Abstract
Prostate cancer prognostication largely relies on visual assessment of a few thinly sectioned biopsy specimens under a microscope to assign a Gleason grade group (GG). Unfortunately, the assigned GG is not always associated with a patient’s outcome in part because of the limited sampling of spatially heterogeneous tumors achieved by 2-dimensional histopathology. In this study, open-top light-sheet microscopy was used to obtain 3-dimensional pathology data sets that were assessed by 4 human readers. Intrabiopsy variability was assessed by asking readers to perform Gleason grading of 5 different levels per biopsy for a total of 20 core needle biopsies (ie, 100 total images). Intrabiopsy variability (Cohen κ) was calculated as the worst pairwise agreement in GG between individual levels within each biopsy and found to be 0.34, 0.34, 0.38, and 0.43 for the 4 pathologists. These preliminary results reveal that even within a 1-mm-diameter needle core, GG based on 2-dimensional images can vary dramatically depending on the location within a biopsy being analyzed. We believe that morphologic assessment of whole biopsies in 3 dimension has the potential to enable more reliable and consistent tumor grading.
Keywords: prostate cancer, Gleason grading, intrabiopsy variability, 3D pathology, Gleason grading variability
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death in the United States and the fifth worldwide.1 The worldwide incidence is 1.3 million with a mortality rate of 359,000 per year.2 Surgical removal of the prostate, ie, radical prostatectomy (RP) and radiation therapy are standard treatments for localized PCa. However, despite such aggressive treatment approaches, 30% to 40% of patients still experience biochemical recurrence,3,4 defined as a rise in the blood level of prostate-specific antigen in patients with PCa. Likewise, many patients with indolent PCa may be overtreated with RP and radiation therapy, which can lead to severe complications, such as urinary incontinence or erectile dysfunction.5,6 This suggests that an improved indicator of tumor aggressiveness is needed, allowing for better personalized treatments.
PCa risk management largely relies on the visual assessment of biopsied tissues with optical microscopy (ie, histopathology) to assign a Gleason grade group (GG). Biopsies with lower scores more closely resemble normal tissue and are thus considered to be more indolent (nonlethal disease). According to the current guidelines, a single biopsy should be sectioned and visualized by pathologists at 3 closely spaced levels (typically ~20 μm apart)7 that collectively represent an ~1% of the whole biopsy.8 These “representative” sections from a part of the biopsy may not adequately capture the morphologic heterogeneity of the tumor9 and can also lead to ambiguities, such as tangential 2-dimensional (2D) sections of fully formed glands (Gleason pattern 3) being misinterpreted as poorly formed glands (Gleason pattern 4).10,11 In terms of spatial heterogeneity, Reyes and Humphrey9 found that “representative” sections missed clinically important atypical structures that were apparent in exhaustively serially sectioned specimens. In another study,12 it was shown that biopsies with atypical glandular proliferation might show focal carcinoma in “additional” sections, even if immunohistochemical analysis of “representative” sections did not identify malignancy. These findings suggest that the tumor grade, as assessed from different 2D slices within a 3-dimensional (3D) tumor volume, is likely to vary between slices (intrabiopsy variability). Exhaustively and serially sectioning a whole specimen for histologic analysis may mitigate such variability but would be destructive of valuable clinical specimens and would require extensive manual effort by histotechnologists, which is impractical.
The recent advent of high-throughput open-top light-sheet (OTLS) microscopy of optically cleared tissues provides an elegant solution for obtaining serial digital 2D sections throughout a 3D biopsy in a nondestructive way. OTLS can rapidly collect images in a z-stack arrangement (ie, 3D pathology) from entire biopsies or surgical excisions without tissue sectioning.13,14 Previous studies10,14,15 have also shown that the tissue processing and imaging methods used in this study are gentle and reversible (ie, the specimens may be returned as formalin-fixed, paraffin-embedded blocks after 3D pathology is performed), having no discernable adverse effects on tissue morphology and molecular expression.
The main focus of this study was to assess the extent of intrabiopsy variability in Gleason grading. In collaboration with 4 human readers (X.F., T.M., P.L.F., L.D.T), this study focused on Gleason grading of individual 2D slices that were virtually sampled from 5 evenly spaced levels spanning the OTLS-generated 3D pathology data sets. Unlike several previous studies that have focused on interreader variability of GG, the focus here was to evaluate intrabiopsy variability for individual human readers in terms of GGs across 5 widely spaced levels (~100 μm apart) from each 1-mm-diameter biopsy. The human readers graded every 2D image (5 levels from each of 20 biopsies) in a randomized sequence (100 total images) using a custom-developed web platform (Fig. 1).
Figure 1.
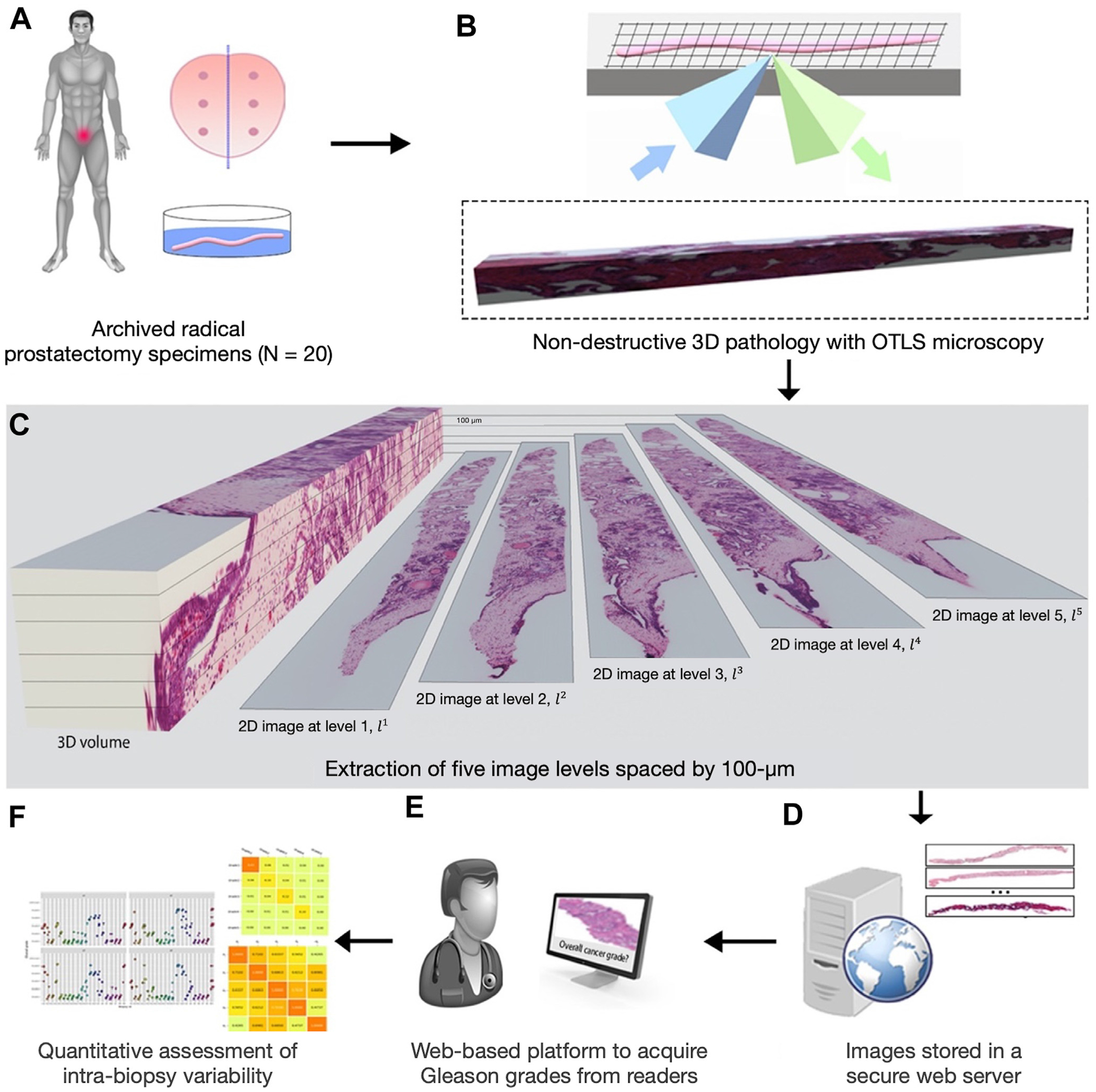
Overview of the present study. (A) Archived formalin-fixed, paraffin-embedded prostatectomy specimens were obtained from a cohort of 20 patients, from which 20 simulated (ex vivo) biopsies were extracted for the analysis. (B) The biopsies, which all contained cancer, were labeled with a fluorescent analog of hematoxylin and eosin (H&E) staining, optically cleared to render the tissues transparent to light, and then comprehensively imaged in 3D with an OTLS microscope. (C) Five 2D images were extracted at 100-μm intervals from each biopsy, resulting in a total of 100 images from the 20 biopsies. (D) 2D images were stored on a university server to allow our collaborating human readers access to the images remotely. (E) The human readers graded all 2D images in a randomized order using an in-house–developed web platform. (F) After collecting the grades from 4 readers, intrabiopsy variability was quantified. OTLS, open-top light-sheet; 2D, 2 dimension; 3D, 3 dimension.
Materials and Methods
Patient Collection and Volumetric Image Acquisition With Open-Top Light-Sheet
The study was reviewed and approved by the Institutional Review Board of the University of Washington (study 00004980), where research specimens were previously obtained from patients with informed consent. Archived formalin-fixed, paraffin-embedded prostatectomy specimens were gathered from ~200 patients with low-risk to intermediate-risk PCa as part of a previous active surveillance study.16 The cases were initially graded during post-RP histopathology as having GGs of 1, 2, or 3, where approximately half of them were low risk (GG1). Of the ~200 cases that were imaged, 20 cases were randomly selected to be used in this study. One cancerous biopsy per case was selected for the present study. Biopsies with evidence of cancer were initially identified by 2 experienced pathologists.8
Each biopsy core was imaged comprehensively in 3D using a published OTLS microscope system14 after the biopsies were stained with a fluorescent analog of hematoxylin and eosin (H&E) and optically cleared with ethanol dehydration and immersion in ethyl cinnamate. As described previously,8,14 the H&E analog consisted of a nuclear dye, TO-PRO3, and eosin, with sampling of ~0.44 μm/voxel, roughly equivalent to what is achieved with a 10× objective on a standard transillumination bright-field light microscope.10 The volumetric imaging took ~0.5 min/mm3 of tissue for each illumination wavelength, resulting in ~50 GB of raw data per biopsy. However, these data sets were downsampled by a factor of 2 in all dimensions (8× reduction in file size) for this human observer study.
Data Set Preparation
A total of 5 cross-sectional images were extracted at 100-μm intervals spanning nearly the entire diameter of each biopsy (~0.9 mm diameter), resulting in a total of 100 digital images {1 ≤ i ≤ 5, 1 ≤ b ≤ 20}, where is an image extracted from the ith level of a biopsy identified as biopsy number b. Two-channel fluorescence images were false colored to mimic H&E staining using a previously published method.17 2D images were stored in a secure university server, in which external human readers were provided secure access.
Human Readers Involved in This Study
Four human readers (r1, r2, r3, and r4) were involved in this study. r1 is a genitourinary (GU) pathologist with 24 years of experience. r2 is a GU pathologist with 33 years of experience. r3 is a general pathologist with 8 years of experience, and r4 is a GU pathologist with 17 years of experience. Each human reader was given a task of grading all images using a custom-developed interactive web tool.
Collecting Grades From Human Readers Using an In-House Interactive Web Tool
We developed an interactive web tool to facilitate reviewing the 2D images. The tool was developed to support large high-resolution images and provide several important functionalities necessary for analyzing high-resolution images, such as zooming and panning. An account was created for each reader to access the platform with their username and password. The tool shuffled all images in a way that no 2 images from the same biopsy would be displayed to the reader within 3 consecutive grading events. This was done to minimize the likelihood that the reader would be able to recall previous images originating from the same biopsy. The tool allowed the reader to stop/resume their analysis at any time. When they logged back in, the tool would allow for resumption of assessment from the last image reviewed.
For each image, the reader was asked to assess GG as defined by the International Society of Urological Pathology.18 This involved identifying the 2 predominant Gleason patterns (which could be identical if only 1 pattern was seen). Grades were collected from all human readers, resulting in a total of 400 predictions. All predictions and comments were saved into a structured database created with SQLAlchemy, a Python SQL toolkit for database object mapping.19 The web tool was developed in Python 3.8 and JavaScript with “Flask,”20 “Open-Slide,”21 and “OpenSeadragon” libraries.22
Exploring Intrabiopsy Variability in Grading Based on 2-Dimensional Histology Images
Intrabiopsy variability was analyzed quantitatively and qualitatively. It was calculated as the worst pairwise agreement in GG between individual levels. Each level was considered an independent observation. The pairwise agreement between any 2 levels in a biopsy was measured using Cohen κ.23 We used 5 individual levels from a biopsy, and therefore, there would be 10 different pairwise comparisons: {, 1 ≤ i ≤ 5, 1 ≤ j ≤ 5, i ≠j}. This resulted in 10 different Cohen κ scores for a reader. The worst pairwise κ score was then identified as the smallest one among the 10 scores.
Interpretation of Cohen κ score was based on the study by Cohen,23 where values ≤0.20 were taken to indicate no agreement, 0.21 to 0.39 as “minimal,” 0.40 to 0.59 as “weak,” 0.60 to 0.79 as “moderate,” 0.80 to 0.90 as “strong,” and >0.90 as “almost perfect” agreement. Violin and histogram plots were used to visualize distributions of GGs. All analyses were performed in Python 3.8 with the “scikit-learn,”24 “Matplotlib,”25 “plotnine,”26 and “Pingouin”27 libraries.
Results
Distributions of Grade Groups Collected Across 4 Human Readers
The distributions of GGs assigned by all human readers are reported in Table 1. The most frequently assigned grade was GG1, which appears to align with the original study. r1, r2, r3, and r4 assigned 50%, 44%, 56%, and 59% of images as GG1, respectively. r4 did not assign any GG for 1 image because of challenges with poor image quality. Overall, across all readers, 52% of the grades were assigned as GG1, and 18%, 14%, 9%, and 7% of the grades were assigned as GG2, GG3, GG4, and GG5, respectively. The average and SD of the GGs were 1.92 ± 1.08, 2.16 ± 1.33, 2.07 ± 1.41, and 1.85 ± 1.26 for r1, r2, r3, and r4, respectively.
Table 1.
Distributions of GGs assigned by all human readers
| r 1 | r 2 | r 3 | r 4 | Total (%) | |
|---|---|---|---|---|---|
| GG1 (%) | 50 | 44 | 56 | 59 | 52 |
| GG2 (%) | 20 | 22 | 12 | 16 | 18 |
| GG3 (%) | 18 | 19 | 9 | 11 | 14 |
| GG4 (%) | 12 | 4 | 15 | 6 | 9 |
| GG5 (%) | 0 | 11 | 8 | 7 | 7 |
| Total (%) | 100 | 100 | 100 | 99 | 100 |
GG, grade group.
Among the 20 samples, the GG determinations that fluctuated the most were between GG1 and GG2 for r1 (n = 5, tied with the flipped grades between GG2 and GG3), r2 (n = 5), and r3 (n = 6), whereas the second most common flipped grades were between GG2 and GG3 for r2 (n = 4) and r3 (n = 3) (Table 2). For r4, the most common flipped grades were between GG2 and GG3 (n = 5), and the second most common was between GG1 and GG2 (n = 4, tied with the flipped grades between GG4 and GG5).
Table 2.
Number of samples (of 20) where 2 GGs were coassigned to the same sample by (a) r1, (b) r2, (c) r3, and (d) r4
| (a) r 1 | ||||
|---|---|---|---|---|
| GG2 | GG3 | GG4 | GG5 | |
| GG1 | 5 | 3 | 0 | 0 |
| GG2 | — | 5 | 1 | 0 |
| GG3 | — | — | 1 | 0 |
| GG4 | — | — | — | 0 |
| (b) r 3 | ||||
| GG2 | GG3 | GG4 | GG5 | |
| GG1 | 6 | 2 | 2 | 0 |
| GG2 | — | 3 | 2 | 0 |
| GG3 | — | — | 2 | 3 |
| GG4 | — | — | — | 2 |
| (c) r 2 | ||||
| GG2 | GG3 | GG4 | GG5 | |
| GG1 | 5 | 1 | 0 | 1 |
| GG2 | — | 4 | 1 | 2 |
| GG3 | — | — | 3 | 3 |
| GG4 | — | — | — | 3 |
| (d) r 4 | ||||
| GG2 | GG3 | GG4 | GG5 | |
| GG1 | 4 | 2 | 1 | 0 |
| GG2 | — | 5 | 1 | 1 |
| GG3 | — | — | 1 | 2 |
| GG4 | — | — | — | 4 |
GG, grade group.
Experiment: Pairwise Agreement in Grade Groups When Grading Different Levels Within a Biopsy
In this experiment, the agreement in GG for all combinations of 2 levels within the same biopsy was measured for each reader. The worst pairwise agreement between levels was calculated using the Cohen κ method to quantify intrabiopsy agreement and found to be κ = 0.43 (“weak agreement”) for r1, 0.38 (“minimal agreement”) for r2, 0.34 (“minimal agreement”) for r3, and 0.34 (“minimal agreement”) for r4 (Fig. 2). All readers assigned at least 2 different grades for 50% of the biopsies. Overall, the pairwise agreement between levels within a biopsy tended to decrease (worsen) as the distance between the levels increased, implying that the grade is spatially heterogeneous within the tumor.
Figure 2.
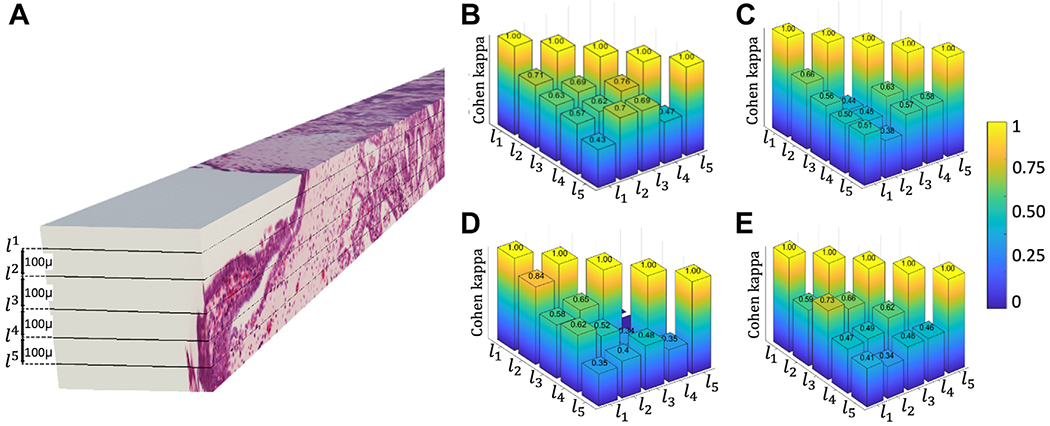
Intrabiopsy agreement. (A) An example of 3-dimensional volume illustrating the 2-dimensional images extracted at 100-μm intervals through the volume of a biopsy. Pairwise agreements in grade groups (Cohen κ) at different biopsy levels for different readers (B) r1, (C) r2, (D) r3, and (E) r4. As the distance between levels increases, the agreement tends to decrease.
Figure 3 illustrates an example showing variability in tumor morphology across 2 different levels within the same biopsy and its impact on grading. The zoomed-in region in Figure 3B was extracted from the first level of biopsy 19, , with small, closely packed tumor glands (pattern 3 or 4). However, in the same zoomed-in region at the third level of the biopsy, , which was 200 μm away in depth (Fig. 3C), the Gleason pattern 3 glands disappear, and benign glands are observed instead. Several Gleason pattern 3 glands are still observed in (outside the zoomed-in region). Overall, according to all readers, was assigned GG3, whereas was assigned GG1.
Figure 3.
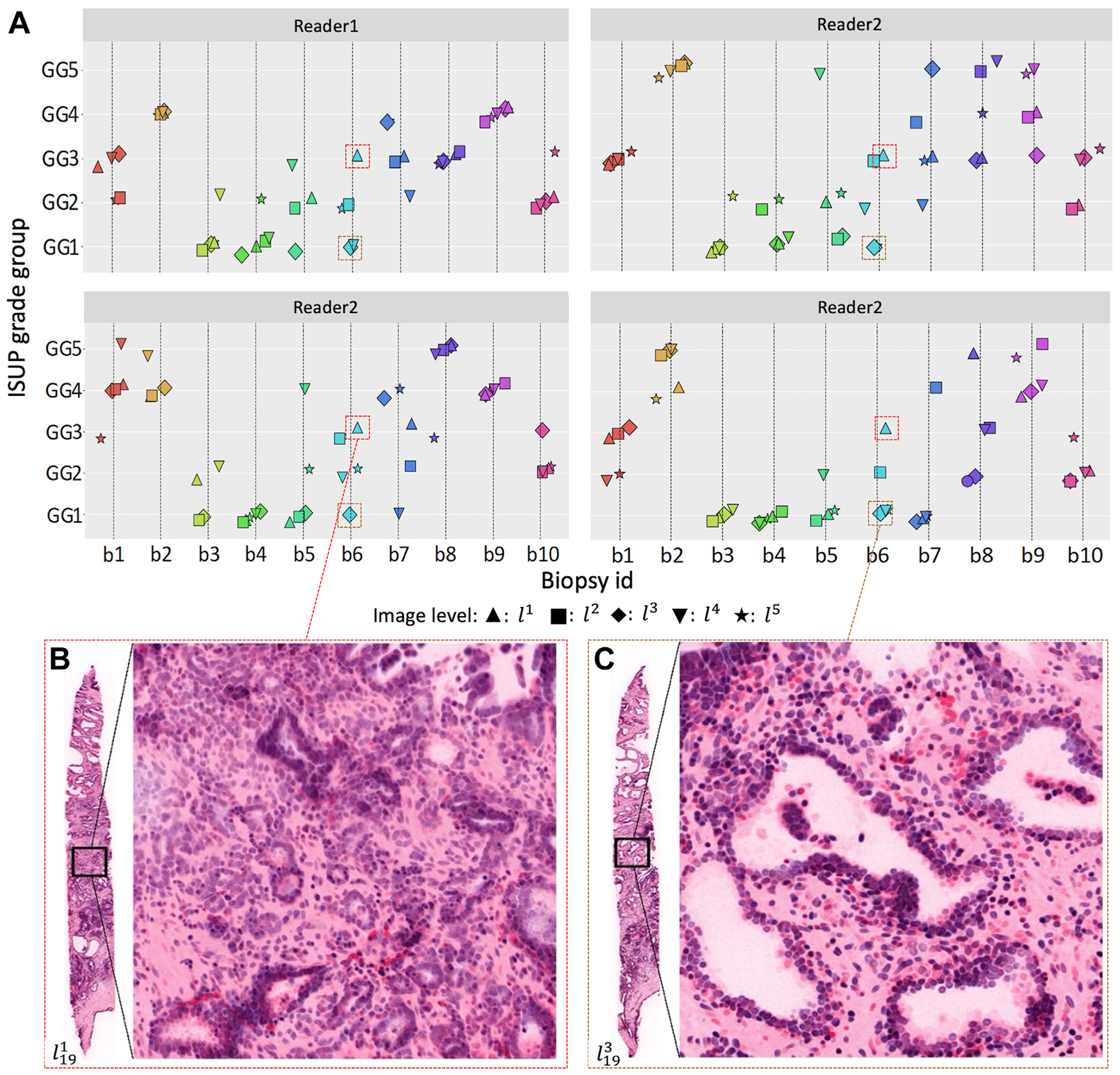
Distributions of GGs assigned by the human readers. (A) Scatter plots of the GGs assigned by each reader for 10 of 20 biopsies. To emphasize the range of variations, we specifically chose those 10, with different GGs for visualization purposes. Each subplot shows the grades assigned by 1 reader. Each data marker represents a GG assigned by looking at a 2-dimensional image at a particular spatial level within a biopsy. Each level is represented by a specific marker shape (see legend below the plots). (B, C) Two example images extracted from different levels within the same biopsy, l119 and l319. (B) The zoomed region of l119 contains mostly poorly formed glands, the presence of which indicates higher GGs, whereas (C) in the same axial region of l319, 200 μm far away from l119 in depth, the glands appear well formed and have recognizable patterns, with clear boundaries, mostly associated with lower GGs. GG, grade group; ISUP, International Society of Urological Pathology.
Discussion
This study focused on evaluating the extent of variability in tumor grading when human readers were given the task of grading individual 2D sections obtained from a 3D tumor volume. Using a global collaboration involving 4 pathologists, we attempted to ascertain intrabiopsy variability for each individual human reader in terms of Gleason grading across multiple levels spanning a 3D biopsy data set. Unlike previous studies28–32 that have focused on the quantitative assessment of the extent of interreader variability in GG, this study focused on intrabiopsy variability because of tumor spatial heterogeneities.
Our experiment revealed low intrabiopsy agreement on GG grading. We found that for the majority of biopsies, readers assigned different GGs to different levels within individual biopsies. Moreover, the degree of intrabiopsy variability found in this study is significant in relation to other sources of “uncertainty” in Gleason grading, such as intrareader variability.29–31,33,34 For instance, r1 regraded a subset of the samples (n = 25) after a washout period of ~10 months. Intraobserver agreement for the 25 samples for r1 was 0.57, which was much higher than the intrabiopsy agreement for the same reader (κ = 0.43). Similarly, Melia et al31 reported a κ score κ of 0.66 when an observer graded the same slides at different time points. In this study, significantly lower κ values for intrabiopsy variability were seen, suggesting that this variability is due at least in part to intrinsic morphologic differences within each biopsy rather than variations in interpretation by a single reader over time.
To better understand the implications of histology sectioning in clinical decision making, Reyes and Humphrey9 found that some of the diagnostic needle biopsies tended to be undergraded because of incomplete sectioning. They exhaustively serially sectioned specimens and defined the following 2 sets of slice groups: “diagnostic” slices, which represented slices used in clinical practice for diagnostic purposes, and “residual” slices, which represented the slices other than the diagnostic ones. It was observed that 4 needle biopsies, which were identified as focal glandular atypia in their diagnostic slices, were identified definitively as carcinoma in the residual slices. The clinical implications of such findings are profound as diagnosis of carcinoma likely prompts RP, whereas diagnosis of atypia typically results in only clinical follow-up. Similarly, for all pathologist readers in our study, the first level of biopsy 19 was evaluated GG3, falling into the intermediate-risk group (Fig. 3A) typically requiring RP, whereas the third level of the same biopsy (200 μm away) was graded GG1 by the same readers, a low-risk group (Fig. 3B) typically assigned to active surveillance. Such discrepancies observed in different 2D images of the same tumor support the value of 3D analysis. Looking at the entirety of a 3D tumor volume to derive a composite 3D-based GG, instead of looking at individual 2D slices, may yield better predictions of disease outcome.
As mentioned previously, the main finding of this study is that there are significant morphologic variations within prostate needle biopsies as a function of spatial location within the biopsy volume. This level of variability is significantly greater than what can be attributed to the uncertainty in grade based on the visual assessment by pathologists (intraobserver variability). Although the focus of the study was not on interobserver variability between pathologists, κ metrics revealed weak agreement between human readers (κ = 0.46). This finding appears to align with previous studies,32,35 which have found that although agreement between pathologists improves as their experience increases, significant variability between readers remains.31,36,37
A limitation of this study is that the intact prostate biopsies were imaged using a lower-resolution prototype of OTLS microscopy, equivalent to what is achieved with a ~10× objective on a standard transillumination bright-field light microscope.10 Thus, pseudo-OTLS images lack the higher spatial resolution to see the nucleoli and make the diagnoses of cancer difficult for GG5 cancerous cells. These cells can potentially be confused with inflammatory cells, small cell carcinoma, or other cell types. Additionally, differential diagnosis between cancerous and noncancerous lesions is sometimes dependent on the assessment of nuclear morphology, such as adenosis versus well-differentiated prostate adenocarcinoma, hyperplasia versus well-differentiated prostate adenocarcinoma, and atypical small acinar proliferation lesions. This situation may contribute to some degree of uncertainty by the pathologists. However, all biopsies were preselected to contain low-risk to intermediate-risk PCa (GG1-GG3) and Gleason grading is based on gland morphology rather than high-resolution cytologic features, and therefore, the lower resolution of our data sets should have had a minimal effect on the grading process. Additionally, although pseudo-H&E images have the advantage of being familiar to practicing pathologists, there may be slight deviations from conventional H&E images.17 These factors may also contribute to some degree of uncertainty by the pathologists. Higher-resolution data sets will be obtained in the future (40× equivalent), leveraging the more recent OTLS systems.38,39
Since Gleason grading evolved many decades ago from the visual assessment of 2D histology slides, there is a potential need for a more accurate grading system based on 3D pathology that better correlates with clinical outcome. Additionally, the integration of computational pathology and machine learning within 3D pathology could pave the way for a new generation of spatial biomarkers to predict disease outcome or treatment response more accurately and efficiently.8,11 Our previous study40 provided evidence along these lines, suggesting that 3D histomorphometric analysis was superior to analogous 2D analyses for determining PCa aggressiveness.
In conclusion, this preliminary study presents evidence of a high degree of intrabiopsy variability for a 2D-based tumor grading system. Our findings reveal that Gleason grading based on 2D images varies dramatically as a function of spatial position within a 3D biopsy volume that is being analyzed. Consequently, clinical decision making and patient management may be affected. We believe that morphologic assessments of whole biopsy specimens in 3D can enable more reliable and consistent tumor grading than the standard of care that is based on the limited numbers of 2D histology sections.
Funding
This study received funding support from the National Institutes of Health National Center for Advancing Translational Sciences through TL1 TR002318 (R.S.) and the Department of Defense Prostate Cancer Research Program through W81WH-18-10358 (J.T.C.L. and L.D.T.), W81XWH-19-1-0589 (N.P.R.), and W81XWH-20-1-0851 (A.M. and J.T.C.L.). Support was also provided by the National Cancer Institute through R01CA268207 (J.T.C.L. and A.M.), K99 CA240681 (A.K.G.), U24CA199374 (A.M.), R01CA249992 (A.M.), R01CA202752 (A.M.), R01CA208236 (A.M.), R01CA216579 (A.M.), R01CA220581 (A.M.), R01CA257612 (A.M.), U01CA239055 (A.M.), U01CA248226 (A.M.), U54CA254566 (A.M.), R01 LM013864 (A.J.), U01 DK133090 (A.J.), U01 CA248226 (A.J.), U01 CA239055 (A.J.), and the Pacific Northwest Prostate Cancer Specialized Programs of Research Excellence (SPORE)—P50CA097186 (L.D.T.). Additional support was provided by the National Heart, Lung, and Blood Institute through R01HL151277 (A.M.), the National Institute of Biomedical Imaging and Bioengineering through R01EB031002 (J.T.C.L.) and R43EB028736 (A.M.), the VA Merit Review Award IBX004121A from the US Department of Veterans Affairs (A.M.), the Prostate Cancer Foundation Young Investigator Award (N.P.R.), the Prostate Cancer UK charity (J.T.C.L.), the National Science Foundation 1934292 HDR: I-DIRSE-FW (J.T.C.L. and R.S.), the Canary Foundation, Academy of Finland, Cancer Foundation Finland, and Governmental Research Funding (VTR) of HUS Helsinki University Hospital (T.M.).
Declaration of Competing Interest
A.K.Glaser is a cofounder of Alpenglow Biosciences, Inc. N.P. Reder is a cofounder and CEO of Alpenglow Biosciences, Inc. A. Janowczyk reports personal fees from Roche and Merck and personal fees from Lunaphore outside the submitted work. L.D. True is a cofounder of Alpenglow Biosciences, Inc. A. Madabhushi is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata, Inc. Currently, he serves on the advisory board of Picture Health, Aiforia, Inc, and SimBioSys. He also currently consults for Biohme, SimBioSys, and Castle Biosciences. He also has sponsored research agreements with AstraZeneca, Boehringer Ingelheim, Eli-Lilly, and Bristol Myers Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. He is also involved in 3 different R01 grants with Inspirata, Inc. J.T.C. Liu is a cofounder and board member of Alpenglow Biosciences, Inc, which has licensed the open-top light-sheet microscopy portfolio developed in his laboratory at the University of Washington. The other authors report no relevant conflicts of interest.
Footnotes
Ethics Approval and Consent to Participate
The study was reviewed and approved by the Institutional Review Board of the University of Washington (study 00004980), where research specimens were previously obtained from patients with informed consent.
Data Availability
The data sets used during the current study are available from the corresponding author on reasonable request. The original 3-dimensional pathology data sets used in this study are available publicly through The Cancer Imaging Archive—https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=145754446
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 2.Shi Z, Platz EA, Wei J, et al. Performance of three inherited risk measures for predicting prostate cancer incidence and mortality: a population-based prospective analysis. Eur Urol. 2021;79(3):419–426. [DOI] [PubMed] [Google Scholar]
- 3.Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294(4):433–439. [DOI] [PubMed] [Google Scholar]
- 4.Zincke H, Oesterling JE, Blute ML, Bergstralh EJ, Myers RP, Barrett DM. Longterm (15 years) results after radical prostatectomy for clinically localized (stage T2c or lower) prostate cancer. J Urol. 1994;152(5 pt 2):1850–1857. [DOI] [PubMed] [Google Scholar]
- 5.Nolsøe AB, Jensen CFS, Østergren PB, Fode M. Neglected side effects to curative prostate cancer treatments. Int J Impot Res. 2021;33(4):428–438. [DOI] [PubMed] [Google Scholar]
- 6.Link RE, Morton RA. Indications for pelvic lymphadenectomy in prostate cancer. Urol Clin North Am. 2001;28(3):491–498. [DOI] [PubMed] [Google Scholar]
- 7.van der Kwast TH, Lopes C, Santonja C, et al. Guidelines for processing and reporting of prostatic needle biopsies. J Clin Pathol. 2003;56(5):336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie W, Reder NP, Koyuncu C, et al. Prostate cancer risk stratification via nondestructive 3D pathology with deep learning-assisted gland analysis. Cancer Res. 2022;82(2):334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes AO, Humphrey PA. Diagnostic effect of complete histologic sampling of prostate needle biopsy specimens. Am J Clin Pathol. 1998;109(4):416–422. [DOI] [PubMed] [Google Scholar]
- 10.Reder NP, Glaser AK, McCarty EF, Chen Y, True LD, Liu JTC. Open-top light-sheet microscopy image atlas of prostate core needle biopsies. Arch Pathol Lab Med. 2019;143(9):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JTC, Glaser AK, Bera K, et al. Harnessing non-destructive 3D pathology. Nat Biomed Eng. 2021;5(3):203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arista-Nasr J, Martínez-Mijangos O, Martínez-Benítez B, Bornstein-Quevedo L, Lino-Silva S, Urbina-Ramírez S. Atypical small acinar proliferation: utility of additional sections and immunohistochemical analysis of prostatic needle biopsies. Nephrourol Mon. 2012;4(2):443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser AK, Reder NP, Chen Y, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng. 2017;1(7):0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser AK, Reder NP, Chen Y, et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat Commun. 2019;10(1):2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Xie W, Glaser AK, et al. Rapid pathology of lumpectomy margins with open-top light-sheet (OTLS) microscopy. Biomed Opt Express. 2019;10(3):1257–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley S, Fazli L, McKenney JK, et al. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: the Canary Prostate Cancer Tissue Microarray. Adv Anat Pathol. 2013;20(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serafin R, Xie W, Glaser AK, Liu JTC. FalseColor-Python: a rapid intensity-leveling and digital-staining package for fluorescence-based slide-free digital pathology. PLoS One. 2020;15(10):e0233198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40(2):244–252. [DOI] [PubMed] [Google Scholar]
- 19.Bayer M. SQLAlchemy. In: Brown A, Wilson G, eds. The Architecture of Open Source Applications Volume II: Structure, Scale, and a Few More Fearless Hacks. 2012. Accessed April 1, 2022. http://aosabook.org/en/sqlalchemy.html
- 20.Grinberg M. Flask Web Development: Developing Web Applications with Python. O’Reilly Media, Inc; 2018. [Google Scholar]
- 21.Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.OpenSeadragon. Accessed April 1, 2022. https://openseadragon.github.io/
- 23.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Measure. 1960;20(1):37–46. [Google Scholar]
- 24.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22(3):276–282. [PMC free article] [PubMed] [Google Scholar]
- 25.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 26.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
- 27.Kibirige H, Lamp G, Katins J, et al. has2k1/plotnine: See the [changelog]. https://plotnine.readthedocs.io/en/stable/changelog.html#v0-9-0. Published online September 29, 2022.
- 28.Vallat R. Pingouin: statistics in Python. J Open Source Softw. 2018;3(31):1026. [Google Scholar]
- 29.Rodriguez-Urrego PA, Cronin AM, Al-Ahmadie HA, et al. Interobserver and intraobserver reproducibility in digital and routine microscopic assessment of prostate needle biopsies. Hum Pathol. 2011;42(1):68–74. [DOI] [PubMed] [Google Scholar]
- 30.Abdollahi A, Meysamie A, Sheikhbahaei S, et al. Inter/intra-observer reproducibility of Gleason scoring in prostate adenocarcinoma in Iranian pathologists. Urol J. 2012;9(2):486–490. [PubMed] [Google Scholar]
- 31.Melia J, Moseley R, Ball RY, et al. A UK-based investigation of inter- and intra-observer reproducibility of Gleason grading of prostatic biopsies. Histopathology. 2006;48(6):644–654. [DOI] [PubMed] [Google Scholar]
- 32.Rutgers JJ, Bánki T, van der Kamp A, et al. Interobserver variability between experienced and inexperienced observers in the histopathological analysis of Wilms tumors: a pilot study for future algorithmic approach. Diagn Pathol. 2021;16(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths DFR, Melia J, McWilliam LJ, et al. A study of Gleason score interpretation in different groups of UK pathologists; techniques for improving reproducibility. Histopathology. 2006;48(6):655–662. [DOI] [PubMed] [Google Scholar]
- 34.McKenney JK, Simko J, Bonham M, et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol. 2011;186(2):465–469. [DOI] [PubMed] [Google Scholar]
- 35.Nakai Y, Tanaka N, Shimada K, et al. Review by urological pathologists improves the accuracy of Gleason grading by general pathologists. BMC Urol. 2015;15(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Yoshida K, Kawashima A, et al. Impact of measurement method on interobserver variability of apparent diffusion coefficient of lesions in prostate MRI. PLoS One. 2022;17(5):e0268829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh RV, Agashe SR, Gosavi AV, Sulhyan KR. Interobserver reproducibility of Gleason grading of prostatic adenocarcinoma among general pathologists. Indian J Cancer. 2011;48(4):488–495. [DOI] [PubMed] [Google Scholar]
- 38.Glaser AK, Bishop KW, Barner LA, et al. A hybrid open-top light-sheet microscope for versatile multi-scale imaging of cleared tissues. Nat Methods. 2022;19(5):613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barner LA, Glaser AK, Huang H, True LD, Liu JTC. Multi-resolution open-top light-sheet microscopy to enable efficient 3D pathology workflows. Biomed Opt Express. 2020;11(11):6605–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie W, Reder NP, Koyuncu CF, et al. Prostate cancer risk stratification via non-destructive 3D pathology with deep learning-assisted gland analysis. Cancer Res. Published online December 1, 2021; canres.2843.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used during the current study are available from the corresponding author on reasonable request. The original 3-dimensional pathology data sets used in this study are available publicly through The Cancer Imaging Archive—https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=145754446


