeneLinear_diene_1_3-butene |

|
|
| 1.23E-02 | bond:C(=O)N_carboxamide_(NH2) |

|
|
| 1.41E-02 | bond:C(=O)O_carboxylicAcid_alkenyl |

|
|
| 2.07E-02 | bond:CC(=O)C_ketone_alkene_generic |

|
|
| 2.22E-04 | bond:P~S_generic |
|
|
| 1.19E-03 | bond:P=O_phosphorus_oxo |
|
|
| peripheral nerve and sensation | 2.53E-03 | bond:P=O_phosphate) |

|
| 5.13E-03 | bond:NN_hydrazine_acyclic_(connect_no Z) |
|
|
| 1.46E-02 | ring:hetero_[6]_N_pyridine |

|
|
| sense organs and special senses | 1.27E-03 | chain:alkeneLinear_monoene_ehtylene_terminal |
|
| 6.10E-03 | bond:CN_amine_aromatic_generic |
|
|
| 1.05E-02 | bond:CN_amine_ter-N_aromatic_aliphatic |

|
|
| 1.41E-02 | ring:hetero_[6]_Z_generic |
|
|
| 1.53E-02 | bond:CX_halide_alkyl-Cl_trichloro_(1_1_1-) |

|
|
| skin and appendages skin | 9.21E-04 | bond:C(=O)O_carboxylicAcid_generic |
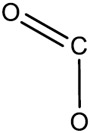
|
| 2.01E-03 | bond:CC(=O)C_ketone_aromatic_aliphatic |

|
|
| 6.08E-03 | ring:aromatic_phenyl |

|
|
| 9.03E-03 | ring:hetero_[6]_N_diazine_(1_3-)_generic |

|
|
| 9.35E-03 | group:carbohydrate_pentofuranose_2-deoxy |

|
|
| vascular | 1.14E-03 | ring:aromatic_benzene |

|
| 3.89E-03 | chain:alkeneCyclic_diene_cyclohexene |

|
|
| 7.51E-03 | bond:CN_amine_aromatic_generic |
|
|
| 1.53E-02 | bond:CN_amine_sec-NH_aromatic |

|
|
| 1.79E-02 | bond:CX_halide_aromatic-X_generic |
|
In addition to the top-performing model for each endpoint, also listed in Table 1 are the results for the optimal model in the 2 other feature set categories, such that the top models are listed for structure-only, assay-only, and structure+assay feature sets (all model results are provided in Table S5). These results indicate that the structure-only model performance falls within the 0.01 error range of the optimal model for 10 out of 14 endpoints, and within 0.02 of the optimal model for the remaining 4 endpoints. In contrast, the assay-only models perform more poorly than either the structure-only or structure+assay models, deviating by 0.05 (kidney ureter and bladder, peripheral nerve and sensation) to 0.19 (brain and coverings), averaging 0.1 less than the optimal model across the 14 modeled endpoints.
Discussion
Machine learning provides a promising tool for toxicity prediction because of advantages such as high efficiency, low cost, and high accuracy. Machine learning has been used to predict chemical - induced toxicity in animals based on structure or biological activity 15, 34, 35. In this study, we applied five machine learning algorithms (NB, RF, SVM, NNET and XGboost) with multiple performance metrics (AUC-ROC, BA and MCC) to build prediction models for 14 human toxicity endpoints. We obtained human in vivo toxicity data on 732 chemicals and built prediction models for each of the 14 endpoints using assay data (assay-only), chemical structure data (structure-only), or a combination of both (structure+assay). For some endpoints, the dataset was balanced with a roughly equal number of toxic and nontoxic chemicals, whereas others had imbalanced datasets with one type of chemicals, in most cases nontoxic, outnumbering the other type, e.g., toxic, by several folds. Despite of this diversity in the dataset composition, we were able to generate models with good performance (Figure 1). To test if data balancing approaches could further improve model performance, we applied four data balancing methods to the musculoskeletal endpoint, and the resulting AUC-ROC values are as follows: Down-sampling (0.77±0.04), Up-sampling (0.85±0.02), ROSE (0.84±0.02) and SMOTE (0.80±0.03). In this case, data balancing did not yield any model that outperformed the original model (0.88±0.02).
Feature selection, the process of selecting a subset of relevant features, has played an important role in building robust machine learning models for classification 36, 37. The data employed in this study for modeling were high-dimensional (729-bit ToxPrints, 147 assay readouts, and 1024-bit ECFP4 fingerprints). Three feature selection methods, including Fisher’s exact test with p value, importance scores from XGboost and RF algorithm, were used to count the number of chemicals with or without a certain feature that fall into the toxic or nontoxic category. When different feature sets based on different selection methods were applied to build the models, we found that the feature set with the best model performance was not necessarily the largest one (Figure 2 and Supplemental Table S5), and an optimal subset of features could be identified that could produce the best performing model. These results suggested that feature selection could help avoid overfitting, improve model performance and increase the speed of learning. In addition to alleviating the effect of dimensionality, the feature selection process also identified features that contributed the most to predicting each in vivo toxicity endpoint 37, 38.
The predictive performances of the models were different for the 14 endpoints, but models for most of the endpoints were accurate and robust (12 of the 14 endpoints with AUC-ROC ≥ 0.75, Table 1). The endpoints with slightly less predictive models, i.e., cardiac (0.74±0.01) and gastrointestinal (0.72±0.01), may have more complex toxicity mechanisms and need data on additional targets that are not covered by the current Tox21 assays to improve model performance 10. Moreover, our results indicated that the performance of a classifier depends on the specific dataset, model type and feature selection method (Figure 2, Table 1 and Supplemental Table S5), implying that pre-selecting classifiers based on evidence from related datasets may help increase discriminative performance 39.
The feature selection process identified features that contributed the most to the prediction of each toxicity endpoint. The top five most significant Tox21 assays are listed in Table 2 and the extended list can be found in Table S8. The biological targets and pathways covered by these assays could provide clues to potential mechanisms leading to the particular type of human toxicity. For example, peroxisome proliferator-activated receptor gamma (PPARγ) was identified as the most significant contributor to liver toxicity. As a main isotype of the PPAR family, PPARγ is a nuclear receptor that plays a key role in liver homeostasis and regulating adipogenesis 40, 41. The hypoxia response element (HRE) assay was found to significantly contribute to vascular toxicity. The hypoxia inducible factor 1 alpha (HIF-1α) can bind to hypoxia response elements (HRE) in the enhancer of vascular endothelial growth factor (VEGF) to increase its transcription, which is an important growth factor for angiogenesis and vascularization 42, 43. Androgen receptor (AR) signaling and estrogen receptor (ER) signaling were the top two most significant contributors for peripheral nerve and sensation toxicity. Consistent with these results, classical intracellular steroid receptors, such as AR and ER, have been detected in the glial and neuronal compartments of the peripheral nervous system 44, 45. Similarly, the modeling process identified ToxPrint structure features that contributed the most to each in vivo toxicity endpoint. The top five most significant ToxPrint structure features are listed in Table 3 with the extended list provided in Table S9. For example, organophosphorus compounds are known to cause neural toxicity, and the top three structure features identified for “peripheral nerve and sensation” all contain phosphorus groups. These features can serve as structure alerts in new chemical design to minimize or avoid toxicity during the chemical review process for industrial chemicals, pesticides or drugs.
In a prospective evaluation, each of the top-performing models was applied to predicting each of the 14 human toxicity endpoints for the structurable portion of the full Tox21 library (including the training set). The detailed results in terms of probabilities (ranging from 1-toxic, to 0-non-toxic) are presented in Table S6 and summarized in Table 4. According to Table 4, some endpoints are predicted to be highly represented across the entire Tox21 library, even at the 0.8 probability threshold (e.g., brain and coverings – 75%, cardiac – 53%, skin and appendages skin – 38%, behavioral – 37%, and vascular – 31%), whereas other endpoints are predicted at much lower frequency (liver – 0.2%, peripheral nerve and sensation – 0.2%, blood – 1%, sense organs and special senses –2%, and lungs thorax or respiration – 2%). Overall, the total number of chemicals with averaged probabilities >0.5 over the 14 endpoints is 297, indicating that approximately 4% of the Tox21 chemicals have significant potential for adverse outcomes across multiple human toxicity endpoints.
Table 4.
The summary of probabilities for predicting the full Tox21 library (including training set).
| Endpoint | >0.5 | >0.8 | % reduction from 0.5 to 0.8 |
|---|---|---|---|
| behavioral | 2770 | 2637 | 5% |
| brain and coverings | 5798 | 5311 | 8% |
| endocrine | 640 | 596 | 7% |
| musculoskeletal | 765 | 720 | 6% |
| skin and appendages skin | 2882 | 2695 | 6% |
| vascular | 2391 | 2169 | 9% |
| gastrointestinal | 2831 | 1472 | 48% |
| kidney ureter and bladder | 706 | 596 | 16% |
| blood | 640 | 83 | 87% |
| cardiac | 6829 | 3736 | 45% |
| liver | 332 | 13 | 96% |
| lungs thorax or respiration | 1631 | 123 | 92% |
| peripheral nerve and sensation | 66 | 11 | 83% |
| sense organs and special senses | 906 | 125 | 86% |
A particularly noteworthy result of the present study is the finding that the structure-only models were equal or nearly equal in performance to models based on both structure and assay features. This finding has important implications for future screening applications since it implies that Tox21 assay data used in many of the current models, which would be costly and difficult, or even impossible (if the chemical were unavailable or insoluble/unstable in Dimethyl sulfoxide (DMSO)) to generate for non-Tox21 library chemicals, would not be needed to produce a model prediction. Hence, the structure-only models could be applied, in principle, to predict toxicity for any chemical for which a unique structure could be represented, extending beyond the Tox21 library to other large libraries or data sets. On the other hand, assay-only models could, in principle, be applied to chemicals for which a unique structure was unavailable, such as for those non-structurable chemicals in the Tox21 library. In addition, as discussed above, the assay features shown to be influential in model performance can potentially illuminate mechanistic elements of an in vivo toxicity endpoint.
In summary, we applied five machine learning algorithms to build predictive models for 14 human toxicity endpoints using chemical structure, in vitro assay data, and a combination of both types of data. We found that model performance depended on the specific dataset type, feature selection method and learning algorithm used. Overall, we constructed models with surprisingly good performance for most of the in vivo human toxicity endpoints in this study (12 out of 14, AUC-ROC >0.75), particularly considering the level of aggregation, variability, and uncertainties in the quality of data used to construct the training sets. These models, particularly those based on structure-only, can be used to screen large sets of new chemicals for potential toxicity with the understanding that the predictions would benefit from further support from nearest neighbor data and examination of features contributing to the prediction. Future work that could potentially improve the current models could include a broader range of assay data, such has been generated in EPA’s ToxCast program (we estimate that approximately one third of the current training set of chemicals with toxicity endpoint data have also been screened in a broad panel of ToxCast assays). The modeling process has also been shown to be useful in identifying structure features and/or assay targets that are helpful in improving model performance and, thus, can serve as a guide to understand the mechanism of chemical-induced toxicity. Additionally, given that animal in vivo data remain the standard for guideline toxicity studies, it might be of interest to compare the predictions of the present human-based toxicity models to the results for similar rodent endpoints to assess the level of concordance. In comparing the present results to those published previously by Liu et al., their use of ToxCast assay data, their relatively small dataset sizes, and their prediction of rodent endpoints from guideline toxicity studies might help account for differences in model performances. Finally, as the models constructed in this study were trained on a collection of chemicals containing a significant number of drugs, the models may be more suitable for making predictions on drug-like molecules than other types of chemicals, such as environmental contaminants. Given the high rate of failure in clinical trials and the perennial challenge of predicting human toxicity of prospective drug candidates, this presumption is worthy of further exploration.
Supplementary Material
Acknowledgements
We would like to thank Srilatha Sakamuru, Jinghua Zhao, Li Zhang, and Dr. Caitlin Lynch for technical assistance with the qHTS assays. This work was supported by the Intramural Research Programs of the National Toxicology Program (Interagency agreement #Y2-ES-7020–01), National Institute of Environmental Health Sciences, the U.S. Environmental Protection Agency (Interagency Agreement #Y3-HG-7026–03), and the National Center for Advancing Translational Sciences, National Institutes of Health.
The views expressed in this article are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the National Center for Advancing Translational Sciences, the National Institutes of Health, or the United States government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Supporting Information
Table S1 Summary of the in vivo toxicity data used in the study; Table S2 Raw in vivo toxicity data records from ChemIDplus; Table S3 Full listing of binarized human in vivo toxicity results; Table S4 Unique chemicals and aggregated data used for modeling; Table S5 Optimal AUC-ROC value, together with BA and MCC for each model; Table S6 Predicted toxic potential of Tox21 compounds; Table S7 Subset of features that achieved the best predictive performance measured by AUC-ROC; Table S8 Significance of Tox21 assays measured by Fisher's exact test with p value; Table S9 Significance of chemical structure features measured by Fisher's exact test with p value. (XLSX)
Figure S1 Clustering of the various human toxicity endpoints based on toxic (red)/nontoxic (white) chemicals derived from the ChemIDPlus Advanced database; Figure S2 Clustering of the various human toxicity endpoints based on significant structure feature. (PDF)
References
- (1).Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, and Sayre P (2008) The toxicity data landscape for environmental chemicals. Environmental health perspectives 117, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).DeNicola N, Zlatnik MMG, and Conry MJ (2018) Toxic environmental exposures in maternal, fetal, and reproductive health. Obstetrics-Gynecology. [Google Scholar]
- (3).Akhtar A. (2015) The flaws and human harms of animal experimentation. Cambridge quarterly of healthcare ethics : CQ : the international journal of healthcare ethics committees 24, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Houck KA, and Kavlock RJ (2008) Understanding mechanisms of toxicity: insights from drug discovery research. Toxicology and applied pharmacology 227, 163–178. [DOI] [PubMed] [Google Scholar]
- (5).Collins FS, Gray GM, and Bucher JR (2008) Toxicology. Transforming environmental health protection. Science 319, 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kavlock RJ, Austin CP, and Tice RR (2009) Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal 29, 485–487; discussion 492-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tice RR, Austin CP, Kavlock RJ, and Bucher JR (2013) Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Krewski D, Acosta D Jr., Andersen M, Anderson H, Bailar JC 3rd, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S Jr., Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, and Zeise L (2010) Toxicity testing in the 21st century: a vision and a strategy. Journal of toxicology and environmental health. Part B, Critical reviews 13, 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR, and Xia M (2013) The Tox21 robotic platform for the assessment of environmental chemicals - from vision to reality. Drug Discov Today 18, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Huang R, Xia M, Sakamuru S, Zhao J, Lynch C, Zhao T, Zhu H, Austin CP, and Simeonov A (2018) Expanding biological space coverage enhances the prediction of drug adverse effects in human using in vitro activity profiles. Sci Rep 8, 3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Raies AB, and Bajic VB (2016) In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip Rev Comput Mol Sci 6, 147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Deeb O, and Goodarzi M (2012) In silico quantitative structure toxicity relationship of chemical compounds: some case studies. Current drug safety 7, 289–297. [DOI] [PubMed] [Google Scholar]
- (13).Liu J, Mansouri K, Judson RS, Martin MT, Hong H, Chen M, Xu X, Thomas RS, and Shah I (2015) Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure. Chem Res Toxicol 28, 738–751. [DOI] [PubMed] [Google Scholar]
- (14).Koutsoukas A, St. Amand J, Mishra M, and Huan J (2016) Predictive Toxicology: Modeling Chemical Induced Toxicological Response Combining Circular Fingerprints with Random Forest and Support Vector Machine. Frontiers in Environmental Science 4. [Google Scholar]
- (15).Shah I, Liu J, Judson RS, Thomas RS, and Patlewicz G (2016) Systematically evaluating read-across prediction and performance using a local validity approach characterized by chemical structure and bioactivity information. Regul Toxicol Pharmacol 79, 12–24. [DOI] [PubMed] [Google Scholar]
- (16).Huang R, Xia M, Sakamuru S, Zhao J, Shahane SA, Attene-Ramos M, Zhao T, Austin CP, and Simeonov A (2016) Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun 7, 10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu J, Patlewicz G, Williams AJ, Thomas RS, and Shah I (2017) Predicting Organ Toxicity Using in Vitro Bioactivity Data and Chemical Structure. Chem Res Toxicol 30, 2046–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sosnin S, Karlov D, Tetko IV, and Fedorov MV (2018) Comparative Study of Multitask Toxicity Modeling on a Broad Chemical Space. Journal of chemical information and modeling 59, 1062–1072. [DOI] [PubMed] [Google Scholar]
- (19).Sheridan RP, Wang WM, Liaw A, Ma J, and Gifford EM (2016) Extreme gradient boosting as a method for quantitative structure–activity relationships. Journal of chemical information and modeling 56, 2353–2360. [DOI] [PubMed] [Google Scholar]
- (20).PubChem. (2018) Tox21 phase II data. http://www.ncbi.nlm.nih.gov/pcassay?term=tox21.
- (21).Huang R, Xia M, Cho M-H, Sakamuru S, Shinn P, Houck KA, Dix DJ, Judson RS, Witt KL, and Kavlock RJ (2011) Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environmental health perspectives 119, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Huang R. (2016) A Quantitative High-Throughput Screening Data Analysis Pipeline for Activity Profiling. Methods Mol Biol 1473, 111–122. [DOI] [PubMed] [Google Scholar]
- (23).Yang C, Tarkhov A, Marusczyk J, Bienfait B, Gasteiger J, Kleinoeder T, Magdziarz T, Sacher O, Schwab CH, Schwoebel J, Terfloth L, Arvidson K, Richard A, Worth A, and Rathman J (2015) New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model 55, 510–528. [DOI] [PubMed] [Google Scholar]
- (24).Steinbeck C, Han Y, Kuhn S, Horlacher O, Luttmann E, and Willighagen E (2003) The Chemistry Development Kit (CDK): An open-source Java library for chemo-and bioinformatics. Journal of chemical information and computer sciences 43, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, and Steinbeck C (2013) KNIME-CDK: Workflow-driven cheminformatics. BMC bioinformatics 14, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chen T, He T, Benesty M, Khotilovich V, and Tang Y (2015) Xgboost: extreme gradient boosting. R package version 0.4-2, 1–4. [Google Scholar]
- (27).Liaw A, and Wiener M (2002) Classification and regression by randomForest. R news 2, 18–22. [Google Scholar]
- (28).Dimitriadou E, Hornik K, Leisch F, Meyer D, and Weingessel A (2005) Misc Functions of the Department of Statistics (e1071), TU Wien. R package version, 1.5–7. [Google Scholar]
- (29).Ripley B. (2002) Modern applied statistics with S, Springer, New York. [Google Scholar]
- (30).Sing T, Sander O, Beerenwinkel N, and Lengauer T (2005) ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941. [DOI] [PubMed] [Google Scholar]
- (31).Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, and Müller M (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, and Judson RS (2017) The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of cheminformatics 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, and Sayeeda Z (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic acids research 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Baskin II. (2018) Machine Learning Methods in Computational Toxicology. Methods Mol Biol 1800, 119–139. [DOI] [PubMed] [Google Scholar]
- (35).Idakwo G, Luttrell J, Chen M, Hong H, Zhou Z, Gong P, and Zhang C (2019) A review on machine learning methods for in silico toxicity prediction. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev, 1–23. [DOI] [PubMed] [Google Scholar]
- (36).Nie F, Huang H, Cai X, and Ding CH (2010) Efficient and robust feature selection via joint ℓ2, 1-norms minimization, In Advances in neural information processing systems pp 1813–1821. [Google Scholar]
- (37).Jović A, Brkić K, and Bogunović N (2015) A review of feature selection methods with applications, In 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) pp 1200–1205, IEEE. [Google Scholar]
- (38).Cai J, Luo J, Wang S, and Yang S (2018) Feature selection in machine learning: A new perspective. Neurocomputing 300, 70–79. [Google Scholar]
- (39).Deist TM, Dankers FJ, Valdes G, Wijsman R, Hsu IC, Oberije C, Lustberg T, van Soest J, Hoebers F, and Jochems A (2018) Machine learning algorithms for outcome prediction in (chemo) radiotherapy: An empirical comparison of classifiers. Medical physics 45, 3449–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).M Zardi E, Navarini L, Sambataro G, Piccinni P, M Sambataro F, Spina C, and Dobrina A (2013) Hepatic PPARs: their role in liver physiology, fibrosis and treatment. Current Medicinal Chemistry 20, 3370–3396. [DOI] [PubMed] [Google Scholar]
- (41).Liss KH, and Finck BN (2017) PPARs and nonalcoholic fatty liver disease. Biochimie 136, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Burroughs SK, Kaluz S, Wang D, Wang K, Van Meir EG, and Wang B (2013) Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future medicinal chemistry 5, 553–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wu J.-b., Tang Y.-l., and Liang X.-h. (2018) Targeting VEGF pathway to normalize the vasculature: an emerging insight in cancer therapy. OncoTargets and therapy 11, 6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Giatti S, Romano S, Pesaresi M, Cermenati G, Mitro N, Caruso D, Tetel MJ, Garcia-Segura LM, and Melcangi RC (2015) Neuroactive steroids and the peripheral nervous system: an update. Steroids 103, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Melcangi R, Magnaghi V, Galbiati M, and Martini L (2001) Glial cells: a target for steroid hormones, In Progress in brain research pp 31–40, Elsevier. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.