Abstract
Participants in mixed-species bird flocks (MSFs) have been shown to associate with species that are similar in body size, diet, and evolutionary history, suggesting that facilitation structures these assemblages. In addition, several studies have suggested that species in MSFs resemble each other in their plumage, but this question has not been systematically investigated for any MSF system. During the nonbreeding season of 2020 and 2021, we sampled 585 MSFs on 14 transects in 2 habitats of Tongbiguang Nature Reserve in western Yunnan Province, China. We performed social network analysis and the Multiple Regression Quadratic Assignment Procedure to evaluate the effect of 4 species traits (body size, overall plumage color, distinctive plumage patterns, and diet) and evolutionary history on species association strength at the whole-MSF and within-MSF levels. All 41 significant relationships showed that species with stronger associations were more similar in their various traits. Body size had the strongest effect on association strength, followed by phylogeny, plumage patterns, and plumage color; diet had the weakest effect. Our results are consistent with the hypotheses that the benefits of associating with phenotypically similar species outweigh the potential costs of interspecific competition, and that trait matching can occur in plumage characteristics, albeit more weakly than in other traits. Several explanations exist as to why similarities in plumage may occur in MSFs, including that they could reduce predators’ ability to target phenotypically “odd” individuals. Whether trait matching in plumage occurs through assortative processes in ecological time or is influenced by co-evolution requires further study.
Keywords: co-evolution, interaction networks, mixed-species animal groups, phenotypic similarity, trait convergence
Mixed-species bird flocks (MSFs hereafter) are important components of bird communities that can be found globally in forested habitats (Greenberg 2000; Zou et al. 2018). MSFs occur seasonally in the nonbreeding season in temperate regions, but they can be found throughout the year in the tropics, where in some forests, more than half the individual birds are in MSFs at any one time (Latta and Wunderle 1996; Zhou et al. 2019). Some species only join MSFs temporarily, but other species forage inside MSFs most of the time, with the pinnacle of stability being in lowland tropical forests in South America, where individuals of different species defend the same territory, and territorial borders may remain the same for decades (Martínez and Gomez 2013). Participating in MSFs can reduce predation risk and increase foraging efficiency (Morse 1977; Sridhar et al. 2009; Goodale et al. 2017). Therefore, mixed-species flocking is one of the most well-known examples of how facilitation can structure bird communities.
Given that many bird species in MSFs interact in close proximity, evidence for competition between species has long been searched for. Some researchers have found evidence for “checkerboard” relationships in MSFs, whereby close congeners that potentially compete with each other do not co-occur (Diamond 1975; Graves and Gotelli 1993; Colorado and Rodewald 2015). However, if competition were the major factor structuring these communities, we would expect co-occurring species to be dissimilar in their traits (Webb et al. 2002), but that does not seem to be the general finding. Rather, species in MSFs have been found to be more similar to each other than expected in diet and size (Sridhar et al. 2012; Mammides et al. 2018; Cestari et al. 2020) and evolutionary history (Gómez et al. 2010; Péron 2017; Mammides et al. 2018; Zhang et al. 2020). Species are even more similar in their foraging niche than would be randomly expected, although they may forage on different substrates (Jones et al. 2020). In fact, it was hypothesized that MSFs, as well as mixed-species groups in other taxa, are likely to be based on supplemental benefits, in which all individuals offer or receive a similar benefit regardless of their species identity, such as when birds dilute the risk of predation among other species of similar sizes that are vulnerable to the same predators (Goodale et al. 2020). Species that have general similarities in diet and size may also display “activity matching.” For example, they may travel at the same speeds (Hutto 1988; Darrah and Smith 2013; Sridhar and Guttal 2018). While matching each other in some aspects, species in MSFs can also reduce competition by niche partitioning in other axes of their ecology, such as their foraging techniques and microhabitats (Sridhar and Guttal 2018; Jones et al. 2020). General similarities among species in MSFs are, therefore, evidence that facilitation may be a stronger force than the competition in shaping their assembly (Sridhar et al. 2012).
One type of matching in MSFs that has rarely been systematically or quantitatively addressed is the matching of plumage characteristics. The forces driving this have been thought to include facilitation of interspecific communication when birds use the same food resources or share predators (i.e., the species would recognize the same signals: Moynihan 1968; Wheatcroft and Price 2013), or confusion of predators with similar appearance (Barnard 1979). The latter hypothesis is related to the idea that predators can focus on 1 individual to attack when it is phenotypically dissimilar from the majority of the group members (known as the “oddity effect” and best studied in fish and invertebrates: Landeau and Terborgh 1986; Jeschke and Tollrian 2007). Empirically, several observers have remarked that similar-looking bird species tend to participate together in MSFs (Moynihan 1962, 1968; Bell 1983; Willis 1989; Sazima 2010). Beauchamp and Goodale (2011) tested 22 species pairs reported to show mimicry in MSFs and found evidence for plumage resemblance as perceived by human eyes for 14 pairs. These similarities could represent mimicry or more general similarities such as similar body color or plumage patterns (e.g., eye lines, contrasting color patterns in wings or tail). Moreover, they could either represent convergence over evolutionary time or simply result from assortment between similar species in time and space (as would seem to be the case for body size matching in MSFs).
Earlier research has shown that some MSF benefits require the participating species to be close together inside MSFs. For example, flycatching birds adjust their positions to be close to gleaning birds, which disturb insects in the air (Hino 1998; Sridhar and Shanker 2014). Likewise, if similar species are benefitting from the oddity effect in MSFs, we would expect them to not only co-occur in MSFs but also associate spatially close within MSFs because predators are expected to be confused when they see many similar individuals within their field of view (Jeschke and Tollrian 2007; Rodgers et al. 2013). In forests, the most lethal predators would be Accipiter hawks that fly through the forest under the canopy (Krams et al. 2020); as their field of vision is restricted because of the dense vegetation present, we predict that similarities would operate in a smaller space (e.g., a cube of 4 m to 1 side) than the whole MSF (a cube that can be of 20 m to a side). Such spatial associations between similar or dissimilar species might be expected to influence phylogenetic patterns in modules of species that associate together within MSFs, or in the MSF as a whole (Péron 2017). Hence studies of trait matching in MSFs should be evaluated both among and within MSFs.
We present a study of phenotypic matching in MSFs in 2 different habitats of southwestern China, including plumage traits (overall color and distinct patterns), as well as body size, diet, and phylogeny. We investigated associations both at the whole-MSF and within-MSF levels. In the area of southwestern China in which we worked, previous observers have noted that there appear to be distinct MSF types that are distinguishable by the plumage colors of their members; specifically, there appear to be a group of birds with yellow underparts that associate together, and other groups that are composed mainly of birds with rufous and white, or dull green, plumage (SKR and YL, per. obs). In this region, and southern China more generally, the dominant “nuclear” bird species in MSFs (i.e., species important to MSF formation and cohesion, sensuMoynihan 1962) are closely related species of fulvetta in the Alcippe genus (Zou et al. 2007; Zhou et al. 2019). These species are often highly gregarious, sometimes averaging more than 30 individuals per MSF (Chen and Hsieh 2002). This is important because when 1 species represents a large proportion of the MSF, any evolutionary process of color convergence could accelerate, because the numerically dominant species could act as a model onto which other species could converge. Nuclear species are often very active, making conspicuous wing and tail movements (Hutto 1994), especially in mobbing contexts (Jiang et al. 2020). Therefore, plumage matching could also be in relation to specific plumage patterns such as on the wings or tails.
We hypothesized that if competition is the dominant force structuring MSFs, species with similar traits would avoid each other within and among MSFs and that the species that associate together most strongly would be phylogenetically distant. The competition hypothesis makes no predictions about plumage traits. In contrast, if MSFs are structured mainly by facilitative interactions, then we predicted that species would match each other in various traits, including those related to plumage, and phenotypically similar species would associate with each other both within and among MSFs. As to plumage matching specifically, we hypothesized that if it was driven by the oddity effect, it would apply mostly to birds that aggregate closely together within MSFs and would be thus more strongly captured in the within-MSF data.
Materials and Methods
Study site and field data collection
We conducted our fieldwork inside and close to Tongbiguang Nature Reserve (24.62°N, 97.63°E), in Yinjiang County, western Yunnan Province, China. Elevation in the reserve ranges from 210 to 3,400 m asl, with the vegetation being primary and secondary monsoon evergreen broad-leaved forest. Secondary forest was lightly logged in the 1990s and early 2000s, including some forests in which natural forest was mixed with planted timber species (Betula alnoides, Cunninghamia lanceolata), now 10–20 years old. In our study, we worked in both primary and secondary forest, but only from 800 to 1,700 m asl, where preliminary observations suggested that some of the plumage traits noticed by SKR and YL appeared to be prevalent.
We placed 14 one-km transects inside and outside the nature reserve along existing trails, primarily on dirt roads, but some with some stretches of tarred roads, 8 in primary forest and 6 in secondary forest, with each transect at least 300 m away from all others at all points. One notetaker and 1 observer walked together along these transects once per day, either in the morning (7:00–11:00) or in the afternoon (14:30–18:30), with sampling balanced between these 2 times. We visited each transect an average of 11.7 ± 4.59 (mean ± SD) times from December 2020 to March 2021. We concentrated on this time during the nonbreeding season because MSFs are most common then; subtropical MSFs can break down in the breeding season (Jiang et al. 2020). While walking the transects, we noted every bird we saw or heard within 30 m (estimated by the observer) and noted whether it was in an MSF or not. We defined an MSF as 2 or more species foraging together and moving in the same direction for at least 5 min (Goodale et al. 2009). We considered MSFs collected on the same transect on different days as independent from each other, because MSFs dissolve at night and reassemble each morning, an assumption that has been made in other studies of Asian MSFs (Zhang et al. 2013; Goodale et al. 2014).
When an MSF was encountered, we began 2 different kinds of observations. The first was at the whole-MSF level: we noted all the species and individuals in the MSF for more than 5 min but no longer than 15 min. When these data were collected, we then measured which species were associated together spatially within MSFs. For these data, we recorded all the birds inside a cube, visually estimated, in a moment in time (i.e., a scan of 5 s). In preliminary observations, we experimented with the size of this cube and found that cubes smaller than 4 m on 1 side (64 m3) often had only 1 species. We, therefore, fixed the cube to be 64 m3 so as to increase the chances it held multiple species, while at the same time being considerably smaller than the size of the whole MSF. We first collected the data at the front edge of the MSF where most birds were located. If the MSF was large enough, we took more than 1 within-MSF observation, and to detect new individuals, we avoided the parts of the MSF where the previous observations were made. For any 1 MSF, there was a maximum of 4 within-MSF observations (the average MSF had 2.97 ± 2.0 within-MSF observations; sometimes we were unable to follow the MSF and made no within-MSF observations). Because we sampled different individuals in each within-MSF observation, we considered all observations to be independent of each other.
Bird trait data
Some studies have suggested that rare species might have disproportionate impact on multivariate analyses (Marchant 2002). To reduce such impact, we deleted species that were recorded less than 3 times in MSFs during our entire fieldwork. This procedure reduced the number of MSF species in the analyses from 123 to 95.
For each species, we extracted (1) body length, (2) diet, (3) plumage color, and (4) plumage patterns from Birds of the World (https://birdsoftheworld.org/bow/home). We only extracted information for female adults and for the subspecies present in the Tongbiguan region. We chose to score females because dimorphic males tend to resemble females in the nonbreeding season. We extracted body length in centimeters and classified diets into 4 categories: insectivores and frugivores, defined by eating a majority of insects and fruits, respectively; omnivores, for which it was difficult to decide which category was a majority; and an “others” category that consisted of nectarivores or carnivores (both of which were rare).
Our original color system, based on the previous observations of SKR and YL in the region between 2016 and 2019, included 3 categories: yellow-belly species, rufous-white species (overall body color dominated by both rufous and white), greenish species (the whole body dominated by green), and an “others” category. However, in this system, a high number of species (30 of 95) were categorized as others. Therefore, we created a more extensive categorization system that included not only the original 3 color categories but also white-black, gray-brown, and black (all assessed at the whole body level), as well as “others” (now including only 12 species). For the plumage patterns analysis, we scored a total of 9 patterns: crest, eye stripe, eye ring, eyebrow, coronal stripe, wing bar(s), contrasting rump, contrasting tail, and contrasting tail edge. In scoring, we assigned 1 when a species clearly had the pattern, 0 if it did not, and 0.5 if the trait was not clear. LZ performed all color and plumage pattern categorization.
Phylogenetic data
Because similarities and dissimilarities between species could be due to phylogeny, we downloaded 10,000 phylogenies from Bird Tree (http://birdtree.org/) with an Ericson backbone (Jetz et al. 2012) for the 95 species. Then, following the general approach of Felsenstein (2004), we used the function consensus from package “ape” in software R 4.1.0, and generated a strict consensus tree.
Statistical analysis
We conducted all analyses in the R statistical environment (version 4.1.0), considering results significant when P ≤ 0.05. We first used Mann–Whitney tests to calculate the differences between habitats (primary vs. secondary forest) in species richness and number of individuals per MSF, or per within-MSF observation. We then analyzed the data for each of the 14 transects separately, for both within-MSF and whole-MSF data (i.e., 28 analyses in total). Using the function get_network from package “asnipe” (Farine 2013), we calculated the simple ratio index (hereafter SRI) for all the species pairs on each transect, provided that both species were seen in MSFs on that transect (a provision which means that a lack of association cannot be simply caused by species absences). SRI is calculated as
where X represents the number of times species a and b were observed together in an MSF on that transect, Ya represents the number of times species a was observed without b in MSFs, Yb represents the number of times species b was observed without a in MSFs, and Yab represents the number of times species a and b were observed together at a transect but did not associate, and is defined here as 0 (Farine and Whitehead 2015). SRI ranges from 0 to 1; the higher the SRI, the more the 2 species associated with each other.
To evaluate the effects of species traits and phylogenetic relationships on association strength, we used the Multiple Regression Quadratic Assignment Procedure (MRQAP) method, following Farine (2013, 2017) and Dekker et al. (2007). MRQAP is a method that uses multiple matrices as independent variables, and is robust to some nonindependence in the data (a potential problem for this project, as we investigated similarities between species pairs, and the same species could be in multiple pairs). In our case, the association strength (SRI) between 2 species was the response variable, and the similarities/dissimilarities in body length, diet, plumage color, plumage patterns, and phylogenetic distances were the explanatory variables. We expressed similarity/dissimilarity in the following ways: (1) body length differences were expressed as dissimilarities, the absolute body length difference between the species in centimeters, normalized to a range of 0–1, using the function data.Normalization from package “clusterSim”; (2) diet matrixes represented similarities between the species pair, assigned 1 if their diets were the same, 0 if they were not, and 0 if they were both “other”; (3) color was represented by similarities like diet; (4) the plumage patterns were calculated as the proportion of traits the species pair shared, only counting a shared trait if they had the exact same value (i.e., 0.5 and 0.5 was considered the same, but 0.5 and 0, or 0.5 and 1, were not); (5) phylogenetic distances between species pairs were calculated using the function cophenetic.phylo from the package “ape,” and then scaled by the multiple 10307 to make the distances range from 0 to 1, with higher phylogenetic distance representing a more remote common ancestor. After preparation of the matrixes for each of the 28 datasets, we used the mrqap.dsp function in the “asnipe” package to run the MRQAP and ran 999 node permutations for each network (transect) and kind of sampling (within-MSF vs. whole-MSF). Note that in conducting multiple tests, as the 14 we performed here for the different transects, some researchers advocate reducing the P-value that is considered significant (e.g., Bonferroni corrections), to reduce Type I errors. However, such tests are known to be overly conservative (Moran 2003), and increase the chance of Type II errors (Nakagawa 2004). As our 14 tests were each based on a different dataset, we took the approach of considering P ≤ 0.05 unadjusted as significant, although we also show Bonferroni-corrected results in the main table.
To investigate if species pairs that had similar plumage color had higher associations than pairs that were different in color, for those datasets in which color was a significant factor, we conducted a generalized linear model with a quasi-binomial distribution, in which SRI was compared between species pairs of the same color versus pairs of different colors. A similar follow-up analysis focused only on those pairs of the same color, and compared their SRIs among the different color categories found in the dataset that had at least 3 samples, followed by multiple comparisons, using Tukey’s honest significant differences method.
Results
We recorded 578 whole-MSF observations and 11,016 individual detections during fieldwork; we detected 95 species in MSFs at least 3 times, and MSFs averaged 4.89 ± 3.52 species (mean ± SD) and 18.79 ± 16.05 individuals. There was no significant difference in the species richness of MSFs between habitats (P > 0.05), but the number of individuals per MSF was slightly higher in secondary forest than in primary forest (W = 36655, P = 0.01; primary forest: 17.10 ± 15.06; secondary forest: 20.65 ± 16.89). We also collected 583 within-MSF observations (ensuring they included more than one species); we recorded 59 species at least 3 times in these observations. The species richness of the within-MSF observations was overall very low (2.2 ± 0.58), and there was no difference between the 2 habitats (primary vs. secondary forest) in species richness or numbers of individuals.
Mean values for species traits, at both the whole-MSF and the within-MSF levels, are shown in Table 1. The species that were represented have generally small body size and are mostly insectivorous or omnivorous. The color categories of yellow-belly, greenish, and rufous-white species were most common, and the most common of the plumage patterns were eye rings, wing bars, and eyebrow stripes.
Table 1.
The distribution of traits among the 95 species observed at least 3 times in whole-MSF level data and the 59 species observed in the within-MSF level data
| Traits | Number of species | ||
|---|---|---|---|
| Whole-MSF level (n = 95) | Within-MSF level (n = 59) | ||
| Body length (cm) (mean ± SD) | 18.38 ± 7.9 | 15.24 ± 5.43 | |
| Diet | Omnivorous | 47 | 30 |
| Insectivorous | 41 | 27 | |
| Frugivorous | 4 | 1 | |
| Others | 3 | 1 | |
| Plumage color | Yellow-belly | 27 | 21 |
| Greenish | 23 | 12 | |
| Rufous-white | 14 | 10 | |
| Gray-brown | 9 | 6 | |
| Black | 7 | 0 | |
| White-black | 3 | 3 | |
| Others | 12 | 7 | |
| Plumage pattern | Crest | 24 | 14 |
| Eye stripe | 28 | 22 | |
| Eyebrow stripe | 36 | 30 | |
| Coronal stripe | 11 | 9 | |
| Eye ring | 56 | 38 | |
| Wing bars | 39 | 26 | |
| Contrasting rump | 9 | 8 | |
| Contrasting tail | 9 | 7 | |
| Contrasting tail edge | 27 | 22 | |
The number shown in the plumage patterns category represents the number of species that had the trait.
As for the MRQAP results, transects varied in the number of relationships that were significant between traits and associations, but the direction of the effects was strikingly consistent: In all 41 significant results, association strength increased with similarity (Table 2). Body length dissimilarity had the most consistent effect: 4 transects (3 in primary forest and 1 in secondary forest) at the within-MSF level and 12 transects (all 8 transects in primary forest and 4 in secondary forest) at the whole-MSF level showed negative effects on association strength (smaller differences showed stronger associations). Significant negative relationships between association strength and phylogenetic distance were also found on many transects: 8 transects at the within-MSF level (4 in primary forest and 4 in secondary forest) and 3 transects at the whole-MSF level (1 in primary and 2 in secondary) showed negative effects on association strength. Plumage pattern similarity was the next most consistent effect: 4 transects at the within-MSF level (1 in primary forest and 3 in secondary forest) and 4 transects (1 in primary forest and 3 in secondary forest) at the whole-MSF level showed association strength increasing with similarities. In contrast, plumage color similarity was not as strong: 2 transects in the secondary forest at the within-MSF level, and 2 transects at the whole-MSF level (1 in primary forest and 1 in secondary forest) showed significant increases in association strength (Figure 1). Finally, diet had the weakest influence: Only 2 transects in the primary forest at the whole-MSF level showed that association strength increased with diet similarity.
Table 2.
Results of the influence of traits on species association strength calculated by the social network analysis
| Habitats | Variables | Within-MSF level | Whole-MSF level | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of significant transects | Number of Bonferroni-corrected significant transects | Mean Estimate | P(β ≥ r) | P(β ≤ r) | P(|β| ≤ |r|) | Number of significant transects | Number of Bonferroni-corrected significant transects | Mean estimate | P(β ≥ r) | P(β ≤ r) | P(|β| ≤ |r|) | ||
| Primary forest (n = 8) | Body length dissimilarity | 3 | 1 | −0.75 | ≥0.003 | ≥0.98 | ≤0.03 | 8 | 5 | −0.29 | ≥ 0 | ≥ 0.99 | ≤ 0.03 |
| Diet similarity | 0 | 0 | / | / | / | / | 2 | 0 | 0.02 | ≥ 0.98 | ≥ 0.02 | ≤ 0.02 | |
| Plumage color similarity | 0 | 0 | / | / | / | / | 1 | 1 | 0.02 | 0.99 | 0.001 | 0.001 | |
| Plumage pattern similarity | 1 | 0 | 0.04 | 0.97 | 0.04 | 0.05 | 1 | 0 | 0.02 | 0.98 | 0.02 | 0.02 | |
| Phylogenetic distance | 4 | 3 | −1.78 | ≥0 | ≥0.99 | ≤0.03 | 1 | 0 | −0.43 | 0.005 | 1 | 0.005 | |
| Secondary forest (n = 6) | Body length dissimilarity | 1 | 0 | −0.14 | 0.02 | 0.99 | 0.04 | 4 | 4 | −0.28 | ≥ 0 | ≥ 0.99 | ≤ 0.002 |
| Diet similarity | 0 | 0 | / | / | / | / | 0 | 0 | / | / | / | / | |
| Plumage color similarity | 2 | 0 | 0.03 | ≥0.98 | ≥0.005 | ≤0.02 | 1 | 0 | 0.02 | 0.97 | 0.03 | 0.04 | |
| Plumage pattern similarity | 3 | 2 | 0.01 | ≥0.98 | ≥0 | ≤0.03 | 3 | 1 | 0.01 | ≥ 0.99 | ≥ 0 | ≤ 0.02 | |
| Phylogenetic distance | 4 | 3 | −0.47 | ≥0.001 | ≥0.99 | ≤0.02 | 2 | 0 | −0.53 | 0.004 | 1 | 0.007 | |
Each transect was analyzed at the whole-MSF and the within-MSF levels. “P(β ≥ r)” represents the probability of the regression coefficient in the model being equal or larger than the permuted value; “P(β ≤ r)” represents the probability of the regression coefficient being equal or less than the permuted value; “P(|β| ≤ |r|)” represents the probability that the absolute value of the regression coefficient (β) being equal or less than the absolute permuted value. P(|β| ≤ |r|) was considered significant if it was ≤0.05. We also show how many results would be considered significant if the dataset was Bonferroni-corrected, given that we made 14 tests, one for each transect.
Figure 1.
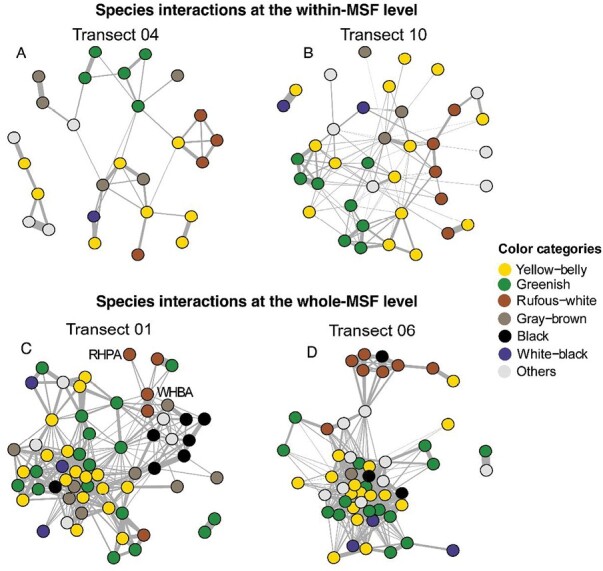
Species interactions in the separate network analysis for the within-MSF (A, B) and whole-MSF (C, D) data, showing the transects in which color had significant effects illustrating the general plumage of the nodes. Each node is a species and was assigned 1 of 7 general color patterns (at the within-MSF level, only 6 colors were recorded). Edges represent the association strength between 2 species, with thicker edges having higher association strength. On transect 01, RHPA represents rufous-headed parrotbill Psittiparus bakeri and WHBA represents white-hooded babbler Gampsorhynchus rufulus; these 2 species could be a case of mimicry.
For the 4 transects in which color was a significant factor, we found that the association strengths within species pairs of the same color species were significantly higher than within different colors (all t ≥ 2.68, all P ≤ 0.008, Figure 2). As to which colors had the highest associations, a significant difference between colors was found on transect 10 at the within-MSF level, where greenish species pairs had significantly higher associations than yellow-bellied species pairs (Z = 2.14, P < 0.001, Figures 1B and 2B), and also found on transect 06 at the whole-MSF level, where rufous-white species pairs had significantly higher associations than yellow-belly, greenish, and “others” species pairs (all Z ≥ 2.95, all P ≤ 0.03, Figures 1D and 2D).
Figure 2.
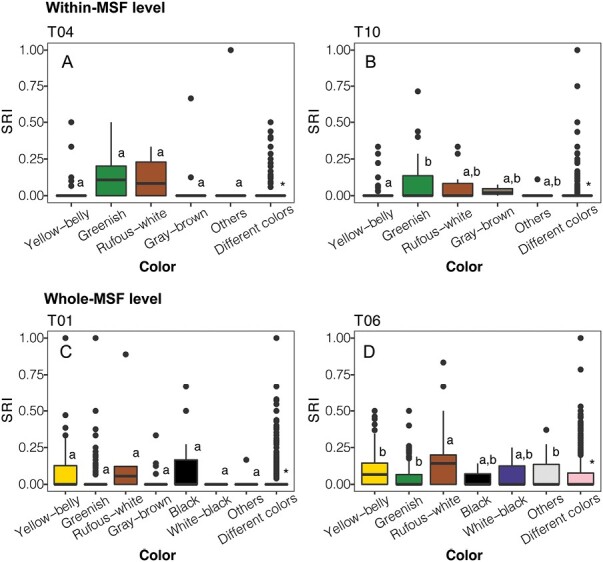
The association strength of species pairs by color, at the within-MSF level (A, B) and whole-MSF level (C, D), for the transects that were significant for color in the MRQAP analyses. In all 3 cases, associations between species pairs of the same color were stronger than associations between pairs of different colors, as represented by the asterisk. Differences between different color categories are shown by letters, with categories that share letters not being significantly different from each other. The y axis includes SRIs that were equal to 0.
Discussion
In this study, we aimed to determine which is the more important force organizing MSFs: competition, in which we would predict associations between species with divergent evolutionary history and traits, other than plumage characteristics, or facilitation, which would predict similarities across traits, including in plumage. We specifically hypothesized that species would share plumage similarities at the within-MSF level due to a predator’s preference for odd phenotypes when attacking many individuals within its visual field. Our results provided strong support for the hypothesis that facilitative interactions play an important role in structuring MSFs (Sridhar et al. 2012). The results were remarkably consistent: Every single relationship that was found to be significant in the analysis—18 at the within-MSF level and 23 at the whole-MSF level; 21 in primary forests and 20 in secondary forests—was in the direction of greater similarity increasing association strength. The strength of the relationships with association strength was intermediate for plumage similarity among the other traits in their influence: not as strong as body size and phylogeny, but not as weak as diet. However, in contrast to what was originally hypothesized, we did not find plumage similarity at the within-MSF level to be stronger than at the whole-MSF level. Therefore, while we gathered the first systematic evidence for plumage resemblances in an MSF system, we have limited ability to determine the mechanism behind the result.
We acknowledge some limitations to our project and conclusions. First, we assumed independence among MSFs seen on the same transect, and among within-MSF observations made on the same MSF; MRQAP is not a mixed model, and so aspects of the sampling design could not be incorporated into the modeling. As we minimized the repeated observations of the same individuals in the within-MSF data, we do not think these data violated the assumption of independence.
Second, we did not quantitatively assess plumage coloration in our plumage analyses. As we were testing a color categorization scheme based on pre-existing observations by human observers, we believe our methods were appropriate for our purposes. Several studies have shown that conclusions reached when scoring using human eyes are similar to those from spectrophotometry (Armenta et al. 2008; Seddon et al. 2010; Bergeron and Fuller 2017). To validate and extend our findings, future studies could incorporate quantitative assessments of the colors of bird plumage from museum specimens (Delhey 2015), photographs, or illustrations (Zhi and Guo 2018), and integrate birds’ color sensitivity, including UV colors (Cuthill et al. 2000).
Third, also regarding our plumage analyses, we only used female plumage because these MSFs mostly occur in the nonbreeding season when males resemble females more than they do in the breeding season. Only 8.4% of the species that were recorded in the MSFs in this study were clearly sexually dimorphic in plumage in the nonbreeding season, so this aspect of the methods should not have affected the result strongly.
Overall, we expected the within-MSF data to show more matching than the whole-MSF analysis in plumage as well as other traits, because the MSFs are large and some members on the periphery of the MSFs may be moving without a lot of coordination with other participants. Indeed, previous studies have suggested that particular associations between species in MSFs (i.e., that could be detected by modularity analyses) may be difficult to see when analyzing large species-rich MSFs as opposed to smaller ones (Bangal et al. 2021, 2022). Yet contrary to these expectations, the 2 kinds of analyses had roughly the same number of significant results, although they had somewhat of a different mixture of traits that were represented (body size was dominant for the whole-MSF analyses, phylogeny more prominent in the within-MSF analyses, see discussion below). The failure of the within-MSF data to show stronger results might be because of sample size issues: On average, there were only 2 species per 64 m3 cube, and thus there were many fewer species included in the within-MSF analysis (59 species compared to the 95 in the whole-MSF analysis).
There were also few differences between analyses of data collected in primary forest versus that collected in secondary forest. Secondary forests had almost as many significant results as did primary forests, despite having fewer transects (6 compared to 8 for primary forests). Hence, there was no evidence for human activity causing a decrease in matching among MSF participants, although it produces in general a decline in the size and encounter rate of MSFs (Zou et al. 2018).
Several mechanisms could explain plumage resemblances among MSF members. Predators may be confused by many individuals that look alike and have difficulty targeting 1 individual, as predicted by the oddity effect (Barnard 1979), and shown in a system of hawks attacking pigeons (Rutz 2012). This confusion might occur particularly when individuals move together, either in the same direction (Ioannou et al. 2012), or scattering independently (Ruxton et al. 2007), as often occurs in an MSF after an alarm call (Gaddis 1980; Goodale and Kotagama 2005). Predators could also be the target of plumage mimicry, if some of the species are poisonous or distasteful, as appears the case in a system of Papua New Guinea (Diamond 1992). Alternatively, similarities between birds could lessen aggressive attacks in groups (Stawarczyk 1984), especially on subordinate species, akin to social dominance mimicry outside of MSFs (Prum 2014). Or similarities could increase interspecific communication (Moynihan 1968), particularly in the case of plumage patterns on tails and wings that are used in visual communication (e.g., mobbing; Curio et al. 1978; Carlson et al. 2017).
Our data do not allow us to differentiate among these different mechanisms behind plumage similarity. A strong pattern for similarity at the within-MSF spatial scale might have been some evidence for oddity as a driving factor, because similar-looking prey within a predator’s field of vision might confuse it (Jeschke and Tollrian 2007), but this was not found. The similarities between plumage patterns observed in these MSFs support the idea of interspecific communication, because some of these traits can be used in dynamic visual signaling, such as wing or tail flicking. However, the way we scored these traits (looking for similarity between 2 species in multiple traits) does not allow us to distinguish whether the particular traits themselves were important (e.g., wing bars), or rather if it was the overall impression of similarity that was of essence. Some hypotheses such as distastefulness remain completely uninvestigated. Thus, research on the benefits of plumage similarities in MSFs should be extended in multiple directions in the future.
Some of the mechanisms for color resemblance might just require similar birds to choose to associate together, whereas others may be driven by co-evolution. Particularly, distastefulness (Dumbacher et al. 2008) and social dominance (Prum 2014) have been shown to lead to mimicry—that is, very close resemblances in plumage and pattern—in birds (reviewed by Hedley and Caro 2022). Some mimicry appears to be occurring in this system. In our rufous-white color system, for example, White-hooded Babbler (Gampsorhynchus rufulus), in which juveniles have rufous heads (Figure 3A) and were often seen in MSFs close to Rufous-headed Parrotbills (Psittiparus bakeri) (Figure 3B). The White-hooded Babbler is a leader of the rufous-white system, and occurs in monospecific groups within MSFs, which makes them a good potential model on which other species could converge. The species is also conspicuous in its active behavior and often initiated mobbing responses when we played the vocalizations of the Collared Owlet (Taenioptynx brodiei), a common predatory bird that preys on small- to medium-sized birds (Dutour et al. 2017). The reason that these Rufous-headed Parrotbills mimic this species when they are young could be either that they are social subordinates, or that the larger bird is a formidable adversary to predators, an idea recently suggested among the woodpeckers (Miller et al. 2019).
Figure 3.

An example of bird species that may be mimicking each other in the MSF systems described in this study, under the “rufous-white” color category. Juvenile white-hooded babbler (A, Gampsorhynchus rufulus) and Rufous-headed Parrotbill (B, Psittiparus bakeri). Photographs by Xiangle Zeng and used by permission.
In addition to plumage traits, we also found strong evidence that birds prefer to associate with species that are similar in size and evolutionary history, confirming earlier field studies (e.g., Mammides et al. 2015, 2018), and meta-analyses (Sridhar et al. 2012). Body size had the largest effect of any explanatory variable (16 significant relationships including both whole-MSF level and within-MSF analyses). There have been several field studies that have suggested that multiple MSF systems can exist in the same area, segregating by size, and 2 of these (King and Rappole [2001] in Myanmar; Nimnuan et al. [2004] in Thailand) are near our study region (see also Bell [1983] for a study in Papua New Guinea). The importance of body size in MSF structure is likely to be mainly driven by predation. Small birds are more vulnerable to predators when alone, so joining with other species or individuals could dilute their predation risk (Turner and Pitcher 1986; Zhou et al. 2021); indeed, small birds are over-represented in MSFs globally (Sainz-Borgo et al. 2018). Raptors, and specifically bird-eating Accipiter hawks, are also sized-structured communities (Reynolds and Meslow 1984; Rebollo et al. 2017), and hence associating with similar birds equates to greater time spent with other species vulnerable to the same predator. The fact that body size was particularly dominant at the whole-MSF level, means that the rules about body size are strong for all participants in MSFs, including those that seem peripherally attached to the center of the MSF.
Phylogeny was the second-most consistent factor on the transect scale (found in 11 transects, combining results whole-MSF and within-MSF analyses). Our results confirm the result of previous work that mixed-species MSFs tend to be phylogenetically clustered (Gómez et al. 2010; Sridhar et al. 2012; Péron 2017; Mammides et al. 2018). In contrast to studies that have looked for “checkerboard relationships” (Graves and Gotelli 1993; Colorado and Rodewald 2015), our analyses included all species and not just close congeners. Given that many traits are evolutionarily conserved (Losos 2008), the strength of phylogeny suggests that similar species tend to associate together in MSFs and that this similarity may extend to traits we did not measure that have a strong phylogenetic component.
Diet was the least influential trait, despite it having been found in past studies to be important to MSF community assembly (Sridhar et al. 2012). Perhaps in our study, this was due mostly to a lack of variation in this trait: The large majority of species were insectivores or omnivores, and these 2 groups might actually not differ strongly in the composition of their diets. In the future, it would be interesting to do more detailed observations of foraging techniques (e.g., Jones et al. 2020) to assess species matching or niche differentiation on this axis.
In conclusion, we found evidence that similarities between species increase associations among species in MSFs, providing evidence that these communities are structured by facilitation more than competition. All significant relationships were in the direction of similarities increasing association, and evidence for matching was found across a range of traits. Results were consistent at both whole- and within-MSF levels, and in both primary and secondary forests. Body size and phylogeny had the most consistent influence at the transect level, followed by plumage patterns and coloration, with diet having the weakest influence. We provided the first systematic, community-wide evidence that the plumage similarity of MSF participants is higher than would be randomly expected.
Acknowledgments
We are grateful for the permission to conduct this study in Tongbiguan Nature Reserve from the Forestry Bureaus of Dehong Prefecture and Yingjiang County. We thank Changsheng Zuo and Yunhui Zhang, who helped us with the permission to enter the villages in our study site. We also appreciate the help of Jiaxing Li with the fieldwork and Xiaolei Zeng with the scoring of the plumage traits. Finally, we thank Xiangle Zen for providing photographs, and 5 anonymous reviewers for their comments that helped improve the manuscript.
Contributor Information
Liping Zhou, Guangxi Key Laboratory of Forest Ecology and Conservation, College of Forestry, Guangxi University, Nanning 530004, China; Kunming Natural History Museum of Zoology, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China.
Christos Mammides, Nature Conservation Unit, Frederick University, 7, Yianni Frederickou Street, Pallouriotissa, Nicosia 1036, Cyprus.
Youfang Chen, State Key Laboratory of Vegetation and Environmental Change, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Wenyi Zhou, Florida Museum of Natural History, University of Florida, Gainesville, FL 34201, USA; Department of Biology, University of Florida, Gainesville, FL 34201, USA.
Wenzhang Dai, School of Life Science and Institute of Wetland Ecology, Nanjing University, Nanjing 210000, China.
Edward L Braun, Department of Biology, University of Florida, Gainesville, FL 34201, USA.
Rebecca T Kimball, Department of Biology, University of Florida, Gainesville, FL 34201, USA.
Yang Liu, State Key Laboratory of Biocontrol, School of Ecology, Sun-Yatsen University, Guangzhou 510275, China.
Scott K Robinson, Florida Museum of Natural History, University of Florida, Gainesville, FL 34201, USA.
Eben Goodale, Guangxi Key Laboratory of Forest Ecology and Conservation, College of Forestry, Guangxi University, Nanning 530004, China; Department of Health and Environmental Science, Xi’an Jiaotong Liverpool University, Suzhou 215123, China.
Funding
This study was funded by postdoctoral funding to LZ from Guangxi University (A3360051014, A3360051010) and from the Guangxi Key Laboratory of Forest Ecology and Conservation (AE33600102).
Author Contributions
The idea of this study was conceptualized by LP, EG, SR, YL, EB, and RK. Field data were gathered by LZ, YC, WZ, and WD. Data analysis was conducted by LZ, CM, and YC. The original draft of the manuscript was written by LP and EG, with all authors contributing to editing and all authors read and approved the draft.
Conflict of Interest Statement
The authors declare that there is no conflict of interest.
References
- Armenta JK, Dunn PO, Whittingham LA, 2008. Quantifying avian sexual dichromatism: a comparison of methods. J Exp Biol 211(15):2423–2430. [DOI] [PubMed] [Google Scholar]
- Bangal P, Sridhar H, Shanker K, 2021. Phenotypic clumping decreases with flock richness in mixed-species bird flocks. Front Ecol Evol 8:537816. [Google Scholar]
- Bangal P, Sridhar H, Shizuka D, vander Meiden LN, Shanker K, 2022. Flock-species richness influences node importance and modularity in mixed-species flock networks. Oecologia 198(2):431–440. [DOI] [PubMed] [Google Scholar]
- Barnard CJ, 1979. Predation and the evolution of social mimicry in birds. Am Nat 113:613–618. [Google Scholar]
- Beauchamp G, Goodale E, 2011. Plumage mimicry in avian mixed-species flocks: More or less than meets the eye? Auk 128:487–496. [Google Scholar]
- Bell HL, 1983. A bird community of lowland rainforest in New Guinea. 5. Mixed-species feeding flocks. Emu 82:256–275. [Google Scholar]
- Bergeron ZT, Fuller RC, 2017. Using human vision to detect variation in avian coloration: How bad is it? Am Nat 191(2):269–276. [DOI] [PubMed] [Google Scholar]
- Carlson NV, Pargeter HM, Templeton CN, 2017. Sparrowhawk movement, calling, and presence of dead conspecifics differentially impact blue tit Cyanistes caeruleus vocal and behavioral mobbing responses. Behav Ecol Sociobiol 71:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari C, da Silva Gonçalves C, de Melo C, 2020. Disentangling abiotic and biotic mechanisms behind the formation of heterospecific Nearctic-Neotropical shorebird flocks. Evol Ecol 34:1047–1061. [Google Scholar]
- Chen CC, Hsieh F, 2002. Composition and foraging behaviour of mixed-species flocks led by the Grey-cheeked Fulvetta in Fushan Experimental Forest, Taiwan. Ibis 144:317–330. [Google Scholar]
- Colorado GJ, Rodewald AD, 2015. Assembly patterns of mixed-species avian flocks in the Andes. J Anim Ecol 84(2):386–395. [DOI] [PubMed] [Google Scholar]
- Curio E, Ernst U, Vieth W, 1978. The adaptive significance of avian mobbing. Z Tierpsychol 48:184–202. [Google Scholar]
- Cuthill IC, Partridge JC, Bennett ATD, Church SC, Hart NSet al. , 2000. Ultraviolet vision in birds. Advan Study Behav 29(C):159–214. [Google Scholar]
- Darrah AJ, Smith KG, 2013. Comparison of foraging behaviors and movement patterns of the wedge-billed woodcreeper Glyphorynchus spirurus traveling alone and in mixed-species flocks in Amazonian Ecuador. Auk 130(4):629–636. [Google Scholar]
- Dekker D, Krackhardt D, Snijders TAB, 2007. Sensitivity of MRQAP tests to collinearity and autocorrelation conditions. Psychometrika 72:563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhey K, 2015. The colour of an avifauna: A quantitative analysis of the colour of Australian birds. Sci Rep 5:18514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JM, 1975. Assembly of species communities. In: Diamond JM, Cody ML, editors. Ecology and Evolution of Communities. Boston (MA): Harvard University Press. 42–344. [Google Scholar]
- Diamond JM, 1992. Rubbish birds are poisonous. Nature 360:19–20. [DOI] [PubMed] [Google Scholar]
- Dumbacher JP, Deiner K, Thompson L, Fleischer RC, 2008. Phylogeny of the avian genus Pitohui and the evolution of toxicity in birds. Mol Phylogenet Evol 49:774–781. [DOI] [PubMed] [Google Scholar]
- Dutour M, Lena JP, Lengagne T, 2017. Mobbing behaviour in a passerine community increases with prevalence in predator diet. Ibis 159(2):324–330. [Google Scholar]
- Farine DR, 2013. Animal social network inference and permutations for ecologists in R using asnipe. Methods Ecol Evol 4(12):1187–1194. [Google Scholar]
- Farine DR, 2017. asnipe: Animal social network inference and permutations for ecologists. Available from: https://rdrr.io/cran/asnipe/.
- Farine DR, Whitehead H, 2015. Constructing, conducting and interpreting animal social network analysis. J Anim Ecol 84(5):1144–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, 2004. Inferring Phylogenies. Sunderland: Sinauer Associates. [Google Scholar]
- Gaddis P, 1980. Mixed flocks, accipiters, and antipredator behavior. Condor 82(3):348–349. [Google Scholar]
- Gómez JP, Bravo GA, Brumfield RT, Tello JG, Cadena CD, 2010. A phylogenetic approach to disentangling the role of competition and habitat filtering in community assembly of Neotropical forest birds. J Anim Ecol 79:1181–1192. [DOI] [PubMed] [Google Scholar]
- Goodale E, Beauchamp G, Ruxton GD, 2017. Mixed-Species Groups of Animals: Behavior, Community Structure, and Conservation. London: Academic Press. [Google Scholar]
- Goodale E, Kotagama SW, 2005. Alarm calling in Sri Lankan mixed-species bird flocks. Auk 122(1):108–120. [Google Scholar]
- Goodale E, Kotagama SW, Raman TRS, Sidhu S, Goodale Uet al. , 2014. The response of birds and mixed-species bird flocks to human-modified landscapes in Sri Lanka and southern India. For Ecol Manag 329:384–392. [Google Scholar]
- Goodale E, Nizam BZ, Robin VV, Sridhar H, Trivedi Pet al. , 2009. Regional variation in the composition and structure of mixed-species bird flocks in the Western Ghats and Sri Lanka. Curr Sci 97:648–663. [Google Scholar]
- Goodale E, Sridhar H, Sieving KE, Bangal P, Colorado ZGJet al. , 2020. Mixed company: A framework for understanding the composition and organization of mixed-species animal groups. Biol Rev 95(4):889–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves GR, Gotelli NJ, 1993. Assembly of avian mixed-species flocks in Amazonia. Proc Natl Acad Sci USA 90(4):1388–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RS, 2000. Birds of many feathers: the formation and structure of mixed-species flocks of forest birds. In: Boinski S, Garber PA, editors. On the Move: How and Why Animals Travel in Groups. Chicago (IL), USA: University of Chicago Press. [Google Scholar]
- Hedley E, Caro T, 2022. Aposematism and mimicry in birds. Ibis 164:606–617. [Google Scholar]
- Hino T, 1998. Mutualistic and commensal organization of avian mixed-species foraging flocks in a forest of western Madagascar. J Avian Bio 29:17–24. [Google Scholar]
- Hutto RL, 1988. Foraging behavior patterns suggest a possible cost associated with participation in mixed-species bird flocks. Oikos 51(1):79–83. [Google Scholar]
- Hutto RL, 1994. The composition and social-organization of mixed-species flocks in a tropical deciduous forest in western Mexico. Condor 96:105–118. [Google Scholar]
- Ioannou CC, Guttal V, Couzin ID, 2012. Predatory fish select for coordinated collective motion in virtual prey. Science 337:1212–1215. [DOI] [PubMed] [Google Scholar]
- Jeschke JM, Tollrian R, 2007. Prey swarming: Which predators become confused and why? Anim Behav 74:387–393. [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO, 2012. The global diversity of birds in space and time. Nature 491(7424):444–448. [DOI] [PubMed] [Google Scholar]
- Jiang D, Sieving KE, Meaux E, Goodale E, 2020. Seasonal changes in mixed-species bird flocks and antipredator information. Ecol Evol 10:5368–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HH, Walters MJ, Robinson SK, 2020. Do similar foragers flock together? Nonbreeding foraging behavior and its impact on mixed-species flocking associations in a subtropical region. Auk 137:1–16. [Google Scholar]
- King DI, Rappole JH, 2001. Mixed specks bird flocks its dipterocarp forest of north central Burma (Myanmar). Ibis 143:380–390. [Google Scholar]
- Krams IA, Krama T, Freeberg TM, Krams R, Sieving KE, 2020. Attacks of songbirds in mixed-species flocks by Eurasian Sparrowhawks: Strategies of predators and potential prey. J Field Ornithol 91(4):367–374. [Google Scholar]
- Landeau L, Terborgh J, 1986. Oddity and the ‘confusion effect’ in predation. Anim Behav 34(5):1372–1380. [Google Scholar]
- Latta SC, Wunderle JM, 1996. The composition and foraging ecology of mixed-species flocks in pine forests of Hispaniola. Condor 98(3):595–607. [Google Scholar]
- Losos JB, 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett 11:995–1003. [DOI] [PubMed] [Google Scholar]
- Mammides C, Chen J, Goodale UM, Kotagama SW, Goodale E, 2018. Measurement of species associations in mixed-species bird flocks across environmental and human disturbance gradients. Ecosphere 9(7):e02324. [Google Scholar]
- Mammides C, Chen J, Goodale UM, Kotagama SW, Sidhu Set al. , 2015. Does mixed-species flocking influence how birds respond to a gradient of land-use intensity? Proc R Soc B 282:1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant R, 2002. Do rare species have any place in multivariate analysis for bioassessment? J N Am Benthol Soc 21(2):311313. [Google Scholar]
- Martínez AE, Gomez JP, 2013. Are mixed-species bird flocks stable through two decades? Am Nat 181(3):E5353–E5E59. [DOI] [PubMed] [Google Scholar]
- Miller ET, Leighton GM, Freeman BG, Lees AC, Ligon RA, 2019. Ecological and geographical overlap drive plumage evolution and mimicry in woodpeckers. Nat Commun 10:1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MD, 2003. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos 100(2):403–405. [Google Scholar]
- Morse DH, 1977. Feeding behavior and predator avoidance in heterospecific groups. BioScience 27:332–339. [Google Scholar]
- Moynihan M, 1962. The organization and probable evolution of some mixed species flocks of Neotropical birds. Smithson Misc Collect 143:1–140. [Google Scholar]
- Moynihan M, 1968. Social mimicry: Character convergence versus character displacement. Evolution 22:315–331. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, 2004. A farewell to Bonferroni: The problems of low statistical power and publication bias. Behav Ecol 15(6):1044–1045. [Google Scholar]
- Nimnuan S, Round PD, Gale GA, 2004. Structure and composition of mixed-species insectivorous bird flocks in Khao Yai National Park. Nat Hist Bull Siam Soc 52:71–79. [Google Scholar]
- Péron G, 2017. Multicontinental community phylogenetics of avian mixed-species flocks reveal the role of the stability of associations and of kleptoparasitism. Ecography 40:1267–1273. [Google Scholar]
- Prum RO, 2014. Interspecific social dominance mimicry in birds. Zool J Linn Soc 172(4):910–941. [Google Scholar]
- Rebollo S, Martínez-Hesterkamp S, García-Salgado G, Pérez-Camacho L, Fernández-Pereira JMet al. , 2017. Spatial relationships and mechanisms of coexistence between dominant and subordinate top predators. J Avian Biol 48(9):1226–1237. [Google Scholar]
- Reynolds RT, Meslow EC, 1984. Partitioning of food and niche characteristics of coexisting Accipiter during breeding. Auk 101:761–779. [Google Scholar]
- Rodgers GM, Kimbell H, Morrell LJ, 2013. Mixed-phenotype grouping: The interaction between oddity and crypsis. Oecologia 172(1):59–68. [DOI] [PubMed] [Google Scholar]
- Rutz C, 2012. Predator fitness increases with selectivity for odd prey. Curr Biol 22(9):820–824. [DOI] [PubMed] [Google Scholar]
- Ruxton GD, Jackson AL, Tosh CR, 2007. Confusion of predators does not rely on specialist coordinated behavior. Beha Ecol 18(3):590–596. [Google Scholar]
- Sainz-Borgo C, Koffler S, Jaffe K, 2018. On the adaptive characteristics of bird flocks: Small birds form mixed flocks. Ornital Neotrop 29:289–296. [Google Scholar]
- Sazima I, 2010. Five instances of bird mimicry suggested for Neotropical birds: A brief reappraisal. Rev Bras Ornitol 18(4):328–335. [Google Scholar]
- Seddon N, Tobias JA, Eaton M, Ödeen A, 2010. Human vision can provide a valid proxy for Avian perception of sexual dichromatism. Auk 127(2):283–292. [Google Scholar]
- Sridhar H, Beauchamp G, Shanker K, 2009. Why do birds participate in mixed-species foraging flocks? A large-scale synthesis. Anim Behav 78(2):337–347. [Google Scholar]
- Sridhar H, Guttal V, 2018. Friendship across species borders: Factors that facilitate and constrain heterospecific sociality. Philos Trans R Soc B Biol Sci 373(1746):20170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar H, Shanker K, 2014. Using intra-flock association patterns to understand why birds participate in mixed-species foraging flocks in terrestrial habitats. Behav Ecol Sociobiol 68:185–196. [Google Scholar]
- Sridhar H, Srinivasan U, Askins RA, Canales-Delgadillo JC, Chen CCet al. , 2012. Positive relationships between association strength and phenotypic similarity characterize the assembly of mixed-species bird flocks worldwide. Am Nat 180:777–790. [DOI] [PubMed] [Google Scholar]
- Stawarczyk T, 1984. Aggression and its suppression in mixed-species wader flocks. Ornis Scandinavica 15(1):23–37. [Google Scholar]
- Turner GF, Pitcher TJ, 1986. Attack abatement: A model for group protection by combined avoidance and dilution. Am Nat 128:228–240. [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ, 2002. Phylogenies and community ecology. Annu Rev Ecol Syst 33(1):475–505. [Google Scholar]
- Wheatcroft D, Price TD, 2013. Learning and signal copying facilitate communication among bird species. Proc R Soc B 280(1757):20123070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis EO, 1989. Mimicry in bird flocks of cloud forests in southeastern Brazil. Rev Bras Biol 49:615–619. [Google Scholar]
- Zhang Q, Han R, Huang Z, Zou F, 2013. Linking vegetation structure and bird organization: Response of mixed-species bird flocks to forest succession in subtropical China. Biodivers Conserv 22:1965–1989. [Google Scholar]
- Zhang Q, Holyoak M, Goodale E, Liu Z, Shen Yet al. , 2020. Trait-environment relationships differ between mixed-species flocking and nonflocking bird assemblages. Ecology 101(1):e03124. [DOI] [PubMed] [Google Scholar]
- Zhi X, Guo C, 2018. Bird species recognition based on deep learning and decision fusion. In: Huang T, Lv J, Sun C, Tuzikov AV, editors. Advances in Neural Networks. Cham: Springer International Publishing, 568–577. [Google Scholar]
- Zhou L, Peabotuwage I, Gu H, Jiang D, Hu Get al. , 2019. The response of mixed-species bird flocks to anthropogenic disturbance and elevational variation in southwest China. Condor 121(3):1–13. [Google Scholar]
- Zhou L, Peabotuwage I, Luo K, Quan R-C, Goodale E, 2021. Using playback to test leadership in mixed-species flocks and compare flocking with mobbing. Anim Behav 180:151–166. [Google Scholar]
- Zou F, Chuan Lim H, Marks BD, Moyle RG, Sheldon FH, 2007. Molecular phylogenetic analysis of the grey-cheeked fulvetta Alcippe morrisonia of China and Indochina: A case of remarkable genetic divergence in a “species”. Mol Phylogenet Evol 44(1):165–174. [DOI] [PubMed] [Google Scholar]
- Zou F, Jones H, Colorado ZGJ, Jiang D, Lee T-Met al. , 2018. The conservation implications of mixed-species flocking in terrestrial birds, a globally-distributed species interaction network. Biol Conserv 224:267–276. [Google Scholar]


