Abstract
Troponin-based Ca2+ regulation of striated muscle contraction emerged approximately 700 million years ago with largely conserved functions during evolution. Troponin I (TnI) is the inhibitory subunit of troponin and has evolved into three muscle type-specific isoforms in vertebrates. Cardiac TnI is specifically expressed in the adult heart and has a unique N-terminal extension implicating a specific value during natural selection. The N-terminal extension of cardiac TnI in higher vertebrates contains β-adrenergic regulated protein kinase A (PKA) phosphorylation sites as a mechanism to enhance cardiac muscle relaxation and facilitate ventricular filling. Phylogenic studies showed that the N-terminal extension of cardiac TnI first emerged in the genomes of early tetrapods as well as primordial lobe-finned fishes such as the coelacanth whereas it is absent in ray-finned fish. This apparently rapid evolution of β-adrenergic regulation of cardiac function suggests a high selection value for the heart of vertebrate animals on land to work under higher metabolic demands. Sequencing and PKA phosphorylation data showed that lungfish cardiac TnI has evolved with an amphibian like N-terminal extension with prototype PKA phosphorylation sites while its overall structure remained fish-like. The data demonstrate that the submolecular structure of TnI may evolve ahead of the whole protein for cardiac muscle contractility to adapt to new environmental conditions. To understand the evolution of the β-adrenergic regulation of TnI and cardiac adaptation to the increased energetic demands of life on land adds knowledge for the treatment of human heart diseases and failure.
Keywords: Troponin, heart, proteins, selection value
Introduction
The contraction of vertebrate skeletal and cardiac muscles is powered by actin-activated myosin ATPase motors under Ca2+ regulation via troponin associated with sarcomeric actin thin filaments. The troponin complex is composed of three protein subunits: The Ca2+-binding subunit troponin C (TnC), the tropomyosin-binding subunit troponin T (TnT), and the actomyosin ATPase inhibitory subunit troponin I (TnI). Muscle contraction is activated by an increase of cytosolic Ca2+ concentration resulting in binding to TnC and inducing conformational changes in troponin and the thin filament to release the inhibition of TnI and allow for strong myosin crossbridge formation, which activates myosin ATPase and generate power strokes to contract the sarcomere (Gordon et al. 2000; Katrukha 2013).
Each troponin subunit is encoded by a separate gene. TnC shares structural and functional similarities with the calmodulin-family of proteins (Baba et al. 1984), while TnI and TnT genes have co-evolved as striated muscle-specific genes. Genes encoding TnI and TnT can be traced at least 700 million years ago, coinciding with the emergence of striated muscles in a pre-Bilateria ancestor (Brunet et al. 2016; Cao et al. 2019). As striated muscle diverged into skeletal and cardiac types in vertebrates, troponin also diverged into homologous fiber type-specific isoforms. While TnC has two isoforms, a fast skeletal muscle form and a cardiac/slow skeletal muscle form, TnI and TnT each has evolved into three fiber-type specific isoforms in fast skeletal, slow skeletal, and cardiac muscles (Jin et al. 2008), encoded by closely linked pairs of isoform genes indicating their origin from gene duplications (Chong & Jin 2009).
Phylogenetic and protein conformational analysis indicated that fast skeletal muscle TnI and TnT were the ancestral types from which cardiac and slow skeletal muscle isoforms emerged (Chong and Jin 2009), consistent with data supporting fast skeletal muscle evolving prior to slow and cardiac muscles (OOta and Saitou 1999). While the three isoforms of vertebrate TnI have highly conserved homologous core structures, cardiac TnI of higher vertebrate species has a unique N-terminal extension (Wilkinson and Grand 1978).
The N-terminal extension of cardiac TnI is a conserved trait of tetrapods, i.e., amphibians, reptiles, birds, marsupials and eutherian mammals (Hastings et al. 1991; Ausoni et al. 1994; Warkman and Atkinson 2004; Feng et al. 2012). In sharp contrast, it is completely absent in cardiac TnI of lower classes of Chordates, including Hyperotreti (hagfishes), Hyperoartia (lampreys), Chondrichthyes (sharks and rays), and Actinopterygii (ray finned fishes) (Shaffer and Gillis 2010; Gross and Lehman 2016). Sequence analysis noted that cardiac TnI of ray-finned fishes groups with slow skeletal muscle TnI of other bilaterians (Shaffer and Gillis 2010; Gross and Lehman 2016), indicating that fish do not possess as clearly diverged cardiac TnI isoform as seen in tetrapods. Thus, emergence of the N-terminal extension of cardiac TnI likely occurred during the evolution of tetrapods with a novel function and selection value. Studies of mammalian and avian species have shown that embryonic hearts express exclusively slow skeletal muscle TnI that is replaced by cardiac TnI during perinatal development (Jin 1996). Adult hearts of higher vertebrates exclusively express cardiac TnI, supporting a hypothesis that the N-terminal extension of cardiac TnI is selected for adult heart functions of air-breathing tetrapods.
Extensively documented from mammalian heart studies, the N-terminal extension of tetrapod cardiac TnI is a substrate of protein kinase A (PKA). It contains PKA phosphorylation sites (Ser23 and Ser24 in human cardiac TnI) which are phosphorylated in response to β-adrenergic stimulation (Solaro et al. 1976; Kranias & Solaro 1982). PKA phosphorylation of cardiac TnI results in a reduction in Ca2+ affinity of TnC to increase cardiac muscle relaxation and enhance diastolic function of the heart (Zhang et al. 1995; Kentish et al. 2001; Rao et al. 2013).
β-adrenergic regulation emerged in fish. Teleost hearts express β-adrenergic receptors and are innervated by adrenergic neurons (Fabbri et al. 1998; Reid et al. 1998; Wang et al. 2009) although some fish species may be more reliant on circulating catecholamines than direct neuronal innervation (Mendonça and Gamperl 2009). The heart of ray-finned fish had evolved with responsiveness to β-adrenergic stimulation. Enhanced β-adrenergic response was seen in fish hearts in adaptation to cold acclimation (Aho and Vornanen 2001), acidosis (Farrell 1985), or hypoxia (Hanson et al. 2006).
Zebrafish β-adrenergic receptor transfectively expressed in HEK293 cells was capable of associating with Gs and initiating cAMP production (Steele et al. 2011), indicating an ability to stimulate PKA and downstream signaling. However, circulating catecholamine levels vary in fish and may be insufficient in some species to adequately stimulate cardiac β-adrenergic receptors (Kawasaki et al. 2008; Mendonça and Gamperl 2009). Knocking out the β1-adrenergic receptor in zebrafish did not significantly change heart rate, distinct from that in mice (Ecker et al. 2006; Steele et al. 2011), indicating that β-adrenergic signaling in fish is preliminary in comparison with that of tetrapods. In skinned trout cardiac muscle, PKA treatment resulted in a decrease in force generation at maximal activation but did not alter Ca2+-sensitivity, passive tension, or cooperativity of activation (Gillis and Klaiman 2011; Kirkpatrick et al. 2011), implicating a likely correlation with the absence of the N-terminal extension in cardiac TnI.
To understand the physiological function and adaptive value of the N-terminal extension in cardiac TnI, here we studied its evolutionary emergence in primordial lobe-finned fish. Phylogenetic analyses, sequencing and PKA phosphorylation studies showed that lungfish cardiac TnI has evolved with an amphibian like N-terminal extension with prototype PKA phosphorylation sites while its overall structure remained fish-like, demonstrating that this submolecular structure of TnI had evolved ahead of the whole protein for cardiac muscle contractility to adapt to new environmental conditions. The data of molecular evolution provide novel evidence for understanding the β-adrenergic regulation of TnI and cardiac adaptation to energetic demands and adds knowledge for the treatment of human heart diseases and failure.
Materials and Methods
Phylogenetic Analysis of TnI Proteins
Amino acid sequences of TnI from representative species were retrieved from the GenBank database. NIH/NCBI Basic Local Alignment Search Tool (BLAST) was used to search troponin I sequences previously unidentified in GenBank. The standard protein BLAST program was employed using clownfish (Amphiprion ocellaris: XM_023267923.1), zebrafish (Danio rerio: XP_691548.2), and clawed frog (Xenopus laevis: L25721.1) cTnI as query sequences, each selected as representative species from bony fishes and amphibians to probe for species closely related to lungfishes. For each search, algorithm parameters were modified to include the top 200 results.
Amino acid sequences were aligned in MegAlign Pro software (DNAStar) employing the MUSCLE multiple sequence alignment algorithm using the default UPGMB cluster method (Edgar 2004). Alignment settings were adjusted to align the N-terminal extension of cardiac TnI. As mammalian cardiac TnI’s are known to have a N-terminal extension, their sequences were first aligned under a gap penalty of magnitude 10, which allows this segment group. This setting was then used in subsequent alignments. The alignments were also assessed to ensure the conserved regions were properly aligned. Table 1 in the Supplement lists the sequences studied and their GenBank accession numbers.
SDS-PAGE and Western Blotting
SDS-polyacrylamide gel electrophoresis (PAGE) and Western immunoblotting were carried out as previously described (Feng et al. 2008) to analyze protein samples from lungfish and other representative species. Briefly, total protein of cardiac and skeletal muscle tissues was extracted by high-speed homogenization in SDS-PAGE sample buffer containing 50 mM Tris-HCl, 2% SDS, 1% 2-mercaptoethanol, 10% glycerol and 0.1% bromophenol blue. The tissue homogenates were heated at 80°C for 5 min and centrifuged in a microcentrifuge at 20,000 x g for 5 min to remove insoluble materials. The SDS-gel samples were resolved on 14% Laemmli gels with an acrylamide:bisacrylamide ratio of 180:1 and visualized by staining with Coomassie blue R250.
Duplicate gels were electrically transferred to nitrocellulose membrane using a semi-dry apparatus (Bio-Rad). The membranes were first stained with Amido Black to visualize all protein bands. The positions of myosin heavy chain (MHC) and actin were used to align against that in Coomassie blue-stained SDS-gel copy to add in the identification of proteins detected in the Western blots. The nitrocellulose membranes were blocked with Tris-buffered saline (TBS) containing 1% bovine serum albumin (BSA) for 30 min, followed by incubation with monoclonal antibody (mAb) TnI-1 against all three TnI isoforms in all vertebrate species (Jin et al., 2001), mAb CT3 against cardiac and slow TnT in all vertebrate species (Chong & Jin, 2009), or mAb 2C8 against all three TnT isoforms (Chong & Jin, 2009) in TBS containing 0.05% Tween-20 (TBS-T) and 0.1% BSA at 4°C overnight. The membranes were washed 3 times 7 min each in TBS containing 0.5% Triton X-100 and 0.1% SDS, followed by 3 TBS washes 3 min each, before incubation with an anti-mouse IgG secondary antibody (Santa Cruz) in TBS-T containing 0.1% BSA at room temperature for 1 hour. Washed again as above, the membranes were developed in 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (BCIP/NBT) to visualize bands recognized by the mAbs.
RT-PCR Amplification and Sequencing of Lungfish Cardiac TnI cDNA
Frozen hearts of spotted African lungfish (Protopterus dolloi) were provided by Dr. Irene Salinas, University of New Mexico. Total RNA was isolated from the frozen cardiac muscle using the Trizol reagent as described in the manufacturer’s instruction. 2 μg of lungfish total cardiac RNA were incubated with 30 pmole of an anchored oligo(dT) primer (5’-TTTTTTTTTTTTTTTTTTTTTV-3’, TV20) in a 30 μL reaction at 70°C for 5 min before chilling on ice for 5 min to allow primer annealing to the beginning of the poly-A tail of mRNAs. Reverse transcription (RT) was performed using AMV reverse transcriptase (Promega) at 42°C for 1 hour.
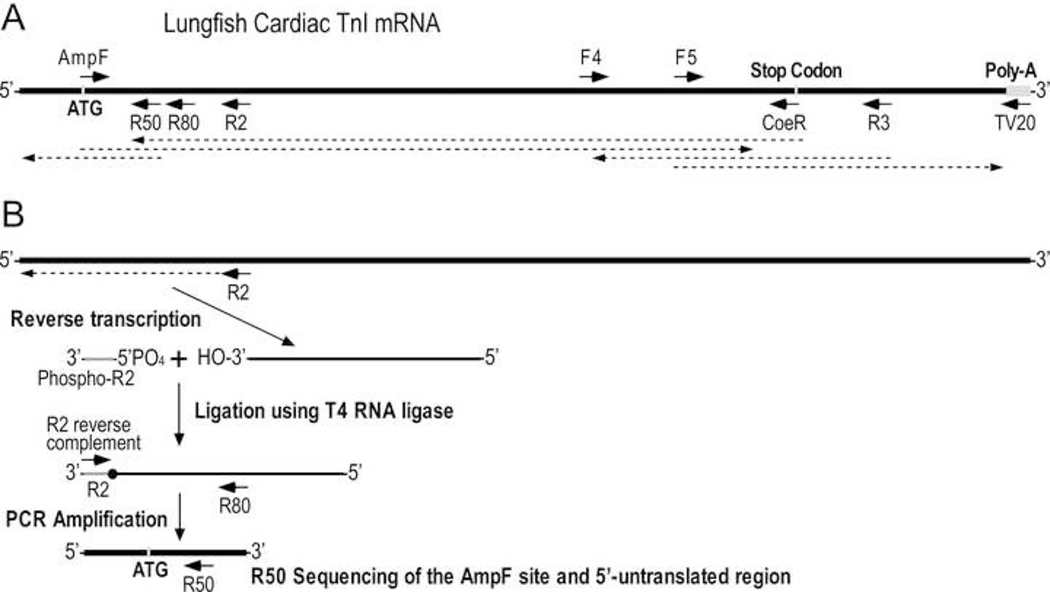
Outlined in Figure 1A, degenerate forward (AmpF) and reverse (CoeR) primers were designed based on the sequences of the coding regions of the translational initiation site of tropical frog (Xenopus tropicalis) cardiac TnI (BC088784.1) and the translational stop codon of coelacanth (Latimeria chalumnae) cardiac TnI (XP_014353977.1) for PCR amplification of lungfish cardiac TnI (94°C for 5 min, 35 cycles of [94°C for 30 seconds, 56°C for 30 seconds, 72°C for 30 seconds], 72°C for 7 min). The PCR product was directly sequenced using the forward and reverse primers using a commercial service (Genewiz).
Fig. 1. Strategy to sequence the entire coding region of lungfish cardiac TnI mRNA.
A, To obtain the sequence of lungfish cardiac TnI, cDNA was made from heart mRNA using an anchored oligo dT primer TV20 for reverse transcription from the beginning of poly-A tail to synthesize total cardiac cDNA. Two degenerate primers AmpF and CoeR were designed based on known sequences at the translational initiation site of frog cardiac TnI and stop codon of coelacanth cardiac TnI for PCR amplify cDNA encoding lungfish cardiac TnI. Internal primers F4, F5, and R3 were designed using the initial sequence data for sequencing reactions and PCR amplification and sequencing of the 3’-untranslated region as well as verify the CoeR segment (see Material and Methods for details). The arrowed dash lines indicate segments determined by the sequencing reactions. B, To sequence the 5’-untranslated region and verify the AmpF segment, an R2 internal reverse primer was used for reverse transcription of lungfish cardiac TnI mRNA. The single stranded cDNA was then ligated to 5’-phosphorylated R2 primer using T4 RNA ligase. PCR amplification of double stranded cDNA was carried out using a reverse complement primer of R2 sequence and an internal reverse primer R80. The PCR product was sequenced using another internal primer R50.
PCR amplification of the authentic sequence of the CoeR primer site and 3’-untranslated region was carried out using a forward primer (F6) matching approximately 250 bases upstream of the translation stop codon paired with the TV20 reverse primer (Figure 1A). The PCR product was directly sequenced using naïve internal forward primer F5 and reverse primer R3.
To obtain the 5’-untranslated region and the authentic sequence of the AmpF primer site using a strategy outlined in Figure 1B (modified from Zhang & Chiang, 1996), a reverse primer approximately 200 bases downstream of the translational initiation codon (R2) was used for reverse transcription of lungfish cardiac TnI mRNA as described above. The single stranded cDNAs synthesized were isolated by precipitating in 0.3M NaCl and 75% ethanol on dry ice for 10 min, followed by centrifugation in a microcentrifuge at 20,000 x g at 4°C for 10 min. The pellet was air dried before resuspension for ligation to a 5’-phosphorylated R2 primer in 20 μL of 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM adenosine triphosphate, 15% (w/v) PEG 8000 and containing 20 units of Murine RNase inhibitor and 10 units of T4 RNA ligase (New England Biolabs). PCR amplification was performed using an R2-reverse complementary primer (R2-RC) and a naïve internal reverse primer (R80). The resulting PCR product was sent for direct sequencing by Genewiz using another naïve reverse primer (R50).
E. coli Expression and Immunological Confirmation of Cloned Lungfish Cardiac TnI cDNA
The amino acid sequence deduced from the cloned full length cDNA encoding lungfish cardiac TnI was reverse-translated into E. coli preferred codons. A cDNA was synthesized and constructed into pET17b expression plasmid at a commercial service (GeneUniversal) for protein expression in transformed BL21(DE3)pLysS E. coli culture as described previously (Ogut and Jin, 1998). Total protein extracts from induced and uninduced E. coli cultures were analyzed using SDS-PAGE and Western blotting as described above.
Skinned Preparations of Lungfish Cardiac Muscle
Adapted from a previously described method (Feng & Jin 2020), skinned muscle fibers were prepared from cryosections of South American lungfish (Lepidosiren paradoxa) heart. 40 μm thick and 150–200 μm wide lungfish cardiac muscle strips were cut using a cryostat at −20°C and stored in a relaxation solution containing 50% glycerol (v/v), 40 mM BES, 10 mM EGTA, 6.86 mM MgCl2, 5.96 mM ATP, 1 mM dithiothreitol (DTT), 3.28 mM potassium propionate, pH 7.0, 33 mM creatine phosphate, 200 U/mL creatine kinase and protease inhibitor cocktail at −20°C. The muscle strips were permeabilized at 4 – 6°C in the relaxation solution plus 1% Triton X-100 for 30 min to remove membrane structures to obtain skinned preparations containing preserved myofibrils suitable for the study of myofilament functions.
The skinned preparations were transferred to 200 μL of relaxing solution containing 1 mM DTT and 100U/mL of the catalytic subunit of PKA from bovine heart (Sigma-Aldrich, St Louis, MO, USA). This concentration of PKA was selected to mimic conditions used by Gillis et al. (Gillis and Klaiman 2011). The PKA treatment was carried out at 15°C for 1 hour. Control muscle strips were incubated in the same solution under the same conditions without PKA. After the incubation, the preparations were immediately frozen in liquid nitrogen for SDS-PAGE and phosphoprotein analysis.
Pro-Q Diamond Phosphoprotein Staining
Phosphorylated proteins resolved in SDS-gel were detected via Pro-Q Diamond stain (Invitrogen). The SDS-gels were pre-fixed in 50% methanol, 10% acetic acid with two changes 30 min each, followed by three washes 10 min each in deionized water. The gels were stained with Pro-Q Diamond stain in a dark container with vigorous shaking for 90 min, and then de-stained in 20% acetonitrile, 50 mM sodium acetate, pH 5.0 with 5 changes 30 min each. After final washes in deionized water for two changes 5 min each, the gels were scanned on a Typhoon 9410 fluorescence imager (GE Healthcare) using the fluorescence mode (550V, high sensitivity, green laser 532 nm for excitation and 560 nm long pass for emission).
Results
Early Emergence of the Three Muscle Type TnI Isoform Genes in All Vertebrates
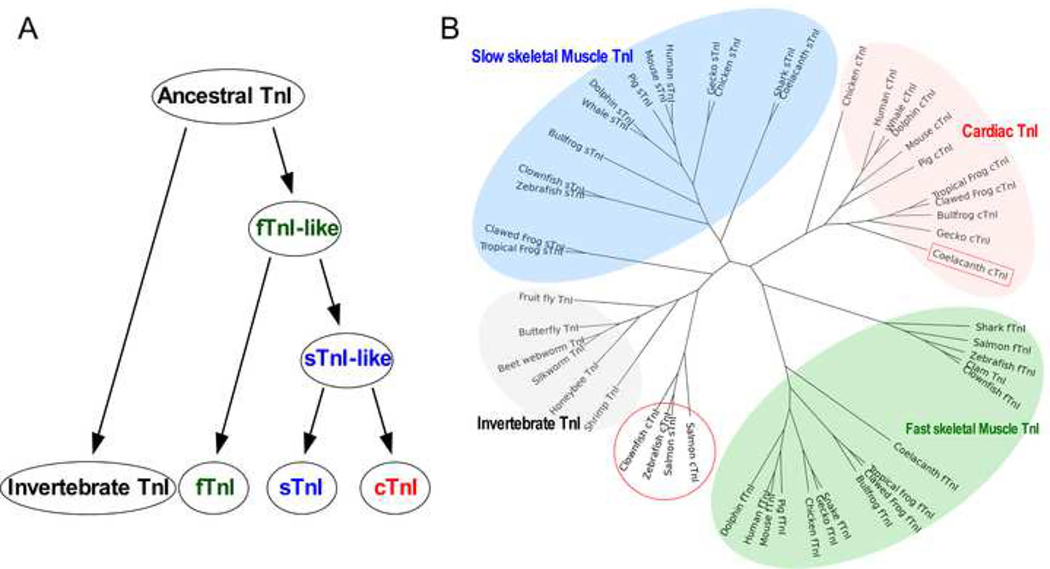
Our protein level phylogenetic analysis confirms a previous notion that fast TnI likely emerged first early in vertebrate muscle from the single TnI gene in the common ancestor of vertebrates and invertebrates. From the fast TnI gene, homologous genes encoding slow skeletal muscle and cardiac TnI isoforms emerged by gene duplications. The three TnI isoform genes are present in all lower order vertebrates such as hagfish, sharks, and bony fishes (Chong and Jin 2009), indicating the early divergence of muscle fiber-type specific TnI isoforms at least 330 million years ago (Figure 2A). Consistent with previous observations of the evolution of TnT, another subunit of the troponin complex which co-evolved with TnI (Jin et al. 1998), amino acid sequence alignment of TnI isoforms in representative vertebrate and invertebrate species reveals that each muscle type TnI isoform is conserved across vertebrate species but clearly diverged from each other and from invertebrate TnI (Figure 2B).
Fig. 2. Evolution of TnI isoforms.
A, The evolutionary lineage demonstrates that fast skeletal muscle TnI (fTnI) was the first vertebrate TnI from an ancestor that also gave rise to invertebrate TnI. Slow skeletal muscle TnI and then cardiac TnI emerged subsequently from gene duplications. B, The phylogenetic tree of TnI isoforms of representative vertebrate and invertebrate species reveals that each muscle type-specific isoform is conserved among species whereas the three isoforms are significantly diverged. An exception is the fish slow skeletal muscle and cardiac TnI, indicating their early states after divergence.
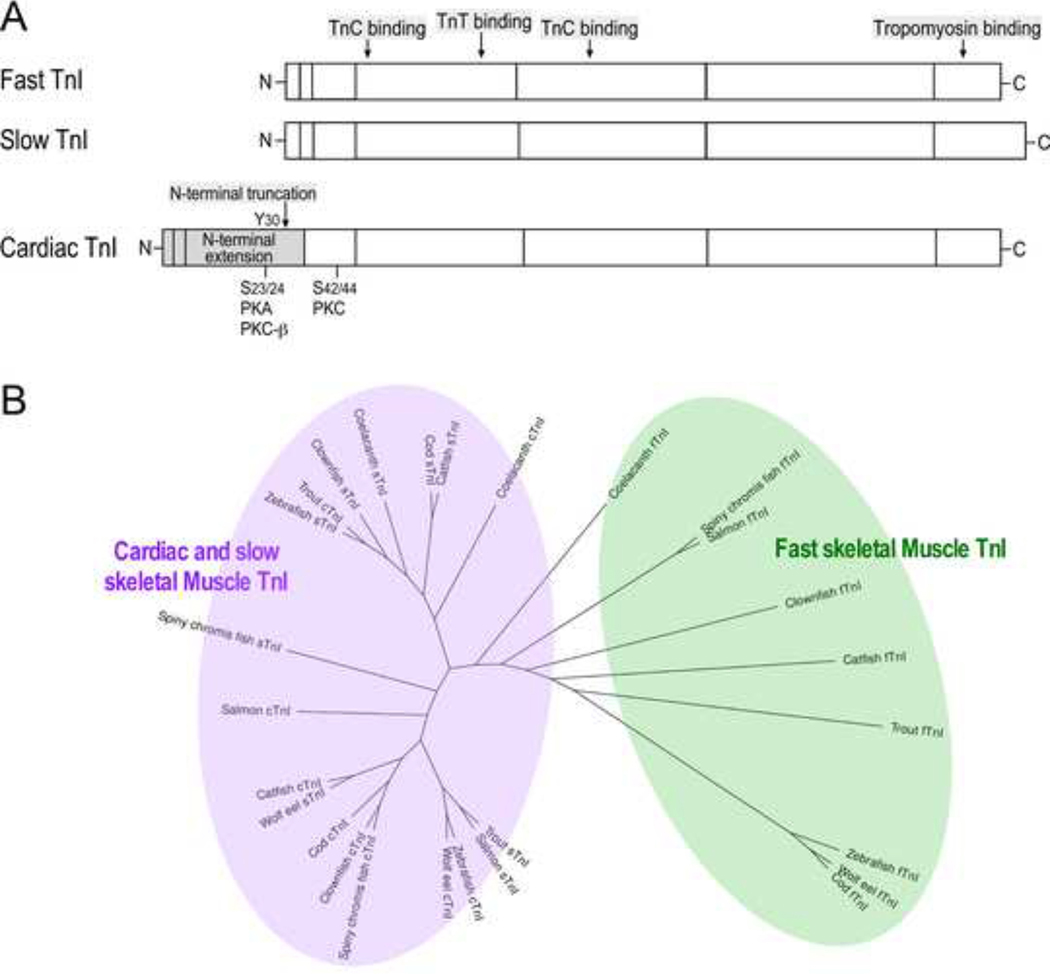
As the Newest TnI Isoform Cardiac TnI I Has Evolved with An Additional N-terminal Extension
The N-terminal extension of cardiac TnI is a unique additional structure in higher order vertebrates (Figure 3A). Within this region are PKA phosphorylation sites (Ser23/Ser24 in mammalian cardiac TnI) which respond to β-adrenergic stimulation to increase the relaxation velocity of cardiac muscle and ventricular filling, resulting in higher stroke volume (Sheng & Jin 2015). As noted previously (Gross and Lehman 2016), our results found that cardiac TnI in lower order vertebrates like ray-finned fishes often grouped together with slow TnI in a phylogenetic tree due to the lack of the N-terminal extension (Figure 3B). Despite the possibility of mis-annotation of sequences within the database due to their high degree of similarity, the data strongly support the emergence of cardiac TnI from duplication of the slow TnI gene (Chong & Jin 2009), which was likely during the evolution of primordial fishes.
Fig. 3. Cardiac TnI has evolved with a unique additional N-terminal extension.
A, The linear protein maps illustrate that while the core structure of the three muscle type TnI isoforms is highly conserved, the N-terminal extension of cardiac TnI is an additional structure that is absent from the skeletal muscle isoforms. The N-terminal extension of cardiac TnI is a regulatory structure containing PKA phosphorylation sites Ser23 and Ser24. B, Phylogenetic lineage analysis showed that in contrast to the fast TnI, cardiac TnI and slow TnI from bony fishes show high degrees of similarity and grouped together, suggesting the emergence of vertebrate cardiac TnI in primordial fishes.
Emergence of the N-terminal Extension of Cardiac TnI and Tetrapod Evolution
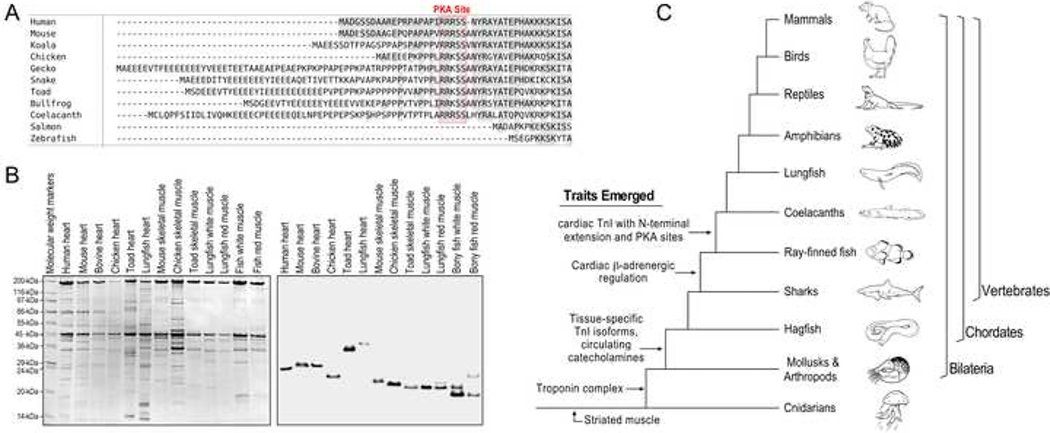
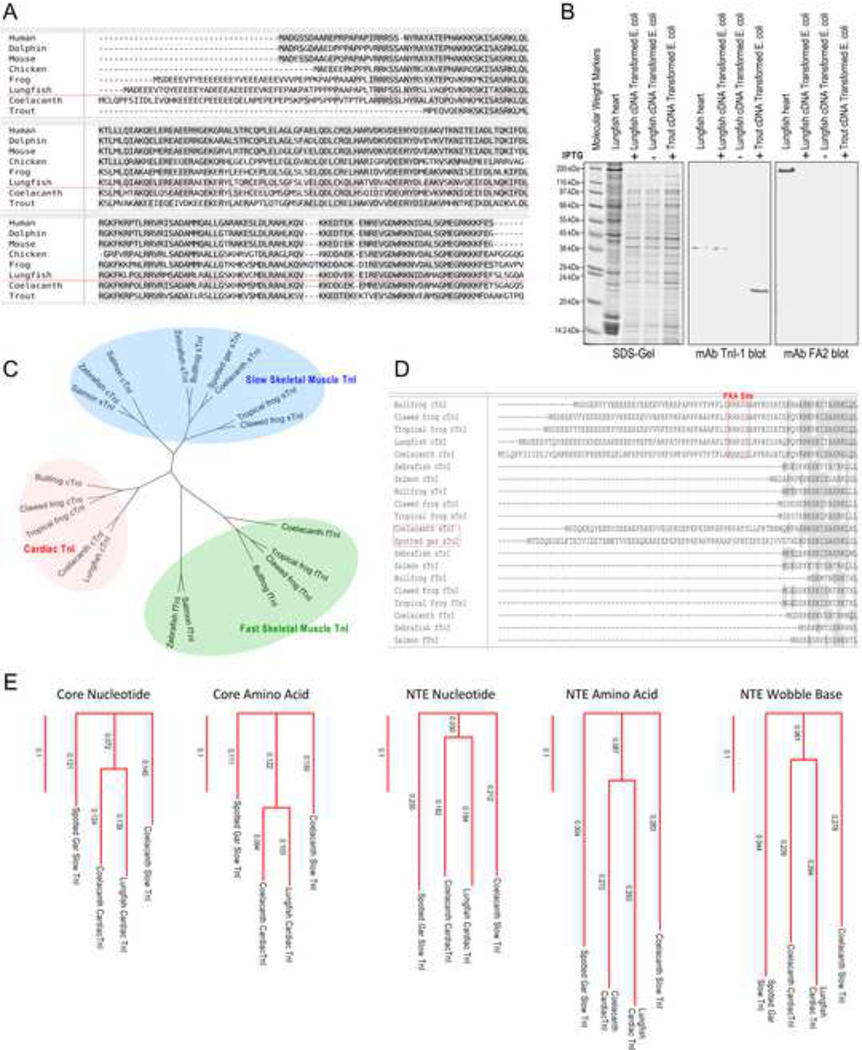
Amino acid sequence alignment of previously known cardiac TnI from representative species reveals that the N-terminal extension, while absent in all ray-finned fishes of class Actinopterygii, is present in coelacanth, a species of lobe-finned fishes of class Sarcopterygii (Figure 4A). The coelacanth is considered to be a member of the lineage pre-dating tetrapods, and they possess fleshy lobed fins likened to proto-limbs (Forey 1988). While this is the earliest known emergence of a cardiac TnI with the N-terminal extension in a fish species, the sequence of coelacanth cardiac TnI N-terminal extension is comparable to that in higher order vertebrates including amphibians, reptiles, birds, marsupials, and eutherian mammals.
Fig. 4. Evolution of the N-terminal extension of cardiac TnI.
A, Amino acid sequence alignment of cardiac TnI of representative vertebrate species demonstrates that the N-terminal extension and the PKA phosphorylation sites are absent in ray-finned bony fishes but present in lobe-finned fishes such as the coelacanth as well as in amphibians, reptiles, birds, marsupials and mammals. B, Coomassie Brilliant Blue-stained SDS-gel (left) and anti-TnI mAb Western blot (right) of total protein extracts from cardiac and skeletal muscles of representative species show that lungfish (South American lungfish, Lepidosiren paradoxa) cardiac TnI has higher molecular weight than that of skeletal muscle TnIs and cardiac TnI of human, mouse, bovine, chicken and toad. Cardiac TnIs of those higher vertebrate species are known to have the N-terminal extension, suggesting lungfish cardiac TnI too contains an N-terminal extension. C, The tree of vertebrate evolution illustrates that striated muscle evolved in cnidarians, and troponin emerged in Bilaterian evolution. The prototype vertebrate hagfish possesses circulating catecholamines, and full β-adrenergic innervation of tissues is seen in some bony fishes where muscle type-specific isoforms of TnI also began to emerge. The cardiac TnI N-terminal extension and S23/S24 appears to have co-evolved in coelacanths and are present in all higher order vertebrates.
Lungfish are the other members of the class of Sarcopterygii lobe-finned fish and are a close relative of coelacanths and modern tetrapods (Zhu and Yu 2002; Wang et al. 2021). Western blotting of cardiac and skeletal muscles from representative vertebrate species demonstrated that lungfish cardiac muscle possesses a high molecular weight TnI significantly larger than the isoforms found in lungfish red and white muscles or skeletal muscle TnI isoforms in other vertebrate species examined which do not contain the N-terminal extension (Figure 4B). It is worth noting that the cardiac isoform of TnI in lungfish heart is also larger than that of cardiac TnI in human, mouse, bovine, chicken and toad, all of which have an N-terminal extension (Figure 4B).
Given the data from sequence and protein analyses, we proposed a possible emergence of the N-terminal extension of cardiac TnI after ray-finned fishes but before lobe-finned fishes (Figure 4C). As coelacanths are a rare species for us to obtain samples for biochemical studies, we turned to the lungfish. While the swim bladders of coelacanths contain fat and do not function to breathe air, lungfish possess fully formed lungs and pulmonary circulation that are considered the precursor to the tetrapod cardiopulmonary system. Therefore, lungfish are considered the closest relative to modern tetrapods (Zhu and Yu 2002) and provide an informative model species for us to investigate the evolution and adaptive value of the N-terminal extension of cardiac TnI in the heart of higher vertebrates.
Lungfish possess three isoforms of TnI and TnT
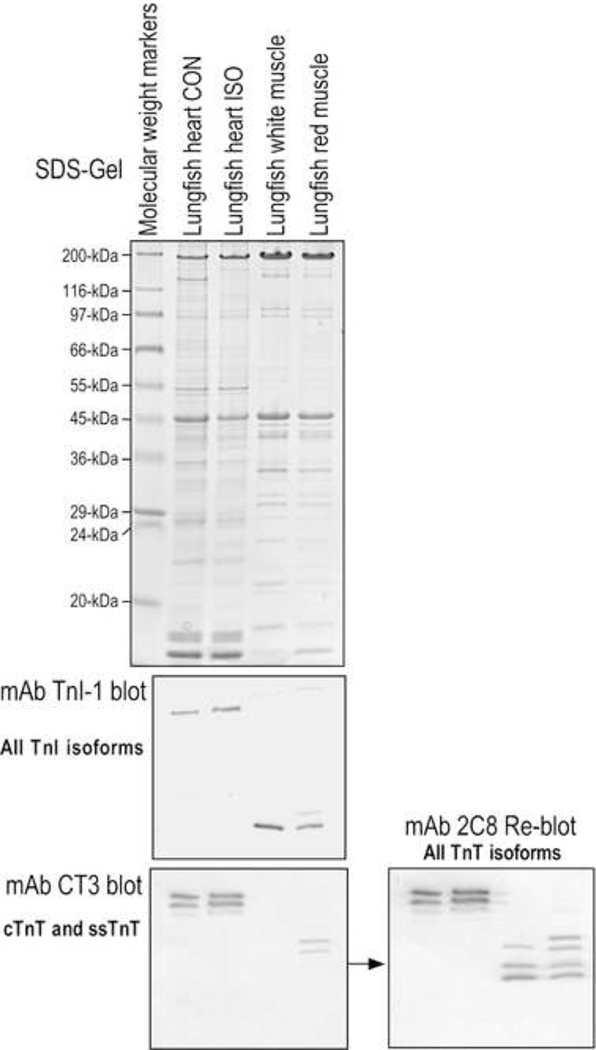
Consistent with previous findings from hagfish, sharks, bony fishes, and coelacanths, we found that lungfish possesses three distinct isoforms of TnI (Figure 5). Pan-TnI mAb TnI-1 Western blots on lungfish cardiac, white and red skeletal muscles detected three isoforms of TnI with the cardiac isoform significantly larger than the skeletal muscle isoforms, consistent with the presence of an N-terminal extension. Immunoblotting with mAb CT3 against cardiac and slow skeletal muscle TnT and pan-TnT mAb 2C8 confirms the co-expression of muscle type-specific isoforms of TnI and TnT in lungfish cardiac, fast and slow skeletal muscles (Figure 5).
Fig. 5. The three TnI isoforms found in lungfish.
Coomassie Blue-stained SDS-gel of South American lungfish (Lepidosiren paradoxa) cardiac and skeletal muscles and Western blots using a pan-TnI mAb TnI-1 which recognizes a highly conserved C-terminal epitope (Jin et al., 2001) showed that lungfish possess three TnI isoforms. A parallel blot with mAb CT3 against cardiac and slow skeletal muscle TnT, followed by reblotting of the same membrane with a pan-TnT mAb 2C8, demonstrates the co-presence of cardiac, fast and slow skeletal muscle TnT isoforms and multiple alternative splice forms in lungfish muscles.
Sequence of Lungfish Cardiac TnI and Its N-terminal Extension
Using RT-PCR approaches (Figure 1), we have determined the coding region sequence of lungfish cardiac TnI with the flanking 5’- and 3’-noncoding regions. The cDNA and amino acid sequences of lungfish cardiac TnI have been deposited to GenBank database with accession number MZ670767.
The sequence data reveal that lungfish cardiac TnI has an N-terminal extension similar to that of coelacanth and amphibians, and clearly different from ray-finned fish cardiac TnIs (Figure 6A). The N-terminal extension of lungfish cardiac TnI is longer than that of amphibians, consistent with the Western blot result in Figure 4B. The sequence of the lungfish cardiac TnI shows a conserved overall structure similar to that in other vertebrate cardiac TnIs, supporting the fidelity of the cDNA sequencing results (Figure 6A). Authenticity of the cloned lungfish cardiac TnI cDNA was further confirmed by expression in E. coli. The Western blot in Figure 6B shows a gel mobility of the encoded protein identical to that of native cardiac TnI in lungfish heart.
Fig. 6. The primary structure of lungfish cardiac TnI and the presence of N-terminal extension containing the two PKA substrate serines.
A, The amino acid sequence of lungfish cardiac TnI is aligned with cardiac TnI from representative vertebrate species. The result shows that lungfish cardiac TnI has a conserved core structure similar to cardiac TnI in the other species. In contrast to cardiac TnI of ray-finned fish, lungfish cardiac TnI has a long N-terminal extension similar to that of coelacanth and amphibian cardiac TnIs. Despite of the significant difference in length, the N-terminal extension of lungfish, coelacanth, amphibian, avian and mammalian cardiac TnIs all have the two PKA substrate serine residues and similar flanking sequences. B, The SDS-gel and mAb TnI-1 Western blot show that the lungfish cardiac TnI protein expressed in E. coli from the cloned cDNA has a gel mobility same as that of native cardiac TnI in the heart of spotted African lungfish (Protopterus dolloi). In comparison, E. coli expressed rainbow trout cardiac TnI that lacks the N-terminal extension (GenBank accession # HM012798.1) showed a significantly faster mobility. The Western blot using mAb FA2 against cardiac MHC further confirmed the mobility position of MHC band in our SDS-PAGE conditions. C, The phylogenetic tree shows that lungfish cardiac TnI is closely related to coelacanth and amphibian cardiac TnIs. In contrast, cardiac and slow skeletal muscle TnIs in ray-finned fishes are not specifically diverged and both lack the N-terminal extension while fish fast TnI is closely related to amphibians. D, Alignment of N-terminal segment of cardiac TnI from multiple fish and amphibian species revealed that besides cardiac TnI, slow skeletal muscle TnI of coelacanth and spotted gar also have a long N-terminal extension but lacking the two PKA phosphorylated serine residues, suggesting a possible transitional state. E, The phylogenetic trees showed that the N-terminal extensions (NTE) found in fish TnIs are less conserved than the core structures and the degree of divergence is higher at amino acid level than that at nucleotide level, indicating a low degree of functional convergence. A tree constructed using the wobble bases of N-terminal extension codons is shown as a reference level of low functional selection.
The amino acid sequence alignment further demonstrates the presence of the two PKA phosphorylated serine residues and flanking sequences in lungfish cardiac TnI like that in the cardiac TnI of tetrapods (Figure 6A). The phylogenetic tree in Figure 6C demonstrates that lungfish cardiac TnI groups with cardiac TnI of higher vertebrates and apart from skeletal muscle TnI and cardiac TnI of other fishes. An interesting finding is that while nearly all fish cardiac and skeletal muscle TnIs lack the N-terminal extension, slow skeletal muscle TnI of coelacanths and spotted gar both contain an N-terminal extension but without the two PKA phosphorylated serine residues (Figure 6D). Both of these species possess rudimentary capacity to breath air (Burggren et al. 2016). Since cardiac TnI emerged from duplication of the slow TnI gene, the finding of such transitional state from which cardiac TnI containing PKA phosphorylation sites may have emerged suggests that the emergence of N-terminal extension of cardiac TnI began prior to the divergence of slow and cardiac TnI genes as a molecular basis to lead the evolution of hearts of air-beathing tetrapod species. Phylogenetic distance analysis showed that the N-terminal extensions of the four fish TnIs are less conserved than the core structures (the region corresponds to that encoded by exons 4–8 of mouse cardiac TnT gene) (Figure 6E). The degree of N-terminal extension similarity is notably lower at amino acid level than that at full nucleotide level (comparable to that of the less functionally relevant codon wobble bases) (Figure 6E). Such low similarity among the N-terminal extension of the fish cardiac TnI indicates a low degree of functional convergence of this submolecular protein structure.
PKA phosphorylation of cardiac TnI
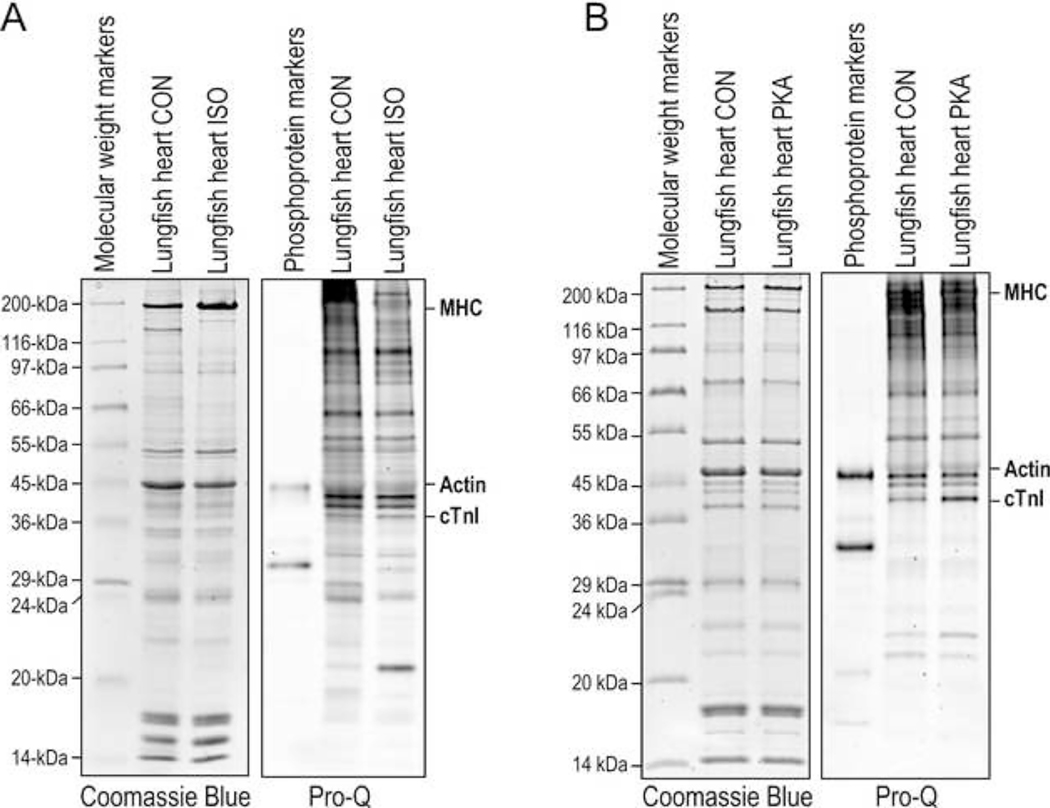
ProQ phosphoprotein staining of total protein extract of lungfish cardiac muscle found that β-adrenergic agonist isoproterenol treatment did not produce significant phosphorylation of cardiac TnI (Figure 7A). As evidence that the isoproterenol treatment was effective, an unidentified 20 kDa protein had significantly increased phosphorylation following treatment (Figure 7A). As β-adrenergic regulation evolved around the time of fishes, this observation suggests that lungfish heart has not evolved a complete β-adrenergic signaling pathway.
Fig. 7. PKA phosphorylation of the N-terminal extension of lungfish cardiac TnI.
A, Coomassie blue-stained (left) and phosphoprotein Pro-Q Diamond stained (right) SDS-gels showed that treatment of lungfish cardiac muscle with β-adrenergic agonist isoproterenol (ISO) did not result in significant phosphorylation of cardiac TnI as compared with control (CON), indicating not fully evolved β-adrenergic responses. B, Coomassie blue-stained (left) and phosphoprotein Pro-Q Diamond stained (right) SDS-gels showed that PKA treatment of skinned lungfish cardiac muscle sections resulted in significant phosphorylation of cardiac TnI, suggesting an emerging capability of a responder to PKA regulation.
Nonetheless, PKA treatment of skinned lungfish cardiac muscle produced significant phosphorylation of cardiac TnI (Figure 7B), indicating that its N-terminal extension has evolved to be a substrate of PKA. This finding suggests that lungfish heart has obtained an emerging capacity of PKA-mediated β-adrenergic regulation of cardiac TnI and heart function, likely a submolecular trait that leads the transition and adaptation to life on land.
Discussion
Troponin has been identified in all clades of Bilateria, including Deuterostomia (Chordata, Hemichordata, Echinodermata) and Protostomia (Mollusca, Annelida, Platyhelminthes, Nematoda, Arthropoda) (Barnes et al. 2016; Yaguchi et al. 2017), while cnidarians and ctenophores that also have striated muscles appear to lack troponin and other molecular hallmarks of bilaterian striated muscles (Steinmetz et al. 2012). The early emergence of troponin regulation in Bilateria evolution highlights its significance in striated muscle function and adaptations.
Cardiac TnI is specifically expressed in adult hearts of higher vertebrates (Jin 1996). The rich database of troponin structure allows us to comprehensively study the intriguing evolution of the N-terminal extension of cardiac TnI and to understand its specific function in the hearts of tetrapods. Pre-tetrapods present an excellent model to understand the cardiac adaptation during the transition to land and how the evolution of protein structure and function served as material basis. Through detailed phylogenetic analysis of vertebrate TnI isoforms revealing the emergence of the N-terminal extension in cardiac TnI, and by sequencing and characterization of cardiac TnI from lungfish as a prototype tetrapod, the present study provides the following novel findings.
Rapid Emergence and Fixation of the N-terminal Extension of Cardiac TnI
Intriguing in our analysis is a rapid emergence and fixation of the N-terminal extension in cardiac TnI of lobe-finned fishes, whereafter it remains conserved in higher order vertebrates. Lobe-finned fishes can be traced back approximately 416 million years ago, and research suggests that the first tetrapods diverged from Sarcopterygii to adapt to land life approximately 385 million years ago (George and Blieck 2011). Contingent upon this adaptation to land, it is clear that molecular structures and protein functions of key organs must pre-adapt to allow for the capacity of living in the new environment. The data that all cardiac TnI sequenced to date, including numerous from lower vertebrates, either have a functional form of N-terminal extension containing the PKA phosphorylation sites or have no N-terminal extension at all with no transitional intermediates reflects a rapid evolution and fixation with high selection value. On the other hand, a likely prototype N-terminal extension was found in slow skeletal muscle TnI, the ancestral gene of cardiac TnI, of coelacanths and spotted gar (Figure 6D). Coelacanths have a cardiac TnI with a functional form of the N-terminal extension and a slow skeletal muscle TnI with an N-terminal extension lacking the PKA phosphorylation sites. The N-terminal extension of spotted gar slow TnI also lacks the PKA phosphorylation sites. Although spotted gar is classified as a ray-finned fish, it has a rudimentary capacity to breath air (Burggren et al. 2016). Therefore, gars may represent the early state of the rapid emergence the N-terminal extension in cardiac TnI for fixation in lobe-finned fishes. Sequencing their cardiac TnI genes would help to look into this hypothesis.
Function of the N-terminal Extension of Cardiac TnI in Tetrapod Evolution to Give Rise to the Ability to Move to Land
In contrast to the lack of N-terminal extension in cardiac TnI in ray-finned fishes, lobe-finned fishes such as coelacanths and lungfishes possess a cardiac TnI with the N-terminal extension containing PKA phosphorylation sites similar to that of amphibians and other higher tetrapods. Lobe-finned fishes of the order Sarcopterygii are thought to be a primary ancestor of present-day tetrapods, and are theorized to have been positively impacted from the increase in atmospheric oxygen during the Early Devonian era (George and Blieck 2011; Meyer et al., 2021; Wang et al. 2021). Therefore, we hypothesize that the addition of the N-terminal regulatory structure in cardiac TnI may have been a crucial gain of function in the hearts of early tetrapods and reflect a key adaptation to increased energetic demands of life on land. This apparently rapid evolution of the β-adrenergic regulation of cardiac function from ancestral ray-finned fishes to lobe-finned fishes suggests a high selection value in the cardiac function of vertebrate animals on land.
The N-terminal extension of TnI might have emerged in fish as a new exon independently in both cardiac and slow TnI genes. This trait was fixed in cardiac TnI of lobe-finned fish and tetrapod hearts for a selection value in air-breathing based higher rate of metabolism and living on land. Although it was not fixed in slow skeletal muscle TnI of tetrapods, the N-terminal extensions of coelacanth and spotted gar slow TnIs provide information for likely ancestral structures. The low degree of functional convergence of lungfish and coelacanth cardiac TnI N-terminal extensions (Figure 6E) suggests that they have not experienced much evolutionary selection pressure, but have laid a foundation for the heart of lobe-finned fish to be ready for moving to land.
PKA Phosphorylation of Cardiac TnI in Heart Function and β-Adrenergic Regulation
Numerous studies have reported the importance of β-adrenergic regulation of cardiac TnI in modulating the function of troponin and the Ca2+-activation of myocardial contraction where cardiac TnI plays a fundamental role in inhibiting actomyosin ATPase and muscle relaxation. Although the heart-specific N-terminal extension of cardiac TnI is not directly involved in interactions with TnC, TnT and other myofilament regulatory proteins, PKA phosphorylation of the N-terminal extension tunes muscle contractility and increases the diastolic function of the heart (Sheng and Jin 2014) via modulating the overall conformation and function of cardiac TnI (Akhter et al. 2012). A restrictive deletion of the N-terminal extension of cardiac TnI occurs in the mammalian heart in adaptation to heart failure conditions (Yu et al. 2001; McConnell et al. 2009), also resulting in increased myocardial relaxation (Barbato et al. 2005; Feng et al. 2008; Gunther et al. 2016). The physiological function of the N-terminal extension of cardiac TnI plays a central role in Ca2+ regulation of cardiac performance and highlights the importance of its evolution and value during natural selection.
The 20 kDa phosphorylated protein we observed in Figure 7A served as evidence that lungfish heart responds to β-adrenergic regulation. Its size suggests that it is likely myosin light chain 2 (MLC2) which we have seen increase in phosphorylation level in mouse hearts as a response to impaired diastolic function (Wei et al, 2015). Cardiac MLC2 is known to be phosphorylated specifically by cardiac myosin light chain kinases but not PKA (Sheikh et al, 2015). Since the N-terminal phosphorylation of cardiac TnI is critical to diastolic cardiac function in higher vertebrates, an increased MLC2 phosphorylation under β-adrenergic stimulation in lungfish heart may implicate that in the absence of PKA regulation of cardiac TnI, MLC2 phosphorylation could play a complementary role. This finding is worth investigating in future studies for understanding the functional significance of MLC2 phosphorylation in mammalian hearts.
Due to the limitation that we only had access to fresh cardiac muscle tissues from two lungfishes for the phosphorylation assay, one for isoproterenol treatment and the other for control, we could only obtain qualitative data without statistical analysis. Future quantitative studies are needed to further establish the biochemical properties of lungfish cardiac TnI.
Value of the Prototype Function of Cardiac TnI in Human Health and Treatment of Heart Failure
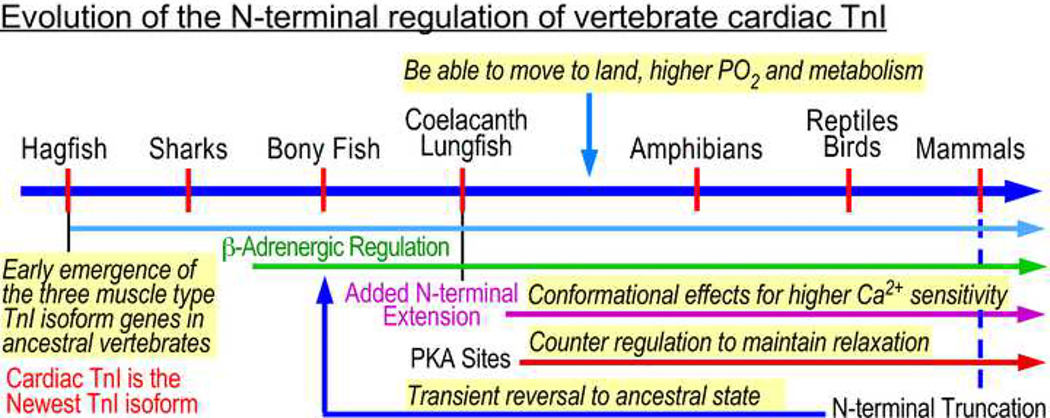
Mammalian embryonic hearts exclusively express slow skeletal muscle TnI lacking the N-terminal extension, with a transition to cardiac TnI occurring weeks after birth (Jin 1996; Feng et al. 2009). Deletion of the N-terminal extension from adult mammalian cardiac TnI results in significant conformational changes to the remaining slow TnI-like core structure and reduces binding affinity for cTnC in a Ca2+-dependent manner (Akhter et al. 2012). Still, mice expressing N-terminal extension truncated cardiac TnI live through adulthood with increased ventricular relaxation velocity and diastolic cardiac function with improved lifespan (Feng et al. 2008; Biesiadecki et al. 2010). A physiologically occurring endogenous restrictive proteolytic truncation of the N-terminal extension of cardiac TnI is present in normal mammalian hearts at low levels and increases in cardiac stresses (Yu et al. 2001). As the N-terminal truncated cardiac TnI may mimic the prototype function of fish cardiac TnI that has no N-terminal extension, the cardiac TnI N-terminal extension-based addition of heart function is a reversable trait with potent effects on cardiac functions. Since cardiac TnI has a rapid turnover rate with a half-life of 3–4 days (Martin 1981), the N-terminal truncated cardiac TnI will be replaced by newly synthesized intact protein when the stress condition is relieved, presenting an attractive mechanism that transiently returns the function of mammalian hearts to an ancestral and compensatory state for the treatment of human heart diseases and failure (Figure 8).
Fig. 8. The evolution and function of the N-terminal extension of cardiac TnI.
The TnI isoforms genes emerged early in the evolution of vertebrate striated muscles. The evolution of a cardiac-specific isoform of TnI and the emergence of the N-terminal extension and PKA phosphorylation sites coincide with the emergence of lungs to increase oxygenation and metabolic rate in cardiac muscle. The N-terminal extension with the PKA phosphorylation sites of cardiac TnI functions to regulate heart function, which could be a key step to enable early tetrapods to move to land where increased energetic demand requires adaptation to allow for higher calcium sensitivity and ultimately stronger contraction. A restrictive proteolytic cleavage that selectively removes the N-terminal extension of cardiac TnI occurs in in mammalian hearts as a physiological adaptation to chronic heart failure, implicating a benefit to transiently reverse the function of cardiac TnI to an ancestral state like that of bony fishes.
Supplementary Material
Acknowledgements
We thank Dr. Irene Salinas and Dr. Ottavia Benedicenti for providing lungfish hearts.
Funding:
This research was supported by grants from the National Institutes of Health HL127691 and HL138007 to JPJ.
Footnotes
Conflicts of interest/Competing interests
The authors declare no conflicts of interest.
Availability of data and material
All data required to evaluate the conclusions of the paper are presented within the paper or supplementary materials, and all genome sequences are publicly available on NCBI GenBank. Any additional data may be requested from the authors.
References
- Aho E, Vornanen M (2001) Cold acclimation increases basal heart rate but decreases its thermal tolerance in rainbow trout (Oncorhynchus mykiss). J Comp Physiol 171(2):173–179. doi: 10.1007/s003600000171. [DOI] [PubMed] [Google Scholar]
- Akhter S, Zhang Z, Jin J-P (2012) The heart-specific NH2-terminal extension regulates the molecular conformation and function of cardiac troponin I. Am J Physiol Cell Physiol 302(4):H923–H933. doi: 10.1152/ajpheart.00637.2011. https://www.physiology.org/doi/10.1152/ajpheart.00637.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausoni S, Campione M, Picard A, Moretti P, Vitadello M, De Nardi C, Schiaffino S (1994) Structure and regulation of the mouse cardiac troponin I gene. J Biol Chem 269(1):339–346. [PubMed] [Google Scholar]
- Baba ML, Goodman M, Berger-Cohn J, Demaille JG, Matsudag G (1984) The early adaptive evolution of calmodulin. Mol Biol Evol 1(6):442–455. doi: 10.1093/oxfordjournals.molbev.a040330. [DOI] [PubMed] [Google Scholar]
- Barbato JC, Huang QQ, Hossain MM, Bond M, Jin JP (2005) Proteolytic N-terminal truncation of cardiac troponin I enhances ventricular diastolic function. J Biol Chem 280(8):6602–6609. doi: 10.1074/jbc.M408525200. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Hwang H, Ono K, Lu H, Ono S (2016) Molecular evolution of troponin I and a role of its N-terminal extension in nematode locomotion. Cytoskeleton 73(3):117–130. doi: 10.1002/cm.21281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiadecki BJ, Tachampa K, Yuan C, Jin J-P, De Tombe PP, Solaro RJ (2010) Removal of the cardiac troponin I N-terminal extension improves cardiac function in aged mice. J Biol Chem 285(25):19688–19698. doi: 10.1074/jbc.M109.086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet T, Fischer AHL, Steinmetz PRH, Lauri A, Bertucci P, Arendt D (2016) The evolutionary origin of bilaterian smooth and striated myocytes. Elife 5:1–24. doi: 10.7554/eLife.19607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren WW, Bautista GM, Coop SC, Couturier GM, Delgadillo SP, García RM, Alvarez González CA (2016) Developmental cardiorespiratory physiology of the air-breathing tropical gar, Atractosteus tropicus. Am J Physiol Regul Integr Comp Physiol 311(4):R689–R701. doi: 10.1152/ajpregu.00022.2016. [DOI] [PubMed] [Google Scholar]
- Cao T, Thongam U, Jin J-P (2019) Invertebrate troponin: Insights into the evolution and regulation of striated muscle contraction. Arch Biochem Biophys 666:40–45. doi: 10.1016/j.abb.2019.03.013. 10.1016/j.abb.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SM, Jin J-P (2009) To Investigate Protein Evolution by Detecting Suppressed Epitope Structures. J Mol Evol 68(5):448–460. doi: 10.1038/jid.2014.371. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker PM, Lin CC, Powers J, Kobilka BK, Dubin AM, Bernstein D (2006) Effect of targeted deletions of β1- and β2-adrenergic-receptor subtypes on heart rate variability. Am J Physiol Hear Circ 290(1):192–199. doi: 10.1152/ajpheart.00032.2005. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5): 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri E, Capuzzo A, Moon TW (1998) The role of circulating catecholamines in the regulation of fish metabolism: An overview. Comp Biochem Physiol C Toxicol Pharmacol 120(2):177–192. doi: 10.1016/S0742-8413(98)10017-8. [DOI] [PubMed] [Google Scholar]
- Farrell AP (1985) A protective effect of adrenalin on the acidotic teleost heart. J Exp Biol 116:503–508. [Google Scholar]
- Feng H-Z, Chen M, Weinstein LS, Jin J-P (2008) Removal of the N-terminal Extension of Cardiac Troponin I as a Functional Compensation for Impaired Myocardial β-Adrenergic Signaling. J Biol Chem 283(48):33384–33393. doi: 10.1074/jbc.M803302200. http://www.jbc.org/lookup/doi/10.1074/jbc.M803302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H-Z, Chen X, Hossain MM, Jin J-P (2012) Toad heart utilizes exclusively slow skeletal muscle troponin T: An evolutionary adaptation with potential functional benefits. J Biol Chem 287(35):29753–29764. doi: 10.1074/jbc.M112.373191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H-Z, Jin J-P (2020) High efficiency preparation of skinned mouse cardiac muscle strips from cryosections for contractility studies. Exp Physiol 105(11):1869–1881. doi: 10.1113/EP088521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forey PL (1988) Golden jubilee for the coelacanth Latimeria chalumnae. Nature 336:727–732. doi: 10.1038/336727a0. [DOI] [Google Scholar]
- George D, Blieck A (2011) Rise of the earliest tetrapods: An Early Devonian origin from marine environment. PLoS One 6(7):1–7. doi: 10.1371/journal.pone.0022136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis TE, Klaiman JM (2011) The influence of PKA treatment on the Ca2+ activation of force generation by trout cardiac muscle. J Exp Biol 214(12):1989–1996. doi: 10.1242/jeb.052084. [DOI] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M (2000) Regulation of contraction in striated muscle. Physiol Rev 80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Gross SM, Lehman SL (2016) Functional phosphorylation sites in cardiac myofilament proteins are evolutionarily conserved in skeletal myofilament proteins. Physiol Gen 48(6):377–387. doi: 10.1152/physiolgenomics.00112.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther LK, Feng H-Z, Wei H, Raupp J, Jin J-P, Sakamoto T (2016) Effect of N-Terminal Extension of Cardiac Troponin I on the Ca2+ Regulation of ATP Binding and ADP Dissociation of Myosin II in Native Cardiac Myofibrils. Biochemistry 55(12):1887–1897. doi: 10.1021/acs.biochem.5b01059. https://pubs.acs.org/doi/10.1021/acs.biochem.5b01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson LM, Obradovich S, Mouniargi J, Farrell AP (2006) The role of adrenergic stimulation in maintaining maximum cardiac performance in rainbow trout (Oncorhynchus mykiss) during hypoxia, hyperkalemia and acidosis at 10°C. J Exp Biol 209(13):2442–2451. doi: 10.1242/jeb.02237. [DOI] [PubMed] [Google Scholar]
- Hastings KEM, Koppe RI, Marmor E, Bader D, Shimada Y, Toyota N (1991) Structure and developmental expression of troponin I isoforms: cDNA clone analysis of avian cardiac troponin I mRNA. J Biol Chem 266(29):19659–19665. [PubMed] [Google Scholar]
- Huang QQ, Jin J-P (1999) Preserved close linkage between the genes encoding troponin I and troponin T, reflecting an evolution of adapter proteins coupling the Ca2+ signaling of contractility. J Mol Evol 49(6):780–788. doi: 10.1007/PL00006600. [DOI] [PubMed] [Google Scholar]
- Jin J-P (1996) Alternative RNA splicing-generated cardiac troponin T isoform switching: A non-heart-restricted genetic programming synchronized in developing cardiac and skeletal muscles. Biochem Biophys Res Commun 225(3):883–889. doi: 10.1006/bbrc.1996.1267. [DOI] [PubMed] [Google Scholar]
- Katrukha IA (2013) Human cardiac troponin complex: structure and functions. Biochemistry 78(13):1447–1465. doi: 10.1134/S0006297913130063. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Saito K, Deguchi T, Fujimori K, Tadokoro M, Yuba S, Ohgushi H, Kawarabayasi Y (2008) Pharmacological characterization of isoproterenol-treated medaka fish. Pharmacol Res 58(5–6):348–355. doi: 10.1016/j.phrs.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Kentish JC, McCloskey DT, Layland J, Palmer S, Leiden JM, Martin AF, Solaro RJ (2001) Phosphorylation of Troponin I by Protein Kinase A Accelerates Relaxation and Crossbridge Cycle Kinetics in Mouse Ventricular Muscle. Circ Res 88(10):1059–1065. doi: 10.1161/hh1001.091640. https://www.ahajournals.org/doi/10.1161/hh1001.091640. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick KP, Robertson AS, Klaiman JM, Gillis TE (2011) The influence of trout cardiac troponin I and PKA phosphorylation on the Ca2+ affinity of the cardiac troponin complex. J Exp Biol 214(12):1981–1988. doi: 10.1242/jeb.052860. [DOI] [PubMed] [Google Scholar]
- Kranias EG, Solaro RJ (1982) Phosphorylation of troponin I and phospholamban during catecholamine stimulation of rabbit heart. Nature 298:182–184. doi: 10.1038/298182a0. 10.1038/298182a0. [DOI] [PubMed] [Google Scholar]
- Mendonça PC, Gamperl AK (2009) Nervous and humoral control of cardiac performance in the winter flounder (Pleuronectes americanus). J Exp Biol 212(7):934–944. doi: 10.1242/jeb.027680. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I, Wong WY, Nowoshilow S, Kneitz S, Kawaguchi A, Fabrizius A, Xiong P, Dechaud C, Spaink HP, Volff JN, Simakov O, Burmester T, Tanaka EM, Schartl M (2021) Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590(7845):284–289. doi: 10.1038/s41586-021-03198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BK, Popovic Z, Mal N, Lee K, Bautista J, Forudi F, Schwartzman R, Jin JP, Penn M, Bond M (2009) Disruption of protein kinase A interaction with A-kinase-anchoring proteins in the heart in vivo: effects on cardiac contractility, protein kinase A phosphorylation, and troponin I proteolysis. J Biol Chem 284(3):1583–1592. doi: 10.1074/jbc.M806321200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogut O, and Jin J-P (1998) Developmentally regulated, alternative RNA splicing-generated pectoral muscle-specific troponin T isoforms and role of the NH2-terminal hypervariable region in the tolerance to acidosis. J. Biol. Chem. 273:27858–27866. [DOI] [PubMed] [Google Scholar]
- OOta S, Saitou N (1999) Phylogenetic relationship of muscle tissues deduced from superimposition of gene trees. Mol Biol Evol 16(6):856–867. doi: 10.1093/oxfordjournals.molbev.a026170. [DOI] [PubMed] [Google Scholar]
- Rao VS, Korte FS, Razumova MV., Feest ER, Hsu H, Irving TC, Regnier M, Martyn DA (2013) N-terminal phosphorylation of cardiac troponin-I reduces length-dependent calcium sensitivity of contraction in cardiac muscle. J Physiol 591(2):475–490. doi: 10.1113/jphysiol.2012.241604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SG, Bernier NJ, Perry SF (1998) The adrenergic stress response in fish: Control of catecholamine storage and release. Comp Biochem Physiol C Toxicol Pharmacol 120(1):1–27. doi: 10.1016/S0742-8413(98)00037-1. [DOI] [PubMed] [Google Scholar]
- Shaffer JF, Gillis TE (2010) Evolution of the regulatory control of vertebrate striated muscle: The roles of troponin I and myosin binding protein-C. Physiol Gen 42(3):406–419. doi: 10.1152/physiolgenomics.00055.2010. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Lyon RC, Chen J (2015) Functions of myosin light chain-2 (MYL2) in cardiac muscle and disease. Gene 569:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J-J, Jin J-P (2014) Gene regulation, alternative splicing, and posttranslational modification of troponin subunits in cardiac development and adaptation: A focused review. Front Physiol 5(165):1–16. doi: 10.3389/fphys.2014.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaro RJ, Moir AJG, Perry SV (1976) Phosphorylation of troponin I and the inotropic effect of adrenaline in the perfused rabbit heart. Nature 262:615–617. doi: 10.1038/262615a0. [DOI] [PubMed] [Google Scholar]
- Steele SL, Yang X, Debiais-Thibaud M, Schwerte T, Pelster B, Ekker M, Tiberi M, Perry SF (2011) In vivo and in vitro assessment of cardiac-adrenergic receptors in larval zebrafish (Danio rerio). J Exp Biol 214(9):1445–1457. doi: 10.1242/jeb.052803. [DOI] [PubMed] [Google Scholar]
- Steinmetz PRH, Kraus JEM, Larroux C, Hammel JU, Amon-Hassenzahl A, Houliston E, Wörheide G, Nickel M, Degnan BM, Technau U (2012) Independent evolution of striated muscles in cnidarians and bilaterians. Nature 487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang J, Zhu C, Yang L, Ren Y, Ruan J, Fan G, Hu J, Xu W, Bi X, et al. (2021) African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 184(5):1362–1376.e18. doi: 10.1016/j.cell.2021.01.047. 10.1016/j.cell.2021.01.047. [DOI] [PubMed] [Google Scholar]
- Wang Z, Nishimura Y, Shimada Y, Umemoto N, Hirano M, Zang L, Oka T, Sakamoto C, Kuroyanagi J, Tanaka T (2009) Zebrafish β-adrenergic receptor mRNA expression and control of pigmentation. Gene 446(1):18–27. doi: 10.1016/j.gene.2009.06.005. 10.1016/j.gene.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Warkman AS, Atkinson BG (2004) Amphibian Cardiac Troponin I Gene’s Organization, Developmental Expression, and Regulatory Properties Are Different from Its Mammalian Homologue. Dev Dyn 229(2):275–288. doi: 10.1002/dvdy.10434. [DOI] [PubMed] [Google Scholar]
- Wei B, Wei H, Jin JP (2015) Dysferlin deficiency blunts β-adrenergic-dependent lusitropic function of mouse heart. J Physiol. 593:5127–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JM, Grand RJA (1978) Comparison of amino acid sequence of troponin I from different striated muscles. Nature 271:31–35. doi: 10.1038/271031a0. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Tanaka H (2017) Troponin-I is present as an essential component of muscles in echinoderm larvae. Sci Rep 7:1–8. doi: 10.1038/srep43563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZB, Zhang LF, Jin JP (2001) A proteolytic NH2-terminal truncation of cardiac troponin I that is up-regulated in simulated microgravity. J Biol Chem 276(19):15753–15760. doi: 10.1074/jbc.M011048200. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhao JJ, Potter JD (1995) Phosphorylation of both serine residues in cardiac troponin I is required to decrease the Ca2+ affinity of cardiac troponin C. J Biol Chem 270(51):30773–30780. doi: 10.1074/jbc.270.51.30773. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chiang VL (1996) Single-stranded DNA ligation by T4 RNA ligase for PCR cloning of 5’-noncoding fragments and coding sequence of a specific gene. Nucleic Acids Res 24(5):990–991. doi: 10.1093/nar/24.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Yu X (2002) A primitive fish close to the common ancestor of tetrapods and lungfish. Nature 418:767–770. doi: 10.1038/nature00871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.