Abstract
We have identified between Mex67p and Mtr2p a complex which is essential for mRNA export. This complex, either isolated from yeast or assembled in Escherichia coli, can bind in vitro to RNA through Mex67p. In vivo, Mex67p requires Mtr2p for association with the nuclear pores, which can be abolished by mutating either MEX67 or MTR2. In all cases, detachment of Mex67p from the pores into the cytoplasm correlates with a strong inhibition of mRNA export. At the nuclear pores, Nup85p represents one of the targets with which the Mex67p-Mtr2p complex interacts. Thus, Mex67p and Mtr2p constitute a novel mRNA export complex which can bind to RNA via Mex67p and which interacts with nuclear pores via Mtr2p.
Transport through nuclear pores requires concerted action between the structural components of the nuclear pore complex (NPC) and the soluble transport factors that bind to the transport substrates and shuttle between the nuclear and cytoplasmic compartments (for reviews, see references 2 and 31). Substantial progress toward an understanding of nuclear protein import has been achieved in the past few years, but very little is known about how RNA is exported from the nucleus into the cytoplasm. Among the factors required for nuclear protein import are the classical nuclear localization signal-receptor complex, consisting of importin/karyopherin α and β, the small GTPase Ran, and several Ran-binding proteins, as well as repeat sequences containing nucleoporins (for reviews, see references 5 and 11). Recently, additional routes of import into the nucleus were discovered, suggesting that major classes of transport substrates use different import pathways. Transportin and Kap123p were identified as novel transport factors that bind directly to their transport substrates, hnRNP protein A1 and ribosomal protein L25, respectively (30, 38). Transportin and Kap123p belong to a growing family of importin β-like proteins which have a Ran GTP-binding domain in their amino-terminal portions (5, 10). Recently, Mtr10p, which is also a member of this protein family, was shown to be the importin for yeast Np13p (34, 41). An essential role for Ran in energy-dependent nuclear protein import has been firmly established, but how Ran and the many Ran activity-modulating proteins participate in the actual translocation process is still controversial.
The Ran system is also involved in transport from the nucleus (9, 14, 18, 36). It has been firmly established that nuclear export sequences (NES), first identified in viral proteins such as human immunodeficiency virus Rev and protein kinase inhibitor, mediate the exit of proteins from the nucleus (for a review, see reference 8). For the Rev protein, which is an RNA-binding protein with a specificity for unspliced or partially spliced viral transcripts, viral mRNA is coexported through association with Rev (3). Initially, it was found that Rev NES interact with Rip (6, 47), which resembles repeat sequence-containing nucleoporins and accordingly was suggested to be a NES receptor at the nuclear pores. Recently, however, vertebrate CRM1 and its yeast homologue, Xpo1p, which also belong to the family of importin β-like proteins which have a Ran GTP-binding domain, were shown to be the receptors for leucine-rich NES signals (4, 7, 32, 46). Similarly, the export of importin α from the nucleus is mediated by another exportin, CAS (23).
The nuclear export of cellular RNA may proceed by a similar mechanism, which would mean that specific NES-containing RNA-binding proteins facilitate nuclear export of different classes of RNA. In fact, a NES called the M9 sequence (which surprisingly also acts like an NLS) was found in hnRNP A1 (27). It was shown that M9 mediates the shuttling of hnRNP A1 between the nucleus and the cytoplasm. Accordingly, it was suggested that hnRNP A1 is involved in the nuclear export of mRNA (17). NES signals which mediate nuclear export have also been found in other putative shuttling transport factors, such as Kap95 (16), RanBP1 (37), Gle1p (29), and Mex67p (40), and some of them are actually involved in mRNA export. Finally, Crm1p/Xpo1p not only may be involved in the nuclear export of NES-containing proteins but also could play a role in mRNA export (46).
Due to the lack of an in vitro system for RNA export, factors directly involved in RNA transport reactions have not been firmly ascribed. Different genetic approaches have identified proteins involved in mRNA export in the yeast Saccharomyces cerevisiae. Both through a genetic screen for mutants defective in poly(A)+ RNA export (1, 20) and by synthetic lethal screens for nucleoporin mutants, many factors involved in poly(A)+ RNA export have been found (2). However, it is yet not clear whether these proteins have a direct role in mRNA export or are pleiotropically linked to the mRNA export machinery. Among the many components, Nup159p (12), Mtr2p (19), Gle1p (29), and Mex67p (40) could play a direct role in the mRNA export process because conditionally lethal mutants exhibit a rapid and strong onset of the mRNA export defect. Furthermore, Npl3p, a yeast hnRNP protein which shuttles between the nucleus and the cytoplasm, has also been implicated in mRNA export (24).
We recently reported that the nuclear pore-associated protein Mex67p is required for nuclear mRNA export in yeast (40). We have now found that Mex67p forms a complex with Mtr2p. The Mex67p-Mtr2p complex, either isolated from yeast or reconstituted in vitro, binds directly to RNA via the Mex67p subunit. Thus, Mex67p and Mtr2p constitute an mRNA export complex which interacts with RNA and gains physical contact with the nuclear pores.
MATERIALS AND METHODS
Strains, plasmids, and microbiological techniques.
The yeast strains used in this work are listed in Table 1. Growth and transformation of yeast and Escherichia coli and yeast genetic procedures were as described earlier (40). The MTR2 gene was cloned as a 1.6-kb SalI/HindIII fragment into centromeric plasmids pRS316-URA3 and pRS315-LEU2 and high-copy-number 2μm plasmid YEP13-LEU2 (15) and as an SnabI/HindIII fragment into pBluescript (cut with SmaI/HindIII). The MEX67 gene was cloned as a 3.5-kb SacI fragment into centromeric plasmid pHT4467-ADE3-URA3 (40) and 2μm plasmid pRS424-URA3. The MEX67-GFP gene contained in a 4.2-kb SacI fragment was blunt ended and subcloned into centromeric vector pASZ11-ADE2 cut with SmaI. The dihydrofolate reductase (DHFR) open reading frame (ORF) contained in a 0.54-kb NdeI/BamHI fragment was subcloned into the PNOP1-ProtA-TEV cassette (13a). Other plasmids used in this study were pHT4467-ADE3-URA3-mex67-5 and pRS314-mex67-5 (40).
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| RS453 | mata/α ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 (38) |
| CH1462 | matα ade2 ade3 his3 leu2 ura3 (38) |
| MEX67-GFP | mata ade2 his3 leu2 trp1 ura3 mex67::HIS3(pUN100-LEU2-MEX67-GFP) (38) |
| mex67-5-GFP | mata ade2 his3 leu2 trp1 ura3 mex67::HIS3(pUN100-LEU2-mex67-5-GFP) (38) |
| MEX67 shuffle | mata or α ade2 his3 leu2 trp1 ura3 mex67::HIS3(pRS316-URA3-MEX67) (38) |
| RW+mex67-5 | matα ade2 ade3 his3 leu2 ura3 trp1 mex67::HIS3(pHT4467-URA3-ADE3-MEX67, pRS314-TRP1-mex67-5) |
| RW+mex67-nes1 | matα ade2 ade3 his3 leu2 ura3 trp1 mex67::HIS3(pHT4467-URA3-ADE3-MEX67, pRS314-TRP1-mex67-nes1) |
| sl190+mex67-5 | matα ade2 ade3 his3 leu2 ura3 trp1 mex67::HIS3 mtr2-190(pHT4467-URA3-ADE3-MEX67, pRS314-TRP1-mex67-5) |
| sl190+mex67-nes1 | matα ade2 ade3 his3 leu2 ura3 trp1 mex67::HIS3 mtr2-190(pHT4467-URA3-ADE3-MEX67, pRS314-TRP1-mex67-nes1) |
| mex67-5 | mata ade2 his3 leu2 ura3 trtp1 mex67::HIS3(pHT4467-URA3-mex67-5) (38) |
| MTR2 shuffle | mata or α ade2 his3 leu2 trp1 ura3 mtr2::HIS3(pRS316-URA3-MTR2) |
| GFP-MTR2 | mata or α ade2 his3 leu2 trp1 ura3 mtr2::HIS3(pRS315-LEU2-GFP-MTR2) |
| MTR2-GFP | mata or α ade2 his3 leu2 trp1 ura3 mtr2::HIS3(pRS315-LEU2-MTR2-GFP) |
| MEX67+/GFP-MTR2 | mata ade2 his3 leu2 trp1 ura3 mex67::HIS3 mtr2::HIS3(pRS316-MEX67, pRS315-GFP-MTR2) |
| mex67-5/GFP-MTR2 | mata ade2 his3 leu2 trp1 ura3 mex67::HIS3 mtr2::HIS3(pRS314-mex67-5, pRS315-GFP-MTR2) |
| MTR2-ProtA | mata ade2 his3 leu2 trp1 ura3 mtr2::HIS3(pRS316-LEU2-MTR2-ProtA) |
| mex67-5/mtr2-190 | mata ade2 his3 leu2 trp1 ura3 mex67::HIS3 mtr2::HIS3(pRS316-URA3-MTR2, pRS315-LEU2-mtr2-190, pRS314-TRP1-mex67-5) |
| nup85Δ/MTR2 shuffle | matα ade2 his3 leu2 trp1 ura3 mtr2::HIS3 nup85Δ::HIS3(pRS316-URA3-MTR2) |
| mtr2-26 | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3(pRS315-mtr2-26) |
| mtr2-26/MEX67-GFP | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3 mex67::HIS3(pRS315-mtr2-26, pASZ11-ADE2-MEX67-GFP) |
| mtr2-9 | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3(pRS315-mtr2-9) |
| mtr2-9/MEX67-GFP | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3 mex67::HIS3(pRS315-mtr2-9, pASZ11-ADE2-MEX67-GFP) |
| GFP-mtr2-9 | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3(pRS315-GFP-mtr2-9) |
| mtr2-21 | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3(pRS315-mtr2-21) |
| mtr2-21/MEX67-GFP | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3 mex67::HIS3(pRS315-mtr2-21, pASZ11-ADE2-MEX67-GFP) |
| GFP-mtr2-21 | mata ura3 ade2 leu2 his3 trp1 mtr2::HIS3(pRS315-GFP-mtr2-21) |
Isolation of MTR2 as a high-copy-number suppressor and in a synthetic lethal screen with thermosensitive mex67-5.
A yeast genomic library in 2μm plasmid YEP13-LEU2 (15) was used to transform a thermosensitive mex67-5 mutant. Transformants were incubated for 8 h at 30°C before plates were shifted to 35°C. In total, about 25,000 transformants were screened. After 2 days, 78 suppressor colonies were picked from the 35°C plates and restreaked on SDC-leu plates at 37°C. Colonies which could also grow at 37°C were analyzed for growth on 5-fluoro-orotic acid (FOA)-containing plates at 30°C. Only colonies not able to grow on FOA may contain an extragenic suppressor of thermosensitive mex67-5. From such suppressor colonies, YEP13 plasmids were recovered. After retransformation into the mex67-5 strain to confirm the complementation at 37°C, the inserts were partly sequenced from both ends. The complementing activity within the genomic inserts was restricted to a single gene by subcloning. For the synthetic lethal screen, a sector-forming strain, RW+mex67-5, was generated (Table 1). UV mutagenesis and isolation of synthetically lethal mutants, including all the tests for specificity, were carried out as recently described (40, 48). For recovery of the mtr2-190 allele, a 2.2-kb SalI/XbaI fragment containing the MTR2 gene was inserted into pRS315. This plasmid was digested with SnabI/BamHI to release the entire MTR2 ORF plus 162 bp upstream of the ATG codon and 147 bp downstream of the stop codon. The isolated linearized plasmid, which contained 5′ (210 bp) and 3′ (221 bp) noncoding sequences of MTR2, was used to transform the sl190 mutant. From the obtained Leu+ transformants, total DNA was prepared, and gap-repaired plasmid pRS315-mtr2-190 was recovered upon transformation of E. coli. mtr2-190 and MTR2 were sequenced.
Construction of MTR2 fusion genes and an MTR2 gene disruption.
Two immunoglobulin G (IgG)-binding domains or the GFP gene (21, 42) was used for the tagging of Mtr2p as previously described (45). To do so, a new BglII restriction site was introduced before the stop codon (in boldface) of MTR2 (AGATCTTAGTGGGAAGATTCC), and a BamHI fragment encoding the protein A (ProtA), ProtA-tobacco edge virus protein site (TEV), or green fluorescent protein (GFP) tag was inserted into this restriction site (40). MTR2-ProtA, MTR2-TEV-ProtA, or MTR2-GFP was then cloned into vector pRS315-LEU2. MTR2 was also tagged with GFP at its amino-terminal end by subcloning of the 0.5-kb XhoI/HindIII fragment containing the MTR2 ORF into the PNOP1-GFP cassette (13a) to yield plasmid pRS315-PNOP1-GFP-MTR2. Mtr2p-GFP but not GFP-Mtr2p in combination with the thermosensitive mex67-5 allele gave synthetic lethality at 30°C (data not shown). The thermosensitive mtr2-26, mtr2-9, and mtr2-21 alleles were also tagged with GFP by subcloning of the corresponding 0.5-kb XhoI/HindIII fragments into the PNOP1-GFP cassette.
For disruption of the MTR2 gene, pBluescript-MTR2 was cut with PstI, which removes an internal 298-bp fragment from the MTR2 gene. The HIS3 gene, isolated as a blunt-ended BamHI fragment, was then inserted into this plasmid, also blunt ended. pBluescript-mtr2::HIS3 was cut with SalI/NotI to release mtr2::HIS3, which was used to transform the diploid strain RS453. Heterozygous His+ transformants were analyzed for correct integration at the MTR2 locus by PCR-Southern analysis and tetrad analysis. A 2:2 segregation for viability was found, confirming earlier data indicating that MTR2 is an essential gene (19).
Isolation of thermosensitive mtr2 mutant alleles.
A collection of thermosensitive mutant alleles of MTR2 was generated as described previously (28). Primers 5′GCAGCCGGTTGGGTGG3′ and 5′GGTGCGAAGCCCTAC3′ were used to amplify the MTR2 gene by PCR under suboptimal conditions (6.5 mM MgCl2, 0.5 mM MnCl2, dGTP, dCTP, and dTTP [1 mM each]; dATP [0.2 mM]; 1 μg of template DNA; 5 U of Taq polymerase). Vector pRS315-MTR2 was digested with SnaBI/BamHI to release the MTR2 ORF, leaving 210 nucleotides 5′ upstream and 221 bp 3′ downstream of the MTR2 ORF which were homologous to both ends of the PCR product. Five micrograms of linearized vector and 10 μg of PCR product were used to transform strain MTR2 shuffle. A total of 5,000 transformants were replated on FOA plates and incubated at 30°C for 4 days. The killing rate on FOA was 25%. Surviving Ura− colonies were tested at 30 and 37°C for a thermosensitive phenotype. A total of 10 thermosensitive alleles were isolated. Plasmids containing thermosensitive mtr2 mutant alleles were recovered from yeast strains as described previously (40).
Expression and localization of GFP-Mtr2p, Mex67p-GFP, GFP–Mtr2-9p, and GFP–mtr2-21p.
The in vivo locations of Mtr2p, Mex67p, and mutant Mtr2-9p and Mtr2-21p proteins were analyzed with strains expressing the corresponding GFP fusion proteins plus the ADE2 gene (plasmid pASZ11-ADE2) as described previously (40). The cells were examined in the fluorescein channel of a Zeiss Axioskop fluorescence microscope. Pictures were taken with a Xillix Microimager charge-coupled device camera. In some cases, digital pictures were further processed by digital confocal imaging by use of the software Openlab (Improvision, Coventry, United Kingdom).
Affinity purification of Mtr2p-TEV-ProtA.
Affinity purification of Mtr2p-TEV-ProtA by IgG-Sepharose chromatography was done as described earlier (45) with modifications and elution from the IgG-Sepharose column with recombinant TEV protease (Life Technologies, Berlin, Germany; Catalog no. 10127-017) as described previously (41). A whole-cell extract was prepared from 4.5 g of yeast spheroplasts, lysed in 20 mM HEPES (pH 7.4)–100 mM potassium acetate–2 mM magnesium acetate–0.5% Tween 20 (lysis buffer), loaded onto a column containing 300 μl of IgG-Sepharose beads (Pharmacia, Freiburg, Germany), equilibrated with lysis buffer, washed with 15 ml of lysis buffer, and incubated at 16°C for 1 h with 50 U of TEV protease in 300 μl of cleavage buffer (20 mM HEPES [pH 7.4], 100 mM potassium acetate, 1 mM dithiothreitol [DTT], 0.1% Tween 20, 0.5 mM EDTA). Elution was performed by transferring the whole mixture onto a spin column. The eluate (25 μl) was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and then by Western blotting. The purification of ProtA-TEV-DHFR was performed in parallel under identical conditions and with the same amount of cells.
Expression and purification of Mex67p and Mtr2p from E. coli.
The MTR2 ORF was cloned into vector pET8c-His6 (39, 43). A recombinant six-histidine (His6)-tagged protein, His6-Mtr2p, was overexpressed in E. coli BL21 cells and purified with Ni-nitrilotriacetic acid (NTA)–agarose as previously described (44). Recombinant Mtr2p was purified through a MonoQ column and injected into rabbits. From the immune serum, antibodies were affinity purified by use of nitrocellulose strips containing recombinant Mtr2p antigen (15). The construction of pET8c-His6-MEX67 was described earlier (40). Recombinant Mex67p without the His6 tag was overexpressed in E. coli by inserting the entire MEX67 ORF into pET9d (Novagen, Madison, Wis.). For coexpression in E. coli, plasmids pET8c-His6-MTR2 and pET9d-MEX67 (full length) were cotransformed into BL21 cells. Protein complexes were purified in a manner similar to that of single proteins (44).
The entire MTR2 ORF was also cloned into plasmid pPROEX1 (Life Technologies), which contains the Trc promoter, ribosome-binding sites, His6 sequences, and the recombinant TEV protease cleavage site. pPROEX1-MTR2 was cotransformed with pET9d-MEX67 into BL21 cells. Expression and purification of the His6-TEV-Mtr2p-Mex67p complex by Ni-NTA affinity chromatography were done as described above. However, the complex was eluted by incubation of the Ni-NTA–agarose beads with recombinant TEV protease as recommended in the manufacturer’s instructions. The eluted Mtr2p-Mex67p complex was further purified by fast protein liquid chromatography (FPLC) on a MonoS column.
In vitro binding to homopolymeric RNA.
For the in vitro RNA-binding assay, E. coli extracts containing Mex67p or His6-tagged Mtr2p were incubated with the homoribopolymers poly(A), poly(U), poly(G), and poly(C) bound to agarose beads (Sigma, Munich, Germany). Ten microliters of E. coli lysate was diluted in 150 μl of 100 mM NaCl or 500 mM NaCl–2.5 mM MgCl2–10 mM Tris-HCl (pH 7.5)–0.8% Triton X-100 before 20 μl of the homoribopolymer-bead mixture was added. The mixture was rotated on a wheel for 60 min at 4°C. The beads were recovered by centrifugation and washed three times with 0.5 ml of buffer before bound proteins were eluted by boiling in 20 μl of SDS sample buffer.
RNA band shift and UV-cross-linking assays.
RNA probes were produced as follows. KS-RNA (a 77-mer) was produced by T7 polymerase runoff transcription of the polylinker area of the HindIII-linearized pBluescript KS vector in the presence of [α-32P]CTP. For the production of homopolymers, a DNA 40-mer [poly(dA) or poly(dG)] was inserted between the NotI and SacI sites in the polylinker of pBluescript KS. A vector containing poly(dC) between the same sites was a gift from D. Ostareck (European Molecular Biology Laboratory). The vectors were linearized with NotI and transcribed with T3 polymerase in the presence of the corresponding α-32P-labeled nucleoside triphosphate. The resulting RNA 62-mers (each containing a stretch of 40 identical nucleotides) were purified by gel electrophoresis. For the band shift assay, purified recombinant Mex67p-Mtr2p complex or Mtr2p alone (0.5 to 1 μg) was incubated for 30 min at 30°C with 3 ng of 32P-labeled KS-RNA (40,000 cpm/ng) or 1 ng of 32P-labeled poly(A)-, poly(G)-, or poly(C)-containing RNA (100,000 cpm/ng) in 20 μl of 20 mM HEPES (pH 7.5)–7.5 mM MgCl2–150 mM NaCl–0.05 mg of bovine serum albumin per ml–10% glycerol–0.2 U of RNasin (Promega) per ml; RNA-protein complexes were detected by electrophoresis on 5% nondenaturing polyacrylamide–0.5× Tris-borate-EDTA gels (120 V, 2.5 h) and then autoradiography. In vitro-transcribed tRNAMet, used for the competition experiment, was produced as previously described (43). For the same experiment, a synthetic poly(dA) 40-mer was used as single-stranded DNA. This was hybridized to a poly(dT) 40-mer in order to obtain double-stranded DNA. To analyze RNA binding by UV cross-linking, the Mex67p-Mtr2p complex isolated from yeast via MTR2-ProtA and bound to IgG-Sepharose beads was mixed with in vitro-transcribed 32P-labeled KS-RNA or homopolymeric RNA for 20 min at 30°C and UV irradiated for 10 min on ice in a UV Stratalinker 1800 (Stratagene); 10 U of RNase T1, 15 μg of RNase A, and 10 U of RNase I were added, and the RNA was digested for 30 min at 37°C before the addition of SDS sample buffer (with or without DTT and boiling), SDS-PAGE, and autoradiography.
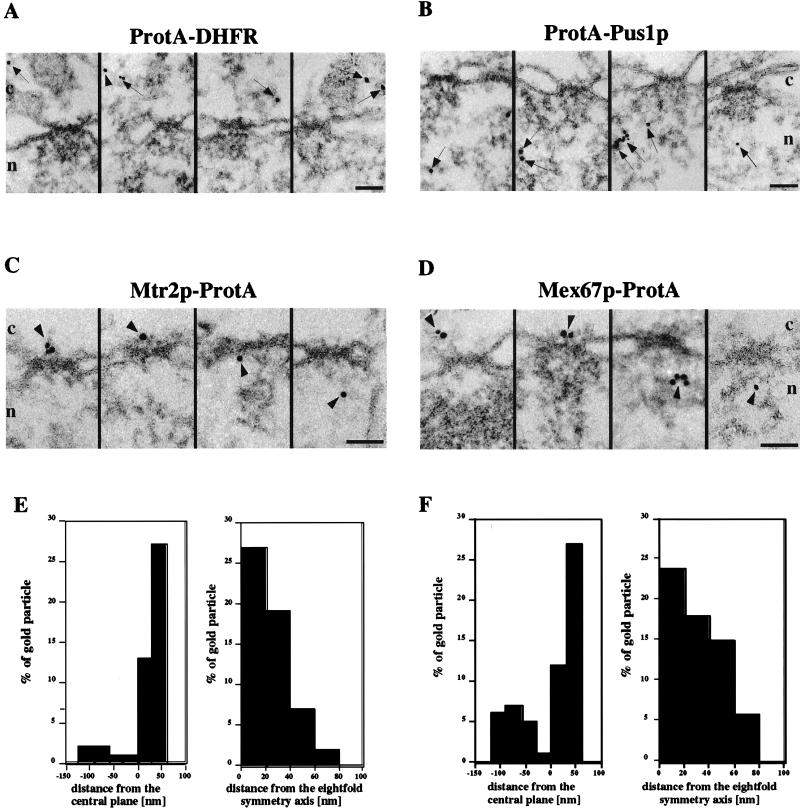
Immunoelectron microscopy.
The preparation of cells for preembedding immunogold labeling is described in detail elsewhere (2a). Briefly, Mex67p-ProtA and MTR2p-ProtA cells grown to an optical density at 600 nm of 0.51 in glucose-containing medium were prefixed with 2% paraformaldehyde before the cell wall was removed with 5 U of Zymolyase 20T (Seikagaku Corporation, Tokyo, Japan) per ml. To allow access of the antibody to both sides of the nuclear envelope, cells were extracted with 0.05% Triton X-100 and incubated with a polyclonal anti-ProtA antibody (Sigma, St. Louis, Mo.) conjugated to 8-nm colloidal gold particles (33). Next, samples were fixed with 2% glutaraldehyde, postfixed with 1% OsO4, dehydrated, and embedded in Epon 812. For controls, strains expressing cytosolic mouse DHFR tagged with ProtA (ProtA-DHFR) and Pus1p, a nuclear protein involved in tRNA biogenesis, tagged with ProtA (ProtA-Pus1p) (43) were used.
For quantitation, postembedding gold labeling was performed. The Triton X-100 extraction step was omitted, cells were embedded in LR White resin (Polyscience Ltd., Northampton, United Kingdom), and sections were immunolabeled with the polyclonal anti-ProtA antibody followed by gold-conjugated goat anti-rabbit IgG (Aurion, Wageningen, Netherlands). Quantitation was performed by a stereological counting method (25).
Miscellaneous methods.
DNA recombinant work (restriction analysis, end filling, ligation, PCR amplification, and DNA sequencing) was performed as described previously (26). SDS-PAGE and Western blotting were done as described by Siniossoglou et al. (45). mRNA in situ hybridization was performed as described by Segref et al. (40). Affinity purification of the Mtr2p-ProtA fusion protein by IgG-Sepharose chromatography was performed as described previously (45).
Cycloheximide (final concentration, 10 μg/ml) was added to exponentially growing yeast cells 10 min before the shift to 37°C. After 10 min of exposure to the restrictive temperature, cells were resuspended in 1 ml of minimal medium containing 10 μg of cycloheximide per ml and incubated at 26°C for 10, 20, and 45 min.
RESULTS
Identification of MTR2 as a high-copy-number suppressor and in a synthetic lethal screen with thermosensitive mex67-5.
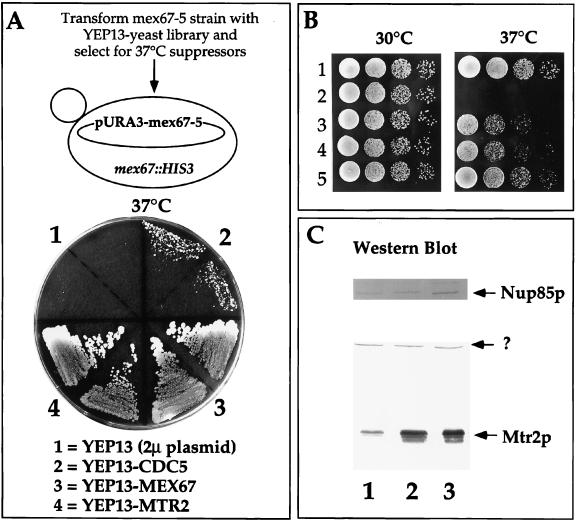
The thermosensitive mex67-5 mutant stops cell growth and accumulates poly(A)+ RNA inside the nucleus shortly after a shift to the restrictive temperature (40). Concomitantly, the mutant Mex67-5p protein dissociates from nuclear pores and mislocalizes to the cytoplasm. To search for Mex67p-interacting proteins involved in NPC association, we screened for high-copy-number suppressors of the thermosensitive mex67-5 allele. Two suppressors, MTR2 and CDC5, were found (Fig. 1A). MTR2 was recently isolated in another genetic screen for poly(A)+ RNA export mutants (19). CDC5 encodes a mitotic kinase frequently found in suppressor screens (22). MTR2 also suppressed the thermosensitive mex67-5 allele when inserted into a low-copy-number plasmid (Fig. 1B). Similar extents of overexpression of Mtr2p were observed with both low- and high-copy-number MTR2-containing plasmids (Fig. 1C).
FIG. 1.
Genetic interaction between MEX67 and MTR2. (A) Isolation of the MTR2 gene as a high-copy-number suppressor of thermosensitive mex67-5. A YPD plate contained the thermosensitive mex67-5 strain transformed with the indicated high-copy-number 2μm (2μ) plasmids and grown for 3 days at 37°C. Note that YEP13-CDC5 does not complement the growth defect of thermosensitive mex67-5 as well as does YEP13-MTR2 at 37°C. (B) Growth of MEX67+ and mex67-5 cells transformed with various plasmids at 30 and 37°C. Rows: 1, haploid RS453; 2 to 5, thermosensitive mex67-5 cells transformed with low-copy-number plasmids pRS316 (2) and pRS316-MTR2 (3) and high-copy-number plasmids YEP13-MTR2 (4) and pRS316-MEX67 (5). (C) Expression of Mtr2p in mex67-5 yeast strains. Whole-cell SDS lysates from mex67-5 cells transformed with low-copy-number plasmids pRS316 (lane 1) and pRS316-MTR2 (lane 2) and high-copy-number plasmid YEP13-MTR2 (lane 3) were analyzed by Western blotting with anti-Mtr2p and anti-Nup85p antibodies. The band marked by a question mark was cross-reactive with anti-Mtr2p antibodies.
In addition, a synthetic lethal screen was performed with the thermosensitive mex67-5 allele. One of the synthetically lethal mutants obtained, sl190, was complemented by MTR2 (data not shown). The mtr2-190 allele carries a single nucleotide exchange giving rise to a leucine (position 140)→ proline mutation within Mtr2p. This mutation does not cause any growth defect by itself but causes lethality when combined with mex67-5. Thus, using two different genetic approaches, we identified MTR2 as a component genetically interacting with MEX67.
Mtr2p forms a complex with Mex67p.
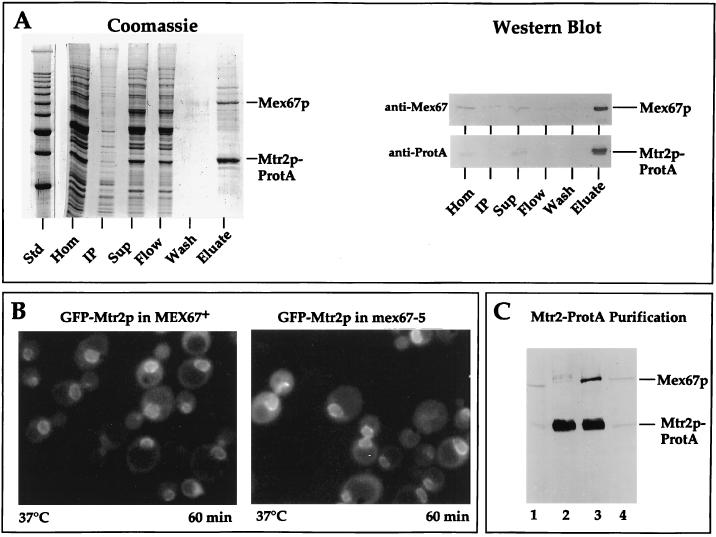
The strong genetic interaction between MTR2 and MEX67 suggested that both encoded proteins physically associate. To facilitate biochemical purification, Mtr2p was tagged at its carboxy-terminal end with two IgG-binding domains derived from Staphylococcus aureus ProtA. The Mtr2p-ProtA fusion protein was functional, since it complemented the otherwise nonviable mtr2::HIS3 null mutant (data not shown). From this complemented strain, Mtr2p-ProtA was affinity purified under nondenaturing conditions by IgG-Sepharose chromatography (Fig. 2A). A prominent band which migrated at 70 kDa copurified with Mtr2p-ProtA and was identified as Mex67p by Western blotting (Fig. 2A, anti-Mex67p) and mass spectrometry (data not shown). Less prominent bands are currently under investigation. Mtr2p and Mex67p thus physically associate in living cells.
FIG. 2.
Mtr2p and Mex67p form a complex at nuclear pores. (A) Affinity purification of Mtr2p-ProtA by IgG-Sepharose chromatography. A cell homogenate (Hom), an insoluble pellet (IP), a soluble supernatant (Sup), the flowthrough (Flow), a pH 5 wash fraction (Wash), and a 300-fold equivalent of the eluted complex (Eluate) were analyzed by SDS-PAGE followed by Coomassie blue staining or by Western blotting with IgG coupled to horseradish peroxidase to detect the ProtA moiety (anti-ProtA) or with anti-Mex67p antibodies (anti-Mex67). The positions of Mex67p and Mtr2p-ProtA are shown. Std, marker proteins (10-kDa ladder with a stronger 50-kDa band). (B) Nuclear envelope location of GFP-Mtr2p in MEX67+ and thermosensitive mex67-5 cells as revealed by fluorescence microscopy. MEX67/GFP-MTR2 and mex67-5/GFP-MTR2 cells (Table 1) were shifted for 60 min to 37°C before pictures were taken. Under the same conditions, thermosensitive cells expressing Mex67-5–GFP showed cytoplasmic mislocalization (40). (C) Affinity purification of Mtr2p-ProtA from thermosensitive mex67-5 (lanes 1 and 2) and MEX67+ (lanes 3 and 4) cells grown at 30°C by IgG-Sepharose chromatography. A whole-cell lysate (lanes 1 and 4) and the eluate fraction from the IgG-Sepharose column (lanes 2 and 3) were analyzed by SDS-PAGE and Western blotting with antibodies against Mtr2p-ProtA and Mex67p.
Mtr2p is a nuclear pore-associated protein which binds nuclear pores independently of Mex67p.
As shown above, Mtr2p forms a complex with Mex67p. We therefore analyzed whether Mtr2p is also a nuclear pore-associated protein under steady-state conditions. The location of Mtr2p was analyzed by fluorescence light microscopy with a GFP-tagged version of Mtr2p which can complement an mtr2 mutant strain. Like Mex67p-GFP, GFP-Mtr2p exhibited a punctate nuclear envelope location in living cells, with signals in the nucleoplasm and cytoplasm as well (Fig. 2B). In addition, GFP-Mtr2p clustered together with the NPC in nup133 mutant cells, arguing that Mtr2p is physically associated with nuclear pores (data not shown).
Next, we analyzed whether the NPC location of tagged Mtr2p is altered in thermosensitive mex67-5 cells. Mutant Mex67-5p–GFP dissociates from nuclear pores at 37°C and is located in dot-like structures throughout the cytoplasm (40). When the location of GFP-Mtr2p was tested in the thermosensitive mex67-5 mutant shifted to the restrictive temperature, GFP-Mtr2p still was associated with the nuclear envelope (Fig. 2B). This result shows that upon dissociation of mutant Mex67-5p into the cytoplasm at the nonpermissive temperature. Mtr2p remains bound to the NPC. Thus, Mtr2p can localize to the NPC independently of Mex67p.
Since mutant Mex67-5p and GFP-Mtr2p are found, respectively, in the cytoplasm and at the nuclear pores at the nonpermissive temperature, Mex67p and Mtr2p may separate under these conditions. To show this idea biochemically, Mtr2p-ProtA was purified from either mex67-5 mutant or wild-type strains (Fig. 2C). Mutant Mex67-5p no longer copurified with Mtr2p-ProtA, demonstrating that the physical interaction between Mtr2p and mutant Mex67-5p was impaired. This lack of interaction was not due to increased proteolytic instability of the mutant protein, since in whole-cell extracts both wild-type Mex67p and mutant Mex67-5p were present in comparable amounts (Fig. 2C, lanes 1 and 4).
mtr2 mutant alleles affect the association of Mtr2p with Mex67p and nuclear pores.
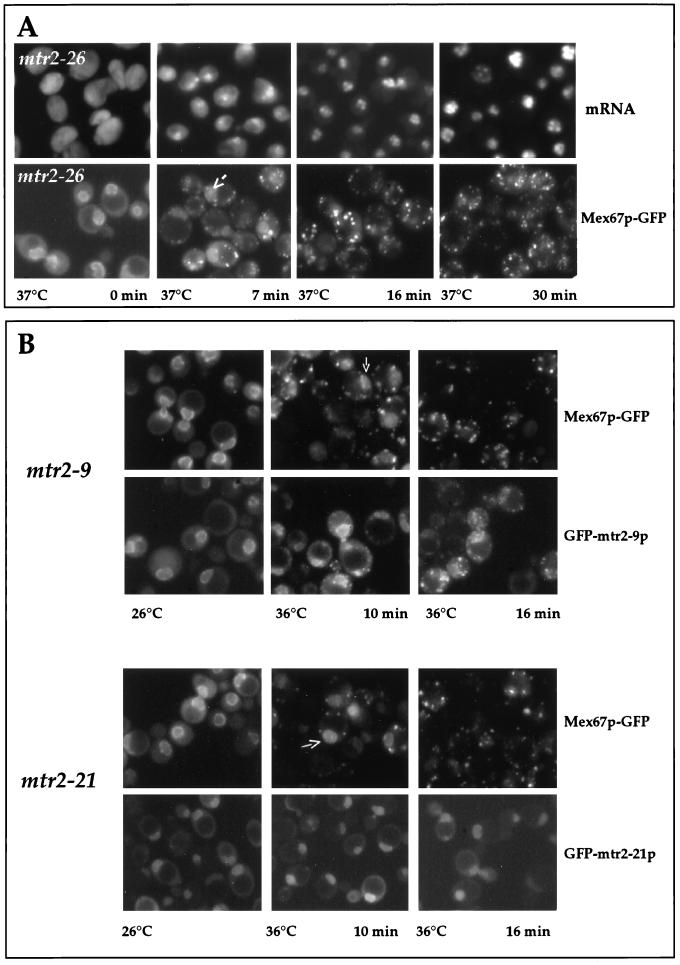
The lack of association of Mex67-5p with Mtr2p, taken together with the mislocalization of Mex67-5p to the cytoplasm, could suggest that Mtr2p is involved in the binding of Mex67p to the nuclear pores. To verify this idea with mtr2 mutants as well, a collection of thermosensitive mtr2 alleles was generated by mutagenic PCR and analyzed for their phenotypic defects (Table 2). All mtr2 mutants obtained exhibited inhibition of nuclear mRNA export at the restrictive temperature, as shown for the mtr2-26 mutant, which started to accumulate poly(A)+ inside the nucleus after a 7-min shift to 37°C (Fig. 3A). Concomitantly, GFP-tagged wild-type Mex67p, when expressed in all of the mtr2 mutant strains, dissociated from the nuclear envelope and accumulated in numerous cytoplasmic foci (Fig. 3). These data show that intact Mex67p can no longer associate with the nuclear pores and mislocalizes to the cytoplasm when Mtr2p is mutated. Furthermore, the extent of cytoplasmic mislocalization of Mex67p correlates with the inhibition of mRNA export. Interestingly, during the early onset of Mex67p release from the nuclear envelope, the intranuclear pool of Mex67p-GFP became more evident in a number of cells (Fig. 3, arrows). The cytoplasmic mislocalization of Mex67p-GFP was reversible, because Mex67p-GFP was retargeted to the nuclear envelope when thermosensitive mtr2 cells were brought back from the restrictive to the permissive temperature (data not shown). Thus, cytoplasmic foci containing Mex67p-GFP do not represent irreversible aggregates.
TABLE 2.
Analysis of mtr2 mutant alleles
| Genotype | mRNA export defecta | Location of:
|
Complemention by MEX67d | Synthetic lethality with nup85Δe | Complemention by NUP85f | |
|---|---|---|---|---|---|---|
| Mtr2pb | Mex67pc | |||||
| MTR2 | − | NPC | NPC | |||
| mtr2-9 | + | Cytoplasm | Cytoplasm | − | + | + |
| mtr2-26 | + | ND | Cytoplasm | − | + | + |
| mtr2-21 | + | NPC | Cytoplasm | + | − | − |
Analyzed after 30 min at 37°C. −, no defect; +, defect.
Wild-type or mutant Mtr2p was localized as a fusion protein with GFP after 16 min at 36°C. ND, not determined.
Wild-type Mex67p was localized as a fusion protein with GFP after 16 min at 36°C.
Analyzed at 37°C. −, no complementation; +, complementation.
−, no synthetic lethality; +, synthetic lethality.
Analyzed at 33°C. −, no complementation; +, complementation.
FIG. 3.
Nuclear mRNA export and localization of GFP-tagged Mtr2p and Mex67p in mtr2 mutant cells. (A) Correlation between inhibition of mRNA export and cytoplasmic mislocalization of Mex67p-GFP in mtr2-26 cells. mtr2-26 cells were grown at 26°C before a shift to 37°C. Samples were taken at the indicated times and analyzed for mRNA export defects [by in situ hybridization with a Cy3-labeled oligo(dT) probe] and for Mex67p-GFP mislocalization. (B) Localization of Mex67p-GFP and GFP-Mtr2p mutant proteins in living cells. mtr2-9/MEX67-GFP, GFP-mtr2-9, mtr2-21/MEX67-GFP, and GFP-mtr2-21 cells (Table 1) were grown at 26°C before a shift to 36°C for 0, 10, and 16 min. The corresponding GFP-tagged proteins were localized in the cells by fluorescence microscopy.
To address the location of mutant Mtr2p in living cells, several of the corresponding thermosensitive alleles were tagged with GFP. Except for Mtr2-26p, all the other mutant Mtr2p-GFP fusion proteins were functional, could complement the mtr2 null strain at permissive temperatures, and exhibited a thermosensitive phenotype at 37°C. Two different phenotypes were observed in this collection of thermosensitive mtr2 mutants (Fig. 3B and Table 2). In one class, represented by mtr2-9, both mutant Mtr2-9p and Mex67p mislocalized to cytoplasmic foci at the restrictive temperature (Fig. 3B and Table 2). However, biochemical analysis showed that Mtr2-9p–ProtA did not copurify with Mex67p (data not shown). Therefore, it remains to be determined whether Mtr2-9p still forms with Mex67p in vivo a complex which mislocalizes to the cytoplasm but is unstable during biochemical purification or whether Mtr2-9p and Mex67p dissociate from both the nuclear pores and each other.
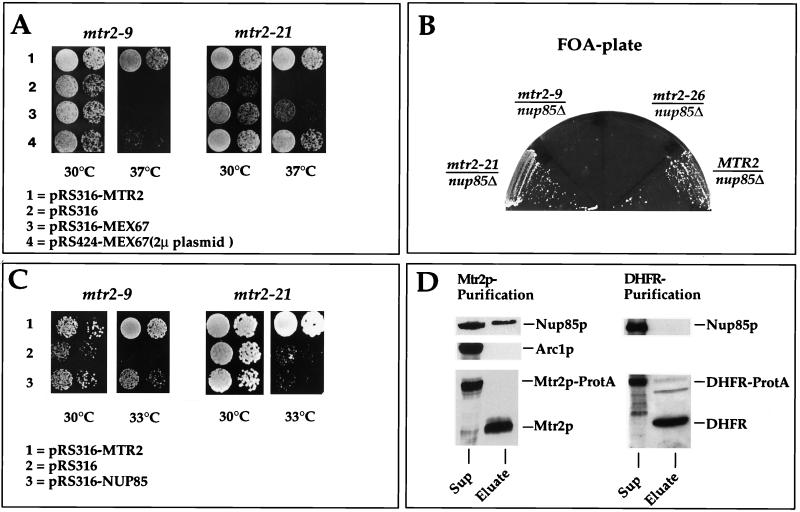
In the second class of mutants, represented by mtr2-21, an intranuclear location of mutant Mtr2-21p was more prominent than a nuclear envelope location at both the permissive and the restrictive temperatures (Fig. 3B). After a shift to the restrictive temperature, mutant Mtr2-21p did not mislocalize to cytoplasmic foci, whereas Mex67p-GFP no longer attached to the pores and accumulated in cytoplasmic foci (Fig. 3B and Table 2). This result suggests that the interaction between Mex67p and Mtr2-21p was impaired, so that Mex67p dissociated into the cytoplasm but Mtr2-21p remained restricted to the nuclear compartment. Biochemical purification of Mtr2-21p–ProtA also showed a lack of association between Mex67p and Mtr2-21p (data not shown). In agreement with the in vivo location data, we observed that the overexpression of Mex67p was not sufficient to restore the growth defect of mtr2-9 cells at the restrictive temperature but efficiently complemented the thermosensitive phenotype of mtr2-21 cells (Fig. 4A and Table 2).
FIG. 4.
Gentic and physical interactions between Mtr2p and Nup85p. (A) Suppression of the thermosensitive mtr2-9 and mtr2-21 growth defect at 37°C by overexpression of MEX67. Precultures were diluted in growth medium, and equivalent amounts of cells (diluted in 10−1 steps) were spotted onto YPD plates. Plates were incubated for 3 days. (B) Genetic interaction between thermosensitive nup85Δ and mtr2 alleles. Strain nup85Δ/MTR2 shuffle (Table 1) was transformed with plasmids containing the MTR2, mtr2-26, mtr2-9, and mtr2-21 genes. The synthetic lethal relationship between the nup85Δ and mtr2 alleles was tested by streaking transformants on FOA-containing plates. Growth inhibition on FOA plates indicates synthetic lethality. (C) Complementation of the thermosensitive mtr2-9 and mtr2-21 growth defect at 33°C by overexpression of NUP85. Precultures were diluted in growth medium, and equivalent amounts of cells (diluted in 10−1 steps) were spotted onto YPD plates. Plates were incubated for 3 days. (D) Western blot analysis of purified Mtr2p reveals an association with Nup85p. Mtr2p-TEV-ProtA and Prot-TEV-DHFR were affinity purified as described in Materials and Methods. Each TEV eluate (25 μl) was analyzed by SDS-PAGE followed by Western blotting with anti-Mtr2p, anti-Nup85p, anti-Arc1p, and anti-DHFR antibodies. The blots were developed with the Amersham ECL kit. Sup, supernatant.
Mtr2p interacts with Nup85p both genetically and physically.
Nup85, a member of the Nup84p nucleoporin complex, is involved in NPC organization and RNA export and is functionally linked to MEX67 (40, 45). We therefore analyzed whether the different thermosensitive mtr2 alleles are synthetically lethal in combination with the nup85Δ allele. Strikingly, mtr2 alleles which caused the cytoplasmic mislocalization of mutant Mtr2p (e.g., mtr2-9) were synthetically lethal in combination with nup85Δ (Fig. 4B). On the contrary, mutant alleles which did not cause the cytoplasmic mislocation of mutant Mtr2p (e.g., mtr2-21) were not synthetically lethal in combination with nup85Δ (Fig. 4B). Consistent with these findings was the fact that the thermosensitive phenotype of mtr2-9 cells but not of mtr2-21 cells was partially complemented at 33°C by the overexpression of NUP85 (Fig. 4C and Table 2).
The interaction between MTR2 and NUP85 was tested biochemically. Mtr2p-TEV-ProtA was affinity purified from yeast spheroplasts (see Materials and Methods), and the Mtr2p eluate (which was obtained by proteolytic TEV cleavage) was analyzed by Western blotting with anti-Nup85p antibodies (Fig. 4D). Although not associated stoichiometrically, Nup85p was detected in the Mtr2p eluate, as revealed by immunoblot analysis, whereas an unrelated protein, such as Arc1p (43), was completely absent. We also tested for the presence of Nup84p and Seh1p in the Mtr2p eluate; only Seh1p was detected, although in small amounts (38a). To test the specificity of the Mtr2p-Nup85p interaction, we affinity purified ProtA-TEV-DHFR and released DHFR from IgG-Sepharose beads by using TEV protease under identical conditions. No Nup85p could be detected (Fig. 4D).
ProtA-tagged Mex67p and Mtr2p can be found on both sides of the NPC.
To determine the location of Mex67p and Mtr2p on an ultrastructural level, we performed preembedding immunoelectron microscopy, a procedure that yields structurally well-preserved yeast NPCs and that has recently been used to localize yeast nucleoporins (2a). We found a significant and statistically relevant number of gold particles at the nuclear pores in cells expressing Mtr2p-ProtA or Mex67p-ProtA (Fig. 5C and D); however, colloidal gold was also seen in the nucleus and cytoplasm. The NPC labeling observed with Mtr2p-ProtA and Mex67p-ProtA was specific, since the gold-conjugated anti-ProtA antibody used did not give NPC labeling when control strains expressing either cytosolic ProtA-DHFR or nuclear ProtA-Pus1p were tested. In this case, the immunolabeling was, as expected, mainly in the nucleus for ProtA-Pus1p and in the cytoplasm for ProtA-DHFR (Fig. 5A and B).
FIG. 5.
Immunoelectron microscopy of Mex67p and Mtr2p. Nuclear envelope cross sections with adjacent nucleoplasm and cytoplasm from Triton X-100-extracted cells expressing ProtA-DHFR (A), ProtA-Pus1p (B), Mtr2p-ProtA (C), and Mex67p-ProtA (D). Cells were preembedding labeled with gold-conjugated anti-ProtA antibody. A gallery of representative photographs is shown. For Mtr2p-ProtA and Mex67p-ProtA, selected examples of labeled NPCs and the quantitative analysis of gold particles associated with NPCs (E and F) are also shown. The distances of gold particles relative to the twofold symmetry axis of the NPC (axis parallel to the nuclear envelope passing through the center of the NPC, with positive distances representing the cytoplasmic side of the NPC) and the eightfold symmetry axis of the NPC (axis perpendicular to the nuclear envelope and passing through the center of the NPC) were determined in cross sections. Cross-sectioned nuclear envelopes from two experiments which yielded 38 cells and 65 gold particles associated with the NPC for Mex67p and 32 cells and 55 gold particles associated with the NPC for Mtr2p were analyzed. The arrows indicate gold particles in the cytoplasm and the nucleus, whereas the arrowheads indicate gold particles associated with NPCs. Cytoplasmic (c) and nuclear (n) sides of the nuclear envelope are indicated. Scale bars, 100 nm.
Visual inspection of representative electron microscopy photographs revealed a distribution of Mtr2p-ProtA and Mex67p-ProtA on both sides of the NPCs. To determine this distribution more precisely, we performed quantitative analysis of the distribution of gold particles associated with the NPCs in cross-sectioned nuclear envelopes. As shown in Fig. 5E and F, although the NPCs were labeled overall for both Mtr2p-ProtA and Mex67p-ProtA, there was a preference for labeling on the cytoplasmic side of the NPCs (i.e., positive values). This finding could have been due to the fact that with the preembedding labeling technique, the antibody first reaches the cytoplasmic epitope, where it is bound, before it can diffuse to the nuclear epitope. However, quantitation of postembedding labeling at the NPCs also revealed preferential labeling on the cytoplasmic side of the NPCs (see below and Table 3).
TABLE 3.
Quantitation of antibody labelinga
| Protein | No. of gold particles/μm2
|
||
|---|---|---|---|
| Cytoplasmic | Nuclear | Nuclear pore | |
| ProtA-Mex67p | 1.5 | 1.4 | 1.6 (1.0 and 0.6) |
| ProtA-Mtr2p | 1.1 | 0.6 | 0.8 (0.6 and 0.2) |
| ProtA-Pus1p | 0.5 | 1.9 | 0 |
| ProtA-DHFR | 2.2 | 0.3 | 0 |
Quantitation was performed as described in Materials and Methods. For ProtA-Mex67p, 22 cells were analyzed and 354 gold particles were counted. For ProtA-Mtr2p, 22 cells were analyzed and 201 gold particles were counted. For ProtA-Pus1p, 20 cells were analyzed and 202 gold particles were counted. For ProtA-DHFR, 20 cells were analyzed and 168 gold particles were counted. Numbers in parentheses indicate cytoplasmic and nuclear sides of the nuclear pore, respectively.
Since through preembedding labeling some cytoplasmic epitopes could be lost (for this technique, yeast cells are treated with Triton X-100 to facilitate labeling on both the cytoplasmic and the nuclear sides of the NPC; see Materials and Methods), gold particle distribution was quantified by immunolabeling of LR White-embedded yeast cells (postembedding labeling). This quantitation revealed an equal distribution of the gold particles in the NPC, the cytoplasm, and the nucleus for Mex67p-ProtA (Table 3). This result is different from the GFP labeling data, which showed an apparent accumulation of Mex67p-GFP at the nuclear envelope. However, the values given in Table 3 cannot be compared directly with the GFP labeling data because the electron microscopy data are from the quantitation of gold particles in cell sections, while the intensity of GFP fluorescence is derived from whole cells. For cell expressing Mtr2p-ProtA, there were fewer gold particles in the nucleus than in the NPC or the cytoplasm. Neither ProtA-DHFR nor ProtA-Pus1p, which was used as a control, was found at the NPC but was located predominantly at the cytoplasm or the nucleus, respectively (Table 3).
The Mex67p-Mtr2p complex binds to RNA in vitro through the Mex67p subunit.
We previously reported that Mex67p can be UV cross-linked to poly(A)+ RNA in vivo (40). We therefore wanted to test whether the newly identified complex, Mex67p-Mtr2p, can bind to RNA in vitro and to analyze if one or both components of the complex are responsible for binding to RNA.
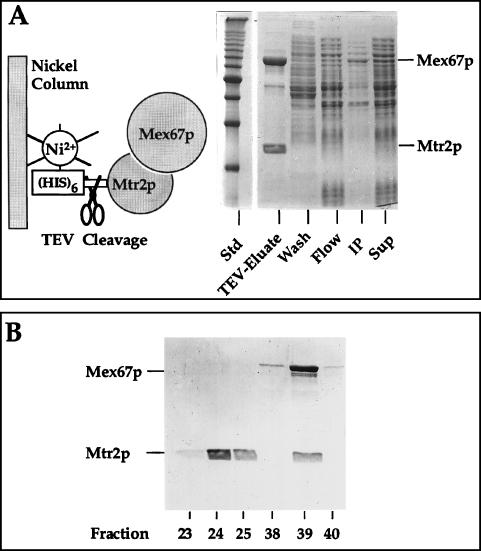
To obtain a recombinant Mex67p-Mtr2p complex for in vitro RNA-binding studies, Mtr2p was His6 tagged, and the proteolytic TEV cleavage site was inserted between the tag and Mtr2p (Fig. 6A, left panel). His6-TEV-Mtr2p was coexpressed with untagged Mex67p and affinity purified from an E. coli lysate by Ni-NTA–agarose chromatography. After TEV cleavage, large amounts of a recombinant Mtr2p-Mex67p complex could be eluted from the Ni-NTA–agarose column (Fig. 6A, TEV-Eluate). Thus, Mtr2p and Mex67p can assemble into a heterodimeric complex upon coexpression in E. coli, without any requirement for other yeast factors. As a control, His6-Mtr10p (an unrelated recombinant protein) was coexpressed with Mex67p in E. coli and purified, but Mex67p did not coenrich (data not shown).
FIG. 6.
Purification of recombinant Mtr2p and Mex67p from E. coli. (A) Coexpression and complex formation of Mtr2p and Mex67p in bacteria. His6-TEV-Mtr2p and untagged Mex67p were expressed and purified from E. coli. A soluble supernatant (Sup), an insoluble pellet (IP), the flowthrough (Flow), a wash fraction (Wash), and the TEV protease-eluted complex (TEV-Eluate) were analyzed by SDS-PAGE and Coomassie blue staining. The positions of Mex67p and Mtr2p are shown. Std, marker proteins (10-kDa ladder). (B) Purification of the recombinant Mex67p-Mtr2p complex by ion-exchange chromatography. Fractions from the MonoS column (23 to 25 and 38 to 40) were analyzed by SDS-PAGE and Coomassie blue staining. The two weakly staining bands below full-length Mex67p were cross-reactive with anti-Mex67p antibodies and thus may correspond to proteolytic breakdown products. Note that Mtr2p tends to migrate as a broad and fuzzy band.
The recombinant Mtr2p-Mex67p complex was further purified, since the TEV eluate exhibited RNase activity, most likely due to contaminating E. coli RNase. Therefore, the TEV eluate was applied to an FPLC MonoS column, and proteins which bound to the resin were eluted with a salt gradient (Fig. 6B). At about 150 mM salt (i.e., fraction 24), free Mtr2p eluted from the column, whereas the Mtr2p-Mex67p complex eluted at about 700 mM salt (i.e., fraction 39) (Fig. 6B).
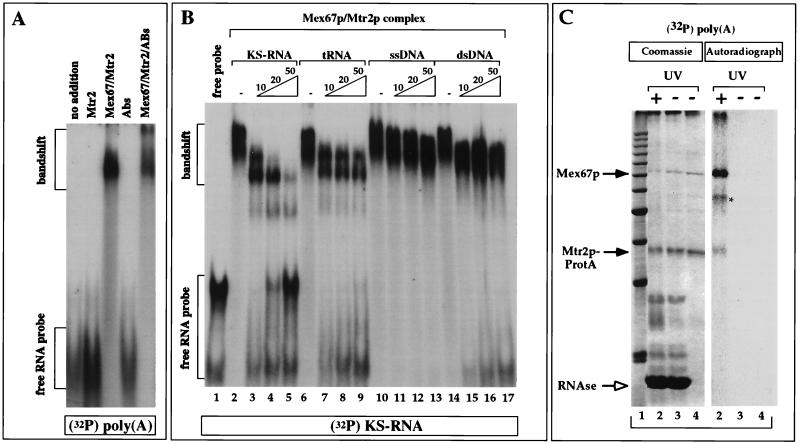
The binding of Mex67p and Mtr2p to RNA was first analyzed by a gel retardation (band shift) assay. The Mex67p-Mtr2p complex and Mtr2p purified by FPLC were incubated with various in vitro-transcribed, 32P-labeled RNAs (for a description of the RNAs, see Materials and Methods), and the protein-RNA complexes formed were analyzed by a gel retardation assay (Fig. 7A). Mtr2p alone did not detectably bind to RNA, while the Mex67p-Mtr2p complex produced a strong band shift with all RNAs tested [shown for poly(A) in Fig. 7A; data not shown for poly(G) and poly(C)]. When affinity-purified anti-Mex67p antibodies were included in the binding reaction, the protein-RNA complex was supershifted, confirming that Mex67p was present in the protein-RNA complex (Fig. 7A).
FIG. 7.
Band shift assays and UV cross-linking to detect the binding of Mex67p to RNA. (A) Binding to RNA was tested in a band shift assay with in vitro-transcribed, 32P-radiolabeled poly(A)+ RNA and purified recombinant proteins in the absence or presence of anti-Mex67p antibodies (Abs or ABs). (B) Competition of the formation of the 32P-labeled KS-RNA–Mex67p–Mtr2p complex by an excess (10-, 20-, or 50-fold) of unlabeled KS-RNA, tRNA, single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA). 32P-labeled KS-RNA (5 ng) was incubated with 1 μg of E. coli-derived, purified Mex67p-Mtr2p complex in the absence (−) or presence of increasing amounts of cold competitor KS-RNA, yeast tRNAMet, single-stranded DNA, and double-stranded DNA. (C) In vitro UV cross-linking of Mex67p to RNA. The Mex67p-Mtr2p-ProtA complex bound to IgG-Sepharose beads was incubated with 1 ng of 32P-labeled poly(A)+ RNA and UV irradiated before analysis by SDS-PAGE and Coomassie blue staining or autoradiography. Lane 1, 10-kDa marker proteins; lanes 2, UV-irradiated beads treated with RNase; lanes 3, non-UV-irradiated beads treated with RNase; lanes 4, input of beads. Proteins were finally released from the beads with SDS. The positions of Mex67p and Mtr2p-ProtA are indicated by arrows. The asterisk indicates an unidentified band.
To test the specificity of the interaction between Mex67p and RNA, the binding of Mex67p to heteropolymeric 32P-labeled KS-RNA (derived from transcription of the pBluescript KS polylinker) was inhibited with an excess of unlabeled KS-RNA, yeast tRNAMet, and single- or double-stranded DNA oligonucleotides of similar lengths (Fig. 7B). When a 10-fold excess of unlabeled KS-RNA was added to the reaction, we observed the formation of faster-migrating complexes as well as unshifted free probe (Fig. 7B, lane 3). This result suggests that the amount of total RNA present in this reaction was large enough to saturate binding to the Mex67p-Mtr2p complex and that the faster-migrating bands represented specific RNA-protein complexes. The intensity of these complexes was further decreased as larger amounts of unlabeled probe were included in the reaction (20- or 50-fold; Fig. 7B, lanes 4 and 5). The same complexes were also observed when an excess of unlabeled yeast tRNAMet was added together with 32P-labeled KS-RNA. However, the formation of these complexes was inhibited less in the presence of 50-fold tRNA than in the presence of 50-fold KS-RNA (Fig. 7B, compare lanes 5 and 9). Therefore, tRNA inhibited readily the formation of the slower-migrating complex, but the faster-migrating complexes were more resistant to tRNA, suggesting a more specific association with KS-RNA. Finally, when single- or double-stranded DNA oligonucleotides of similar lengths were added to the binding reaction, we did not observe a significant decrease in the intensity of the band shifts but observed instead a small increase in the mobility (Fig. 7B, lanes 10 to 17). We conclude from these experiments that Mex67p has an affinity for RNA but not for single- or double-stranded DNA. Furthermore, Mex67p exhibits a preference for binding to homopolymeric or heteropolymeric KS-RNA rather than tRNA.
When His6-tagged Mex67p was isolated from E. coli in the absence of Mtr2p, it had the tendency to aggregate after elution from the Ni-NTA–agarose column and dialysis (data not shown). Therefore, we could not test by band shift assays whether Mex67p alone can bind to RNA. However, His6-Mex67 remained soluble in the E. coli lysate. When this soluble lysate was incubated with agarose beads carrying covalently attached RNA homopolymers, Mex67p bound to poly(A) and poly(G) and, to a lesser extent, to poly(U) and poly(C) (data not shown). The binding of Mex67p to poly(A) and poly(U) was still observed at 500 mM NaCl. When the same in vitro binding assay was performed with recombinant Mtr2p, no significant association with the four homopolymeric RNAs was observed (data not shown). Thus, Mex67p has the capability to bind to homopolymeric RNAs in the absence of Mtr2p.
The property of the native Mex67p-Mtr2p complex isolated from yeast to associate with RNA was verified by UV cross-linking. When the Mex67p-Mtr2p-ProtA complex immobilized on IgG-Sepharose beads was incubated with radiolabeled poly(A)+ RNA and UV irradiated, Mex67p could be efficiently cross-linked to RNA (Fig. 7C). Mtr2p was cross-linked to RNA only very inefficiently. To make sure that the observed radioactive signals were due to the formation of covalent bonds between proteins and RNA, the beads were boiled in the presence of DTT. Although this treatment led to massive dissociation of IgG chains from the IgG-Sepharose beads, the same cross-linked products could be observed, showing that nonspecific cross-linking to, e.g., IgG did not take place (data not shown). Similar results were obtained when other RNAs (poly(C), poly(G), or KS-RNA) or when the recombinant Mex67p-Mtr2p complex was used in the UV-cross-linking assay (data not shown).
In summary, using three different methods (band shift, UV-cross-linking, and binding to immobilized ribohomopolymers), we found that the Mex67p-Mtr2p complex binds to RNA via the Mex67p subunit.
DISCUSSION
We have identified a complex which is essential for mRNA export and which consists of at least two components, Mex67p and Mtr2p. Mex67p can directly bind to RNA in vitro, and Mtr2p is required for the association of Mex67p with the nuclear pores. Furthermore, a genetic and physical interaction of Mtr2p with Nup85p was found, suggesting that Nup85p or the Nup85p complex is (one of) the target(s) at the NPC to which Mtr2p and Mex67p bind. Previously, we have found that a putative RNA-binding protein called Mip6p interacts with Mex67p on the basis of two-hybrid data; however, we could not further confirm this possible interaction, since no genetic and biochemical interactions between Mex67p and Mip6p have yet been found (39a).
The fact that Mex67p and Mtr2p physically interact explains their strong genetic interaction and their common involvement in mRNA export. Whether Mex67p and Mtr2p are in permanent contact or whether they reversibly associate and dissociate remains to be shown. The observed locations of Mex67p and Mtr2p on both sides of the NPC suggest that the complex plays a role during translocation through the pore channel and release of the mRNA transport substrate from the pores into the cytoplasm.
Recently, Xpo1p (Crm1p) was found to be involved in both nuclear protein and mRNA export and was suggested to be the receptor for leucine-rich NES signals (46). We did not find a significant intranuclear accumulation of Mex67p-GFP in xpo1-1 mutant cells at the restrictive temperature (38a). Certain hnRNP proteins which shuttle between the nuclear and cytoplasmic compartments also have been implicated to mediate the nuclear export of cellular mRNA (24, 27, 35). Therefore, hnRNP proteins such as Npl3p could functionally overlap the Mex67p-Mtr2p complex, e.g., by acting upstream in the splicing, assembly, and initial transport of the hnRNPs, whereas Mex67p-Mtr2p functions downstream in nuclear mRNA export, requiring transport-competent mRNPs.
An interaction among Mex67p, Mtr2p, and the nuclear pores appears to be essential for mRNA export. If Mex67p is mutated in such a way that it no longer can bind to Mtr2p, nuclear mRNA export and cell growth are inhibited. Concomitantly, Mex67p mislocalizes to the cytoplasm, but Mtr2p stays at the pores or inside the nucleus. On the contrary, when the binding of Mtr2p to the nuclear pores is impaired (e.g., when mtr2 mutant alleles are synthetically lethal with nup85Δ), both Mex67p and Mtr2p mislocalize to the cytoplasm and mRNA export is blocked. These defective interactions among Mtr2p, Mex67p, and Nup85p can be rescued by the overproduction of the appropriate partner protein; e.g., an impaired interaction between Mex67p and Mtr2p can be complemented by the overexpression of either Mex67p or Mtr2p. Alternatively, MTR2 mutations which affect the interaction with Nup85p can be (partly) rescued by NUP85. It is not clear why the overexpression of NUP85 can partly rescue the thermosensitive mtr2 phenotype, but one possibility is that more binding sites for Mtr2p are provided by overproduced Nup85p.
It is interesting that the mislocalization of either Mex67p alone or Mex67p and Mtr2p always occurs into numerous foci scattered throughout the cytoplasm when cells containing certain mutant alleles of either mex67 or mtr2 are incubated at the restrictive temperature. These foci may represent dynamic structures during mRNA transport. If so, Mex67p-Mtr2p could play a role not only in mRNA export from the nucleus but also in cytoplasmic transport of mRNPs to sites of mRNA consumption.
Although Mex67p does not exhibit motifs indicative of an RNA-binding protein (e.g., the RNP motif, the RGG box, or the KH motif [2]), it was found to bind to RNA in several independent in vitro assays. Mex67p thus contains a novel RNA-binding domain. In vitro, Mex67p binds efficiently to homo- and heteropolymeric RNAs but not to single- or double-stranded DNA. From our in vitro binding experiments, we cannot conclude whether Mex67p has a specific affinity for a particular class of RNA. However, the fact that Mex67p can be UV cross-linked in vivo to poly(A)+ RNA and its requirement for mRNA nuclear export (40) suggest that the binding of Mex67p to RNA in vitro probably reflects the ability of Mex67p to associate in vivo directly with mRNA. Whether the binding of Mex67p to mRNA exhibits sequence specificity or is controlled in a cooperative way by Mtr2p and/or additional factors such as hnRNP proteins will be addressed in the future. It will be also interesting to determine which domain of Mex67p binds to RNA and how RNA binding is made reversible in living cells.
We have reported an evolutionary conservation between Mex67p and putative higher eukaryotic counterparts (40). A human homologue of Mex67p called Tap was found in a two-hybrid screen with herpesvirus saimiri Tip (tyrosine kinase-interacting protein) as bait (49). Interestingly, on human chromosome X at least two further Tap homologues which lack part or all of the conserved carboxy-terminal domain were found (15a). These findings give rise to the speculation that several members of the human Tap family are involved in different aspects of mRNA transport (e.g., regulated mRNA export, tissue-specific mRNA transport, or transport of different mRNAs). Recently, the Tap protein was found to bind to viral CTE RNA and was shown to be the mediator of CTE-dependent RNA export (13). Furthermore, microinjected recombinant Tap overcomes mRNA export inhibition caused by the presence of saturating amounts of CTE RNA. It is thus likely that Tap and Tap-related proteins are involved not only in CTE-dependent viral RNA export but also in mRNA export in higher eukaryotic cells, with a function similar to that of yeast Mex67p. Whether an Mtr2p homologue exists in higher eukarytoic cells is not yet known.
In summary, Mex67p and Mtr2p, which form a complex and have the capability to associate with both mRNA and the nuclear pores, are essential constituents of the mRNA export machinery in yeast. Further biochemical and genetic analyses should reveal how Mex67p in conjunction with Mtr2p interacts with RNA, other RNA-binding proteins, transport factors of the importin/Ran machinery, and NPC components during mRNA export from the nucleus to the cytoplasm.
ACKNOWLEDGMENTS
We thank Juri Rappsilber and Matthias Mann for performing mass spectroscopy of the Mex67p band and Kasten Weis for sending the xpo1-1 mutant. We also thank Kerstin Kloke for excellent technical assistance. Critical proofreading by Craig Hart (University of Geneva, Geneva, Switzerland) is also acknowledged.
E.H. was the recipient of a grant from the Deutsche Forschungsgemeinschaft (SFB352). This work was also supported by grants from the Swiss National Science Foundation (4036-044061 and 3100-053034), by the Kanton Basel-Stadt, and by the M. E. Müller Foundation of Switzerland.
REFERENCES
- 1.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 2.Doye V, Hurt E C. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 2a.Fahrenkrog, B., E. Hurt, U. Aebi, and N. Panté. Submitted for publication.
- 3.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 4.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 5.Fornerod M, Vandeursen J, Vanbaal S, Reynolds A, Davis D, Gopal Murti K, Fransen J, Grosveld G. The human homologue of yeast Crm1 is in a dynamic subcomplex with Can/Nup214 and a novel nuclear pore component, Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 8.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 9.Goldfarb D S. Nuclear transport—whose finger is on the switch? Science. 1997;276:1814–1816. doi: 10.1126/science.276.5320.1814. [DOI] [PubMed] [Google Scholar]
- 10.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görlich D, Mattaj I W. Protein kinesis—nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 12.Gorsch L C, Dockendorff T C, Cole C N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 13a.Hellmuth, K. Unpublished data.
- 14.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 15.Hurt E C. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Hurt, E. C. Unpublished data.
- 16.Iovine M K, Wente S R. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadowaki T, Hitomi M, Chen S, Tartakoff A M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol Biol Cell. 1994;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadowaki T, Zhao Y, Tartakoff A M. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci USA. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahana J, Silver P. Use of the A. victoria green fluorescent protein to study protein dynamics in vivo. Curr Prot Mol Biol. 1996;9:722–728. doi: 10.1002/0471142727.mb0907cs34. [DOI] [PubMed] [Google Scholar]
- 22.Kitada K, Johnson A L, Johnston L H, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Export of importin a from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 24.Lee M S, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 25.Lucocq J. Quantitation of gold labeling and estimation of labeling efficiency with a stereological counting method. J Histochem Cytochem. 1992;40:1929–1936. doi: 10.1177/40.12.1453009. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 27.Michael W M, Choi M Y, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 28.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 29.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 30.Nakielny S, Siomi M C, Siomi H, Michael W M, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- 31.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 32.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 33.Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pemberton L F, Rosenblum J S, Blobel G. A distinct and parallel pathway for nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 36.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 37.Richards S A, Lounsbury K M, Carey K L, Macara I G. A nuclear export signal is essential for the cytosolic localization of the ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 38a.Santos-Rosa, H. Unpublished data.
- 39.Schlaich N L, Hurt E C. Analysis of nucleocytoplasmic transport and nuclear envelope structure in yeast disrupted for the gene encoding the nuclear pore protein Nup1p. Eur J Cell Biol. 1995;67:8–14. [PubMed] [Google Scholar]
- 39a.Segref, A. Unpublished data.
- 40.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Hurt E C. Mex67p, which is an essential factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senger, B., G. Simos, F. R. Bischoff, A. V. Podtelejnikov, M. Mann, and E. C. Hurt. Mtr10p functions as a nuclear import receptor for the mRNA binding protein Np13p. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 42.Shibasaki F, Price E R, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 43.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. The yeast protein Arc1p binds to tRNA and functions as a cofactor for methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 44.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 45.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt E C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 46.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 47.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 48.Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt E. A new subclass of nucleoporins that functionally interacts with nuclear pore protein NSP1. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon D-K, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung J U. Tap: a novel cellular protein that interacts with Tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6:571–582. doi: 10.1016/s1074-7613(00)80345-3. [DOI] [PubMed] [Google Scholar]