Abstract
V(D)J recombination is initiated by a coordinated cleavage reaction that nicks DNA at two sites and then forms a hairpin coding end and blunt signal end at each site. Following cleavage, the DNA ends are joined by a process that is incompletely understood but nevertheless depends on DNA-dependent protein kinase (DNA-PK), which consists of Ku and a 460-kDa catalytic subunit (DNA-PKCS or p460). Ku directs DNA-PKCS to DNA ends to efficiently activate the kinase. In vivo, the mouse SCID mutation in DNA-PKCS disrupts joining of the hairpin coding ends but spares joining of the open signal ends. To better understand the mechanism of V(D)J recombination, we measured the activation of DNA-PK by the three DNA structures formed during the cleavage reaction: open ends, DNA nicks, and hairpin ends. Although open DNA ends strongly activated DNA-PK, nicked DNA substrates and hairpin-ended DNA did not. Therefore, even though efficient processing of hairpin coding ends requires DNA-PKCS, this may occur by activation of the kinase bound to the cogenerated open signal end rather than to the hairpin end itself.
V(D)J recombination is the process in which DNA of lymphoid cells is rearranged to form functional immunoglobulin and T-cell receptor genes. The lymphoid cell-specific proteins RAG1 and RAG2 initiate V(D)J recombination by recognizing recombination signal sequences adjacent to V, D, or J coding sequences and catalyzing a coordinated cleavage at two sites, each at the border between a signal sequence and a coding sequence (19, 31). RAG1 and RAG2 cleave DNA by first nicking the DNA to produce a 3′-hydroxyl and a 5′-phosphate and then mediating a nucleophilic attack by the 3′-hydroxyl at the phosphodiester bond opposite the nick on the second strand, creating a covalently closed hairpin coding end and blunt signal end (30).
After cleavage, the ends are joined by multiple proteins, including those that mediate double-strand break repair: XRCC4 and DNA-dependent protein kinase (DNA-PK) (27). DNA-PK consists of a 460-kDa catalytic subunit (DNA-PKCS or p460), which possesses DNA binding and protein kinase activities, and a smaller subunit (Ku), which directs DNA-PKCS to DNA ends so that it is efficiently activated (11, 12, 33). Ku contains 70- and 86-kDa subunits and binds to several DNA structures, including DNA ends, stem-loop structures, and DNA nicks (3, 9).
Cells from the severe combined immunodeficient (SCID) mouse have a mutation in the DNA-PKCS gene that leads to truncation of the C-terminal kinase domain (4, 5, 7, 14). SCID cells are hypersensitive to ionizing radiation (2, 10, 13) and have a defect in V(D)J recombination that leads to absent coding joints but relatively intact signal joints (18). Cells mutated in Ku are hypersensitive to ionizing radiation and defective for both coding and signal joints (8, 28, 29). Thymocytes from both SCID (25) and Ku86 knockout (34) mice accumulate abnormally high levels of hairpin coding ends, indicating that intact DNA-PK is required for efficient processing of hairpin ends.
These observations suggest that the processing of hairpin coding ends may require activation of the kinase function in DNA-PK. Candidates for the activating DNA include three intermediates created during V(D)J recombination: DNA nicks, open ends, and hairpin ends. Surprisingly, only open-ended DNA efficiently activated the kinase. This result has implications for V(D)J recombination, which will be discussed.
MATERIALS AND METHODS
Oligonucleotide preparation.
A double-stranded 68-bp hairpin-ended DNA molecule was synthesized from three oligonucleotides: oligonucleotide 1, TGCAGCCCAAGCTTGGCGTAATCATCGAATTCAGCTGTCTAGAAG; oligonucleotide 2, CTTCTGCAGGTCGACCTGCAGAAGCTTCTAGACAGCTGAATTCGA; and oligonucleotide 3, TGATTACGCCAAGCTTGGGCTGCAGGTCGACTAGTACTAGTCGACC. Oligonucleotide 1 contains 45 nucleotides, which anneal to the 5′ end of oligonucleotide 3 and the 3′ end of oligonucleotide 2. Oligonucleotide 2 contains 45 nucleotides, of which the 24 nucleotides at the 5′ end are self annealing. Oligonucleotide 3 contains 46 nucleotides, of which the 22 nucleotides at the 3′ end are self annealing. The left hairpin end created by oligonucleotide 3 was identical to a sequence tested in an extrachromosomal V(D)J recombination assay (20).
Each oligonucleotide (100 pmol) was phosphorylated at its 5′ end by incubation in 10 μl of solution with 10 U of T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and either 100 μM ATP for oligonucleotides 2 and 3 or 40 μCi of [γ-32P]ATP (6,000 Ci/mmol) for oligonucleotide 1. Following incubation at 37°C for 45 min, 1 μl of 1 mM ATP was added to the oligonucleotide 1 reaction, and all reaction mixtures were incubated for another 30 min. Reaction mixtures were extracted with phenol once and chloroform twice and precipitated in ethanol. Oligonucleotides 1, 2, and 3 were resuspended in 9 μl of 1× ligase buffer, pooled, heated to 95°C for 5 min, and quick chilled on ice before the addition of 400 U of T4 DNA ligase. After incubation at 16°C overnight, the preparation was resolved by denaturing gel electrophoresis in 6 M urea–40% formamide–6% polyacrylamide. (The gel was prerun for 45 min at 14 W and then run with the DNA samples for 4 h at 14 W.) The band containing the hairpin-ended DNA was excised, and DNA was eluted by dicing the gel slice and incubating it at 37°C overnight in 10 mM Tris (pH 8)–1 mM EDTA–0.4 M NaCl. The eluted DNA was precipitated in ethanol, resuspended in formamide, and resolved on a 7% polyacrylamide denaturing gel. After the two rounds of gel purification, the DNA migrated as a single band on an 8% polyacrylamide denaturing gel. This protocol yielded hairpin preparations less than 1% contaminated with nicked DNA or other species when quantitated on a phosphorimager. Unlabeled hairpin-ended molecules were prepared in parallel, by phosphorylating the oligonucleotides with cold ATP and resolving the samples in lanes adjacent to the labeled preparation. Labeled and unlabeled nicked hairpin-ended DNA molecules were prepared similarly, except that oligonucleotide 3 was not phosphorylated. A single nick was thus positioned centrally, 35 bp from the left end and 33 bp from the right end of the hairpin-ended DNA molecule.
The open-ended 70-bp DNA molecule was excised from pBluescript with KpnI and BamHI, resolved on a 1.5% agarose gel, eluted, and purified with Qiaex beads (Qiagen, Chatsworth, Calif.). Nicked plasmid was prepared by incubating the 3,000-bp pBluescript plasmid (200 μg/ml) with 5 × 10−6 U of DNase I in 20 μl at 37°C for 5 min, heated to 70°C in 25 mM EDTA to stop the reaction, extracted twice with a 1:1 mixture of phenol and chloroform and once with ether, and precipitated in ethanol. Successful nicking was determined by electrophoresis of the products in a 1% agarose gel containing 0.5 μg of ethidium bromide per ml. DNA preparations were stained with PicoGreen dye (Molecular Probes, Eugene, Oreg.) for quantitation on a fluorometer (TD-700; Turner Designs, Sunnyvale, Calif.) which was capable of detecting as little as 100 pg of DNA.
DNA-PK assay.
DNA-PK preparations, which were more than 70% (Promega, Madison, Wis.) or more than 95% (12) pure, were assayed for kinase activity in 10 μl of buffer containing 13 mM spermidine, 25 mM HEPES-KOH (pH 7.5), 15 mM MgCl2, 0.2 mM ATP, 2.5 μCi of [γ-32P]ATP, 20% glycerol, 0.1% Nonidet P-40, 50 mM KCl, 100 mM NaCl, and 1 mM dithiothreitol by incubation at 20°C for 10 min with the specific peptide substrate EPPLSQEAFADLWKK (15). DNA-PK activity was quantitated by measuring phosphorylation of the peptide upon addition of DNA. Reactions were stopped by adding an equal volume of 30% acetic acid, and reaction mixtures were spotted onto Whatman P81 phosphocellulose paper (VWR, San Francisco, Calif.), air dried, washed four times in 15% acetic acid, and counted in a scintillation counter.
DNA-PK electrophoretic mobility shift assay.
DNA-PK or Ku purified to greater than 95% homogeneity (12) was incubated with 0.2 ng of either hairpin-ended or open-ended labeled DNA probe in 10 μl of solution containing 10 mM Tris-HCl (pH 7.4), 20% glycerol, and 200 mM NaCl2 at 20°C for 10 min and then resolved on a 4% nondenaturing polyacrylamide gel in Tris-glycine buffer at 10 V/cm for 30 min.
RESULTS
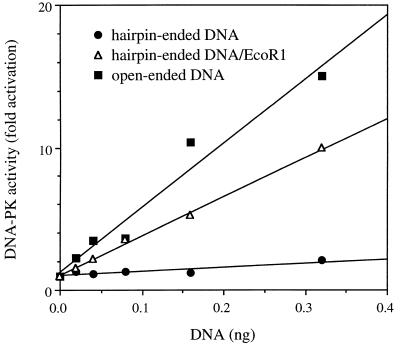
The activation of DNA-PK was tested with a 68-bp DNA molecule containing two hairpin ends (Fig. 1). The hairpin-ended DNA was tested over a range of concentrations by incubation with DNA-PK and found to be strikingly inefficient in activating DNA-PK (Fig. 2). By contrast, a 70-bp open-ended DNA molecule produced a strong activation of DNA-PK. The hairpin-ended DNA preparation did not contain contaminants that inhibited DNA-PK, since cleavage by EcoRI led to strong activation of DNA-PK. The cleavage products consisted of fragments of 28 and 40 bp, which were about 60% as active as the 70-bp open-ended DNA. This degree of activation was about equal to that seen with a molar equivalent of open-ended DNA of 32 bp (data not shown). Hairpin-ended DNA failed to efficiently activate DNA-PK preparations purified to either 70% or greater than 95%. Furthermore, the kinase assay was repeated at the increased temperature of 37°C to promote potential unwinding of the hairpin ends, and again there was no significant activation of DNA-PK (data not shown). In summary, when DNA was rate limiting, activation of DNA-PK by hairpin-ended DNA was less than 5% of the level for open-ended DNA.
FIG. 1.
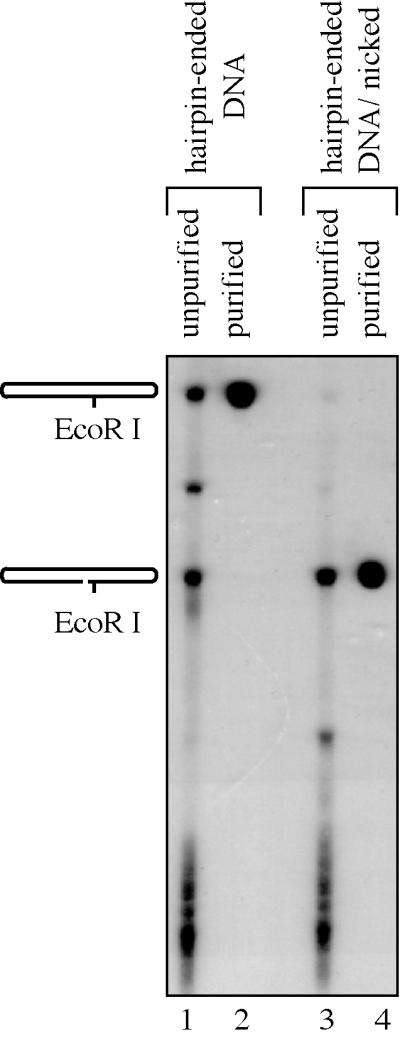
Purification of hairpin-ended and nicked hairpin-ended DNA substrates. Lane 1, ligation products of oligonucleotides used to construct the hairpin-ended DNA. Lane 2, hairpin-ended DNA after gel purification. Lane 3, ligation products of oligonucleotides used to construct the nicked hairpin-ended DNA. Lane 4, nicked hairpin-ended DNA after gel purification. The DNA samples were resolved by denaturing polyacrylamide gel electrophoresis.
FIG. 2.
Inefficient activation of DNA-PK by hairpin-ended DNA. DNA-PK (70% pure) and its peptide substrate were incubated with hairpin-ended DNA (closed circles), hairpin-ended DNA cleaved with EcoRI (open triangles), or open-ended DNA (closed squares). The uncleaved hairpin-ended DNA was treated identically to its cleaved counterpart by incubation with heat-inactivated EcoRI. Kinase activity was measured as the fold increase in counts from phosphorylation of peptide in the presence of DNA over background counts in the absence of DNA. Similar results were obtained with 95% pure DNA-PK.
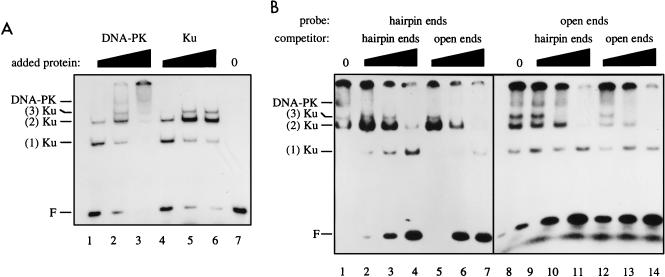
The inefficient activation of DNA-PK by hairpin DNA ends could be due to a failure of Ku to bind hairpin ends or to recruit DNA-PKCS to hairpin ends. To address this issue, we used an electrophoretic mobility shift assay, in which a 70-bp open-ended DNA probe detected protein-DNA complexes consistent with the binding of one, two, or three Ku molecules (22) as well as the binding of both Ku and DNA-PKCS (Fig. 3A). The components of the protein-DNA complexes were verified by showing that the complexes marked as Ku were supershifted by anti-Ku antibodies but not by anti-DNA-PKCS antibodies, and the complex marked as DNA-PK was supershifted by both anti-Ku and anti-DNA-PKCS antibodies (data not shown). We also observed a signal that was retained in the well of the gel. The appearance of this higher-order protein-DNA complex in the well was dependent on the presence of both Ku and DNA-PKCS, but the complex has not been further characterized.
FIG. 3.
Ku and DNA-PK bind to DNA with hairpin ends. (A) Ku and DNA-PK binding to open-ended DNA. A 70-bp open-ended DNA probe was incubated with increasing amounts of 70% pure DNA-PK (0.37, 1.1, and 3.3 Promega units in lanes 1 to 3, respectively) or purified Ku (14, 42, and 126 ng in lanes 4 to 6, respectively) or without added protein (lane 7) and resolved by nondenaturing polyacrylamide gel electrophoresis. Positions are indicated for free DNA probe (F), complexes containing one, two, or three Ku molecules, and a DNA-PK complex containing Ku and DNA-PKCS. (B) DNA-PK binding to hairpin- and open-ended DNA. DNA-PK (1.1 U) was incubated with DNA probes consisting of either a 68-bp hairpin-ended molecule (lanes 1 to 7) or a 70-bp open-ended molecule (lanes 8 to 14). For each probe, incubations were also done with unlabeled competitor DNA consisting of either the hairpin-ended molecule (lanes 2 to 4 and 9 to 11) or the open-ended molecule (lanes 5 to 7 and 12 to 14) in increasing amounts of 0.04, 0.20, and 1.0 ng.
When labeled hairpin-ended DNA was incubated with DNA-PK, it bound to Ku alone or to the DNA-PK complex, but only for high concentrations of DNA-PK (Fig. 3B, lane 1). For lower DNA-PK concentrations, formation of the DNA-PK complex was not observed, suggesting that its affinity might be lower for hairpin ends than for open ends. To directly compare the affinities of DNA-PK for hairpin ends and open ends, competitor DNA consisting of unlabeled hairpin-ended DNA or open-ended DNA was mixed with the labeled hairpin-ended DNA before the DNA-PK preparation was added. The addition of competitor DNA of both types was associated with rapid disappearance of the DNA-PK band (Fig. 3B, lanes 2 to 7). In this experiment, the Ku bands displayed a complex behavior that is difficult to interpret. However, for lower DNA-PK concentrations, at which formation of Ku complexes but not the DNA-PK complex occurred, both hairpin-ended DNA and open-ended DNA competed for Ku binding (data not shown).
To extend this result, the reverse experiment, in which the open-ended DNA was labeled, was performed. Like the hairpin-ended DNA, the open-ended DNA assembled complexes containing either Ku alone or both Ku and DNA-PKCS (Fig. 3B, lanes 1 and 8). Both unlabeled hairpin-ended DNA and open-ended DNA competed for binding activity (Fig. 3B, lanes 9 to 14). In summary, hairpin-ended DNA was capable of assembling a DNA-PK complex, although quantitative determination of the relative affinities of Ku and DNA-PKCS for hairpin- and open-ended DNA must await future experiments.
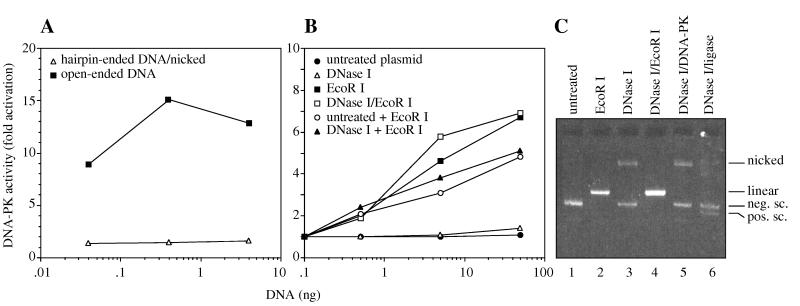
We next tested whether nicked DNA can activate DNA-PK. Nicked hairpin-ended 68-bp DNA failed to activate DNA-PK to any significant degree, while open-ended 70-bp DNA activated DNA-PK strongly (Fig. 4A). To extend this result, a 3,000-bp supercoiled plasmid was nicked with DNase I at a concentration sufficient to produce a preparation free of linear DNA, in which 40% of supercoiled plasmid was converted to nicked plasmid (Fig. 4C, lane 3). Nearly all of the nicked plasmid DNA could be ligated into a covalently closed circle that migrated as a positively supercoiled species when resolved by electrophoresis in agarose containing ethidium bromide (Fig. 4C, lane 6). Thus, the preparation contained 40% simple nicks and at most 8% other structures such as single-stranded gaps. The DNase I-treated plasmid and supercoiled plasmid preparations both failed to activate DNA-PK (Fig. 4B). By contrast, when either of these preparations was linearized with EcoRI, DNA-PK was strongly activated. Similar results were observed for DNA-PK preparations purified to 70% and greater than 95% and for incubation temperatures of 20 and 37°C. We calculated that activation of DNA-PK with the DNase I-treated plasmid preparation was at most 1.5% that of linear plasmid DNA under limiting DNA concentrations. Since about 40% of the preparation was nicked, we estimate that activation by DNA nicks is less than 4% that of open DNA ends.
FIG. 4.
Inefficient activation of DNA-PK by nicked DNA. (A) Nicked hairpin-ended DNA. DNA-PK was tested for activation by 68-bp nicked hairpin-ended DNA or 70-bp open-ended DNA. (B) Nicked plasmid DNA. DNA-PK (70% pure) was tested for activation by untreated supercoiled plasmid DNA (closed circles), plasmid linearized with EcoRI (closed squares), plasmid partially nicked with DNase I (open triangles), plasmid nicked with DNase I and linearized with EcoRI (open squares), an equal mixture of untreated plasmid and plasmid linearized with EcoRI (open circles), or an equal mixture of plasmid nicked with DNase I and plasmid linearized with EcoRI (closed triangles). For the DNA mixtures, each type of DNA was added in the amount indicated on the graph. (C) Analysis of nicked plasmid DNA. DNA preparations were analyzed by electrophoresis in a 1% agarose gel containing ethidium bromide, which resolved nicked, linear, and negatively and positively supercoiled (sc) plasmid DNA at the indicated positions. Negatively supercoiled plasmid DNA was left untreated (lane 1), linearized with EcoRI (lane 2), or partially nicked with DNase I (lane 3). The DNase I-treated plasmid was converted to nicked linear DNA by EcoRI (lane 4), not affected by incubation with DNA-PK (lane 5), and contained very few lesions other than simple nicks, since T4 DNA ligase converted nearly all of the nicked DNA to covalently closed circular DNA that migrated as positively supercoiled DNA (lane 6). Similar results were obtained with 95% pure DNA-PK.
Since the DNase I-treated plasmid preparation contained supercoiled DNA, we tested the effect of supercoiled DNA on kinase activation. A mild inhibition of kinase activation was observed when supercoiled plasmid was mixed with linearized plasmid, but only at high DNA concentrations. Mild inhibition was also seen when the DNase I-treated plasmid was mixed with linearized plasmid. However, even in the mixed samples, activation was strong compared to the results obtained with the DNase I-treated plasmid alone. We also ruled out the possibility that an unforseen ligase activity in the DNA-PK preparation may have repaired the nicked DNA. After incubation of the DNase I-treated plasmid with DNA-PK, no detectable change was observed in the structure of the nicked plasmid (Fig. 4C, lane 5).
DISCUSSION
DNA-PK is required for efficient double-strand break repair and V(D)J recombination (27). Both Ku and DNA-PKCS must be intact for efficient processing of hairpin coding ends during V(D)J recombination (25, 34). Other investigators have reported that DNA molecules ending in loops of 4 to 20 bases will bind to Ku and activate DNA-PK (9, 21). However, they did not examine DNA molecules with perfect hairpin ends such as those created during V(D)J recombination. Surprisingly, we found that hairpin-ended DNA was an extremely poor substrate for activating DNA-PK, even though hairpin-ended DNA was capable of assembling a DNA-PK complex. Thus, hairpin ends and stem-loops may differ with respect to their ability to activate DNA-PK.
DNA nicks bind to Ku (3) and are created as a DNA intermediate during V(D)J recombination (19). However, we found that nicked DNA failed to activate DNA-PK, arguing against the possibility that DNA-PK might be activated at the nicking step, prior to hairpin formation. Previous studies reported conflicting results for the effect of nicked DNA on DNA-PK. Our results concur with those of Weinfeld et al., who found no DNA-PK activation by nicked DNA isolated from irradiated plasmid or generated by DNase I (32). In addition, Gottlieb and Jackson found no activation of DNA-PK with plasmid DNA that had been nicked by DNase I in the presence of ethidium bromide (11). By contrast, Morozov et al. reported full activation of DNA-PK with a gel-purified 350-bp nicked minicircle not exposed to ethidium bromide (21). Our experiments used the same peptide as in those of Morozov et al. for measuring kinase activity, tested DNA substrates both smaller and larger than their minicircle, and created nicked DNA in two different ways: by DNase I treatment and by partial ligation of oligonucleotides in the absence of ethidium bromide. The activity observed by Morozov et al. may be due to features peculiar to the 350-bp minicircle. Indeed, the intrinsic rigidity of DNA inhibits the covalent closure of linear DNA fragments into circles for fragment lengths less than 500 bp (26). The 350-bp minicircle may impose structural abnormalities at the site of the nick, making it an anomalously effective substrate for DNA-PK. By contrast, our experiments used nicked substrates unconstrained by DNA rigidity, and we conclude that simple DNA nicks do not activate DNA-PK.
It is noteworthy that nicked DNA (3, 9) and hairpin-ended DNA bind to Ku, despite failing to activate the kinase. This result is consistent with the recent observation that DNA-PKCS is capable of DNA binding and activation in the absence of Ku at low salt concentrations (12, 33). Therefore, even though a DNA substrate may bind to Ku, it must also bind to DNA-PKCS in a configuration that leads to activation of the kinase. Such a configuration may not occur for hairpin ends or DNA nicks, suggesting that the structure of the DNA ends is critical for activation of the kinase.
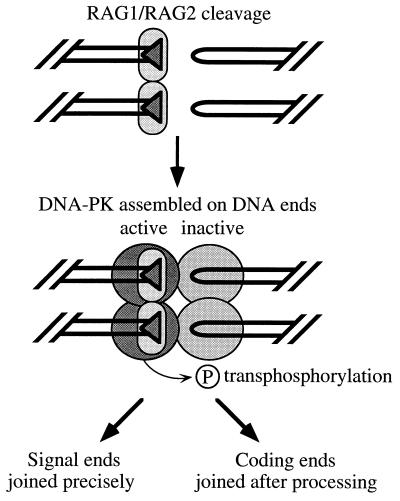
What are the implications of our results for V(D)J recombination? Notably, we obtained results for two different hairpin sequences, including one that was identical to a sequence previously tested in an extrachromosomal V(D)J recombination assay (20). Intact DNA-PK is required for efficient hairpin processing (25, 34) but fails to be efficiently activated by hairpin ends. Curiously, fully intact DNA-PK is not required for signal joining (4, 23), even though it is activated by blunt signal ends. To explain this apparent paradox, we propose a model in which hairpin processing requires assembly and activation of DNA-PK by the cogenerated signal ends and transphosphorylation of target proteins bound to the hairpin end (Fig. 5). The target protein might be Ku, DNA-PKCS, or another protein, which upon transphosphorylation makes the hairpin accessible to a putative hairpin endonuclease. The opened coding ends are then joined by a general double-strand break repair pathway (6). Signal ends are joined precisely, perhaps because proteins bound to the signal ends are protected from phosphorylation by RAG1 and RAG2, which remain bound to the signal ends after cleavage (1). Thus, transphosphorylation would not be involved in signal joining, explaining why signal joining is spared in SCID mice.
FIG. 5.
Model for processing of hairpin ends by activation of DNA-PK. RAG1-RAG2 (small ovals) induce cleavage adjacent to two recombination signal sequences (shaded triangles) to form blunt signal ends and hairpin coding ends. The signal ends activate DNA-PK (large circles) so that phosphorylation in trans will lead to processing of the hairpin end. The target of transphosphorylation could be Ku, DNA-PKCS, or some other protein bound to the hairpin end. By contrast, DNA-PK bound to the hairpin coding end remains inactive as a kinase. Proteins bound to the signal ends may be protected from transphosphorylation by RAG1-RAG2, which remain bound to signal ends after cleavage (1).
Unlike hairpin ends generated by V(D)J recombination, hairpin ends introduced into cells by transfection are joined with equal efficiency in SCID and in wild-type cells (17). This difference may be explained by in vitro V(D)J recombination experiments, which show coupling between cleavage and end joining (16, 24). Components of the end-joining machinery may be assembled in a postcleavage complex that masks the hairpin ends, so that subsequent activation of DNA-PK is required to unmask the ends for the hairpin endonuclease. Such a masking effect would not occur for hairpin ends introduced by transfection.
Our data are consistent with other models as well. Hairpin end joining may require the kinase activity of DNA-PK indirectly, for example, through signaling pathways. Alternatively, coding joints may not require the kinase activity at all but instead require another biochemical activity contained in the enormous DNA-PKCS molecule. Nevertheless, we have demonstrated that, of the three known DNA intermediates in the V(D)J cleavage process, nicked DNA, hairpin-ended DNA, and open-ended DNA, only open-ended DNA is capable of significantly activating DNA-PK, thus adding important constraints to any model for the role of DNA-PK in V(D)J recombination.
ACKNOWLEDGMENTS
We thank O. Hammarsten, B. J. Hwang, D. Brutlag, and J. Danska for helpful conversations and P. Mayerfeld and J. Short for loaning the TD-700 fluorometer.
This research was supported by grant DAMD 17-94-J-4350 from the U.S. Army Medical Research and Materiel Command to G.C. and grant MT-13219 from the Medical Research Council of Canada to S.L., who is a Research Scientist of the National Cancer Institute of Canada supported with funds from the Canadian Cancer Society. V.S. is supported by the Life and Health Insurance Medical Research Fund sponsored by the American United Life Insurance Company. W.K.R. is supported by the Medical Scientist Training Program.
REFERENCES
- 1.Agrawal A, Schatz D. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 2.Biedermann K, Sun J, Giaccia A, Tosto L, Brown J M. Scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blier P, Griffith A, Craft J, Hardin J. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 4.Blunt T, Finnie N, Taccioli G, Smith G, Demengeot J, Gottlieb T, Mizuta R, Varghese A, Alt F, Jeggo P, Jackson S. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 5.Blunt T, Gell D, Fox M, Taccioli G, Jackson S, Lehman A, Jeggo P. Identification of a nonsense mutation in the carboxy-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu G. Double-strand break repair. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 7.Danska J, Holland D, Mariathasan S, Williams K, Guidos C. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errami A, Smider V, Rathmell W K, He D, Hendrickson E A, Zdzienicka M, Chu G. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cells. Mol Cell Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falzon M, Fewell J W, Kuff E L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 10.Fulop G, Phillips R. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 11.Gottlieb T, Jackson S. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 12.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson E, Qin X Q, Bump E, Schatz D, Oettinger M, Weaver D. A link between double-strand break related repair and V(D)J recombination: the scid mutation. Proc Natl Acad Sci USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchgessner C, Patil C, Evans J, Cuomo C, Fried L, Carter T, Oettinger M, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1185. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 15.Lees-Miller S, Sakaguchi K, Ullrich S, Appela E, Anderson C. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leu T, Eastman Q, Schatz D. Coding joint formation in a cell-free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- 17.Lewis S M. P nucleotide insertions, and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 19.McBlane F, van Gent D, Ramsden D, Romeo C, Cuomo C, Gellert M, Oettinger M. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 20.Meier J, Lewis S. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol Cell Biol. 1993;13:1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morozov V, Falzon M, Anderson C, Kuff E. DNA-dependent protein kinase is activated by nicks and larger single-stranded gaps. J Biol Chem. 1994;269:16684–16688. [PubMed] [Google Scholar]
- 22.Paillard S, Strauss F. Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res. 1991;19:5619–5624. doi: 10.1093/nar/19.20.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pergola F, Zdzienicka M Z, Lieber M R. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsden D, Paull T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 25.Roth D, Menetski J, Nakajima P, Bosma M, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 26.Shore D, Langowski J, Baldwin R. DNA flexibility studied by covalent closure of short DNA fragments into circles. Proc Natl Acad Sci USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smider V, Chu G. The end-joining reaction in V(D)J recombination. Semin Immun. 1997;9:189–197. doi: 10.1006/smim.1997.0070. [DOI] [PubMed] [Google Scholar]
- 28.Smider V, Rathmell W K, Lieber M, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 29.Taccioli G, Gottlieb T, Blunt T, Priestly A, Demengeot J, Mizuta R, Lehmann A, Alt F, Jackson S, Jeggo P. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 30.van Gent D, McBlane J F, Ramsden D, Sadofsky M, Hesse J, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 31.van Gent D, Ramsden D, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 32.Weinfeld M, Chaudhry M, D’Amours D, Pelletier J, Poirer G, Povirk L, Lees-Miller S. Interaction of DNA-dependent protein kinase and poly(ADP-ribose) polymerase with radiation-induced DNA strand breaks. Radiat Res. 1998;148:22–28. [PubMed] [Google Scholar]
- 33.Yaneva M, Kowalewski T, Lieber M. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu C, Bogue M, Lim D S, Hasty P, Roth D. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]