Abstract
Proteins bearing a nuclear localization signal (NLS) are targeted to the nucleus by the heterodimeric transporter importin. Importin α binds to the NLS and to importin β, which carries it through the nuclear pore complex (NPC). Importin disassembles in the nucleus, evidently by binding of RanGTP to importin β. The importin subunits are exported separately. We investigated the role of Cse1p, the Saccharomyces cerevisiae homologue of human CAS, in nuclear export of Srp1p (yeast importin α). Cse1p is located predominantly in the nucleus but also is present in the cytoplasm and at the NPC. We analyzed the in vivo localization of the importin subunits fused to the green fluorescent protein in wild-type and cse1-1 mutant cells. Srp1p but not importin β accumulated in nuclei of cse1-1 mutants, which are defective in NLS import but not defective in NLS-independent import pathways. Purified Cse1p binds with high affinity to Srp1p only in the presence of RanGTP. The complex is dissociated by the cytoplasmic RanGTP-binding protein Yrb1p. Combined with the in vivo results, this suggests that a complex containing Srp1p, Cse1p, and RanGTP is exported from the nucleus and is subsequently disassembled in the cytoplasm by Yrb1p. The formation of the trimeric Srp1p-Cse1p-RanGTP complex is inhibited by NLS peptides, indicating that only NLS-free Srp1p will be exported to the cytoplasm.
Transport of proteins and RNAs between the cytoplasm and the nucleus across the nuclear pore complex (NPC) is mediated by shuttling transport receptors. All transporters so far identified belong to the importin β superfamily. Substrate binding and release of these transporters are modulated by the small GTPase Ran, the key regulator of nucleocytoplasmic transport. Several specific transport pathways for different import and export substrates have recently been identified (reviewed in references 52 and 73).
The best understood transport pathway is the import of proteins carrying a classical nuclear localization sequence (NLS), which is characterized by a short segment of basic amino acid residues (20). The NLS is recognized by importin α in the cytoplasm, which in turn binds to importin β. Importin β mediates docking at the NPC and subsequent transport into the nucleus. Docking of NLS substrates to the nuclear envelope and subsequent energy-dependent transfer into the nucleus can be reconstituted with recombinant factors and permeabilized cells in vitro. Two soluble factors, Ran and p10, are sufficient to promote the translocation step (for reviews, see references 16, 32 and 50).
Ran is an abundant, mostly nuclear protein that switches between two conformational states, i.e., bound to GDP (RanGDP) and to GTP (RanGTP). The specific regulators of the Ran GTPase cycle, the cytoplasmic GTPase-activating protein RanGAP1/Rna1p and the nuclear nucleotide exchange factor RCC1/Prp20p, generate RanGDP in the cytoplasm and RanGTP in the nucleus (reviewed in references 32 and 61). RanGTP binds to importin β, which is thereby released from the NLS-importin complex (12, 33, 56). This event is thought to represent import termination in the nucleus (33, 52). Accordingly, a high level of nuclear RanGTP is required for efficient import, whereas cytoplasmic RanGTP prevents nuclear import (33, 40, 62, 74). RanGTP complexed to importin β is completely resistant to GTP exchange and GTP hydrolysis (21, 33, 63). Export of importin β from the nucleus does not require GTP hydrolysis by Ran (40) and most likely occurs in a complex with RanGTP but not in a complex with importin α (29, 75). Once in the cytoplasm, the importin β-RanGTP complex has to be dissociated to allow initiation of a new import cycle. This is achieved by importin α and RanBP1 in vitro (4, 22). Subsequently, the RanGAP converts RanGTP to RanGDP, which prevents rebinding of RanGTP to importin β. RanBP1 contains a RanGTP-binding domain different from that of importin β. Yrb1p, the yeast homologue of RanBP1, is an essential cytoplasmic protein which is required for nucleocytoplasmic transport (reviewed in reference 61).
Recently, a number of importin β-related proteins have been identified which were postulated, and in part have been shown, to represent novel transport receptors (24, 28). They are similar in size (95 to 125 kDa) and secondary structure, possess NPC-binding sites, and are homologous in their N-terminal domains, which mediate binding to RanGTP (13, 28, 45, 63). The members of the importin β superfamily can be divided into import receptors and export receptors. Import receptors different from importin β bind to their substrates directly without using an adapter like importin α (1, 28, 55, 58, 63). Substrates identified so far are mRNA-binding proteins for the import factors transportin/Kap104p and Mtr10p (1, 9, 25, 54, 55, 68), ribosomal proteins for Yrb4p/Kap123p and Pse1p (58, 63), and a protein involved in pre-tRNA processing for Sxm1p (57). Like importin β, other import receptors release their cargo by direct binding to RanGTP. This was shown for transportin, Yrb4p/Kap123p, and Pse1p (40, 63). Efficient dissociation of complexes consisting of RanGTP and importin β-related receptors was shown to require RanBP1/Yrb1p in vitro (4, 18, 22, 28, 48, 63).
In contrast to import receptors, export factors require simultaneous association with RanGTP for high-affinity substrate binding (23, 44, 46). This allows cargo binding in the nucleus, where RanGTP is present, and subsequent release into the cytoplasm, where RanBP1 and RanGAP disassemble the export complex (44, 46). Three export receptors have been described recently. Crm1p from various organisms was identified as the exporter of proteins containing leucine-rich nuclear export signals (NESs) occurring, e.g., in the human immunodeficiency virus Rev protein and in the protein kinase A inhibitor (23, 26, 53, 70). Exportin t, a human protein similar to yeast Los1p (36, 37), was identified as the tRNA exporter (46). The human CAS protein was shown to mediate export of Rch1, a member of the importin α family, from the nucleus. CAS binds to importin α and RanGTP in a cooperative manner and is required for importin α export in vitro (44). Here we characterize the Saccharomyces cerevisiae homologue of CAS, the previously identified Cse1p, which is essential for viability (77). Cse1p forms a trimeric complex with Srp1p and the GTP-bound form of Gsp1p (yeast Ran). Complex formation is prevented by NLS peptides, indicating that only yeast importin α which has released its NLS cargo will be exported to the cytoplasm. Export of Srp1p from the nucleus and NLS-dependent nuclear import are inhibited in cse1 mutants. Our biochemical and in vivo data combined suggest a function of Cse1p as the specific export receptor of yeast importin α.
MATERIALS AND METHODS
Plasmids and strains.
The strains used in this study are listed in Table 1. All green fluorescent protein (GFP) constructs were made with an S65T V163A mutant. To create an SRP1-GFP fusion by homologous recombination by using the pop-in/pop-out strategy (59), GFP was inserted into pRS306 (67) containing SRP1 (47, 78) with engineered restriction sites at the stop codon. This plasmid (pGS288) was linearized with EcoRI and used for transformation of the diploid wild-type strain GSY158 (63), which resulted in GSY412 (pop-in strain). Integration at the SRP1 locus was confirmed by Southern blotting. GSY412 was passed over 5-fluoro-orotic acid plates, and tetrads were dissected to isolate the pop-out strain GSY414. This strain was crossed to the cse1-1 mutant strain Y1709 (77), and haploid mutants containing integrated SRP1-GFP (GSY581) were isolated by tetrad dissection of the diploid. The in vivo localization of Srp1-GFP in the wild type was analyzed in the sister spore GSY591. To construct a GFP-Kap95p fusion, GFP was inserted into pRS306 carrying the RSL1/KAP95 gene (42) with an introduced BamHI site at the start codon of RSL1/KAP95. This plasmid (pGS348), which contains a linker encoding four glycines between GFP and RSL1/KAP95, was linearized in the coding region and used to isolate the pop-in (GSY585) and pop-out (GSY586) strains as described above, except that haploid strains were used. GSY586 was crossed to cse1-1, and a mutant expressing GFP-RSL1/KAP95 (GSY589) was isolated. CSE1 was cloned by PCR from genomic DNA by using the oligonucleotides 5′-CGCGCGGCCGCTTCCAGGATGCTATATTACG-3′ (GS60) and 5′-CGGCTCGAGCAGACCTATGTACTCCGCTGG-3′ (GS61). All PCRs were performed with the proofreading Pwo polymerase (Boehringer Mannheim). The 3,240-bp PCR product was inserted first between the NotI and XhoI sites of pBluescript (Stratagene), generating pGS356. The strategy to obtain pop-in (GSY579) and pop-out (GSY580) strains expressing GFP-CSE1 was as described for GFP-RSL1/KAP95. The coding region of CSE1 plus 146 bp of 3′ sequence was amplified with 5′-GCGGATCCATGTCCGATTTGGAAACCG-3′ (GS62) and GS61, digested with BamHI plus XhoI, and inserted behind the GAL1 promoter of the URA3 plasmids YCpGAL and YEpGAL (62), generating pGAL-CSE1 CEN (pGS364) or pGAL-CSE1 2μm (pGS366). The RSL1/KAP95 coding sequence (42) was similarly subcloned into YCpGAL, generating YCpGAL-KAP95 (pGS305). The CSE1 coding sequence was amplified with GS62 and 5′-GCGGATCCATTACCAACTAATAATTGATT-3′ (GS63), digested with BamHI, and inserted in pGEX-4T (Pharmacia) for expression in Escherichia coli. For expression of a GST-Srp1p fusion in E. coli, the SRP1 coding sequence was inserted into the BamHI site of pGEX-5G (pGS388; a derivative of pGEX-4T encoding a five-glycine linker at the C terminus of glutathione S-transferase [GST]). The reporter plasmid YEpGAL-NLS-GST-GFP (pGS420; 2μm URA3) was derived from pGAL-NLS-GST, pGAL-GST (64), and a BglII/BamHI fragment encoding GFP. YCpGAL-NLS-GST-GFP (pGS422; CEN LEU2) carries the same insert. Plasmid pGS304, encoding the N-terminal 49 amino acid residues of L25 fused to β-galactosidase, was described before (63).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| GS155 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 | 63 |
| GS158 | MATa/α ura3-52/− leu2Δ1/− his3Δ200/ − trp1Δ63/+ | 63 |
| GSY412 | MATa/α ura3-52/− leu2Δ1/− his3Δ200/− trp1Δ63/+ SRP1 SRP1-GFP URA3 (pop-in) | This study |
| GSY414 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 SRP1-GFP (pop-out) | This study |
| Y1709 | MATa ura3-52 his3-11,15 trp1Δ901 ade2-101 cse1-1 | 77 |
| GSY579 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 CSE1 GFP-CSE1 URA3 (pop-in) | This study |
| GSY580 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 GFP-CSE1 (pop-out) | This study |
| GSY581 | MATa ura3 his3 trp1 cse1-1 SRP1-GFP(pRS313) | This study |
| GSY585 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 RSL1/KAP95 GFP-RSL1/KAP95 (pop-in) | This study |
| GSY586 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 GFP-RSL1/KAP95 (pop-out) | This study |
| GSY587 | MATα ura3 leu2 his3 trp1 rat2-1 GFP-CSE1(prS315, prS316) | This study |
| GSY589 | MATα ura3 his3 trp1 cse1-1 GFP-RSL1/KAP95 | This study |
| GS591 | MATa ura3 leu2 his3 trp1 SRP1-GFP(pRS313) | This study |
Protein analysis.
Cse1p and Srp1p were purified from E. coli as GST fusions and cleaved with thrombin to remove GST as described previously (64). The purification of Schizosaccharomyces pombe Rna1p (6), Yrb1p, GST-Gsp1p, and Gsp1p (15, 64) was described before. Labeling of Gsp1p with [γ-32P]GTP (6,000 Ci/mmol) and GTPase assays were carried out as described previously (63). Gsp1p bound to [γ-32P]GTP (Gsp1p[γ-32P]GTP) was supplemented with the additions described in the figure legends and incubated at 25°C. The total assay volume was 50 μl. Released [32P]phosphate was determined by the charcoal method (5). The control (100% hydrolysis) in the GTPase assays was the released [32P]phosphate over the background level in samples containing only Gsp1p[γ-32P]GTP and Rna1p. Solution binding assays were performed at 4°C in PBSKMT (25 mM sodium phosphate, 150 mM NaCl, 3 mM KCl, 1 mM MgCl2, 0.1% Tween 20, pH 7.3). To load Gsp1p with GTP or GDP, Gsp1p or GST-Gsp1p was incubated for 60 min at 0°C with PBSKMT plus 2 mM EDTA in the presence of 1 mM nucleotides, and then 6 mM MgCl2 was added. GST-Gsp1p or GST-Srp1p was bound to 20 μl of glutathione-Sepharose (Pharmacia) per reaction mixture for 1 h. The binding reaction mixtures were then incubated for 1 h with the factors indicated in the figure legends. The beads were washed four times with 1 ml of PBSKMT, and bound proteins were eluted with sodium dodecyl sulfate (SDS) gel loading buffer.
Affinity purification of rabbit antibodies against recombinant Cse1p, immunofluorescence microscopy and detection of poly(A)+ RNA by in situ hybridization were performed as described previously (63). Affinity purification of antibodies against Kap95p and Srp1p was described before (31). Western blot analysis was carried out according to the guidelines for the ECL kit (Amersham) with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Sigma) as the secondary antibody.
The calculation of the cellular Cse1p concentration was based on the observation that 1.2 million cells contain approximately 12 ng of Cse1p (purified Cse1p served as a standard in immunoblots [not shown]) and on previous determinations that a haploid yeast cell contains 6 pg of protein and has a volume of 70 μm3 (34).
RESULTS
In vivo localization of yeast importin α and β.
To analyze the traffic of yeast importin α (Srp1p) and yeast importin β (Rsl1p/Kap95p) across the NPC in living S. cerevisiae cells, we constructed fusion proteins with GFP. We genetically replaced the importin genes by copies encoding Srp1-GFP or GFP-Kap95p. Immunoblot analysis with antibodies against Srp1p, Kap95p, or GFP showed that the respective GFP fusion proteins were present in the resulting strains (Fig. 1A and C), which had growth characteristics identical to those of wild-type strains (not shown). This demonstrates that both fusion proteins are fully functional.
FIG. 1.
Localization of importin α and β in living yeast cells. (A) Lysates from wild-type cells (lanes 1 and 4), cells containing an additional chromosomal copy of SRP1-GFP (pop-in) (lanes 2 and 5), or cells containing only SRP1-GFP (pop-out) (lanes 3 and 6) were analyzed by SDS gel electrophoresis and immunoblotting with anti-Srp1 antibodies (lanes 1 to 3) or anti-GFP antibodies (lanes 4 to 6). The sizes of molecular weight markers (in thousands) are indicated. (B) Haploid cells containing SRP1-GFP instead of SRP1 were grown in liquid medium at 30°C. DAPI (2.5 μg/ml) was added to the culture. After 15 min, cells were viewed by fluorescence microscopy in the fluorescein channel to visualize the GFP signal, in the UV channel to visualize DAPI-stained DNA, or by Nomarski optics. (C) Lysates from wild-type cells (lanes 1 and 4), cells additionally containing GFP-RSL1/KAP95 (lanes 2 and 5), or cells containing only GFP-RSL1/KAP95 (lanes 3 and 6) were analyzed by SDS gel electrophoresis and immunoblotting with anti-Kap95 antibodies (lanes 1 to 3) or anti-GFP antibodies (lanes 4 to 6). (D) Cells expressing only GFP-RSL1/KAP95 were grown in liquid medium. DAPI (2.5 μg/ml) was added to the culture. After 15 min, cells were viewed by fluorescence microscopy to visualize GFP-Kap95p or DNA and by Nomarski optics. Note that we used petite strains in panels B and D to minimize staining of mitochondrial DNA.
Next we examined the in vivo localization of the GFP fusions. Logarithmically grown cells from liquid cultures were directly viewed by fluorescence microscopy. Both chimeras were evenly distributed throughout the cell, concentrated around the nuclear envelope, and excluded from the vacuoles. The nuclei were identified by costaining with the DNA dye DAPI (4′,6-diamidino-2-phenylindole). In contrast to the case for Kap95p, the concentration of Srp1p in the nucleus appeared to be somewhat higher than that in the cytoplasm (Fig. 1B and D). Divergent Srp1p and Kap95p localization data were obtained by indirect fluorescence microscopy before (2, 38, 42, 43, 78). These variations (exclusive localization at the nuclear envelope, the nucleus, or the cytoplasm) are explained by different fixation conditions during sample preparation for immunofluorescence microscopy (63). We compared the localizations of the wild-type importins and the respective GFP fusions and observed identical staining patterns by immunofluorescence microscopy with anti-Srp1 or anti-Kap95 antibodies and different fixation protocols (not shown). Therefore, the GFP moieties do not affect the localization of importin α and β.
cse1 mutants are defective in Srp1p export and NLS import.
Mutations in factors involved in Srp1p transport across the NPC are expected to affect the intracellular distribution of Srp1p. To test this, we crossed the strain containing integrated SRP1-GFP against a number of mutants with defects in factors potentially involved in Srp1p transport. We then analyzed the localization of Srp1p in the resulting double mutants. The previously reported nuclear accumulation of Srp1p in prp20-1 mutants (42) was also observed with Srp1-GFP. Temperature-sensitive rna1-1, npl4-1, npl4-2, rat2-1, xpo1-1, rpb1-1, and Δyrb2 alleles (15, 19, 35, 51, 70, 71) had no effect on the Srp1p localization (not shown). However, cells carrying the cold-sensitive cse1-1 allele (77) showed a complete loss of cytoplasmic Srp1p and a concomitant nuclear accumulation even at the permissive temperature of 30°C, indicating a defect in Srp1p export (Fig. 2A to D). By immunofluorescence microscopy, no significant Srp1p mislocalization in gsp1-1, gsp1-2, npl3-1, pse1-1, rsl1-1, yrb1-1, and Δyrb4 mutants (10, 42, 63–65, 76) was observed (not shown), but again Srp1p was found exclusively in the nuclei of cse1-1 mutants (Fig. 3I to L).
FIG. 2.
Srp1p but not Kap95p accumulates in the nuclei of cse1-1 mutants. Wild-type (wt) (GSY591) or cse1-1 (GSY581) cells expressing SRP1-GFP (A to D) and wild-type (GSY586) or cse1-1 (GSY589) cells expressing GFP-RSL1/KAP95 (E to H), grown at 30°C, were viewed by fluorescence microscopy to detect the GFP signal or by Nomarski optics.
FIG. 3.
Nuclear import of Npl3p and L25 is not affected in cse1-1 mutants. Wild-type (wt) (GSY155) or cse1-1 (Y1709) cells carrying no plasmid (A to D and I to L) or carrying plasmid pGS304 encoding L251–49–β-galactosidase (E to H), grown at 30°C, were prepared for immunofluorescence microscopy by formaldehyde fixation. They were then incubated with antibodies against Npl3p (A to D), or β-galactosidase (βgal) (Promega) (E to H), or Srp1p (I to L) and then with Texas red-conjugated secondary antibodies (Jackson) and with DAPI.
Cse1p (predicted molecular mass, 109.3 kDa) belongs to the importin β superfamily. It has an overall identity of 17.1 to 18.8% with Kap95p, Kap104p, Yrb4p/Kap123p, Pse1p, Mtr10p, Los1p, or Crm1p/Xpo1p (aligned by the FASTA algorithm). Cse1p shows significant homology to the human CAS protein (36% identity and 67% similarity) and is therefore regarded as the yeast homologue of CAS (11). Cse1p was reported earlier to play a role in chromosome segregation during mitosis (77). By quantitative immunoblotting with purified Cse1p as a standard, we calculated that Cse1p represents approximately 0.16% of the total cellular protein; i.e., roughly 55,000 molecules are present in a haploid yeast cell (not shown).
Mutant cse1-1 cells streaked on plates containing rich medium did not grow when incubated at 15°C for 14 days but were rescued by plasmids carrying the CSE1 gene (Fig. 4A). We confirmed the previous observation (cited in reference 77) that the mutant is suppressed by SRP1 overexpression. cse1-1 cells transformed with 2μm plasmids containing the SRP1 gene grew comparably to wild-type cells (Fig. 4A). We next localized various proteins in the cse1-1 mutant to examine nucleocytoplasmic transport. Cells were grown at the permissive temperature of 30°C in liquid medium and then shifted to 15°C for 24 h. Under these conditions, the mutant exhibits a growth rate similar to that of the wild type for more than 48 h (not shown). To test whether Kap95p also mislocalizes in cse1-1 mutants, we used a strain carrying the integrated GFP-KAP95 allele. Fluorescence microscopy showed that the localization of GFP-Kap95p is the same in cse1-1 cells and wild-type cells at 30°C (Fig. 2E to H) or at 15°C (not shown), indicating that nuclear export of Kap95p is not affected. We next examined the intracellular distribution of endogenous proteins by immunofluorescence microscopy. The localizations of nuclear Npl3p (10) (Fig. 3A to D), cytoplasmic Yrb1p (64), and nucleolar Nop1p (72) (not shown) were not changed in cse1-1 mutants at any temperature tested. The mutant also shows a normal distribution of poly(A)+ RNA, which indicates that mRNA export is not inhibited (not shown).
FIG. 4.
NLS import is defective in cse1-1 mutants. (A) cse1-1 cells (Y1709) transformed with a 2μm vector as a control or with SRP1 2μm, CSE1 2μm, or CSE1 CEN plasmids were streaked on YPD plates and incubated for 2 days at 30°C or for 14 days at 15°C. (B) cse1-1 cells carrying YEpGAL-NLS-GST-GFP were transformed with a CEN vector or a CSE1 CEN plasmid. Cultures were grown in selective medium containing raffinose, and then 2% galactose was added and the cultures were split. One half was further incubated at 30°C for 3 h, and one half was incubated at 15°C for 24 h. NLS-GST-GFP was localized by fluorescence microscopy.
Since Srp1p is needed in the cytoplasm for import of NLS proteins, nuclear accumulation of Srp1p could result in an NLS import defect. To test this, we transformed cse1-1 mutants with plasmids encoding a protein containing the simian virus 40 (SV40) large-T-antigen NLS fused to GST and GFP (NLS-GST-GFP; molecular mass, 58.3 kDa). This reporter was located exclusively in the nuclei of wild-type cells or cse1-1 cells complemented with CSE1 plasmids at all temperatures tested (Fig. 4B). However, the reporter accumulated in the cytoplasm of cse1-1 cells, indicating a defect in nuclear import. The NLS import defect was already prominent at 30°C and was profound at 15°C (Fig. 4B). Similar results were obtained with reporters containing the histone H2B NLS fused to β-galactosidase, the SV40 NLS fused to cytoplasmic invertase, or the SV40 NLS fused to β-galactosidase (63) (not shown). We also tested a reporter protein containing the nuclear targeting signal of ribosomal protein L25 fused to β-galactosidase. L25 import was shown to depend on Yrb4p/Kap123p but not on importin (58, 63). The L25 reporter did not mislocalize to the cytoplasm in cse1-1 mutants (Fig. 3E to H). In summary, cse1-1 mutants show specifically a defect in importin α-dependent NLS import. Parallel import pathways and export of mRNA or importin β proceed normally during a shift to the nonpermissive temperature for 24 h.
CSE1 overexpression interferes with mRNA export in wild-type cells.
To examine the effect of CSE1 overexpression on nucleocytoplasmic transport, we transformed wild-type cells with centromeric (CEN) or high-copy (2μm) plasmids containing the CSE1 coding sequence under control of the inducible GAL1 promoter. A single-copy pGAL-KAP95 plasmid encoding galactose-inducible Kap95p, which mediates a dominant-lethal phenotype upon overexpression of RSL1/KAP95 (Fig. 5A), served as a control. Cells transformed with pGAL-CSE1 2μm plasmids did not grow on galactose-containing plates, indicating that CSE1 overexpression is lethal (Fig. 5A). Expression mediated by pGAL-CSE1 CEN plasmids inhibited growth but still allowed colony formation. Wild-type cells transformed with pGAL-CSE1 2μm plasmids or pGAL-KAP95 CEN plasmids were grown in liquid medium, and overexpression was induced by addition of 2% galactose. The growth arrest was established ∼8 h after galactose addition (Fig. 5B). Immunoblot analysis showed that plasmid-encoded Cse1p and Kap95p were expressed approximately fivefold over wild-type levels after 2 to 5 h (Fig. 5C).
FIG. 5.
CSE1 overexpression blocks mRNA export. (A) Wild-type cells (GSY155) were transformed with plasmid pGAL 2μm, pGAL-KAP95 CEN, pGAL-CSE1 CEN, or pGAL-CSE1 2μm, streaked on plates containing glucose (no induction) or galactose (induction), and incubated at 30°C for 2 days. (B) Cells carrying plasmid pGAL, pGAL-CSE1 2μm, or pGAL-KAP95 CEN were grown at 30°C in raffinose-containing medium. Expression was induced by addition of galactose (2%), and growth rates were monitored by measuring absorptions at 600 nm. (C) Extracts from cells overexpressing CSE1 or RSL1/KAP95 for the indicated times were analyzed by SDS gel electrophoresis and immunoblotting with anti-Cse1 or anti-Kap95 antibodies. (D) Cells as described for panel B, carrying YCpGAL-NLS-GST-GFP, were grown in galactose for 2 h (pGAL-CSE1) or 3 h (pGAL and pGAL-KAP95). The cells were either directly viewed by fluorescence microscopy to detect NLS-GST-GFP or prepared for in situ hybridization to localize mRNA and stained with DAPI.
We then investigated whether CSE1 and RSL1/KAP95 overexpression affects nucleocytoplasmic traffic. A number of endogenous and plasmid-encoded proteins were localized microscopically at 2, 3, or 5 h after galactose addition as described above. The localization of Srp1p and Kap95p (both analyzed with the respective GFP fusions in vivo), Yrb1p, or Nop1p was unaffected under all conditions tested (not shown). The L25 reporter and Npl3p (not shown), as well as NLS-GST-GFP and all other reporter proteins containing classical NLSs, accumulated strongly in the cytoplasm after RSL1/KAP95 overexpression but were still nuclear after CSE1 overexpression (Fig. 5D). To examine mRNA export, we localized poly(A)+ RNA under the same conditions. As expected, polyadenylated RNA was observed in the cytoplasm of wild-type cells carrying a control plasmid (Fig. 5D). However, all cells overexpressing CSE1 accumulated poly(A)+ RNA completely in the nucleus even 2 h after induction. Overexpression of RSL1/KAP95 for 2 to 5 h caused only an intermediate inhibition of mRNA export (Fig. 5D). We also tested whether CSE1 overexpression affects NES export but observed no mislocalization of a reporter (70) containing the leucine-rich NES of protein kinase A inhibitor (PKI) (not shown). Taken together, the data indicate that excess amounts of Kap95p strongly inhibit NLS import and import of Npl3p and L25, probably by competition for common import receptor binding sites at the NPC, but do not block mRNA export to a similar extent. Overexpression of CSE1, on the other hand, does not interfere with various protein import and export pathways but blocks the mRNA export pathway.
Localization of Cse1p.
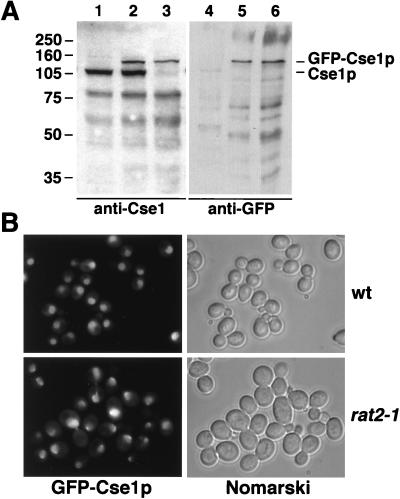
To determine the intracellular distribution of Cse1p, we generated a strain in which CSE1 is replaced by a GFP-CSE1 fusion. This strain grows normally and encodes a functional GFP-Cse1p fusion which was detected by immunoblotting with polyclonal anti-Cse1 antibodies or GFP-specific antibodies (Fig. 6A). The faster-migrating bands in Fig. 6A (lanes 1 to 3) probably represent degradation products of Cse1p. Their intensity varies greatly in different immunoblots, and they can be competed by purified recombinant Cse1p (not shown). GFP-Cse1p is located mainly in the nucleus, with some localization in the cytoplasm (Fig. 6B). Nuclear rim staining was prominent in some wild-type cells. To test whether this staining corresponds to NPCs, we introduced the integrated GFP-CSE1 allele in rat2-1 mutants, which cluster their NPCs at one side of the nuclear envelope (35). Most cells of this strain indeed show a concentration of GFP-Cse1p at a small region of the nuclear envelope (Fig. 6B). The presence of Cse1p at the NPC was also shown by double-labeling immunofluorescence experiments. Here, the cells were prepared by methanol fixation, which results in a loss of most of the cytoplasmic content (63). Using the monoclonal antibody 414, which recognizes NPC antigens (17), and anti-GFP, we observed colocalization of NPCs and GFP-Cse1p in wild-type cells and rat2-1 cells (Fig. 7A).
FIG. 6.
In vivo localization of Cse1p. (A) Lysates from wild-type cells (lanes 1 and 4), cells additionally containing GFP-CSE1 (lanes 2 and 5), or cells containing only GFP-CSE1 (lanes 3 and 6) were analyzed by SDS gel electrophoresis and immunoblotting with affinity-purified anti-Cse1 antibodies (lanes 1 to 3) or anti-GFP antibodies (lanes 4 to 6). The sizes of molecular weight markers (in thousands) are indicated. (B) Wild-type (wt) or rat2-1 cells expressing GFP-Cse1p (GSY580 and GSY587, respectively) were grown at 25°C in liquid medium and viewed by fluorescence microscopy and by Nomarski optics.
FIG. 7.
Immunolocalization of Cse1p. (A) Wild-type (wt) or rat2-1 cells expressing GFP-Cse1p, grown at 25°C, were prepared for immunofluorescence microscopy by methanol fixation, probed with rabbit anti-GFP antibodies (Clontech) and mouse monoclonal antibody 414 (Berkeley Antibodies Co.), and then incubated with fluorescein isothiocyanate-conjugated anti-rabbit IgG and Texas Red–anti-mouse IgG secondary antibodies (Jackson) and stained with DAPI. (B) Wild-type cells expressing Cse1p or GFP-Cse1p were grown at 30°C and prepared for immunofluorescence microscopy by formaldehyde fixation. The cells were incubated with affinity-purified anti-Cse1 antibodies and then incubated with Texas Red–anti-rabbit IgG and stained with DAPI.
We also used affinity-purified anti-Cse1 antibodies to localize Cse1p and GFP-Cse1p in wild-type cells by immunofluorescence microscopy. In methanol-fixed cells, we observed punctate nuclear rim staining, indicating a nucleoporin-like localization (not shown). This staining was identical to the anti-GFP staining of GFP-Cse1p shown in Fig. 7A. In formaldehyde-fixed cells, with anti-GFP antibodies (not shown) or anti-Cse1 antibodies (Fig. 7B), both proteins gave cytoplasmic staining and a stronger labeling of the nucleus, but staining of the nuclear envelope was observed only occasionally. The localization of Cse1p in the cse1-1 mutant was similar to that of wild-type Cse1p, but the mutant protein was less nuclear (Fig. 7B).
Cse1p forms a complex with RanGTP and Srp1p.
The Srp1p export and NLS import defects of the cse1-1 mutant, the localization of Cse1p, the presence of an importin β-like Ran-binding domain at the N terminus of Cse1p (28), and the homology to CAS together suggest that Cse1p represents the specific nuclear export receptor of Srp1p. Human CAS was recently shown to mediate export of importin α in vitro and to form a trimeric complex with RanGTP and importin α (44) which probably represents the complex exported from the nucleus. For Kap95p, Yrb4p/Kap123p, and Pse1p, it was previously shown that binding to the GTP-bound form of Gsp1p (yeast Ran) blocks hydrolysis and exchange of the Gsp1p-bound GTP (21, 63). Therefore, we used the inhibition of Rna1p-induced GTPase activation as an assay to characterize interactions between Gsp1p, Cse1p, and Srp1p.
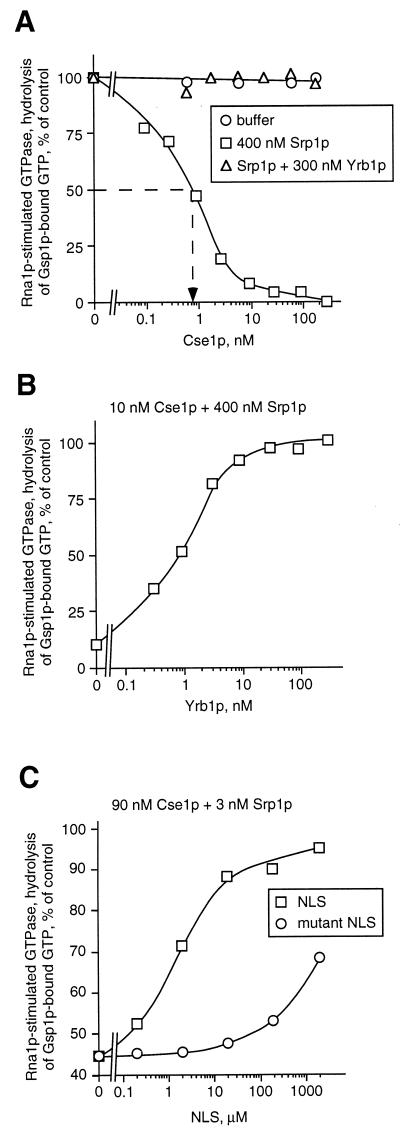
When Gsp1p loaded with [γ-32P]GTP was incubated with recombinant Cse1p, no GTPase inhibition was observed, indicating that the two proteins do not tightly interact under these conditions. However, in the presence of a high Srp1p concentration, Cse1p inhibited the GTPase activity of Gsp1p in a concentration-dependent manner (Fig. 8A). This inhibition was also observed when the Srp1p concentration was varied at a constant concentration of Cse1p (not shown). No GTPase block was obtained when Srp1p and Yrb1p were added simultaneously (see below). We conclude that a trimeric complex consisting of Gsp1p, Cse1p, and Srp1p was formed and that the binding of Gsp1pGTP and Srp1p to Cse1p is highly cooperative. We suggest a constant of approximately 1 nM for the dissociation of Gsp1pGTP from the Gsp1pGTP-Cse1p-Srp1p complex. In parallel experiments, the dissociation constant for a complex consisting of Srp1p, Cse1p, and human RanGTP was approximately 15 nM (not shown).
FIG. 8.
Gsp1pGTP complexed to Cse1p and Srp1p is resistant to Rna1p-induced GTP hydrolysis. (A) Srp1p is required for efficient binding of Gsp1pGTP to Cse1p. Gsp1p[γ-32P]GTP (50 pM) was incubated for 30 min with buffer, with Cse1p at the final concentrations indicated, or with Cse1p and Srp1p (400 nM). Yrb1p (300 nM) was then added to the indicated samples. After 1 min, the GTPase reaction was started by addition of Rna1p (20 nM). Hydrolysis of GTP by Gsp1p was determined as released [32P]phosphate after 2 min by the charcoal method. A 50% inhibition of GTP hydrolysis required 0.7 nM Cse1p (arrow). (B) Yrb1p dissociates the Cse1p-Gsp1pGTP-Srp1p complex in the presence of Rna1p. Gsp1p[γ-32P]GTP (50 pM) was incubated for 30 min with Cse1p (10 nM) and Srp1p (400 nM). Rna1p (20 nM) and then Yrb1p at the final concentrations indicated were added. Released [32P]phosphate was determined after further incubation for 2 min. (C) NLS peptides inhibit the formation of the trimeric complex. Gsp1p[γ-32P]GTP (50 pM) was preincubated for 30 min with Srp1p (3 nM) and peptides corresponding to the SV40 large-T-antigen NLS (CTPPKKKRKV) or mutant NLS peptides (CTPPKTKRKV) at the concentrations indicated. Cse1p (90 nM) was then added, and the reaction mixtures were further incubated for 5 min. The GTPase reaction was started by addition of Rna1p (20 nM). After 2 min, released [32P]phosphate was determined.
We have previously reported that Gsp1pGTP-Yrb4p and Gsp1pGTP-Pse1p complexes are disassembled by Yrb1p in the presence of Rna1p. Because Yrb1p and Rna1p are both cytoplasmic proteins, this was interpreted as a cytoplasmic event allowing recycling of the transport factors after export through the NPC (63). Figure 8B shows that Yrb1p also dissociates the Gsp1p-Cse1p-Srp1p complex in the presence of Rna1p. We incubated Gsp1p[γ-32P]GTP with concentrations of Cse1p and Srp1p allowing complex formation, which is indicated by complete GTPase inhibition. Subsequent addition of Yrb1p resulted in an efficient stimulation of GTP hydrolysis by Gsp1p. Since Cse1p (see below) and Yrb1p (63, 64) have no detectable affinity for Gsp1pGDP, Yrb1p will cause a complete disassembly of the complex when Rna1p is also present.
Since it can be expected that Srp1p will return to the cytoplasm after it has released the NLS substrate into the nucleus, we next investigated whether Srp1p, like the vertebrate importin α (44), preferentially binds to Cse1p in its NLS-free form. To test this, we preincubated Gsp1pGTP and Srp1p with synthetic peptides corresponding to the SV40 large-T-antigen NLS or a nonfunctional mutated NLS as a control (27). In this experiment, we used limiting concentrations of Srp1p. Addition of Cse1p resulted in complete GTPase inhibition in the absence of peptides. NLS peptides, however, prevented the GTPase block in a concentration-dependent manner by sequestering Srp1p (Fig. 8C). Relatively high concentrations of NLS peptides were needed to drive Srp1p quantitatively to the NLS-bound form. This might be due to the low NLS binding affinity of importin α in the absence of importin β (14, 30, 56). A 1,000-fold-higher concentration of mutant NLS peptides was required to achieve a similar extent of GTPase activation (Fig. 8C). We conclude that Cse1p preferentially binds to NLS-free Srp1p. The observed inhibition of the complex formation could also arise from direct binding of NLS peptides to Cse1p. We consider this very unlikely, since the mutant NLS has nearly no effect.
To investigate the formation of the trimeric Gsp1p-Cse1p-Srp1p complex in the absence of Rna1p, we performed solution binding assays. A fusion protein consisting of GST and Gsp1p was immobilized via the GST tag to glutathione-Sepharose. We then added Cse1p and/or Srp1p and tested for binding to Gsp1p. GST-Gsp1pGTP did not efficiently bind to either Cse1p or Srp1p alone (Fig. 9A, lanes 6 and 7). However, when Cse1p and Srp1p were added together, they both bound efficiently (Fig. 9A, lane 8). No binding to GST-Gsp1pGDP was observed (Fig. 9A, lane 9), indicating that complex formation is specific for the GTP-bound form of Gsp1p. Preincubation of Srp1p with NLS peptides strongly inhibited binding of Srp1p and Cse1p, whereas mutant NLS peptides did not disturb complex formation (Fig. 9A, lanes 10 and 11). The concentration of Cse1p in these assays was 18 μM (for comparison, we estimated that the concentration of Cse1p in the nuclei of living cells is approximately 12 μM). We then examined the effect of Yrb1p and Rna1p on the dissociation of the trimeric complex. To test this, Cse1p and Srp1p were prebound to immobilized GST-Gsp1pGTP, unbound proteins were removed by washing, and then Rna1p or Yrb1p was added. Remarkably, Yrb1p alone released the majority of Cse1p and Srp1p. As expected, Yrb1p itself bound efficiently to Gsp1pGTP (Fig. 9A, lane 13). Rna1p alone had no effect, which confirms that Gsp1p-bound GTP is inaccessible to the GTPase-activating protein (Fig. 9A, lane 15). When Yrb1p and Rna1p combined were added to the preformed complex, Cse1p and Srp1p were completely released. Yrb1p was also released, indicating that now GTP hydrolysis had occurred (Fig. 9A, lane 14). Very similar results were obtained when immobilized GST-Srp1p was used instead of GST-Gsp1p (Fig. 9B). Cse1p binding required simultaneous binding of Gsp1pGTP but not Gsp1pGDP. NLS peptides abolished trimer formation, and Yrb1p alone disassembled the complex. Complex dissociation by Yrb1p alone required roughly equimolar amounts (Fig. 9B, lane 9). However, in the presence of Rna1p, substoichiometric concentrations of Yrb1p induced complex disassembly (about 10-fold less Yrb1p than Gsp1p was present in lane 13), indicating that Yrb1p acts catalytically.
FIG. 9.
Formation and dissociation of the Cse1p-Gsp1pGTP-Srp1p complex. (A) GST-Gsp1pGTP or GST-Gsp1pGDP (3 μg) immobilized to glutathione-Sepharose was incubated with or without 6 μg of Cse1p and 4 μg of Srp1p in a total volume of 300 μl (lanes 5 to 15). Srp1p in lanes 10 and 11 was preincubated with NLS peptides or mutant (mut.) NLS peptides (0.45 mg/ml), respectively, for 30 min. The reaction mixtures were incubated for 60 min, and unbound material was removed by four washes. In lanes 12 to 15, the preformed Cse1p-Gsp1pGTP-Srp1p complex was reisolated and further incubated for 10 min in a volume of 1 ml with or without 2 μg of Yrb1p and 0.9 μg of Rna1p. Bound proteins were analyzed by SDS gel electrophoresis and Coomassie blue staining. Lanes 2 to 4 contain Cse1p, Srp1p, and Yrb1p, respectively, corresponding to 50% of the load. The sizes of molecular weight markers (lane 1) (in thousands) are indicated. (B) Immobilized GST-Srp1p (3 μg) was incubated with 6 μg of Cse1p or 3 μg of Gsp1p loaded with GTP or GDP and incubated as described for panel A. In lanes 8 to 15, the preformed Cse1p-Gsp1pGTP-Srp1p complex was further incubated for 20 min at 25°C with buffer, with 200 ng of Rna1p, or with the indicated amounts of Yrb1p. Bound proteins were analyzed by SDS gel electrophoresis and Coomassie blue staining. Lane 1, molecular weight markers.
DISCUSSION
The data presented suggest that Cse1p represents the specific export receptor of Srp1p. Our in vivo assays show that Srp1p but not Kap95p accumulates in the nuclei of cse1-1 mutants. Nuclear accumulation is accompanied by an inhibition of NLS-dependent nuclear import, suggesting that Srp1p becomes limiting for NLS import in the cytoplasm. Other nuclear import pathways (import of Npl3p and L25) proceed normally in cse1 mutants. This indicates that Srp1p is a specific transport substrate for Cse1p, but we do not know if other proteins are also exported by Cse1p. Overexpression of SRP1 suppresses the temperature sensitivity of the cse1-1 mutant, which demonstrates that the interaction with Srp1p is an essential function of CSE1. Cse1p is localized mainly in the nucleus but is also found in the cytoplasm and at the NPC. Cse1p belongs to the importin β superfamily and binds to RanGTP, and the interaction with Ran regulates substrate binding and release. Cse1p is the functional homologue of CAS, which mediates export of importin α in higher eukaryotes (44).
Cse1p binds to Srp1p and to RanGTP cooperatively; we did not detect significant binding to Srp1p or RanGTP alone. However, Cse1p was shown to bind to RanGTP in an overlay assay (28). Several observations indicate that the Srp1p-Cse1p-RanGTP complex is formed in the nucleus and is subsequently exported to the cytoplasm. Complex formation requires RanGTP but not RanGDP, i.e., conditions which are found in the nucleus. NLS-bound Srp1p does not bind to Cse1p and RanGTP, probably because NLS-free Srp1p and NLS-bound Srp1p differ in their conformational states. This indicates that the NLS protein is not exported by Srp1p and Cse1p after Srp1p-mediated import and thus explains the unidirectionality of NLS import (49, 66). Ran-bound GTP in the Srp1p-Cse1p-RanGTP complex is protected from hydrolysis. This prevents complex dissociation before the cytoplasmic environment is reached. The trimeric complex is disassembled by the cytoplasmic protein Yrb1p, which was shown to be essential for NLS import, Npl3p import, and mRNA export in vivo (64). Yrb1p and its mammalian homologue RanBP1 bind strongly to RanGTP and act as a coactivator of the Ran GTPase in vitro (7, 64). Accumulating evidence defines the main function of Yrb1p/RanBP1 as the universal cytoplasmic dissociation factor for complexes consisting of RanGTP and importin β-like receptors. Besides importin β and Cse1p, also RanBP5, RanBP7, RanBP8, Yrb4p/Kap123p, Pse1p, transportin, exportin t, and CAS complexed to RanGTP are targets for Yrb1p/RanBP1 (4, 18, 28, 44, 46, 63). We show that Yrb1p alone is sufficient for the disassembly of the Srp1p-Cse1p-RanGTP complex and that Yrb1p acts catalytically when Rna1p is also present. This indicates that Rna1p-mediated GTP hydrolysis serves to prevent reformation of the complex in the cytoplasm.
An inhibition of export of importin α from the nucleus was observed before in mutants other than cse1-1 mutants. Conditions that inhibit the Ran-specific GDP/GTP exchange factor also resulted in a nuclear accumulation of importin α (8, 42). This is explained by a decrease of the nuclear concentration of RanGTP, which impairs the formation of the trimeric export complex. Srp1p export was also inhibited in an RSL1/KAP95 mutant defective in nuclear export (38). We observed that overexpression of CSE1 blocks mRNA export but does not affect Kap95p export, Srp1p export, NES export, or several import pathways. However, overexpression of RSL1/KAP95 primarily inhibits several protein import pathways. This indicates that excess amounts of Kap95p might compete with other import receptors for NPC binding sites. It is unclear why CSE1 overexpression selectively inhibits mRNA export and not export of, e.g., Srp1p. The mRNA export pathway might be more sensitive to certain mutant conditions than other transport pathways. Interestingly, we did not detect nuclear accumulation of Srp1p in the rat2-1 mutant, which was identified in an mRNA mislocalization screen (35). This nucleoporin mutant and the RAT2/NUP120 deletion mutant show very similar phenotypes. However, nuclear accumulation of Srp1p was reported for the deletion mutant (2). This indicates that the RAT2/NUP120 gene product affects Srp1p export and mRNA export at different levels.
In cell lysates, about one-third of Srp1p was found in a stable complex with the shuttling but mainly nucleus-located cap binding complex (CBC), which consists of yCBP80 (Gcr3p) and yCBP20 (Mud13p). The strong Srp1p-CBC interaction is mediated by an NLS in CBP80 (31). This raises the question whether Srp1p is also exported from the nucleus complexed to CBC. Since only NLS-free Srp1p will be exported by Cse1p, a fraction of Srp1p could be exported by another pathway. U snRNA export from the nucleus to the cytoplasm is mediated by an interaction with CBC (41) and by the NES export receptor Crm1p (23). In contrast to export of Rev and U snRNA, however, importin α export is not affected by the potent Crm1 inhibitor leptomycin B in Xenopus oocytes (23). On the other hand, human importin α contains a sequence similar to a leucine-rich NES which functions as an export signal when fused to a reporter (8), but it is unclear whether Crm1p interacts with this NES-like signal. We observed complete nuclear accumulation of yeast importin α in cse1-1 mutants but did not detect even partial nuclear accumulation in xpo1-1 cells (shifted for up to 3 h to the nonpermissive temperature) which are mutated in yeast CRM1 (70). Furthermore, we did not detect binding of Srp1p to Xpo1p/Crm1p in the presence or absence of RanGTP (69). These observations together suggest that in contrast to CBC complexed to U snRNA, importin α is not exported by the CRM1 pathway.
Cse1p was reported to be involved in chromosome segregation during mitosis (77). We did not test whether Cse1p plays a role in spindle association (60) and/or chromosome segregation (77). Cell cycle defects, which were also observed for the srp1-31 mutant (47), can be explained by the failure to import certain proteins into the nucleus. Cse1p was also described to be involved in cyclin degradation. However, we found genetically that the mutant designated cse1-22 in this report (39) carries a temperature-sensitive mutation outside CSE1 (not shown).
Our in vivo and in vitro experiments show that Cse1p is the functional homologue of the human CAS protein, which mediates export of importin α from the nucleus in a reconstituted in vitro system with permeabilized cells and recombinant transport factors (44). Complex formation of Cse1p/CAS with RanGTP and NLS-free importin α as well as disassembly by Yrb1p/RanBP1 have been conserved during evolution. The major factors involved in the importin α transport cycle are importin β, Ran and its regulators, Cse1p/CAS, and a number of nucleoporins. NLS recognition by importin α starts in the cytoplasm. NLS-bound importin α is imported into the nucleus by importin β. RanGTP binds to importin β and thereby releases the importin α-NLS protein complex. It is still unknown how the NLS substrate dissociates from importin α. It seems that not phosphorylation, as previously proposed (3), but again Ran regulates this event. First, RanGTP-mediated release of importin β weakens the NLS-importin α interaction. Second, NLS-free importin α binds cooperatively to Cse1p/CAS and RanGTP, both of which are very abundant in the nucleus. Thus, the trimeric complex constitutes a trap for NLS-free importin α destined for export. In the cytoplasm, disassembly of the trimeric complex is induced by Yrb1p/RanBP1. Rna1p-mediated GTP hydrolysis then guarantees complete dissociation and allows importin α to enter the next import cycle.
ACKNOWLEDGMENTS
We thank Andrew Schroeder for strains, Wolfgang Nastainczyk for peptide synthesis, Dirk Görlich and Richard Zimmermann for helpful discussions, and Ellen Roth and Sandra Ruprecht for expert technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 2.Aitchison J D, Blobel G, Rout M P. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azuma Y, Takio K, Tabb M M, Vu L, Nomura M. Phosphorylation of Srp1p, the yeast nuclear localization signal receptor, in vitro and in vivo. Biochimie. 1997;79:247–259. doi: 10.1016/s0300-9084(97)83512-2. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff F R, Görlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase activating protein RanGap1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff F R, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boche I, Fanning E. Nucleocytoplasmic recycling of the nuclear localization signal receptor α subunit in vivo is dependent on a nuclear export signal, energy, and RCC1. J Cell Biol. 1997;139:313–325. doi: 10.1083/jcb.139.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossie M A, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc Natl Acad Sci USA. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi N C, Adam E J, Visser G D, Adam S A. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi N C, Adam E J H, Adam S A. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 14.Chi N C, Adam J H E, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett A H, Koepp D M, Schlenstedt G, Lee M S, Hopper A K, Silver P A. Rna1p, a Ran/TC4 GTPase activating protein is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbett A H, Silver P A. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis L I, Fink G R. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 1990;61:965–978. doi: 10.1016/0092-8674(90)90062-j. [DOI] [PubMed] [Google Scholar]
- 18.Deane R, Schäfer W, Zimmermann H P, Müller L, Görlich D, Prehn S, Ponstingl H, Bischoff F R. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-β but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeHoratius C, Silver P A. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:178–181. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 21.Floer M, Blobel G. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- 22.Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- 23.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 24.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 27.Goldfarb D S, Gariépy J, Schoolnik G, Kornberg R D. Synthetic peptides as nuclear localization signals. Nature. 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 28.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Görlich D, Henklein P, Laskey R A, Hartmann E. A 41 aa motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 30.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 31.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 32.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 33.Görlich D, Panté N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 34.Fink G R, editor. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–21. [PubMed] [Google Scholar]
- 35.Heath C V, Copeland C S, Amberg D C, de Priore V, Snyder M, Cole C N. Nuclear pore complex clustering and nuclear accumulation of poly(A)+RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopper A K, Schultz L D, Shapiro R A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 37.Hurt D J, Wang S S, Lin Y H, Hopper A K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iovine M K, Wente S R. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 40.Izaurralde E, Kutay U, Kobbe C v, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 42.Koepp D M, Wong D H, Corbett A H, Silver P A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Küssel P, Frasch M. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol Gen Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- 44.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 45.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 47.Loeb J D L, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G R. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lounsbury K M, Macara I G. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- 49.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 50.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 51.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 53.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 54.Pemberton L F, Rosenblum S R, Blobel G. A distinct parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 56.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 57.Rosenblum S R, Pemberton L F, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 59.Scherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scherf U, Pastan I, Willingham M C, Brinkmann U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc Natl Acad Sci USA. 1995;93:2670–2674. doi: 10.1073/pnas.93.7.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlenstedt G. Protein import into the nucleus. FEBS Lett. 1996;389:75–79. doi: 10.1016/0014-5793(96)00583-2. [DOI] [PubMed] [Google Scholar]
- 62.Schlenstedt G, Saavedra C, Loeb J D J, Cole C N, Silver P A. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff F R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlenstedt G, Wong D H, Koepp D M, Silver P A. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shulga N, Roberts P, Gu Z, Spitz L, Tabb M M, Nomura M, Goldfarb D S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1997;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solsbacher, J., and G. Schlenstedt. Unpublished data.
- 70.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 71.Taura T, Schlenstedt G, Silver P A. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 72.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 73.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 74.Weis K, Dingwall C, Lamond A I. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 75.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1α is essential for nuclear import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 76.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P A. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yano R, Oakes M, Tabb M M, Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]