Abstract
As microtubule-organizing centers, centrosomes direct assembly of the bipolar mitotic spindle required for chromosome segregation and genome stability. Centrosome activity requires the dynamic assembly of pericentriolar material (PCM), the composition and organization of which changes throughout the cell cycle. Recent studies highlight the conserved localization of several mRNAs encoded from centrosome-associated genes enriched at centrosomes, including Pericentrin-like protein (Plp) mRNA. However, relatively little is known about how RNAs localize to centrosomes and influence centrosome function. Here, we examine mechanisms underlying the subcellular localization of Plp mRNA. We find that Plp mRNA localization is puromycin-sensitive, and the Plp coding sequence is both necessary and sufficient for RNA localization, consistent with a co-translational transport mechanism. We identify regions within the Plp coding sequence that regulate Plp mRNA localization. Finally, we show that protein-protein interactions critical for elaboration of the PCM scaffold permit RNA localization to centrosomes. Taken together, these findings inform the mechanistic basis of Plp mRNA localization and lend insight into the oscillatory enrichment of RNA at centrosomes.
Keywords: centrosome, RNA localization, co-translational transport, Pericentrin
Introduction
Centrosomes are microtubule-organizing centers (MTOCs) that support cell division, intracellular trafficking, and ciliogenesis. Consequently, centrosome dysfunction is associated with varied diseases and developmental disorders, including cancer and microcephaly [1–3]. Centrosome function is instructed by the organization and composition of the pericentral material (PCM), the composite of proteins and mRNAs that surround the central pair of centrioles [4–7].
Centrosome activity oscillates in phase with the cell cycle. Centrosomes duplicate once and only once per cell cycle, usually during S-phase [8]. Subsequently, the duplicated centrosomes separate and undergo a maturation process, wherein additional PCM is recruited to support microtubule nucleation and organization [9–13]. The coordinated processes of centrosome duplication, separation, and maturation ensure the timely formation of the bipolar mitotic spindle during M-phase. As cells exit mitosis, centrosomes shed PCM [13, 14]. While these cell cycle-dependent fluctuations in PCM recruitment and shedding instruct the microtubule-organizing activity of centrosomes, the underlying mechanisms remain incompletely understood.
Recent work indicates that some mRNAs specifically enrich at centrosomes in a cell cycle-dependent manner [7, 15–18]. Remarkably, RNAs that localize to centrosomes encode centrosome proteins, raising the intriguing possibility that centrosomal mRNAs may contribute to centrosome maturation, structure, or otherwise influence centrosome activity [19–21]. Consistent with these ideas, the localization of some centrosomal mRNAs is directed by a co-translational transport mechanism, whereby RNA localization and protein translation are coupled [15, 22, 16]. Within cultured mammalian cells, for example, ASPM and NUMA1 mRNAs and nascent peptides are co-trafficked to centrosomes followed by additional on-site translation [15]. Co-translational transport was similarly reported for Centrocortin (Cen) mRNA within Drosophila syncytial embryos [22]. Furthermore, the mislocalization of Cen mRNA to the anterior cortex prevents Cen protein from localizing to distal centrosomes, demonstrating the coupling of RNA transport and local translation [17].
Among the most conserved mRNAs localizing to centrosomes is Pericentrin (PCNT) mRNA, as observed in cell culture, zebrafish, and Drosophila models [16–18]. Human PCNT and Drosophila Pcnt-like protein (Plp) share functionally conserved roles in PCM scaffolding and microtubule nucleation [23–28]. In humans, loss-of-function PCNT mutations are associated with microcephalic osteodysplastic primordial dwarfism type II (MOPD II), as well as cardiac and neurovascular abnormalities [29–33]. Loss of Drosophila Plp also leads to pleiotropic effects, including embryonic lethality, neuronal dysfunction, and sterility [24, 25, 28, 34]. While prior work indicates PCNT mRNA localization requires translation and the microtubule minus end-directed motor dynein, relatively little is understood about mechanisms underlying the co-translational transport of centrosomal RNAs or how their localization is coupled to the cell cycle [16].
Drosophila embryos are a valuable model to investigate how and why RNAs localize to centrosomes. Drosophila embryos progress through 14 rounds of synchronous, abridged S-to-M nuclear division cycles (NCs) without gap phases prior to somatic cellularization [35]. During this period of development, the embryo is largely transcriptionally quiescent and supported by maternal stores of RNAs and proteins [36]. As in mammalian cells, RNAs enrich at embryonic centrosomes preceding mitotic onset, and less RNA localizes to centrosomes during mitosis [17, 15]. RNAs also progressively enrich at centrosomes as embryonic development ensures, concomitant with the lengthening of successive NCs [17, 18]. These findings argue that RNA localization to centrosomes is entrained to the cell cycle and developmental progression.
Prior work by our group and others similarly uncovered cell cycle and developmental stage-specific changes in the organization of Drosophila embryonic centrosomes. The organization and structure of the PCM is largely supported by the formation of Centrosomin (Cnn) flares, which extend during interphase, retract during mitosis, and mature as the NCs proceed [24, 37]. Cnn functions as a PCM scaffold important for centrosome maturation and organization [37–39]. Cnn scaffolding activity, in turn, is supported by Plp, which localizes to the tips of Cnn flares and interacts directly with Cnn via two interaction modules. The interaction between Plp-Cnn is critical for PCM scaffolding and early embryo mitotic divisions [24]. Although the oscillations in centrosomal RNA distributions appear to mirror changes in PCM organization, whether the PCM scaffold influences RNA localization has not been examined.
In this study, we examine the mechanisms that support Plp mRNA localization to centrosomes. We show Plp mRNA localization is puromycin-sensitive, consistent with a co-translational transport mechanism. We further identify a requirement for microtubules to direct Plp mRNA to centrosomes. Through a reporter assay, we discovered the Plp untranslated regions are dispensable for Plp mRNA localization. Rather, regions within the Plp coding sequence (CDS) necessary for PCM scaffolding also direct mRNA localization. We further demonstrate genetic perturbation of the PCM scaffold is sufficient to impair centrosomal mRNA localization. Taken together, these data inform mechanisms underlying Plp mRNA localization and the basis of cell cycle-dependent variances in RNA enrichment at centrosomes.
Results
Microtubules support Plp mRNA localization
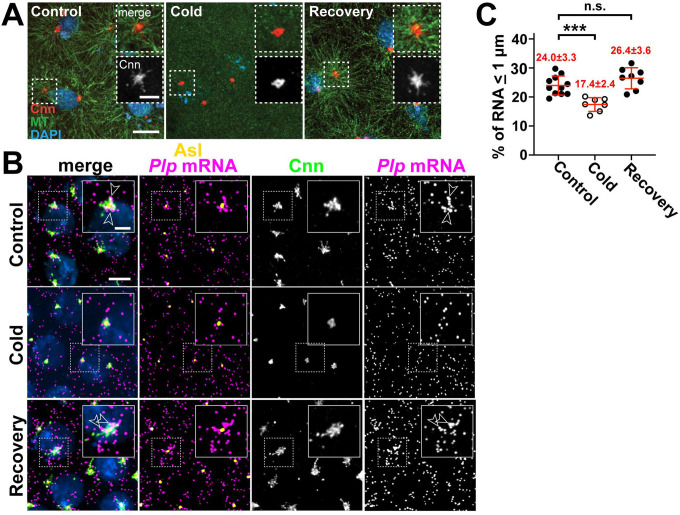
Centrosomes are MTOCs, and RNA localization often utilizes microtubule-based transport, raising the possibility that microtubules help enrich RNA at centrosomes [40, 41]. To investigate the role of microtubules in the subcellular localization of Plp mRNA at centrosomes, we performed microtubule regrowth assays [42]. Microtubule stability is sensitive to temperature; therefore, microtubules were depolymerized by incubating early embryos on ice (see Methods) [43–45]. We first confirmed that cold-shock treatment led to microtubule depolymerization and the loss of Cnn flares, consistent with prior work [24, 46]. To allow microtubule regrowth, we shifted cold-shocked embryos to room-temperature, which also permitted reformation of Cnn flares (Figure 1A).
Figure 1. Plp mRNA localization requires microtubules.
Maximum intensity projections of (A) NC 11 embryos from the indicated conditions labeled with anti-Cnn (PCM; red) and α-Tub antibodies (microtubules; green), and DAPI (DNA; blue). (B) NC 12 P/p-GFP embryos from control, cold-treated, or recovery conditions labeled with GFP smFISH probes to show Plp mRNA distributions (magenta) and labelled with Cnn (green) and Asl (centrioles; yellow) antibodies and DAPI (blue). Dashed box regions are enlarged in insets. Arrowheads show Plp mRNA enrichments at centrosomes. (C) Quantification of GFP mRNA localization (within 1 μm from Asl). Each dot represents a measurement from a single embryo; see Table S1 for N embryos and RNA objects examined. Mean ± S.D. are displayed, n.s. not significant; ***p<0.001 by one-way ANOVA followed by Dunnett’s T3 multiple comparisons test. Scale bars: 5 μm; 2 μm (insets).
Microtubule depolymerization decreased endogenous Plp mRNA localization, as revealed by single molecule fluorescence in situ hybridization (smFISH). This response was reversible, as Plp mRNA localization was restored to WT levels following microtubule regrowth (Figure 1B,C). Thus, microtubules support proper Plp mRNA localization to centrosomes.
Cytoplasmic dynein is a minus-end directed microtubule motor, reviewed in [47]. Prior work established a requirement for dynein to localize PCNT mRNA and protein to centrosomes in cultured human cells [16, 48, 49]. PCNT associates with dynein via a dynein light intermediate chain (DLIC) recognition motif situated in the middle of the PCNT CDS [50]. By sequence analysis, we confirmed that this region contains an AAxxG motif important for DLIC recognition [51]. In contrast, Drosophila Plp and mouse Pcnt lack the AAxxG motif, indicating this region is less well conserved (Figure S1A).
To directly test whether dynein similarly functions in translocating Plp mRNA to centrosomes, we examined RNA distributions in hypomorphic Dynein heavy chain 64C (Dhc) mutant embryos (i.e., DhcLOA homozygous mutants; see Methods). Unexpectedly, we did not observe significant changes to Plp mRNA localization in DhcLOA mutants relative to controls (Figure S1B, C). These findings suggest that either sufficient dynein activity persists in DhcLOA mutants or that other mechanisms support Plp mRNA localization.
Co-translational transport of Plp mRNA to centrosomes
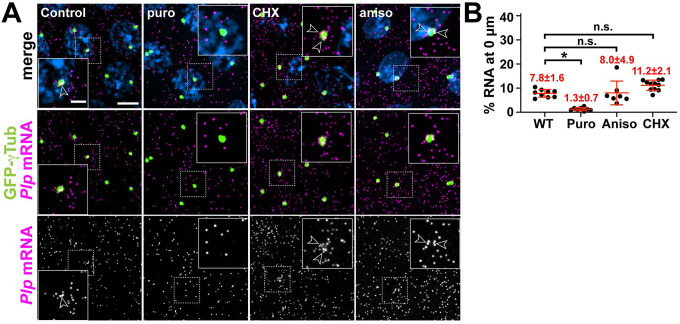
We previously showed some Plp mRNA colocalizes with Plp protein at centrosomes [18]. Consistent with these observations, recent work highlights co-translational transport as a major paradigm for RNA localization to centrosomes [16, 15, 20]. To assess whether translation is required for Plp mRNA localization, we examined Plp distributions following treatment with several translation inhibitors [52].
Puromycin is an A-site tRNA analog that terminates translation and induces ribosome dissociation from the nascent polypeptide [53]. In contrast, anisomycin and cycloheximide (CHX) block translation elongation and freeze ribosomes on mRNAs without releasing the newly synthesized peptide [54–56]. Treatment with these inhibitors revealed Plp mRNA localization is selectively puromycin-sensitive (Figure 2A, B). These results argue that actively engaged ribosomes in association with the nascent peptide are drivers of Plp mRNA localization to centrosomes, similar to human PCNT mRNA [16].
Figure 2. Plp mRNA localization to centrosomes is puromycin-sensitive.
(A) Maximum intensity projections of NC 13 embryos expressing GFP-γTub (green) labeled with Plp smFISH probes (magenta) and DAPI (blue) in controls or following treatment with translation inhibitors: puromycin (puro), cycloheximide (CHX), or anisomycin (aniso). Dashed box regions mark insets. Arrowheads show Plp mRNA enrichments at centrosomes. (B) Percentage of Plp mRNA localizing within 0 μm from the γTub surface. Mean ± S.D. are displayed, n.s. not significant; *p<0.05 by one-way ANOVA followed by Dunnett’s T3 multiple comparisons test. Scale bars: 5 μm; 2 μm (insets).
Domains within the Plp CDS direct Plp mRNA localization
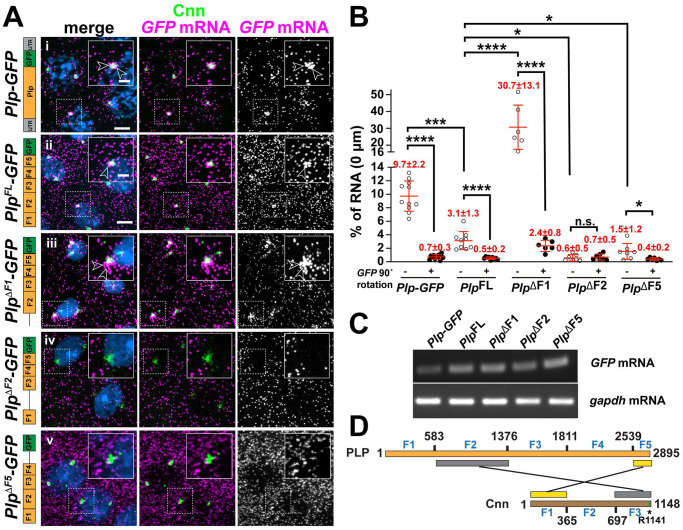
To further investigate the molecular mechanisms of Plp mRNA localization, we utilized a reporter assay to define cis-regulatory elements. As a control, we first examined the localization of endogenous Plp-GFP, an in-frame GFP knock-in at the Plp C-terminus generated via CRISPR (hereafter, Plp-GFP), as schematized in Figure S2A [57]. RNA distributions for Plp-GFP resembled those of untagged Plp mRNA, confirming that the addition of the GFP tag did not alter RNA localization or expression (Figure S2B,C). We then used the maternal α-Tub driver (matGAL4) to direct expression of various GFP reporter transgenes and visualized RNA distributions. Because RNA localization often relies upon sequences and/or structural motifs within the untranslated regions (UTRs) [40], we first examined whether the Plp 5’- and/or 3’-UTRs were sufficient to localize GFP mRNA to centrosomes. Neither the Plp 5’- nor 3’-UTRs directed RNA localization to centrosomes, despite expression levels comparable to controls, suggesting that the localization elements reside within the Plp CDS (Figure S2D, E).
Aligned with this prediction, the Plp CDS was sufficient for RNA localization to centrosomes (Figure 3A,B; PlpFL-GFP). This enrichment was specific and not due to spurious overlap because it was eliminated by rotating the RNA channel by 90° (Figure 3B). Comparing RNA distributions in Plp-GFP versus PlpFL-GFP embryos indicates the Plp CDS mediates localization less efficiently, suggesting that while the Plp CDS encodes sequences necessary and sufficient for RNA localization to centrosomes, other features (e.g., regulatory sequences, splicing events, etc.) influence the extent of RNA enrichment (Figure 3A,B and Figure S2E). Nevertheless, a requirement for the Plp CDS for RNA localization is consistent with the puromycin-sensitivity noted above.
Figure 3. Plp mRNA localization requires sequences within the Plp CDS.
(A) Maximum intensity projections of NC 11 embryos expressing mCherry-Cnn (green) and labeled with GFP smFISH probes (magenta) to mark transgenic Plp mRNA localization and DAPI (blue) in the following genotypes: (i) Plp-GFP, (ii) UAS-PlpFL-GFP, (iii) UAS-PlpΔF1-GFP, (iv) UAS-PlpΔF2-GFP, and (v) UAS-PlpΔF5-GFP. Transgenes in (ii-v) were expressed using matGAL4 in the presence of endogenous Plp. Construct schematics are shown to the left. Arrowheads show RNA enrichments at centrosomes. (B) Quantification of GFP mRNA localization (0 |_im from Cnn surface). Each dot represents a measurement from a single embryo; see Table S1 for N embryos and RNA objects examined. The RNA channel was rotated 90° (+) and images re-quantified to assay the specificity of localization. (C) RT-PCR was used to assay the relative expression of the indicated GFP-tagged constructs from 0–2 hr embryos. (D) Schematic adapted from (Lerit et al., 2015) showing the two direct interaction modules between Plp and Cnn. Asterisk denotes the single point mutation (R1141H) that defines the cnn84 allele and abolishes the direct interaction between Plp F2 and Cnn CM2 (green bar). Mean ± S.D. are displayed, n.s. not significant; *p<0.05; ***p<0.001; ****p<0.0001 by one-way ANOVA followed by Dunnett’s T3 multiple comparisons test. Uncropped gels are available at <10.6084/m9.figshare.24926298>. Scale bars: 5 μm; 2 μm (insets).
To uncover which regions of the Plp CDS direct RNA localization, we leveraged several Plp truncation lines, which divide the ORF into five fragments (F1–F5) and incorporated them into our reporter assay [24, 58] (Figure 3). The truncation lines were all overexpressed relative to Plp-GFP, but comparable to PlpFL-GFP (Figure 3C). Unexpectedly, we found that an N’-terminal truncation of Plp (PlpΔF1-GFP) resulted in significantly more Plp mRNA at centrosomes, suggesting that elements within F1 somehow limit Plp mRNA localization to centrosomes. In contrast, deletion of either F2 (PlpΔF2-GFP) or F5 (PlpΔF5-GFP) resulted in significantly less Plp localized to centrosomes (Figure 3A,B). Taken together, these results suggest that expression levels alone are insufficient to instruct RNA localization to centrosomes. Rather, RNA localization to centrosomes is driven by discrete cis-elements. In particular, PlpΔF2 abolished Plp mRNA localization, indicating that F2 is required for Plp mRNA localization or anchorage at centrosomes.
The PCM scaffold anchors RNAs at centrosomes
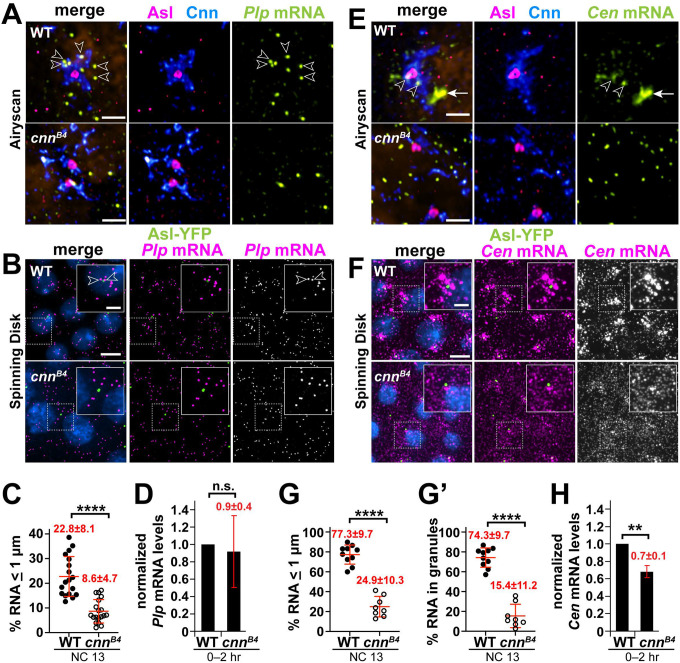
We previously reported that Plp F2 and F5 mediate direct protein-protein interactions with Cnn F3 and Cnn F1, respectively, to maintain the PCM scaffold [24]. The PCM scaffold is impaired in cnnB4 mutants, defined by an R1141H substitution within the highly conserved Cnn Motif 2 (CM2) and sufficient to block the interaction between Plp F2 and Cnn F3 (Figure 3D) [59, 24]. Using super-resolution microscopy, we found that Plp mRNA appeared displaced from the fragmented PCM in cnnB4 mutants, as compared to age-matched controls (Figure 4A). Quantification revealed a significant reduction in Plp mRNA localizing within 1 μm from the centriole (marked with Asterless, Asl) in NC 13 cnnB4 mutants, as compared to controls (22.8±8.1% in WT vs. 8.6±4.7% in cnnB4; Figure 4B,C). A similar reduction was observed in early NCs (Figure S3 A,B). Because total levels of Plp mRNA are similar in 0–2 hr WT and cnnB4 embryos (Figure 4D), we conclude that the PCM scaffold is required to anchor Plp mRNA at centrosomes, likely via protein-protein interactions between Plp and Cnn.
Figure 4. The centrosome scaffold permits mRNA localization.
Maximum intensity projections of NC 13 control and cnnB4 embryos labeled with (A and B) Pip mRNA or (E and F) Cen mRNA smFISH probes. In (A and E), embryos were co-stained with smFISH probes (green), anti-Cnn (blue) and Asl (magenta) antibodies, and DAPI (orange; nuclei), then imaged using a Zeiss LSM 880 Airyscan. For (B and F), embryos expressing Asl-YFP (green) were labeled with smFISH probes (magenta) and DAPI (blue) then imaged by spinning disk confocal microscopy. (C) Quantification shows the percentage of Pip mRNA localizing within 1 μm from the Asl surface. (D) Levels of Pip mRNA or (H) Cen mRNA were normalized to RP49 as detected by qPCR from 0–2 hr WT versus cnn84 embryos and displayed relative to the WT control. (G) The percentage of Cen mRNA localizing and (G’) residing within granules (defined as > 4 RNA molecules per object [17]) within 1 μm from the Asl surface. Each dot represents a measurement from a single embryo; see Table S1 for N embryos and RNA objects examined. Mean ± S.D. are displayed, n.s., not significant or ****p<0.0001 by unpaired student t-test. Scale bars: (A and E) 1 μm; (B and F) 5 μm; 2 μm (insets).
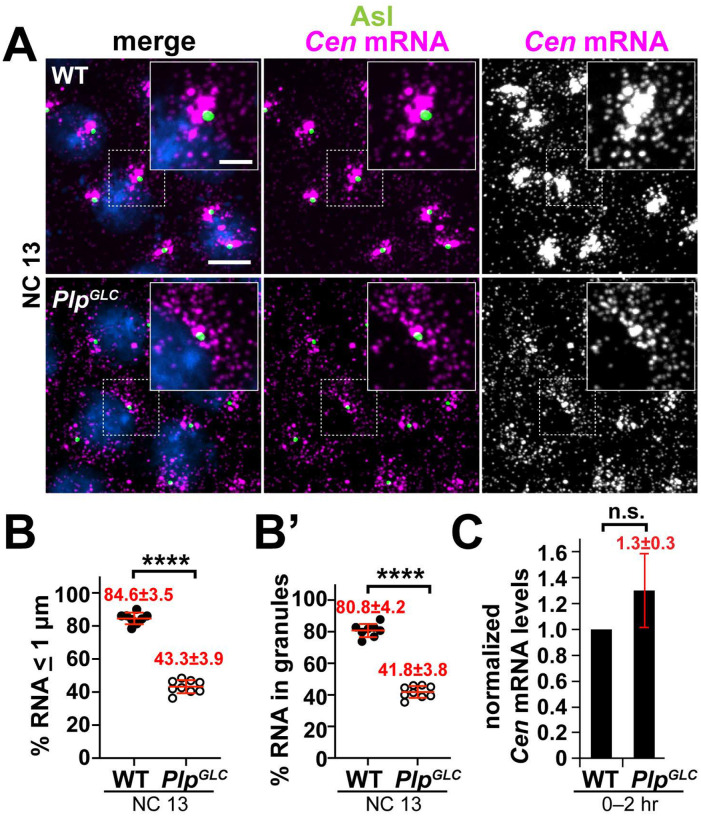
Might the PCM scaffold support the localization of other centrosome-localized RNAs? Normally, Cen mRNA becomes significantly enriched at interphase NC 13 centrosomes within micron-scale granules. However, Cen mRNA granules appear diminished in cnnB4 mutants [17]. Indeed, super-resolution imaging revealed fewer and smaller Cen mRNA granules in cnnB4 embryos, as compared to controls (Figure 4E). Quantitative analysis confirmed significantly less Cen mRNA resides within granules or localizes to centrosomes in cnnB4 versus controls (Figure 4F–G’; Figure S3C–D’).
We next examined whether this reduction in Cen mRNA localization might be attributed to changes in RNA abundance by qPCR. While Cen RNA levels are about 30% reduced in 0–2 hr embryonic extracts from cnnB4 mutants relative to WT, this difference is unlikely to account for the 3-fold reduction in RNA localization to centrosomes (Figure 4G,H). Taken together, these data suggest that an intact PCM scaffold also contributes to Cen mRNA localization, perhaps by stabilizing Cen RNA granules. Future work is required to investigate the relationship between Cnn and Cen RNA granule formation and whether the Cen granule regulates Cen mRNA stability.
As a whole, these studies help establish a generalizable requirement for the PCM scaffold to dock localized RNAs at the centrosome. We sought to further test this model using an independent approach. Plp functionally cooperates with Cnn to ensure PCM scaffolding. Thus, loss of Plp also leads to a PCM fragmentation phenotype [24, 25]. We therefore examined Cen mRNA localization within Plp null embryos derived from germline clones (PlpGLC embryos; see Methods). The significant reduction in Cen mRNA localization to centrosomes and residence within granules in PlpGLC embryos relative to controls support a model wherein the PCM scaffold functions not only in the organization of PCM proteins, but for localized mRNAs as well (Figure 5A–C).
Figure 5. The centrosome scaffold supports Cen mRNA localization and granule formation.
Maximum intensity projections of NC 13 (A) WT or (B) PlpGLC embryos labeled with Cen smFISH probes (magenta), Asl antibodies (green), and DAPI (blue). Charts show the percentage of Cen mRNA (B) localizing or (B’) residing within granules (> 4 RNA molecules per object) within 1 pm from the Asl surface. Each dot represents a measurement from a single embryo; see Table S1 for N embryos and RNA objects examined. (C) Levels of Cen mRNA were normalized to RP49 mRNA as detected by qPCR from 0–2 hr WT versus PlpGLC embryos and displayed relative to the WT control. Mean ± S.D. are displayed, n.s. not significant, and ****p<0.0001 by unpaired student t-test. Scale bars: 5 μm; 2 μm (insets).
Discussion
Although some RNAs localize to centrosomes, relatively little is known about their mechanism of localization and function. In this study, we examined the mechanisms of Plp mRNA localization to centrosomes. We found that Plp mRNA localization requires microtubules, association with the nascent peptide, and defined permissive and restrictive localization elements within the Plp CDS. Our findings are consistent with the idea that Plp mRNA localization is supported by a protein-protein interaction between Plp F2 and Cnn CM2. We propose that emergence of Plp F2 from the ribosome renders the Plp mRNA-protein complex sufficient to associate with Cnn (Figure 6), effectively recruiting Plp mRNA to centrosomes. Finally, we demonstrated a general requirement for the PCM scaffold to support RNA localization at centrosomes.
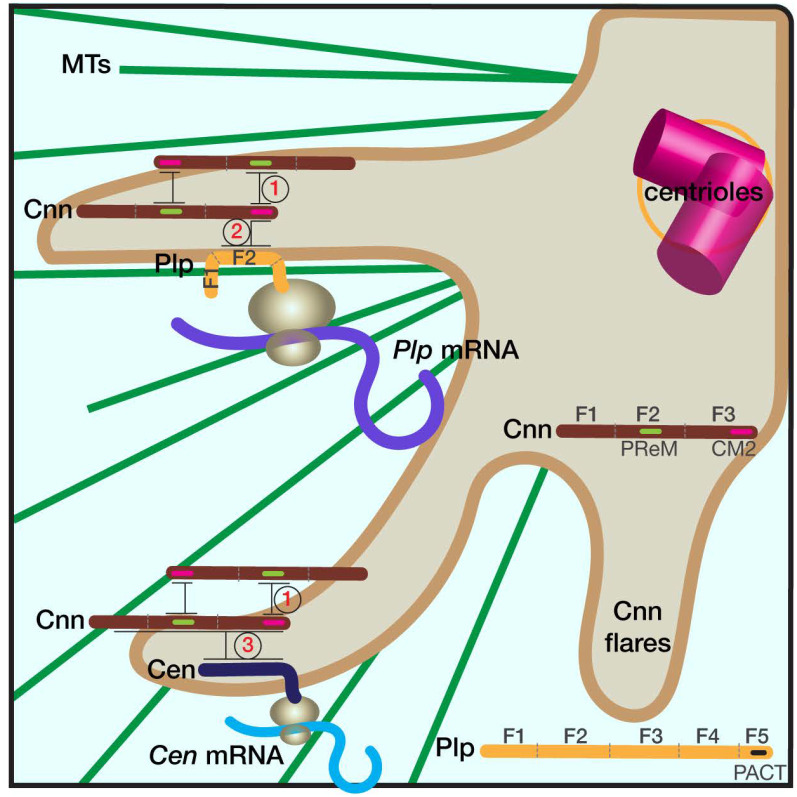
Figure 6. Schematic of RNA localization to centrosomes.
The cartoon shows part of a centrosome with extended Cnn flares (brown), which contribute to PCM scaffolding. Elaboration of the PCM scaffold requires oligomerization of Cnn between its PReM and CM2 motifs (interaction 1), and a direct interaction between CM2 and PLP F2 (interaction 2; [61, 39, 24]). Simplified protein architectures of Cnn and Plp are noted in the figure. We propose that the Plp F2-Cnn CM2 interaction helps transmit and/or anchor the Plp mRNA-protein complex to the centrosome. Accordingly, microtubules (MTs, green), are required both for the extension of Cnn flares and the localization of Plp mRNA to centrosomes [24, 46, 39, 62]. Cen mRNA also localizes to the centrosome via co-translational transport, and Cen protein interacts directly with Cnn (interaction 3). Mutant analysis indicates that an intact PCM scaffold is required for the localization of both Plp and Cen mRNAs. We further propose that the temporal control of PCM scaffold elaboration (i.e., extension of Cnn flares) similarly regulates RNA localization to centrosomes.
Surprisingly, we found an N’-terminal deletion of Plp F1 led to a significant increase in Plp mRNA localization. Recent work demonstrates that the F1 deletion stabilizes Plp, leading to increased protein levels, raising the possibility that the upregulated Plp protein levels in PlpΔF1 mutants might drive Plp mRNA enrichment at centrosomes [60]. However, deletion of F2 led to a significant reduction in Plp mRNA localization to centrosomes, despite a similar uptick in Plp protein levels [60]. These findings argue that protein expression levels alone do not direct RNA enrichment at centrosomes. It is more likely that a specific element in F1 limits Plp mRNA localization. Future investigation will uncover how Plp F1 suppresses the recruitment of Plp mRNA to centrosomes.
In contrast, we found Plp F2 is necessary for Plp mRNA localization. This observation is intriguing given our prior work indicating a direct interaction between Plp F2 and Cnn F3, via the CM2, supports centrosome scaffolding and mitotic fidelity [24]. Cnn CM2 promotes centrosome scaffolding through its interaction with a leucine zipper region within a previously identified phosphoregulated-multimerization (PReM) domain residing in the middle of the Cnn CDS (Figure 6, interaction 1). Phosphorylation of the PReM domain by polo kinase promotes interaction with Cnn CM2, contributing to Cnn oligomerization and scaffold formation [39, 61]. This phosphoregulation likely regulates the timing of centrosome scaffold assembly. Our data indicate Plp mRNA localization requires the Cnn scaffold, suggesting the cell cycle-dependent enrichments of Plp mRNA at centrosomes are likely entrained to centrosome scaffold formation (Figure 6, interaction 2).
We also uncovered a requirement for microtubules to support Plp mRNA localization. Of note, extension of the centrosome scaffold is also microtubule-dependent, as microtubule depolymerization results in the retraction and condensation of Cnn flares (Figure 1A) [24, 46, 39, 62]. In principle, microtubules may be required for Plp mRNA localization because they are necessary for scaffold formation. Alternatively, microtubules may help traffic and/or anchor Plp mRNA to centrosomes. Live imaging the dynamics of Plp mRNA will help decipher these requirements.
Which feature(s) within Plp F2 mediate Plp mRNA localization await identification. The recent development of AlphaFold2 allows us to render predictive models of the Plp F2–Cnn CM2 interface. The CM2 within Cnn F3 is critical for centrosome scaffold formation and the interaction with Plp F2, which can be abolished by the cnnB4 R1141H mutation [24, 59, 61, 39]. Using AlphaFold Multimer, an extension of AlphaFold2, and the COSMIC2 cloud platform, we modeled the interface between Plp F2 and Cnn CM2, which provided five predictive structural models [63, 64]. Cnn exists as a monomer in the cytoplasm [65]. Underscoring the fidelity of the AlphaFold predictions, our Cnn CM2 models are similar to the previously reported 3D crystal structure of the CM2 monomer (PDB: 5MVW), with a root mean square deviation (RMSD) ranging from 0.8 to 1.4, confirming the two superimposed atomic coordinates are similar (Figure S3E) [61]. We centered our analysis of the AlphaFold models on the Plp F2 residues proximal to Cnn CM2. Intriguingly, all models predicted a C-terminal region of Plp F2 (amino acids 1177–1306) apposed Cnn CM2 (Figure S3F). We speculate that this region is important for the Plp F2 and Cnn F3 interaction and key for Plp mRNA localization. While these predictions suggest that Cnn interacts with Plp as a monomer, this requires experimental validation.
Another interacting partner of Cnn is Cen, although the precise interaction interface remains less defined (Figure 6, interaction 3) [59]. We speculate that this protein-protein interaction similarly supports Cen mRNA localization to centrosomes. Given Cen and Plp mRNA localization both require an intact centrosome scaffold, RNA enrichments are probably temporally coordinated with PCM organization (e.g., entrained to elaboration of the Cnn-rich PCM flares). Nonetheless, the distributions of Cen and Plp mRNAs are distinct. Cen mRNA organizes within large RNA granules, whereas Plp mRNA does not. In addition, the localization of the Cen mRNA granule often tends to be more peripheral to the Cnn flares of the mother centrosome [17]. Understanding the mechanisms underlying these differences, and testing their influence with respect to centrosome activity, is a promising area of future research.
Materials and methods
Fly stocks
The following stocks and transgenic lines were used: y1w1118 (Bloomington Drosophila Stock Center (BDSC) #1495) was used as the WT control. Null plp mutant germline clones were generated by the FLP/ovoD method using FRT2A, plp2172 recombinant chromosomes [66, 67]. DhcLOA is a hypomorphic mutation in the dynein heavy chain (Dhc64C) gene defined by an F597Y mutation within Dhc (modeled after the murine Dync1h1 F580Y mutation, legs-at-odd-angles (LOA) [68]. Ubi-GFP-γ-Tub23C expresses GFP- γ Tub under the Ubiquitin (Ubi) promotor [26]; Ubi-Asl-YFP expresses Asl-YFP under the Ubi promoter [69]; PBAC-GFP-Cnn, expresses Cnn tagged at the N-terminus with EGFP under endogenous regulatory elements [24]; mCherry-Cnn expresses Cnn tagged with mCherry with endogenous regulatory elements [70]; Plp-GFP is an in-frame C-terminal GFP knock-in at the Plp endogenous locus generated via CRISPR [57]. UAS-PLPFL-GFP (PlpFL-GFP) expresses full-length PLP isoform PF under the control of upstream activating sequence (UAS) sites [58]; the truncated Plp lines, including ΔF1, ΔF2, ΔF5, all express truncated Plp isoform PF fragments under the UAS promoter and are C-terminally tagged with GFP [58]. The maternal α-Tub GAL4 (mat-GAL4; BDSC #7063) driver was used to drive the expression of all UAS transgenes. To examine maternal effects, mutant or transgenic embryos are progeny derived from mutant or transgenic mothers. Flies were raised on Bloomington formula ‘Fly Food B’ (Lab-Express; Ann Arbor, MI), and crosses were maintained at 25°C in a light and temperature-controlled incubator chamber.
smFISH detection
Stellaris Plp and GFP smFISH probes conjugated to Quasar 570 or 670 dyes (LGC Biosearch Technologies; Middlesex, UK) were designed against the coding region for each gene using the Stellaris RNA FISH probe designer [17, 71, 18]. smFISH probes were dissolved in nuclease-free water at 25 μM and stored at −20°C before use.
smFISH experiments were performed as previously described using RNase-free solutions [17, 71, 18]. Fixed embryos were rehydrated and washed first in 0.1% PBST (PBS plus 0.1% Tween-20), then in wash buffer (WB; 10% formamide and 2× SSC supplemented fresh each experiment with 0.1% Tween-20 and 2 μg/mL nuclease-free BSA)., then incubated with 100 μL of hybridization buffer (HB; 100 mg/mL dextran sulfate and 10% formamide in 2× SSC supplemented fresh each experiment with 0.1% Tween-20, 2 μg/mL nuclease-free BSA, and 10 mM ribonucleoside vanadyl complex (RVC; S1402S; New England Biolabs; Ipswich, MA) for 10–20 min in a 37°C water bath. Embryos were then incubated in 25 μL of HB containing 0.5 μM smFISH probes in a 37°C water bath overnight. Probes used in this study are listed in Table S2. Embryos were washed three times for 30 min in prewarmed WB, stained with DAPI (1:1000) for 1 hr at room temperature, washed with 0.1% PBST, and mounted with Vectashield mounting medium (H-1000; Vector Laboratories; Burlingame, CA). Slides were stored at 4°C and imaged within 1 week.
Dual smFISH and immunofluorescence
Dual smFISH and IF experiments were optimized for maintaining the integrity of RNA signals, as previously described [17, 18]. All the following steps were performed with RNase-free solutions. Fixed embryos were processed exactly as described above for smFISH, except for the addition of primary antibody at the same time embryos were incubated overnight in 25 μL of HB containing 0.5 μM smFISH probes in a 37°C water bath. On the next day, embryos were washed four times for 30 min in prewarmed WB, stained with secondary antibody and DAPI (1:1000) for 2 hr at room temperature, washed with 0.1% PBST, and mounted with Vectashield mounting medium (H-1000; Vector Laboratories). Slides were stored at 4°C and imaged within 1 week.
Microtubule depolymerization and recovery assay
0.5–2.5 hr YFP-Asl embryos were collected and dechorionated with bleach for 30 s. The dechorionated embryos were incubated on ice for 5 min to disrupt the microtubules. Embryos were then either immediately fixed in a 1:1 solution of heptane:37% formaldehyde for 3 min, or, to permit microtubule regrowth (recovery), embryos were incubated in room-temperature PBS for 5 min before the fixation. After fixation, all embryos were rinsed in PBS and manually devitellinized as described [17].
Translational inhibition
To inhibit translation, embryos were treated with inhibitors diluted in Robb’s medium (1 mM calcium chloride, 10 mM glucose, 100 mM HEPES (pH 7.2), 1.2 mM magnesium chloride, 55 mM potassium acetate, 40 mM sodium acetate, and 100 mM sucrose) [72]. To begin, 0.5–2.5 hr embryos were collected and incubated in a 1:1 solution (450 μl: 450 μl) of heptane: Robb’s medium with the appropriate drug or an equivalent volume of DMSO [22]. The concentrations and duration of treatment for each drug are: 3 mM puromycin (Sigma-Aldrich P8833) for 10 min; 0.1 mM anisomycin (Sigma-Aldrich A9789) for 15 min; 0.71 mM cycloheximide (VWR, 97064–724) for 15 min. After drug incubation, Robb’s medium was removed, and 450 μl of 4% paraformaldehyde in PBS was added, and embryos were fixed for 20 min before devitellinization.
Immunofluorescence
For immunofluorescence with Asl and Cnn antibodies, embryos were fixed in a 1:1 solution of anhydrous methanol (Sigma, #322415): heptane for 15 s and devitellinized in methanol by shaking. For visualization of MTs, embryos were prepared as previously described [73]. Briefly, embryos were fixed in a 1:1 mixture of 37% paraformaldehyde: heptane for 3 min, rinsed in PBS, and manually devitellinized using 30G PrecisionGlide needles (BD). Fixed embryos were rehydrated, blocked in BBT buffer (PBS supplemented with 0.1% Tween-20 and 0.1% BSA), and incubated overnight at 4°C with primary antibodies diluted in BBT. After washing, embryos were further blocked in BBT supplemented with 2% normal goat serum and incubated for 2 hr at room temperature with secondary antibodies and DAPI (10 ng/ml, ThermoFisher). Embryos were mounted in Aqua-Poly/Mount (Polysciences, Inc.) prior to imaging.
The following primary antibodies were used: guinea pig anti-Asl (1:4000, gift from G. Rogers, University of Arizona), rabbit anti-Cnn (1:4000, gift from T. Megraw, Florida State University), mouse anti-α-Tubulin DM1α (1:500, Sigma-Aldrich T6199). Secondary antibodies: Alexa Fluor 488, 568, or 647 (1:500, Molecular Probes), and DAPI (10 ng/ml, ThermoFisher).
Microscopy and image analysis
Images were acquired on a Nikon Ti-E system fitted with a Yokagawa CSU-X1 spinning disk head, Hamamatsu Orca Flash 4.0 v2 digital complementary metal oxide-semiconductor (CMOS) camera, Perfect Focus system (Nikon), Nikon LU-N4 solid state laser launch (15 mW 405, 488, 561, and 647 nm) using a Nikon 100x, 1.49 NA Apo TIRF oil immersion objective. The microscope was powered through Nikon Elements AR software on a 64-bit HP Z440 workstation.
Images in Figure 4A and 4E were acquired on a Zeiss LSM 880 AiryScan microscope with a 63× 1.4 NA oil objective (“SR” mode; 2x averaging; 1.32 μs pixel dwell). Raw images were processed with Airyscan joint deconvolution in Zen Blue with varying iterations per channel (15 iterations for Plp or Cen mRNA, 15 iterations for Cnn, 20 iterations for Asl).
smFISH signals were detected and single molecule normalization was performed as described [17, 71, 18]. Briefly, single-channel .tif raw images were segmented in three dimensions using Python scripts adapted from the Allen Institute for Cell Science Cell Segmenter [74]. Each segmented image was compared with the raw image to validate accurate segmentation. RNA objects of ≥50 pixels in segmented images were identified, and object features were extracted, which included surface coordinates. Distances were measured from the surface of each RNA object to the surface of the closest centrosome. We calculated the percentage of total RNA within 1 μm from the centriole (Asl) or 0 μm from the PCM (Cnn or γTub) surface and selected 10, 8, 6 and 4 μm as the upper boundary for the pseudo-cell radius for NC 10, NC 11, NC 12, and NC 13; respectively, based on measuring the centrosome-to-centrosome distances from a set of representative images. Later interphase/prophase embryos were selected by their large, round nuclei and separated centrosomes.
Fiji (National Institutes of Health; [75]) was used to rotate, split, or merge channels. Images were cropped and brightness/contrast adjusted using Adobe Photoshop. Figures were assembled in Adobe Illustrator.
RT-PCR
RNA was extracted from ~2–5 mg of dechorionated 0–2 hr embryos per biological replicate using TRIzol Reagent (#15596026, ThermoFisher Scientific) and treated with1 μL TURBO Dnase (2 U/μL, # AM1907, ThermoFisher Scientific) prior to RT-PCR. 500 ng of RNA was reverse transcribed to cDNA with the iScript cDNA Synthesis Kit following the manufacturer’s protocol (Bio-Rad, #1708891).
qPCR was performed on a Bio-Rad CFX96 Real-time system with iTaq Universal SYBR Green Supermix (#1725121, Bio-Rad; Hercules, CA). Values were normalized to RpL32 (rp49) expression levels. Ct values from the qPCR results were analyzed and the relative expression levels for each condition were calculated using Microsoft Excel. Three biological replicates and three technical replicates were performed on a single 96-well plate using the following primers:
rp49 Forward: CATACAGGCCCAAGATCGTG
rp49 Reverse: ACAGCTTAGCATATCGATCCG
Plp Foward: CGCAGCAAGGAGGAGATAAC
Plp Reverse: TCAGCCTGCAGTTTGTTCAC
Cen Forward: AAAGTACCCCCGGTAACACC
Cen Reverse: TGAGGATACGACGCTCTGTG
To detect the relative RNA expression level for Plp reporter assays, 50 ng cDNA was amplified by PCR for 30 cycles using Phusion High Fidelity DNA Polymerase (M0530L; New England Biolabs) using the following primers:
Plp Forward: CACAAACAGCTCGATCAGGA;
Plp Reverse: TCATTTTGAGCAACCAGCAG;
GFP Forward: ACGTAAACGGCCACAAGTTC;
GFP Reverse: AAGTCGTGCTGCTTCATGTG;
gapdh Forward: CACCCATTCGTCTGTGTTCG;
gapdh Reverse: CAACAGTGATTCCCGACCAG
Statistical methods
Data were plotted and statistical analysis was performed using GraphPad Prism (v. 9) software. To calculate significance, distribution normality was first confirmed with a D’Agnostino and Pearson normality test. Data were then analyzed by unpaired t-test, one-way ANOVA test, or the appropriate non-parametric test and are displayed as mean ± SD. Data shown are representative results from at least two independent experiments.
Protein-protein Complex Prediction
To model the interaction between these Plp and Cnn, we ran AlphaFold2 (2.3.2) using the multimer model on the COSMIC2 cloud platform with the amino acid sequences of Plp F2: 584–1357 (isoform RF) and Cnn CM2: 1082–1148. AlphaFold2 generated five predicted models. We used PyMOL (version 2.5.7) to visualize and process images of these predicted models. We compared the similarity between 3D protein structures by calculating the Root Mean Square Deviation (RMSD) using the align function in PyMOL by running the command: align object1, object2; where object 1 was the CM2 model predicted by AlphaFold, and object 2 was the published 3D crystal structure of CM2 motif (PDB: 5MVW, chain A).
Supplementary Material
Figure S1. Dynein is not essential for Plp mRNA localization. (A) Amino acid alignment of Drosophila melanogaster (Dmel) Plp, mouse (Mmus) PCNT, and human (Hsap) PCNT (Clustal Omega; https://www.ebi.ac.uk/Tools/msa/clustalo/). The amino acid numbers of Plp and PCNT are listed above and fully conserved (*), strongly similar (:), and weakly similar (.) residues are indicated. The dynein light intermediate chain (DLIC) binding motif in human PCNT is noted (blue). (B) Maximum intensity projections of NC 11 embryos of WT and homozygous DhcLOA mutants labeled with Plp smFISH probes (magenta), Asl antibodies (green), and DAPI (blue). Dashed box regions mark insets. (C) The percentage of Plp mRNA localizing within 1 μm from the surface of Asl. Each dot represents a measurement from N=8 WT and N=7 DhcLOA NC 11 embryos; see Table S1. Mean ± S.D. are displayed. n.s. not significant by unpaired student t-test. Scale bars: 5 μm (main panels); 2 μm (insets).
Figure S2. Plp mRNA localization requires the Plp CDS. (A) Maximum intensity projections of NC 11 embryos labeled with anti-Asl antibodies (green), Plp smFISH probes in WT, or GFP smFISH probes in Plp-GFP (magenta). Schematic diagrams of labelled RNAs are shown to the left. (B) The percentage of Plp mRNA localizing within 1 μm of Asl from N=7 WT and 10 Plp-GFP NC 11 embryos. (C) Relative expression level of endogenous Plp RNA in 0–2 hr embryos of the indicated genotypes assayed by RT-PCR. (D) Maximum intensity projections of NC 11 embryos labeled with anti-Cnn antibodies (green), GFP smFISH probes (magenta) and DAPI (blue) in the following genotypes: (i) UAS-Plp5’UTR-GFP-Plp3’UTR, (ii) UAS-GFP-Plp3’UTR, (iii) UAS-GFP, and (iv) UAS-PlpFL-GFP. Transgenes in (ii–v) were expressed using matGAL4 in the presence of endogenous Plp. Insets are enlarged in the upper-right corners. Arrowheads mark Plp mRNA enriched at centrosomes. Schematic diagrams of GFP-tagged constructs are shown on the left. (E) Relative expression level of the GFP-tagged reporter RNAs in 0–2 hr embryos of the indicated genotypes was assayed by RT-PCR. Uncropped gels are available at < https://figshare.com/s/360dfc97047235a2b18a and https://figshare.com/s/71f35163efc18e879e7b >. Scale bars: 5 μm (main panels); 2 μm (insets).
Figure S3. The PCM scaffold permits mRNA localization in early embryos. Maximum intensity projections of NC 11 control and cnnB4 embryos expressing Asl-YFP and labeled with (A) Plp or (C) Cen smFISH probes (magenta) and DAPI (blue). (B) The percentage of Plp mRNA localizing within 1 μm from the Asl surface from N=11 WT and 9 cnnB NC 11 embryos. The percentage of Cen mRNA (D) localizing and (D’) residing within granules (defined as ≥ 4 RNA molecules per granule) within 1 μm from the Asl surface from N=11 WT and 10 cnnB4 NC 11 embryos. (E) The AlphaFold Cnn CM2 predicted structure (gray) was superimposed on the 3D crystal structure of Cnn CM2 (PDB: 5MVW; green) [61]. RMSD = 1.4111, (433 to 433 atoms) out of 490 atoms. (F) Side view and top view images of the top 3-ranked AlphaFold models of the Plp F2–Cnn CM2 interaction. Shown are Plp amino acids 1177–1306 (yellow) and Cnn CM2 (gray). Mean ± S.D. is displayed. ****p<0.0001 by unpaired student t-test. Scale bars: 5 μm (main panels); 2 μm (insets).
Table S1. List of objects quantified in the figures. Tabulation of genotypes, embryos, centrosomes, and RNA objects quantified in this study.
Table S2. smFISH probe sequences. List of Plp, Cen, and EGFP mRNA probes used in this study.
Acknowledgements
For gifts of reagents, we thank Drs. Clemens Cabernard, Timothy Megraw, Nasser Rusan, Greg Rogers, and Simon Bullock. For technical advice and assistance, we are grateful to Dr. Paul Donlin-Asp, Dr. Carolina Eliscovich, Ms. Shuristeen Joubert, Ms. Jordan Goldy, and Mr. Jovan Brockett. We thank Drs. Brian Galletta and Matthew Hannaford for insightful conversations and Dr. Girish Deshpande for constructive feedback on the manuscript.
This work was supported by the Maximizing Investigators’ Research Award (MIRA R35) from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) R35GM138183 to JRB, National Science Foundation Graduate Research Fellowship (NSF GRFP) DGE-1842190 to MAQ, and NIH grants K99GM143517 to JF and R01GM138544 to DAL.
Footnotes
Competing interest statement
The authors have no competing interests to declare.
Data Availability Statement
Uncropped gels from Fig. 3 and S2 are available on FigShare:
https://figshare.com/s/103951922143448b05d2
References:
- [1].Nigg E. A. and Raff J. W., “Centrioles, Centrosomes, and Cilia in Health and Disease,” Cell, 2009. 139 (4): 663–678. [DOI] [PubMed] [Google Scholar]
- [2].Doxsey S., Zimmerman W., and Mikule K., “Centrosome control of the cell cycle,” Trends in Cell Biology, 2005. 15 (6): 303–311. [DOI] [PubMed] [Google Scholar]
- [3].Vertii A., Hehnly H., and Doxsey S., “The Centrosome, a Multitalented Renaissance Organelle,” Cold Spring Harb Perspect Biol, 2016. 8 (12): a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Müller H. et al. , “Proteomic and functional analysis of the mitotic Drosophila centrosome,” The EMBO journal, 2010. 29 (19): 3344–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gupta G. D. et al. , “A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface,” Cell, 2015. 163 (6): 1484–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carden S. et al. , “Proteomic profiling of centrosomes across multiple mammalian cell and tissue types by an affinity capture method,” Dev Cell, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lécuyer E. et al. , “Global Analysis of mRNA Localization Reveals a Prominent Role in Organizing Cellular Architecture and Function,” Cell, 2007. 131 (1): 174–187. [DOI] [PubMed] [Google Scholar]
- [8].Robbins E., Jentzsch G., and Micali A., “The centriole cycle in synchronized HeLa cells,” J Cell Biol, 1968. 36 (2): 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moritz M., Braunfeld M. B., Sedat J. W., Alberts B., and Agard D. A., “Microtubule nucleation by γ-tubulin-containing rings in the centrosome,” Nature, 1995. 378 (6557): 638–640. [DOI] [PubMed] [Google Scholar]
- [10].Gould R. R. and Borisy G. G., “The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation,” J Cell Biol, 1977. 73 (3): 601–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moritz M., Braunfeld M. B., Guénebaut V., Heuser J., and Agard D. A., “Structure of the γ-tubulin ring complex: a template for microtubule nucleation,” Nature cell biology, 2000. 2 (6): 365–370. [DOI] [PubMed] [Google Scholar]
- [12].Mennella V. et al. , “Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization,” Nature cell biology, 2012. 14 (11): 1159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Khodjakov A. and Rieder C. L., “The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules,” J Cell Biol, 1999. 146 (3): 585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mittasch M. et al. , “Regulated changes in material properties underlie centrosome disassembly during mitotic exit,” J Cell Biol, 2020. 219 (4): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Safieddine A. et al. , “A choreography of centrosomal mRNAs reveals a conserved localization mechanism involving active polysome transport,” Nat Commun, 2021. 12 (1): 1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sepulveda G. et al. , “Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates,” eLife, 2018. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ryder P. V., Fang J., and Lerit D. A., “centrocortin RNA localization to centrosomes is regulated by FMRP and facilitates error-free mitosis,” J Cell Biol, 2020. 219 (12): e202004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fang J. and Lerit D. A., “Orb-dependent polyadenylation contributes to PLP expression and centrosome scaffold assembly,” Development, 2022. 149 (13): dev200426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ryder P. V. and Lerit D. A., “RNA localization regulates diverse and dynamic cellular processes,” Traffic, 2018. 19 (7): 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zein-Sabatto H. and Lerit D. A., “The Identification and Functional Analysis of mRNA Localizing to Centrosomes,” Frontiers in Cell and Developmental Biology, 2021. 9 (3174): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Camargo Ortega G. and Gotz M., “Centrosome heterogeneity in stem cells regulates cell diversity,” Trends Cell Biol, 2022. 32 (8): 707–719. [DOI] [PubMed] [Google Scholar]
- [22].Bergalet J. et al. , “Inter-dependent Centrosomal Co-localization of the cen and ik2 cis Natural Antisense mRNAs in Drosophila,” Cell Reports, 2020. 30 (10): 3339–3352.e6. [DOI] [PubMed] [Google Scholar]
- [23].Doxsey S. J., Stein P., Evans L., Calarco P. D., and Kirschner M., “Pericentrin, a highly conserved centrosome protein involved in microtubule organization,” Cell, 1994. 76 (4): 639–650. [DOI] [PubMed] [Google Scholar]
- [24].Lerit D. A. et al. , “Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function,” J Cell Biol, 2015. 210 (1): 79–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Richens J. H. et al. , “The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM,” Biology Open, 2015. 4 (8): 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lerit D. A. and Rusan N. M., “PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells,” The Journal of Cell Biology, 2013. 202 (7): 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fang J. and Lerit D. A., “Drosophila pericentrin-like protein promotes the formation of primordial germ cells,” genesis, 2020. 58 (3–4): e23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martinez-Campos M., Basto R., Baker J., Kernan M., and Raff J. W., “The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis,” The Journal of Cell Biology, 2004. 165 (5): 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lorenzo-Betancor O. et al. , “PCNT point mutations and familial intracranial aneurysms,” Neurology, 2018. 91 (23): e2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aoyama K.-I. et al. , “New PCNT candidate missense variant in a patient with oral and maxillofacial osteodysplasia: a case report,” BMC Medical Genetics, 2019. 20 (1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chun Y. W. et al. , “Impaired Reorganization of Centrosome Structure Underlies Human Infantile Dilated Cardiomyopathy,” Circulation, 2023. 147 (17): 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Delaval B. and Doxsey S. J., “Pericentrin in cellular function and disease,” The Journal of Cell Biology, 2010. 188 (2): 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rauch A. et al. , “Mutations in the Pericentrin (PCNT) Gene Cause Primordial Dwarfism,” Science, 2008. 319 (5864): 816. [DOI] [PubMed] [Google Scholar]
- [34].Galletta B. J. et al. , “Sperm Head-Tail Linkage Requires Restriction of Pericentriolar Material to the Proximal Centriole End,” Developmental Cell, 2020. 53 (1): 86–101.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karr T. L. and Alberts B. M., “Organization of the cytoskeleton in early Drosophila embryos,” The Journal of cell biology, 1986. 102 (4): 1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Vastenhouw N. L., Cao W. X., and Lipshitz H. D., “The maternal-to-zygotic transition revisited,” Development, 2019. 146 (11): [DOI] [PubMed] [Google Scholar]
- [37].Megraw T. L., Li K., Kao L. R., and Kaufman T. C., “The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila,” Development, 1999. 126 (13): 2829. [DOI] [PubMed] [Google Scholar]
- [38].Lucas E. P. and Raff J. W., “Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila Centrosomin,” Journal of Cell Biology, 2007. 178 (5): 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Conduit Paul T. et al. , “The Centrosome-Specific Phosphorylation of Cnn by Polo/Plk1 Drives Cnn Scaffold Assembly and Centrosome Maturation,” Developmental Cell, 2014. 28 (6): 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Buxbaum A. R., Haimovich G., and Singer R. H., “In the right place at the right time: visualizing and understanding mRNA localization,” Nature Reviews Molecular Cell Biology, 2014. 16 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mofatteh M. and Bullock S. L., “SnapShot: Subcellular mRNA Localization,” Cell, 2017. 169 (1): 178–178.e1. [DOI] [PubMed] [Google Scholar]
- [42].Lattao R., Rangone H., Llamazares S., and Glover D. M., “Mauve/LYST limits fusion of lysosome-related organelles and promotes centrosomal recruitment of microtubule nucleating proteins,” Dev Cell, 2021. 56 (7): 1000–1013 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Desai A. and Mitchison T. J., “MICROTUBULE POLYMERIZATION DYNAMICS,” Annual Review of Cell and Developmental Biology, 1997. 13 (1): 83–117. [DOI] [PubMed] [Google Scholar]
- [44].Müller-Reichert T., Chrétien D., Severin F., and Hyman A. A., “Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate,” Proc Natl Acad Sci U S A, 1998. 95 (7): 3661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li G. and Moore J. K., “Microtubule dynamics at low temperature: evidence that tubulin recycling limits assembly,” Mol Biol Cell, 2020. 31 (11): 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Megraw T. L., Kilaru S., Turner F. R., and Kaufman T. C., “The centrosome is a dynamic structure that ejects PCM flares,” Journal of Cell Science, 2002. 115 (23): 4707. [DOI] [PubMed] [Google Scholar]
- [47].Reck-Peterson S. L., Redwine W. B., Vale R. D., and Carter A. P., “The cytoplasmic dynein transport machinery and its many cargoes,” Nat Rev Mol Cell Biol, 2018. 19 (6): 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Purohit A., Tynan S. H., and Doxsey S. J., “Direct Interaction of Pericentrin with Cytoplasmic Dynein Light Intermediate Chain Contributes to Mitotic Spindle Organization,” 1999. [DOI] [PMC free article] [PubMed]
- [49].Young A., Dictenberg J. B., and Doxsey S. J., “Cytoplasmic Dynein-mediated Assembly of Pericentrin and g Tubulin onto Centrosomes,” 2000. [DOI] [PMC free article] [PubMed]
- [50].Tynan S. H., Purohit A., Doxsey S. J., and Vallee R. B., “Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin,” J Biol Chem, 2000. 275 (42): 32763–8. [DOI] [PubMed] [Google Scholar]
- [51].Chaaban S. and Carter A. P., “Structure of dynein–dynactin on microtubules shows tandem adaptor binding,” Nature, 2022. 610 (7930): 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lin J., Zhou D., Steitz T. A., Polikanov Y. S., and Gagnon M. G., “Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design,” Annual Review of Biochemistry, 2018. 87 (1): 451–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nathans D., “Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains,” Proceedings of the National Academy of Sciences, 1964. 51 (4): 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Grollman A. P., “Inhibitors of protein biosynthesis. II. Mode of action of anisomycin,” J Biol Chem, 1967. 242 (13): 3226–33. [PubMed] [Google Scholar]
- [55].Schneider-Poetsch T. et al. , “Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin,” Nature Chemical Biology, 2010. 6 (3): 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Santos D. A., Shi L., Tu B. P., and Weissman J. S., “Cycloheximide can distort measurements of mRNA levels and translation efficiency,” Nucleic Acids Res, 2019. 47 (10): 4974–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gallaud E. et al. , “Dynamic centriolar localization of Polo and Centrobin in early mitosis primes centrosome asymmetry,” PLoS Biol, 2020. 18 (8): e3000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Galletta B. J. et al. , “Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies,” Molecular Biology of the Cell, 2014. 25 (18): 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kao L.-R. and Megraw T. L., “Centrocortin Cooperates with Centrosomin to Organize Drosophila Embryonic Cleavage Furrows,” Current Biology, 2009. 19 (11): 937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Galletta B. J. et al. , “The E3 ligase Poe promotes Pericentrin degradation,” Molecular Biology of the Cell, 2023. 34 (9): br15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feng Z. et al. , “Structural Basis for Mitotic Centrosome Assembly in Flies,” Cell, 2017. 169 (6): 1078–1089.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lee M. J., Gergely F., Jeffers K., Peak-Chew S. Y., and Raff J. W., “Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour,” Nature cell biology, 2001. 3 643. [DOI] [PubMed] [Google Scholar]
- [63].Richard E. et al. , “Protein complex prediction with AlphaFold-Multimer,” bioRxiv, 2022. 2021.10.04.463034. [Google Scholar]
- [64].Cianfrocco M. A., Wong-Barnum M., Youn C., Wagner R., and Leschziner A., “COSMIC2: A Science Gateway for Cryo-Electron Microscopy Structure Determination,” presented at the Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, New Orleans, LA, USA, 2017. [Online]. Available: 10.1145/3093338.3093390. [DOI] [Google Scholar]
- [65].Li K. and Kaufman T. C., “The Homeotic Target Gene centrosomin Encodes an Essential Centrosomal Component,” Cell, 1996. 85 (4): 585–596. [DOI] [PubMed] [Google Scholar]
- [66].Chou T.-b. and Perrimon N., “The Autosomal FLP-DFS Technique for Generating Germline Mosaics in Drosophila melanogaster,” Genetics, 1996. 144 (4): 1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lerit D. A. et al. , “Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function,” The Journal of cell biology, 2015. 210 (1): 79–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Salvador-Garcia D. et al. , “A force-sensitive mutation reveals a spindle assembly checkpoint-independent role for dynein in anaphase progression,” bioRxiv, 2023. 2023.08.03.551815. [Google Scholar]
- [69].Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., and González C., “Functionally Unequal Centrosomes Drive Spindle Orientation in Asymmetrically Dividing Drosophila Neural Stem Cells,” Developmental Cell, 2007. 12 (3): 467–474. [DOI] [PubMed] [Google Scholar]
- [70].Singh P., Ramdas Nair A., and Cabernard C., “The Centriolar Protein Bld10/Cep135 Is Required to Establish Centrosome Asymmetry in Drosophila Neuroblasts,” Current Biology, 2014. 24 (13): 1548–1555. [DOI] [PubMed] [Google Scholar]
- [71].Ryder P. V. and Lerit D. A., “Quantitative analysis of subcellular distributions with an open-source, object-based tool,” Biology Open, 2020. bio.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].“Modified Robb’s medium,” Cold Spring Harbor Protocols, 2011. 2011 (8): pdb.rec12483. [Google Scholar]
- [73].Theurkauf W. E., “Chapter 25 Immunofluorescence Analysis of the Cytoskeleton during Oogenesis and Early Embryogenesis,” in Methods in Cell Biology, vol. 44, Goldstein L. S. B. and Fyrberg E. A. Eds.: Academic Press, 1994, pp. 489–505. [DOI] [PubMed] [Google Scholar]
- [74].Chen J. et al. , “The Allen Cell Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images,” bioRxiv, 2018. 491035. [Google Scholar]
- [75].Schindelin J. et al. , “Fiji: an open-source platform for biological-image analysis,” Nature Methods, 2012. 9 (7): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Dynein is not essential for Plp mRNA localization. (A) Amino acid alignment of Drosophila melanogaster (Dmel) Plp, mouse (Mmus) PCNT, and human (Hsap) PCNT (Clustal Omega; https://www.ebi.ac.uk/Tools/msa/clustalo/). The amino acid numbers of Plp and PCNT are listed above and fully conserved (*), strongly similar (:), and weakly similar (.) residues are indicated. The dynein light intermediate chain (DLIC) binding motif in human PCNT is noted (blue). (B) Maximum intensity projections of NC 11 embryos of WT and homozygous DhcLOA mutants labeled with Plp smFISH probes (magenta), Asl antibodies (green), and DAPI (blue). Dashed box regions mark insets. (C) The percentage of Plp mRNA localizing within 1 μm from the surface of Asl. Each dot represents a measurement from N=8 WT and N=7 DhcLOA NC 11 embryos; see Table S1. Mean ± S.D. are displayed. n.s. not significant by unpaired student t-test. Scale bars: 5 μm (main panels); 2 μm (insets).
Figure S2. Plp mRNA localization requires the Plp CDS. (A) Maximum intensity projections of NC 11 embryos labeled with anti-Asl antibodies (green), Plp smFISH probes in WT, or GFP smFISH probes in Plp-GFP (magenta). Schematic diagrams of labelled RNAs are shown to the left. (B) The percentage of Plp mRNA localizing within 1 μm of Asl from N=7 WT and 10 Plp-GFP NC 11 embryos. (C) Relative expression level of endogenous Plp RNA in 0–2 hr embryos of the indicated genotypes assayed by RT-PCR. (D) Maximum intensity projections of NC 11 embryos labeled with anti-Cnn antibodies (green), GFP smFISH probes (magenta) and DAPI (blue) in the following genotypes: (i) UAS-Plp5’UTR-GFP-Plp3’UTR, (ii) UAS-GFP-Plp3’UTR, (iii) UAS-GFP, and (iv) UAS-PlpFL-GFP. Transgenes in (ii–v) were expressed using matGAL4 in the presence of endogenous Plp. Insets are enlarged in the upper-right corners. Arrowheads mark Plp mRNA enriched at centrosomes. Schematic diagrams of GFP-tagged constructs are shown on the left. (E) Relative expression level of the GFP-tagged reporter RNAs in 0–2 hr embryos of the indicated genotypes was assayed by RT-PCR. Uncropped gels are available at < https://figshare.com/s/360dfc97047235a2b18a and https://figshare.com/s/71f35163efc18e879e7b >. Scale bars: 5 μm (main panels); 2 μm (insets).
Figure S3. The PCM scaffold permits mRNA localization in early embryos. Maximum intensity projections of NC 11 control and cnnB4 embryos expressing Asl-YFP and labeled with (A) Plp or (C) Cen smFISH probes (magenta) and DAPI (blue). (B) The percentage of Plp mRNA localizing within 1 μm from the Asl surface from N=11 WT and 9 cnnB NC 11 embryos. The percentage of Cen mRNA (D) localizing and (D’) residing within granules (defined as ≥ 4 RNA molecules per granule) within 1 μm from the Asl surface from N=11 WT and 10 cnnB4 NC 11 embryos. (E) The AlphaFold Cnn CM2 predicted structure (gray) was superimposed on the 3D crystal structure of Cnn CM2 (PDB: 5MVW; green) [61]. RMSD = 1.4111, (433 to 433 atoms) out of 490 atoms. (F) Side view and top view images of the top 3-ranked AlphaFold models of the Plp F2–Cnn CM2 interaction. Shown are Plp amino acids 1177–1306 (yellow) and Cnn CM2 (gray). Mean ± S.D. is displayed. ****p<0.0001 by unpaired student t-test. Scale bars: 5 μm (main panels); 2 μm (insets).
Table S1. List of objects quantified in the figures. Tabulation of genotypes, embryos, centrosomes, and RNA objects quantified in this study.
Table S2. smFISH probe sequences. List of Plp, Cen, and EGFP mRNA probes used in this study.
Data Availability Statement
Uncropped gels from Fig. 3 and S2 are available on FigShare:
https://figshare.com/s/103951922143448b05d2