Feringa and van der Kant discuss novel findings from the Cohen group implicating a role for the Alzheimer’s risk gene ApoE at the lipid droplet surface.
Abstract
The secreted ApoE protein is a major regulator of lipid transport between brain cells. In this issue, Windham et al. (https://doi.org/10.1083/jcb.202305003) uncover a novel intracellular role for ApoE at the lipid droplet surface, where it regulates lipid droplet size and composition.
Lipid droplets (LDs) are highly dynamic organelles that play important roles in cellular signaling, energy metabolism, and immune activity. LDs consist of a hydrophobic core of neutral storage lipids such as triacyl glycerides (TG) and cholesteryl esters (CE), surrounded by a monolayer of phospholipids coated by a multitude of regulatory proteins (1). Alois Alzheimer had already described lipid saccules as a characteristic of the Alzheimer’s Disease (AD) brain (2), and more recently, LDs have also been shown to accumulate in the aging brain (3, 4). Polymorphisms in the APOE gene are both the strongest genetic risk factor for AD (APOE4) as well as the strongest genetic protective factor for AD (APOE2) compared to the neutral variant (APOE3).
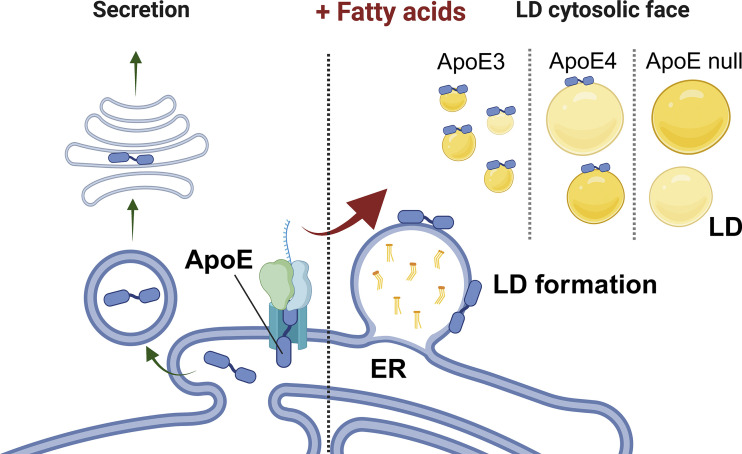
While ApoE has been linked to AD over 20 years ago, a mechanistic link on its impact on AD pathogenesis is still missing. Much research has focused on the role of ApoE as a secreted protein, which binds lipoprotein particles to regulate lipid transport between different brain cell types. Furthermore, secreted ApoE has been shown to bind Amyloid beta (Aβ) and regulate its compaction and clearance thus regulating Amyloid plaque pathology (5). Here Windham et al. (6) define an interesting novel intracellular role for ApoE in shaping lipid droplets, through localization of ApoE at the cytosolic leaflets of these organelles (Fig. 1). ApoE had previously been detected on LD surfaces (7, 8), but little attention was brought to these early observations. However, Windham and colleagues provide convincing evidence that ApoE is present on LDs and plays an important role in the regulation of their size and triglyceride metabolism. The authors describe how ApoE is transferred from the endoplasmic reticulum (ER) to the LD via membrane bridges when cells are challenged with exogenous fatty acids. To do so, ApoE (which contains an N-terminal ER lumen targeting domain) escapes translocation to the ER lumen. Other LD proteins can also avoid translocation to the ER lumen via different mechanisms, as referenced by the authors, and understanding how this occurs for ApoE would be another important next step in elucidating this intriguing novel aspect of ApoE biology.
Figure 1.
ApoE regulates LD dynamics. ApoE undergoes co-translational translocation into the ER lumen, where it enters the secretory route. Once secreted, ApoE regulates lipid transport between different brain cells. Windham and colleagues now show that upon a fatty acid challenge, ApoE escapes translocation into the ER and is recruited to newly formed LDs. In absence of ApoE, or in the presence of the AD risk variant ApoE4, LDs become larger and have increased levels of unsaturated triglycerides that are more sensitive to lipid peroxidation.
The authors also show a physiological role for ApoE at the LD surface. After translocation to newly formed LDs, ApoE restricted LD growth thereby preventing the formation of large LDs with an excess in unsaturated TGs. Importantly, both the presence of AD risk variant ApoE4, as well as ApoE loss, resulted in the formation of larger LDs with higher levels of unsaturated TG species, which are also more vulnerable to lipid peroxidation. By mutating different ApoE domains, Windham et al. (6) present compelling evidence for a previously unknown intracellular role for ApoE at the LD. Furthermore, their finding that ApoE4 shows a loss of function of this mechanism is important for understanding AD but also raises several important open follow-up questions.
For example, to which extent do the extracellular versus intracellular pools of ApoE contribute to AD pathogenesis? Previously, the effects of ApoE4 on many brain cell types have been attributed to a difference in binding of the ApoE isoforms to lipids/Aβ or lipoprotein receptors (5). If ApoE also regulates an intracellular pool of lipids, does this contribute to AD pathogenesis? This is not unlikely, as altered glial lipid metabolism has been shown to contribute to pathogenesis in ApoE4/Tau mouse models of AD (9). Targeting secreted ApoE, or ApoE-receptor interactions, e.g., with antibodies, is one direction of preclinical drug development currently explored for AD, although such interventions are unlikely to affect the intracellular pool of ApoE shown to be important here. Another therapeutic strategy to reduce ApoE levels by antisense oligonucleotides might lead to glial lipid dyshomeostasis based on this novel role for ApoE.
It is still a major unresolved question in the field whether ApoE4 is a loss of function variant, or a toxic gain of function variant, with good evidence for both from multiple groups (5). Could differences in functions of the intra- versus extracellular pools of ApoE explain some of the seemingly discrepant findings? To really understand the impact of the findings by Windham et al. for AD pathogenesis, many of these questions will have to be addressed in mouse models where pathology and behavioral outcomes are part of the equation. A host of novel and exciting ApoE mouse models have recently been developed including models that can switch ApoE on and off in a cell-type specific manner and models that can induce a switch from APOE4 and APOE2 genotypes. It will be a challenge to develop these further to also test how intracellular versus extracellular pools of ApoE contribute to AD pathology, but these new data indicate that this needs to be considered.
Maybe the most exiting in terms of ApoE biology, and in understanding the importance of these findings, is the question of what ApoE is doing on the LDs? The authors show that without ApoE (or with ApoE4), LDs contain more unsaturated fatty acids, which is consistent with other recent findings in ApoE4 iPSC-derived astrocytes (10). But what is the functional consequence of this if it occurs in vivo? What happens with protective variants such as ApoE2 and ApoE Christchurch? The authors show increased lipid peroxidation is an effect of ApoE4 expression, and this might be one potentially pathogenic pathway. However other downstream effects will also need to be explored. For example, how does LD targeted ApoE affect cellular metabolism, and particularly mitochondria which use LD-derived fatty acids to produce energy? With which other proteins (or lipids) does ApoE interact when on the LD surface, and can ApoE travel through the cytosol to regulate other organelles from the inside of the cell? As any good discovery, this work from the Cohen lab raises numerous new questions which many in the field will be eager to address in the future.
Overall, the findings here add a whole new level of ApoE biology on top of what is already known by showing ApoE has a function within the cytosol at the LD surface. While this does not necessarily make the ApoE-Alzheimer puzzle any easier, these findings provide further insight into the complex role ApoE plays in the brain and open several important research routes to understand and eventually treat ApoE4-dependent AD.
Acknowledgments
We thank Aiko Robert for proofreading the manuscript. The figure was created with http://BioRender.com.
This work was supported by a ZonMW Veni (09150162210237) to F.M. Feringa, a ZonMW Vidi (08150172110086) and grant number 2022-250480 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community to R. van der Kant.
References
- 1.Olzmann, J.A., and Carvalho P.. 2019. Nat. Rev. Mol. Cell Biol. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer, A., et al. 1995. Clin. Anat. 10.1002/ca.980080612 [DOI] [PubMed] [Google Scholar]
- 3.Marschallinger, J., et al. 2020. Nat. Neurosci. 10.1038/s41593-019-0566-1 [DOI] [Google Scholar]
- 4.Ralhan, I., et al. 2021. J. Cell Biol. 10.1083/jcb.202102136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenfeld, J., et al. 2024. Nat. Rev. Neurosci. 10.1038/s41583-023-00776-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Windham, I.A., et al. 2024. J. Cell Biol. 10.1083/jcb.202305003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bersuker, K., et al. 2018. Dev. Cell. 10.1016/j.devcel.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejhert, N., et al. 2020. Mol. Cell. 10.1016/j.molcel.2020.01.014 [DOI] [Google Scholar]
- 9.Litvinchuk, A., et al. 2024. Neuron. 10.1016/j.neuron.2023.10.023 [DOI] [Google Scholar]
- 10.Sienski, G., et al. 2021. Sci. Transl. Med. 10.1126/scitranslmed.aaz4564 [DOI] [PMC free article] [PubMed] [Google Scholar]