Abstract
An inverted repeat (IR) within the U5 region of the Rous sarcoma virus (RSV) mRNA forms a structure composed of a 7-bp stem and a 5-nucleotide (nt) loop. This U5-IR structure has been shown to be required for the initiation of reverse transcription. The mRNA of Tf1, long terminal repeat-containing retrotransposon from fission yeast (Schizosaccharomyces pombe) contains nucleotides with the potential to form a U5-IR stem-loop that is strikingly similar to that of RSV. The putative U5-IR stem-loop of Tf1 consists of a 7-bp stem and a 25-nt loop. Results from mutagenesis studies indicate that the U5-IR stem-loop in the mRNA of Tf1 does form and that it is required for Tf1 transposition. Although the loop is required for transposition, we were surprised that the specific sequence of the nucleotides within the loop was unimportant for function. Additional investigation indicates that the loss of transposition activity due to a reduction in the loop size to 6 nt could be rescued by increasing the GC content of the stem. This result indicates that the large loop in the Tf1 mRNA relative to that of the RSV allows the formation of the relatively weak U5-IR stem. The levels of Tf1 proteins expressed and the amounts of Tf1 RNA packaged into the virus-like particles were not affected by mutations in the U5-IR structure. However, all of the mutations in the U5-IR structure that caused defects in transposition produced low amounts of reverse transcripts. A unique feature in the initiation of Tf1 reverse transcription is that, instead of a tRNA, the first 11 nt of the Tf1 mRNA serve as the minus-strand primer. Analysis of the 5′ end of Tf1 mRNA revealed that the mutations in the U5-IR stem-loop that resulted in defects in reverse transcription caused a reduction in the cleavage activity required to generate the Tf1 primer. Our results indicate that the U5-IR stems of Tf1 and RSV are conserved in size, position, and function.
Long terminal repeat (LTR)-containing retrotransposons are mobile elements that possess similarities to retroviruses in both structure and mechanism (4, 25). The genomes of LTR-containing retrotransposons are positive single-stranded RNA molecules that encode proteins analogous to retroviral gag and pol gene products. These include the structural protein, Gag, and the catalytic proteins, protease (PR), reverse transcriptase (RT), and integrase (IN). In a process that is similar to the assembly of retroviral virions, the structural and catalytic proteins of retrotransposons assemble the transposon RNA into virus-like particles (VLPs). Within the context of VLPs, the RT converts the RNA into double-stranded DNA that includes flanking LTRs. The IN then integrates this DNA into the host genome. Extensive similarities also exist between retroviruses and LTR-containing retrotransposons in their specific mechanisms of reverse transcription and integration. Despite the extensive similarities to retroviruses, LTR-containing retrotransposons integrate their reverse transcripts only into the genomes of host cells that have expressed the transposon.
The reverse transcription of RNAs encoded by several retroviruses and LTR-containing retrotransposons has been shown to require specific tRNA molecules to function as primers (7, 9, 12, 27). The 3′-terminal 10 to 18 nucleotides (nt) of the tRNA anneal to the viral mRNA at the primer binding site (PBS), a region immediately downstream of the 5′ LTR. The virus-encoded RT is able to initiate DNA synthesis from the 3′ terminus of the tRNA (25). Besides the annealing of the tRNA to the PBS, the initiation of reverse transcription of Rous sarcoma virus (RSV) has been shown to require the formation of an additional RNA structure termed the U5-inverted repeat (IR) stem-loop (8). The sequence of the LTRs are composed of the U3 (unique in the 3′ end of mRNA), U5 (unique in the 5′ end of mRNA), and R (repeated in both ends of mRNA) regions based on their locations in the genomic RNA. The term U5-IR stem-loop is used because the structure is composed of an IR within the nucleotides of the U5 region of the LTR. The U5-IR stem-loop of RSV is located immediately upstream of the PBS and consists of a 7-bp stem and a 5-nt loop. Mutations that alter the formation of the U5-IR stem and loop reduced the initiation of reverse transcription without causing major defects in packaging the RSV RNA into virions (8, 19). The presence of sequences with the potential to form similar U5-IR structures in the RNA of several retroviruses has been observed (1). These results suggest that this RNA structure may participate in the reverse transcription of many retroviruses (1, 12).
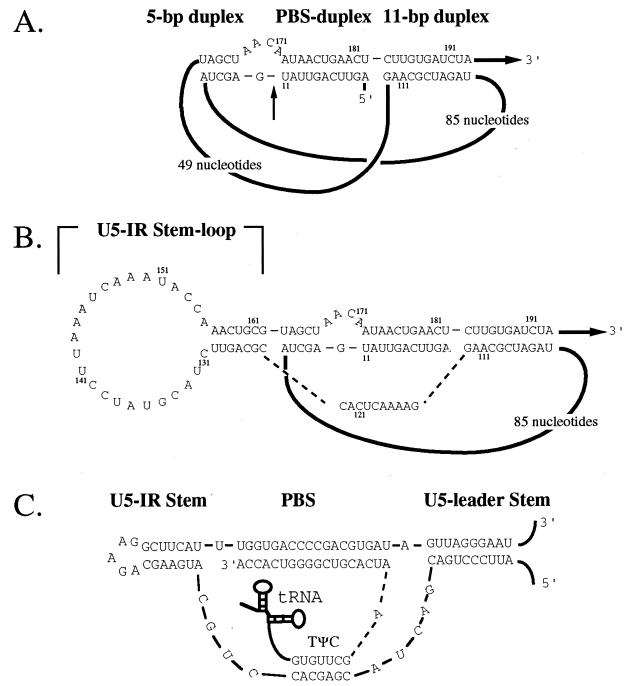
The reverse transcription of the LTR-containing retrotransposon Tf1 of Schizosaccharomyces pombe is exceptional in that the primer for reverse transcription is not derived from a tRNA. Perfect complementarity between the PBS and the first 11 nt of the Tf1 mRNA allows these 11 nt to serve as the primer of minus-strand reverse transcription (13). The results of mutagenesis studies and RNA analysis indicate that the primer is generated by a cleavage that occurs between nt 11 and 12 of the Tf1 mRNA (14). We reported previously that the first 11 nt and the PBS are included in a three-duplex RNA structure (Fig. 1A) that is required for the cleavage reaction (17). Recently, it was found that this unique mechanism of self-primed reverse transcription is likely shared by several LTR-containing retrotransposons (18, 23). These self-primed retrotransposons lack tRNA primers and possess complementarity between their PBSs and sequences near the start of their mRNAs. The potential to form RNA structures significantly similar to that of Tf1 was observed in five of these retrotransposons (18).
FIG. 1.
The putative U5-IR stem-loop of Tf1. (A) An RNA structure consisting of three duplexes is proposed. The regions of duplex structure are the 5-bp duplex, the PBS duplex, and the 11-bp duplex. The 5′ end of the RNA is labeled, and the dark black lines represent extended stretches of sequence. The arrow labeled 3′ indicates that the majority of the transcript is 3′ of the sequence shown. The numbers shown in small type indicate the nucleotide position relative to the first base of the transcript. (B) Base pairing is predicted for the nucleotides (nt 124 to 130 and nt 162 to 156) within the 49-nt loop. The resulting structure is a U5-IR stem-loop with a 7-bp stem and a 25-nt loop. The dashed lines indicate that the nucleotides shown connect directly to the indicated regions of the diagram. (C) U5-IR stem-loop of RSV. The three regions of duplex structure in the transcript of RSV are the U5-IR, the PBS, and the U5-leader stem. The tRNA primer is shown in diagrammatic form, and all the lines between nucleotides indicate direct connections to adjacent nucleotides. The 3′ and 5′ labels indicate the polarity of the RNA and the position of the transcript sequences not shown.
We noticed the potential for additional base pairing within one of the loops of the three-duplex RNA structure of Tf1. This base pairing would result in a larger RNA structure (Fig. 1B) that would have profound similarity to the U5-IR and U5- leader structures of RSV (Fig. 1C). We investigated the existence and function of the U5-IR stem-loop in Tf1 RNA by assessing the effect on transposition of mutations that either disrupted or restored the formation of the RNA structure. Here, we report that the U5-IR stem-loop does form in the Tf1 mRNA in vivo and we present evidence that indicates it consists of a 7-bp stem and a 25-nt loop. These conclusions were based on in vivo results that showed that the U5-IR stem-loop of Tf1 is required for Tf1 transposition. Similar to the essential role that this RNA structure plays in the reverse transcription of RSV, the U5-IR stem-loop of Tf1 was also required for the initiation of reverse transcription. In addition, our data indicates that the formation of the U5-IR stem-loop of Tf1 is not required for the packaging of RNA into VLPs. The results of this study indicate that the U5-IR stems of RSV and Tf1 are conserved in size, position, and function.
MATERIALS AND METHODS
Media.
The minimal liquid and plate media for S. pombe were composed of EMM (21) and were supplemented with amino acids, vitamin B1, 5-fluoroorotic acid (5-FOA) and G418 (Geneticin) at concentrations as previously described (14).
Strains.
The yeast strains and the plasmids used in this study are listed in Table 1. All plasmids were transformed into S. pombe YHL912 (h− ura4-294 leu1-32). Escherichia coli TG1 was used for propagation of plasmids.
TABLE 1.
Yeast strains used in this study
| Yeast straina | Plasmid description
|
||
|---|---|---|---|
| Designationc | Tf1 allele | Region | |
| YHL4093b | pHL891-19 | WTd | |
| YHL4179b | pHL900-1 | PR-fs | |
| YHL5019b | pHL1099-2 | U11A | |
| YHL6062 | pHL1291-1 | A172U | |
| YHL6074 | pHL1297-2 | U11A/A172U | |
| YHL5893 | pHL1264-1 | Gag-Δ166-173 | |
| YHL6082 | pHL1303-1 | C124G | Stem |
| YHL6084 | pHL1305-1 | C126G | Stem |
| YHL6272 | pHL1449-2 | G128C | Stem |
| YHL6263 | pHL1445-1 | G135C | Loop |
| YHL6172 | pHL1359-2 | A137U | Loop |
| YHL6086 | pHL1307-1 | U151A | Loop |
| YHL6259 | pHL1443-1 | C153G | Loop |
| YHL6267 | pHL1447-1 | C158G | Stem |
| YHL6088 | pHL1309-1 | G160C | Stem |
| YHL6090 | pHL1311-1 | G162C | Stem |
| YHL6152 | pHL1347-1 | C124G/G162C | Stem |
| YHL6156 | pHL1349-1 | C126G/G160C | Stem |
| YHL6394 | pHL1566-1 | G128C/C158G | Stem |
| YHL6398 | pHL1568-1 | G135C/C153G | Loop |
| YHL6200 | pHL1373-1 | A137U/U151A | Loop |
| YHL6252 | pHL1439-2 | MSL1 | Loop |
| YHL6176 | pHL1361-1 | MSL2 | Loop |
| YHL6180 | pHL1363-2 | MSL3 | Loop |
| YHL6184 | pHL1365-1 | MSL4 | Loop |
| YHL6188 | pHL1367-1 | MSL5 | Loop |
| YHL6192 | pHL1369-1 | MSL6 | Loop |
| YHL6196 | pHL1371-1 | MSL7 | Loop |
| YHL6255 | pHL1441-1 | MSL8 | Loop |
| YHL6070 | pHL1295-1 | Δ3 (nt 124 to 162 deleted) | Stem-loop |
| YHL6081 | pHL1302-1 | Δ2 (nt 135 to 153 deleted) | Loop |
| YHL6248 | pHL1437-2 | Δ1 (nt 139 to 149 deleted) | Loop |
| YHL6384 | pHL1560-1 | CG-stem | Stem-loop |
| YHL6388 | pHL1563-1 | AG-loop | Stem-loop |
| YHL6390 | pHL1564-1 | RSV-loop | Stem-loop |
| YHL6275 | pHL891-19 | WT | |
| pHL1451-2 | WT | ||
| YHL6331 | pHL1302-1 | Δ2 (nt 135 to 153 deleted) | Loop |
| pHL1451-2 | WT | ||
| YHL6332 | pHL1295-1 | Δ3 (nt 124 to 162 deleted) | Stem-loop |
| pHL1451-2 | WT | ||
| YHL6333 | pHL1303-1 | C124G | Stem |
| pHL1451-2 | WT | ||
| YHL6334 | pHL1305-1 | C126G | Stem |
| pHL1451-2 | WT | ||
| YHL6335 | pHL1309-1 | G160C | Stem |
| pHL1451-2 | WT | ||
| YHL6336 | pHL1311-1 | G162C | Stem |
| pHL1451-2 | WT | ||
| YHL6337 | pHL1449-2 | G128C | Stem |
| pHL1451-2 | WT | ||
| YHL6338 | pHL1447-1 | C158G | Stem |
| pHL1451-2 | WT | ||
All plasmids were transformed into YHL912 (h− ura4-294 leu1-32).
Constructed in previous study (17).
All plasmids contained a URA3 gene except pHL1451-2, which contained a Leu2 gene.
WT, wild type.
Construction of plasmids with site-directed mutations.
All plasmids were constructed by directional cloning of fusion PCR (13) products into a Tf1-containing plasmid with a URA3 selectable marker. The plasmids containing site-directed single- and multiple-point mutations as well as deletion mutations were constructed by cloning into pHL888-3 the XhoI-AvrII fragment amplified from pHL891-19 (17) with oligonucleotides that included the specific mutations as primers (17). The plasmids containing compensating double-point mutations in the 5′ untranslated region of Tf1 were created by using DNA from the plasmids with single-point mutations as a template in fusion PCRs that used the corresponding oligonucleotide primers to generate the additional mutation. All PCRs were performed with the native Pfu enzyme (Stratagene). The specific oligonucleotides used in the construction of each mutation are shown in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a | Mutation (strandb) |
|---|---|---|
| General | ||
| HL38 | GGAAGAGGAATCCTGGC | All (T) |
| HL39 | CCAATTGCTTCCAGTCTTTG | All (B) |
| Single-point mutation | ||
| HL260 | CCAAACTGCCTAGCTAACAATAACTG | G162C (T) |
| HL261 | CAGTTATTGTTAGCTAGGCAGTTTGG | G162C (B) |
| HL262 | CCTTAAATCAAAAACCAAACTGCG | U151A (T) |
| HL263 | CGCAGTTTGGTTTTTGATTTAAGG | U151A (B) |
| HL264 | CAAATACCAAACTCCGTAGCTAAC | G160C (T) |
| HL265 | GTTAGCTACGGAGTTTGGTATTTG | G160C (B) |
| HL266 | GGAAAACTCACCGGAGTTCTACGTATCC | C126G (T) |
| HL267 | GGATACGTAGAACTCCGGTGAGTTTTCC | C126G (B) |
| HL268 | GGAAAACTCACGGCAGTTCTACG | C124G (T) |
| HL269 | CGTAGAACTGCCGTGAGTTTTCC | C124G (B) |
| HL303 | GCAGTTCTACGTTTCCTTAAATC | A137U (T) |
| HL304 | GATTTAAGGAAACGTAGAACTGC | A137U (B) |
| HL380 | AAATCAAATAGCAAACTGCGTAGC | C153G (T) |
| HL381 | CGCAGTTTGCTATTTGATTTAAGG | C153G (B) |
| HL382 | GTTCTACCTATCCTTAAATCAAATACC | G135C (T) |
| HL383 | AAGGATAGGTAGAACTGCGGTGAG | G135C (B) |
| HL384 | TACCAAAGTGCGTAGCTAACAATAACTG | C158G (T) |
| HL385 | GCTACGCACTTTGGTATTTGATTTAAGG | C158G (B) |
| HL386 | CACCGCACTTCTACGTATCCTTAAATC | G128C (T) |
| HL387 | GTAGAAGTGCGGTGAGTTTTCCTTGCG | G128C (B) |
| Multiple-point mutation | ||
| HL305 | CTACGTATGGAAAAATCAAATACC | MSL2 (T) |
| HL306 | TTTGATTTTTCCATACGTAGAACTGCGG | MSL2 (B) |
| HL307 | CTACGTATTTCCAAATCAAATACC | MSL3 (T) |
| HL308 | TTTGATTTGGAAATACGTAGAACTGCGG | MSL3 (B) |
| HL309 | CGTATCCTTTTTTCAAATACCAAACTGCG | MSL4 (T) |
| HL310 | GGTATTTGAAAAAAGGATACGTAGAACTGC | MSL4 (B) |
| HL311 | CGTATCCTTGGGTCAAATACCAAACTGCG | MSL5 (T) |
| HL312 | GGTATTTGACCCAAGGATACGTAGAACTGC | MSL5 (B) |
| HL313 | CGTATCCTTAAAAGTTATACCAAACTGCG | MSL6 (T) |
| HL314 | CGCAGTTTGGTATAACTTTTAAGGATACG | MSL6 (B) |
| HL315 | CGTATCCTTAAACTGGATACCAAACTGCG | MSL7 (T) |
| HL316 | CGCAGTTTGGTATCCAGTTTAAGGATACG | MSL7 (B) |
| HL376 | AGTTGATGGTATCCTTAAATCAAATACC | MSL1 (T) |
| HL377 | GATACCATCAACTGCGGTGAGTTTTCC | MSL1 (B) |
| HL378 | TCAAATACGTAACTGCGTAGCTAAC | MSL8 (T) |
| HL379 | CGCAGTTACGTATTTGATTTAAGGATACG | MSL8 (B) |
| Deletion mutation | ||
| HL253 | GGAAAACTCACΔTAGCTAACAATAACTG | Deletion of nt 124 to 162 (T) |
| HL254 | TTATTGTTAGCTAΔGTGAGTTTTCCTTGCG | Deletion of nt 124 to 162 (B) |
| HL256 | GCTACGCAGTTTGΔGTAGAACTGCGG | Deletion of nt 135 to 153 (B) |
| HL270 | AGTTCTACΔCAAACTGCGTAGCTAAC | Deletion of nt 135 to 153 (T) |
| HL374 | TTCTACGTATΔATACCAAACTGCG | Deletion of nt 139 to 149 (T) |
| HL375 | TTTGGTATΔATACGTAGAACTGCGG | Deletion of nt 139 to 149 (B) |
| HL430 | AGTCCTACΔCAGACTGCGTAGCTAAC | GC-stem (T) |
| HL431 | AGTCTGΔGTAGGACTGCGGTGAGTTTTCC | GC-stem (B) |
| HL432 | AGTCATACΔCGGACTGCGTAGCTAAC | AG-loop (T) |
| HL433 | AGTCCGΔGTATGACTGCGGTGAGTTTTCC | AG-loop (B) |
| HL434 | AGTCAGAAGGACTGCGTAGCTAAC | RSV-loop (T) |
| HL435 | AGTCCTTCTGACTGCGGTGAGTTTTCC | RSV-loop (B) |
Mutated nucleotides are shown in boldface type. Δ, deleted nucleotides.
T, top; B, bottom.
Verification of mutations in constructed plasmids.
The mutation sites in each plasmid were verified by dideoxyribonucleotide termination sequencing reactions (USB Sequenase version 2.0 DNA sequencing kit). In addition, each PCR-generated construct was made in duplicate by using an independent PCR to preclude false assay results caused by nontemplated PCR-generated mutations. The correct constructs were confirmed when each duplicated plasmid gave results in transposition and primer extension assays equivalent to those of its partner.
Immunoblot detection of Tf1 Gag and IN.
Total cellular proteins were collected from 5 ml of cell cultures at an OD600 of 2.0 harvested and broken with glass beads in 400 μl of extraction buffer (10 mM HEPES-KOH [pH 7.8], 15 mM KCl, 5 mM EDTA) containing PR inhibitors (aprotinin [1 μg/ml], leupeptin [0.5 μg/ml], pepstatin [0.7 μg/ml], 3 mM dithiothreitol, and 2 mM phenylmethylsulfonyl fluoride) as previously described (2). The whole-cell extracts were collected, and an equal volume of 2× sample buffer was added. The mixture was boiled for 3 min and 25 μg of total protein from each sample was loaded on sodium dodecyl sulfate–10% polyacrylamide electrophoresis gels for immunoblot analysis. Standard electrotransfer techniques were used (26) with Immobilon P (Millipore) as the membrane. The detection method used was the enhanced chemiluminescence system as described by the manufacturer (Amersham), except that the secondary antibody, horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin, was used at a 10,000-fold dilution. The primary polyclonal antisera used for each filter were from production bleeds 660 (anti-Gag) and 657 (anti-IN) (16).
Transposition assay.
S. pombe strains containing Tf1 plasmids were first grown as patches on EMM plates in the absence of vitamin B1 to induce the nmt1 promoter that was fused to the neo-marked Tf1. The neo gene was inserted into the noncoding region of Tf1 as a marker for transposition. After 4 days of incubation at 32°C, the cells were replica printed onto 5-FOA-containing EMM plates to select against cells retaining the Tf1-containing plasmid which possesses a URA3 marker gene (5). Cells on these plates were then replica printed onto 5-FOA- and G418-containing YES medium (21) and incubated at 32°C for 2 days to detect cells with copies of Tf1-neo that had transposed into the genome (15, 16). The presence of the bacterial neo gene allowed S. pombe to grow in G418 (500 mg/ml).
Isolation of RNA and DNA from whole-cell extracts.
Ten milliliters of stationary-phase cells at an optical density at 600 nm (OD600) of 10 were harvested for nucleic acid isolation. The procedure for making these preparations was modified from previously published procedures (14) and included the steps briefly described below. Nucleic acids were directly isolated from cells suspended in extraction buffer (50 mM Tris-HCl [pH 7.4], 0.15 M NaCl, 10 mM EDTA, and 5% sodium dodecyl sulfate) by extracting with PICA (phenol-chloroform-isoamyl alcohol; 50:50:1, vol/vol/vol) while breaking cells under acid-washed glass beads. After four additional extractions with PICA, the samples were ethanol precipitated and used as material for either DNA blot analysis (2) or RNA-templated primer extension.
DNA blot analysis.
Five micrograms of DNA sample was restriction digested with BstXI and subjected to agarose gel electrophoresis. The DNA was transferred to GeneScreen Plus, and the filter was hybridized with a 32P-labeled neo probe (a 1-kb fragment derived from a BamHI digest of pGH54 [11]).
Primer extension.
The RNA (∼10 μg) isolated from cells in early stationary phase was resuspended in 15 μl of 1× avian myeloblastosis virus (AMV) reverse transcription buffer (Boehringer Mannheim) that contained 1 pmol of oligonucleotide JB890 that was 5′ end 32P labeled. Extension reactions with AMV RT (Boehringer Mannheim) were performed as previously described (14).
Tf1 RNA protection assays.
Cells were inoculated at an OD600 of 0.05 in 50 ml of drop out medium (EMM minus uracil) and grown for 36 h to an OD600 of 10.0. Twenty milliliters of cells was harvested, washed, and then broken with glass beads in 1 ml of extraction buffer (15 mM KCl, 10 mM HEPES-KOH [pH 7.8], 5 mM EDTA, 0.2% Triton X-100) containing PR inhibitors (aprotinin [1 μg/ml], leupeptin [0.5 μg/ml], pepstatin [0.7 μg/ml], 3 mM dithiothreitol, and 2 mM phenylmethylsulfonyl fluoride and 60 U of RNase inhibitor (RNAguard; Pharmacia Biotech). Supernatants of the cell extracts were recovered after centrifugation at 1,000 × g for 5 min. Fifteen microliters of 10× buffer M (100 mM Tris-HCl [pH 7.5], 100 mM MgCl2, 500 mM NaCl) with or without Benzonase (Sigma catalog no. E-8263) at specified concentrations was added to 135 μl of the supernatant and incubated at room temperature for 6 min. The reactions were stopped by the addition of 5 μl of 0.5 M EDTA. Nucleic acids were extracted immediately by the addition of 300 μl each of the nucleic acid extraction buffer and PICA as described above in the isolation of RNA and DNA. After four additional extractions with PICA, the samples were ethanol precipitated, resuspended in 25 μl of H2O, and kept at −20°C until RNA blot analysis. The levels of RNase-resistant RNA were compared to the amount of transcripts present in cells by isolating the total accumulated RNA (isolated by extraction from cells with glass beads and phenol-chloroform as described above).
RNA blot analysis.
Five microliters of RNA sample was mixed with 2 μl of 37% formaldehyde, 10 μl of deionized formamide, 2 μl of 10× MOPS (morpholinepropanesulfonic acid) buffer (0.2 M MOPS, 0.05 M sodium acetate, 5 mM EDTA), and 1 μl of ethidium bromide (0.5 μg/ml). The prepared samples were incubated at 65°C for 5 min and subjected to agarose gel (1%) electrophoresis in 1× MOPS. The RNA was transferred to GeneScreen Plus, and the filter was hybridized with 32P-labeled probes specific for Gag or actin.
RESULTS
We reported previously that retrotransposon Tf1 requires the formation of a three-duplex RNA structure to initiate its self-primed reverse transcription (Fig. 1A) (17). This three-duplex RNA structure includes intervening loops of 49 and 85 nt. Recently, we observed the potential for extended base pairing in the 49-nt loop (Fig. 1B) that would result in a structure analogous to the U5-IR stem-loop of retroviruses (1, 12). This putative U5-IR stem-loop of Tf1 is located upstream of the Tf1 PBS and consists of a 7-bp stem and a 25-nt loop (Fig. 1B). The U5-IR stem-loop of RSV (Fig. 1C), also located immediately upstream of the PBS of RSV, is required for efficient initiation of reverse transcription (8). In addition to the importance of the U5-IR structure in the reverse transcription of RSV, the close proximity of the U5-IR stem-loop of Tf1 to its PBS led us to investigate whether this extended structure of Tf1 exists in vivo and to examine its role in the process of self-primed reverse transcription.
The U5-IR stem was required for Tf1 transposition.
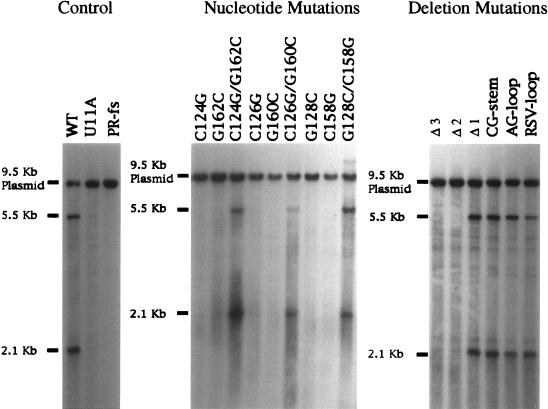
To test whether the sequence proposed to form the U5-IR stem of Tf1 contributed to transposition, several point mutations spanning both strands were constructed (Fig. 2A). Strains carrying these mutations were subjected to an assay that measured within S. pombe cells the transposition of a neo-marked copy of Tf1 from a plasmid into the host genome. Expression of Tf1-neo was regulated by a repressible promoter that was active in the absence of vitamin B1. After the eviction of the Tf1-containing plasmids from the S. pombe cells, transposition events were detected as growth on G418-containing medium (16). The single-nucleotide substitutions generated were C to G at position 124 (C124G), C126G, G128C, C158G, G160C, and G162C (Fig. 2A, substitutions 1 to 6 respectively). We also generated three versions of Tf1 with pairs of compensating mutations that altered nucleotides in the stem but preserved the potential for base pairing. These compensating mutations are designated C124G/G162C, C126G/G160C, and G128C/C158G (Fig. 2A, 1 and 6, 2 and 5, and 3 and 4, respectively). All six single-point mutations drastically reduced transposition activity of Tf1 relative to the level produced by a strain that contained wild-type Tf1. The reduced transposition activities caused by substitutions 1 through 6 were equal to or less than the activity of IN-fs, a strain previously shown to possess levels of transposition activity 100-fold less than that of the wild type (17). The strain labeled PR-fs contained a copy of Tf1 with a frameshift in the PR sequence, and as a result, the lack of RT and IN expression lowered transposition to levels that were undetectable. The transposition defects caused by the single-point mutations were rescued to wild-type levels by the addition of mutations that restored the potential for base pairing (Fig. 2B). The rescue of the low transposition activities to wild-type levels was observed for all three combinations tested. These results indicate that the U5-IR stem did form in vivo and was required for Tf1 transposition.
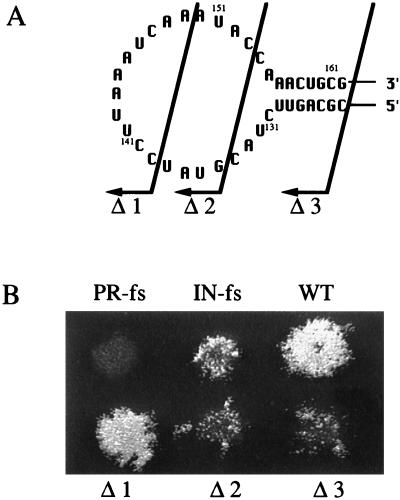
FIG. 2.
The results of transposition assays indicate that the U5-IR stem was required for Tf1 transposition. (A) Substitutions of nucleotides and their positions in the U5-IR stem loop of Tf1 mRNA. The single-point mutations are 1, C124G; 2, C126G; 3, G128C; 4, C158G; 5, G160C; and 6, G162C. The double-point mutations are combinations of mutations 1 and 6, 2 and 5, and 3 and 6. (B) Transposition activity as indicated by growth on G418-containing plates. The agar plate shown contained the transposition assay patches of strains that contained the mutations shown in panel A. WT, strain with a wild-type version of Tf1.
FIG. 6.
Mutants that are defective in transposition produced small or undetectable amounts of Tf1 reverse transcripts. Extracts of total DNA from the strains in this study were digested with the restriction enzyme BstXI and subjected to DNA blot analysis in which Tf1 cDNA was visualized by hybridization with a probe specific for neo. The 2.1-kb band represents the 3′ fragment of the Tf1 cDNA, and the 9.5-kb band represents the Tf1-containing plasmid. The 5.5-kb band is a species of cDNA thought to be derived from single-LTR circles. WT, strain with a wild-type version of Tf1.
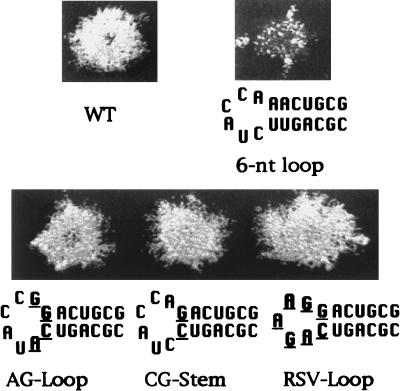
FIG. 5.
Mutations that increase the stability of the U5-IR stem rescued the transposition defect of the 6-nt loop. Shown here are the sequences and the transposition activities of strains that contained modifications in the 6-nt loop version of the U5-IR stem-loop. The base pair in the stem adjacent to the loop was changed from AU to GC in the versions of Tf1 designated AG loop, CG stem, and RSV loop. WT, strain with a wild-type version of Tf1.
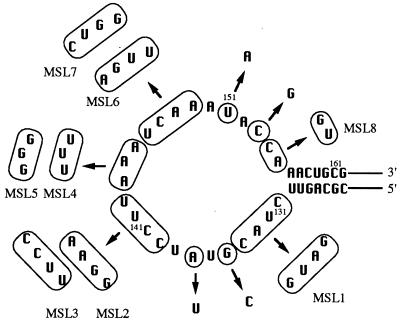
FIG. 3.
Nucleotide substitutions in the U5-IR loop exhibited no effect on Tf1 transposition. The circled nucleotides were altered to the sequences indicated by the arrows. None of these sequence alterations caused a reduction in the transposition activity of Tf1. The numbers represent the nucleotide positions within the Tf1 transcript.
FIG. 4.
Transposition assay results of strains with deletions in the U5-IR structure. (A) Arrows indicate the positions of the deletions. Nucleotides left of the arrows are deleted. Deletion 1 resulted in the formation of a 14-nt loop. Deletion 2 resulted in the formation of a 6-nt loop. Deletion 3 eliminated the U5-IR stem-loop. (B) Growth on G418-containing plates indicates transposition activity. The extent of growth on the G418-containing plate is a direct measure of transposition activity. WT, strain with a wild-type version of Tf1.
Nucleotide substitutions in the U5-IR loop did not affect Tf1 transposition activity.
To address whether nucleotides in the U5-IR loop contribute to Tf1 transposition, we generated a total of 14 mutations distributed throughout this sequence (Fig. 3). These substitutions include four single-point mutations (G135C, A137U, U151A, and C153G), two double-point mutations (G135C/C153G and A137U/U151A) and eight mutations with multiply substituted loops (MSLs) (MSL1 to MSL8). We detected no visible transposition defects caused by any of these mutations in the U5-IR loop (data not shown). Thus, the specific sequence of the nucleotides within the loop was unimportant for transposition.
The U5-IR loop stabilized the base pairing of the U5-IR stem.
Three Tf1-containing plasmids with deletions in the U5-IR stem-loop were made to investigate whether the size of the loop affected Tf1 transposition. The sequences deleted in these constructs were nt 139 through 149 (Δ1), nt 135 through 153 (Δ2), and nt 124 through 162 (Δ3) in the Tf1 RNA (Fig. 4A). Deletion of the entire U5-IR stem-loop (Δ3) greatly reduced Tf1 transposition activity (Fig. 4B). This result was consistent with the requirement of the U5-IR stem in Tf1 transposition. Reducing the size of the loop to 6 nt (Δ2) also led to severe defects in Tf1 transposition, while a 14-nt loop (Δ1) allowed wild-type levels of transposition activity. Thus, a U5-IR loop of 14 nt or more was sufficient for Tf1 transposition.
Additional mutations were made in the version of Tf1 with the 6-nt loop (Δ2) to test whether enhancing the stability of the U5-IR stem by increasing its GC content would rescue the transposition defect caused by this reduction in loop size. We changed the base pair of the stem closest to the loop from UA to CG and designated this version of Tf1 the CG stem. The introduction of the CG base pair restored transposition activity to nearly wild-type levels (Fig. 5). To test whether or not the transposition activity observed in the strain with the CG stem is dependent on specific nucleotides in the loop, we created two versions of Tf1 with substitutions within the loop sequence of the CG stem mutation. The version designated the AG loop carried substitutions of the two nucleotides in the loop adjacent to the stem, and the mutation designated the RSV loop possessed substitutions of the loop sequences with the nucleotides derived from the loop of RSV. Neither of these modifications in the U5-IR loop significantly affected the wild-type transposition activity of the CG stem. The near wild-type levels of transposition activities obtained from these strains indicated that specific nucleotide sequences were not required in the loop even after the size was reduced to 5 nt. Increasing the stability of the U5-IR stem was sufficient to rescue the transposition defect caused by reducing the loop to 6 nt. These results suggest that the role of the 25-nt loop of Tf1 is to allow the formation of a relatively unstable U5-IR stem.
The U5-IR stem-loop was required for efficient generation of the Tf1 primer. (i) Detection of Tf1 reverse transcripts by DNA blot analysis.
We detected normal levels of Tf1 Gag and IN proteins expressed by all the strains that carried mutated versions of Tf1 generated for this study (data not shown). Thus, the transposition defects caused by the mutations described here could be due to defects in the synthesis of the reverse transcripts. This possibility would be expected if the U5-IR stem-loop of Tf1 functioned, like that of RSV, by contributing to the initiation of reverse transcription. The levels of Tf1 reverse transcripts were measured by DNA blot analysis of total DNA extracts digested with the restriction enzyme BstXI and visualized with a probe specific for the neo gene (Fig. 6). The 2.1-kb band resulted from the neo-containing fragment of the Tf1 cDNA, and the 9.5-kb band was derived from the Tf1-containing plasmid. Results shown in Fig. 6 indicate that the transposition-defective mutants produced almost no detectable Tf1 cDNA. These mutant strains included the six single-point mutations in the U5-IR stem and the two deletion mutants Δ2 and Δ3. All of the transposition-competent versions of Tf1, including the three compensating double-point mutations, the 14-nt loop construct (Δ1), and the three elements whose stems had a higher GC content, exhibited visible and near wild-type amounts of Tf1 cDNA. Hence, the defect in transposition correlated with a lack of Tf1 cDNA.
(ii) Detection of Tf1 primer generation by in vitro primer extension.
Because the production of Tf1 cDNA was lower in the strains with reduced transposition activity, we tested whether any of the mutations in the U5-IR stem-loop of Tf1 affected the efficiency of the cleavage reaction that produces the primer of reverse transcription. Since the Tf1 primer is the first 11 nt of the Tf1 mRNA, the cleavage that creates the primer also shortens the Tf1 mRNA by 11 nt at its 5′ end (14). By analyzing the 5′ end of Tf1 mRNA using in vitro primer extension reactions, we detected the presence of the shortened Tf1 mRNA. The appearance of the primer extension product that was 11 nt shorter than the full-length species indicated that cleavage of the Tf1 primer had occurred (14).
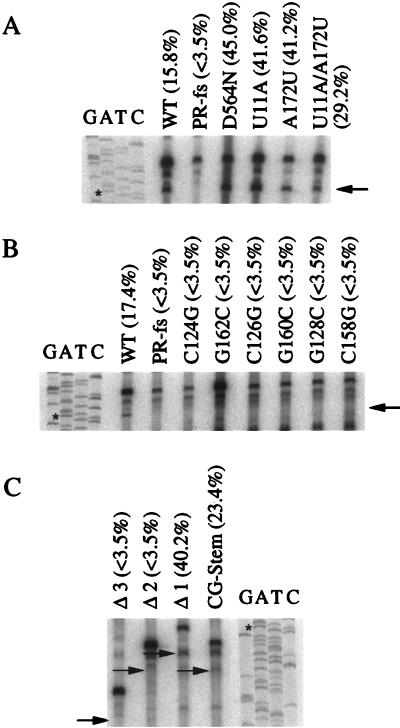
In previous studies it was difficult to detect significant levels of the shortened mRNA in the wild-type strain because the cleaved mRNA was rapidly converted into cDNA. We engineered a mutation (D564N) in the polymerase domain of Tf1 RT to prevent the conversion of the Tf1 RNA into cDNA and to facilitate the detection of the shortened Tf1 mRNA (14, 17). However, to eliminate the possibility that this mutant RT alters the recognition of the RNA structure or the primer, we used the wild-type RT in all the strains for in vitro primer extension reactions in this study. We did, however, include two control strains that contained mutations in the PBS duplex, the U11A and the A172U, because these mutations inhibited reverse transcription and therefore allowed the accumulation of the primer-cleaved Tf1 mRNA. The strain that contained the U11A mutation in Tf1 was previously demonstrated to be able to generate the Tf1 primer (Fig. 7A) yet unable to produce Tf1 reverse transcripts (Fig. 6). As seen in Fig. 7A, both of these mutations in Tf1 allow the accumulation of cleaved Tf1 mRNA that can be readily detected by primer extension. In addition, we speculated that the nucleotide U11 is the last nucleotide of the Tf1 primer of reverse transcription. The mismatch created by the U11A mutation would therefore prevent the addition of deoxyribonucleotides by the Tf1 RT. If this were true, a mutation in the nucleotide opposite U11, A172U, would produce a similar effect and the double mutation that restored complementarity would allow normal reverse transcription to occur. Indeed, the strain that contained the A172U mutation was able to generate the Tf1 primer (Fig. 7A) but failed to produce wild-type levels of Tf1 reverse transcripts and possessed only 2.8% of the transposition activity of the wild type. The strain with the double-point mutation (U11A/A172U) produced near wild-type (95.4%) transposition activity. In addition to providing useful controls for the primer extension assays, these data provide additional evidence that the first 11 nt of Tf1 mRNA serve as the primer of reverse transcription.
FIG. 7.
Detection of cleaved Tf1 mRNA via primer extension analysis. Total RNA was incubated with 32P-labeled oligonucleotide JB890, and AMV RT was added to synthesize DNA from the oligonucleotide. The arrows indicate the positions in the gel where the primer extension products of the cleaved mRNAs produced by the altered versions of Tf1 were expected to migrate. The sequencing ladders were generated with the same oligonucleotide that was used in the primer extension reactions. Percentages are the total Tf1 RNA that was cleaved. The asterisks indicate the positions of primer extension products of the cleaved mRNA generated by wild-type Tf1. The major band between those templated by cleaved and uncleaved mRNA was a nonspecific extension product. Results of primer extension analysis of RNA from the control strains (A), from the six strains with single-point mutations in the U5-IR stem (B), and from the four strains with loop deletions (C) are shown. WT, strain with a wild-type version of Tf1.
We found that all of the transposition-defective strains that contain versions of Tf1 with mutations in the U5-IR stem-loop lacked cleaved Tf1 mRNA (Fig. 7B and C) and therefore lacked the ability to generate the Tf1 primer. In addition, the strains with deletions in the U5-IR stem-loop that transposed normally produced mRNA that had been cleaved between bases 11 and 12 (Fig. 7C). Thus, the U5-IR stem-loop was required for the generation of Tf1 primer.
The U5-IR stem-loop is not required for the packaging of Tf1 mRNA into VLPs.
Since the packaging of the RNA of retroviruses into particles is known to precede the process of reverse transcription, one key question is whether the requirement of the U5-IR stem-loop for reverse transcription of Tf1 is because the formation of the structure is required for the packaging of the Tf1 RNA into the VLPs. Since retrotransposons do not release their VLPs into culture medium, measuring packaged mRNA in purified VLPs has been hindered by the difficulties of isolating particles free of cellular transcripts. Two approaches were taken in this study to address whether mutations in the U5-IR stem-loop affected packaging of Tf1 RNA into VLPs.
(i) Two-plasmid transposition assay.
We conducted transposition assays on strains that contained two plasmids such that wild-type Tf1 was provided without the neo marker gene and the second plasmid contained a version of Tf1-neo with alterations in the sequence of the U5-IR structure. Because the mutations in the U5-IR structure blocked the initiation of reverse transcription, the production of a neo-marked Tf1 cDNA required that both the marked and unmarked Tf1 mRNA be packaged into the VLPs. We obtained normal transposition activities for all the strains containing the mutated U5-IR stem-loops in two-plasmid transposition assays (data not shown). These results indicate that Tf1 RNA with a mutant U5-IR structure can be copackaged with wild-type RNA into VLPs.
(ii) Tf1 RNA protection assay.
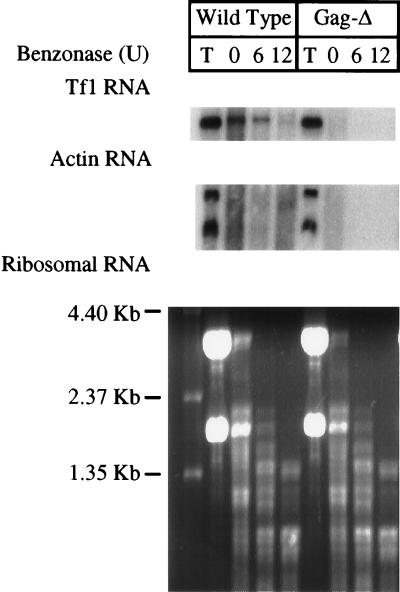
To monitor the packaging of Tf1 RNA, a protection assay was developed to detect the amount of Tf1 RNA that was protected by the Gag protein of the VLPs from degradation by Benzonase (Sigma), an endonuclease that nonspecifically degrades both RNA and DNA. Cellular extracts containing VLPs were subjected to digestion with various concentrations of Benzonase for 6 min. The resistant RNA was isolated from each digested sample and analyzed by RNA blot analysis. In addition to using the wild-type strain that served as the positive control, we used a strain containing Tf1 with an 8-amino-acid deletion in the Gag protein (Gag-Δ lacked amino acid residues 166 to 173) as the negative control strain. This version of Tf1 was used as a negative control for the packaging of Tf1 mRNA because the Gag protein is unstable and ultimately was undetected on immunoblots. Translation of the single primary polypeptide was known to be unaltered by the 8-amino-acid deletion, because high levels of the most-downstream protein, IN (8a), were detected. Using this strain as a negative control for packaging allowed us to avoid the rapid degradation of Tf1 mRNA associated with mutations that inhibit the full translation of the open reading frame. To determine the total level of transcripts that accumulate within cells, preparations of total cellular RNA were subjected to RNA blot analysis and hybridization with probes specific for Gag and actin sequences. The same cultures were subjected to extraction under native conditions to generate supernatant fractions that contained intact VLPs. These samples were subjected to Benzonase treatment followed by phenol extraction in order to characterize the resistant RNA by blot analysis. The T lanes (total cellular RNA) of Fig. 8 show that the strain that expressed wild-type Tf1 and the strain that produced Gag-Δ contained comparable levels of Tf1 and actin mRNA. The Tf1 mRNA from the strain that contained Gag-Δ was sensitive to RNase degradation even before Benzonase treatment, whereas the Tf1 mRNA from the wild-type strain showed significant resistance to Benzonase treatment. This protection from degradation was specific for the Tf1 mRNA, since the actin transcripts as well as the rRNAs were degraded. The actin mRNA was degraded by cellular RNases even before Benzonase was added (lane 0). These results indicate that the Gag protein assembles into a complex or VLP that specifically protects the Tf1 mRNA, but not cellular RNAs, from degradation. As we have previously shown that the bulk of the Tf1 Gag, IN, mRNA, and cDNA assemble into VLPs that cosediment in sucrose gradients (16), this specific protection of the Tf1 transcript from degradation is likely due to VLP packaging. Tf1 mRNA signals were also detected in all strains with mutations in the U5-IR stem-loop, even after Benzonase treatment (Fig. 9A). The total RNA extracts of the strains that expressed mutant versions of Tf1 were also found to contain levels of Tf1 and actin mRNA comparable to those of the wild-type strain (Fig. 9B). These results indicate that Tf1 RNA of the strains containing the mutations in the U5-IR stem-loop were protected by the presence of the Gag protein.
FIG. 8.
RNase protection assay to detect packaging of Tf1 RNA into VLPs. Cellular extracts containing VLPs were treated with various amounts of Benzonase for 6 min. The RNAs extracted from the Benzonase-treated samples and from whole cells were analyzed on RNA blots. Tf1 and actin mRNA were detected by hybridization with probes specific for Tf1 Gag and actin of S. pombe. The rRNAs were visualized by ethidium bromide staining of the agarose gel under UV light. Wild Type, strain containing the wild-type sequence of Tf1; Gag-Δ, strain with deletion in the Tf1 Gag that destabilizes Gag without affecting translation of IN; T, total RNA extracted from whole cells with glass beads and phenol-chloroform; 0, 6, and 12, RNA samples extracted from the native extracts containing VLPs after treatment with 0, 6, and 12 U of Benzonase, respectively.
FIG. 9.
Results of RNA protection assays indicate that the U5-IR stem-loop of Tf1 was not required for the packaging of Tf1 mRNA into VLPs. (A) Tf1 RNAs were protected from nuclease (Benzonase) degradation in all strains that contained versions of Tf1 with mutations in the U5-IR stem-loop. The blotting techniques and designations described in the legend to Fig. 8 were used. Tf1 RNAs were detected after Benzonase treatment in all strains with single-point mutations in the U5-IR stem and with deletion mutations of the U5-IR stem-loop. Tf1 RNAs were not detected in PR-fs and Gag-Δ strains. All rRNAs were degraded by Benzonase. (B) RNA blot analysis of the total accumulated levels of Tf1 and actin RNAs in cells. These total RNA samples were extracted from whole cells with glass beads and phenol-chloroform. WT, strain with a wild-type version of Tf1.
DISCUSSION
The analysis of mutations generated in the 5′ leader of the Tf1 transcript demonstrated that base pairing did occur within the U5-IR stem of Tf1 and formation of the stem was required for Tf1 transposition. The structure of the U5-IR sequence proposed in Fig. 2A also included a 25-nt loop. Surprisingly, none of the substitutions made throughout the loop altered transposition activity. These results indicate that the loop made no sequence-specific contributions to transposition. Nevertheless, deletion analysis indicated that there was a size minimum for the loop, below which transposition would be blocked. Our experiments revealed that this size limit was due to the destabilization of the U5-IR stem associated with small loops. These results support the idea that the function of the loop in the U5-IR structure was not to provide specific contacts with other components but was instead to reduce the steric stress on the base pairs in the stem. These steric forces from the loop that destabilize the stem may have had such dramatic consequences on the structure because additional destabilizing forces were exerted on the U5-IR stem as the result of its direct tethering to the 11-nt duplex just upstream.
The result that the deletion of 19 nt from the U5-IR loop could be rescued by stabilizing the U5-IR stem indicates that, at the level of DNA, the sequences removed from the loop were not required for the insertion of cDNA into target sites by IN. This is significant, as the deleted sequence was predicted to result in a cDNA that lacked a 19-nt segment that was just 18 nt from the 3′ ends of the LTRs. The nonessential nature of this segment of DNA sequence is consistent with analyses of the retrotransposon Ty1 that showed that only the last 4 nt of the LTR are required for efficient integration in in vitro reactions (6, 20).
Strains that contained copies of Tf1 with each of the alterations in the U5-IR stem-loop were tested for defects in reverse transcription. The analysis of DNA blots showed that the U5-IR structure was required for the process of reverse transcription. In an effort to determine whether the role of the U5-IR stem-loop in reverse transcription was related to the initiation of cDNA synthesis, we measured the ability of the mutant strains to cleave the primer of reverse transcription from the Tf1 mRNA. Because each of the strains that exhibited reduced transposition also showed lower levels of cleaved Tf1 mRNA, we concluded that the low levels of reverse transcription were due to a defect that blocked the priming of reverse transcription.
One process that is required for the reverse transcription of retrovirus mRNA is the packaging of the mRNA into virus particles. We speculated that if the mutations in the U5-IR stem-loop of Tf1 inhibited the packaging of the mRNA into VLPs, the RT and any other necessary factors would be unable to cleave the primer from the Tf1 mRNA and to start reverse transcription. Because the isolation of retrotransposon particles from cell extracts is more difficult than harvesting retrovirus particles from culture supernatants, few studies have measured the packaging of a specific transposon mRNA in VLPs. For this reason we developed a genetic approach to detect the presence of the altered Tf1 transcripts in particle complexes that were active for reverse transcription. The evidence that copies of Tf1 mRNA with mutations in the U5-IR sequence could be packaged was the finding that neo-marked Tf1 mRNA with alterations in the U5-IR could be made competent for transposition if coexpressed with wild-type Tf1 that lacked the neo marker. In addition, the rescue of the neo-marked versions of the Tf1 mRNA also indicated that during Tf1 reverse transcription, interstrand transfer of minus-strand strong-stop cDNA likely occurred. Alternatively, the wild-type copy of the Tf1 mRNA could have allowed the neo marker in the altered Tf1 mRNA to be reverse transcribed if the self-priming structure were composed of sequences from separate copies of mRNA. Nevertheless, the ability of the neo marker in the Tf1 mRNAs to be reverse transcribed not only indicated that they were packaged into VLPs but also provided support for our assumption that at least two copies of Tf1 mRNA are packaged into particles.
A physical method of measuring the packaging of Tf1 mRNA was based on the protection from ribonucleases provided by the particle structure. The effectiveness of this approach was demonstrated by the observation that Tf1 mRNA from a wild-type element was specifically protected from degradation while actin mRNA was degraded. That the specific protection of the Tf1 mRNA was due to particle-like structures was indicated by the sensitivity of the Tf1 mRNA to Benzonase, caused by a mutation that reduced the stability of the capsid protein. The mRNAs expressed from the versions of Tf1 with mutations in the U5-IR structure were found to be resistant to Benzonase treatment and therefore assembled into large complexes composed of Gag proteins that were likely VLPs. Interestingly, we observed that the mRNAs with the mutations in the U5-IR sequences possessed even greater stability with regard to ribonuclease degradation than did the wild-type version of the Tf1 mRNA. The eventual degradation of the wild-type mRNA in the incubations with MgCl2 and Benzonase was likely the result of the RNase H (RH)-catalyzed degradation of Tf1 mRNA that occurs during reverse transcription and not the result of sensitivity to Benzonase. This notion is based on the observation that a mutation in a critical residue of RH also resulted in Tf1 mRNA that was significantly more resistant to degradation than wild-type Tf1 mRNA (16a).
The finding that mutations in the U5-IR stem-loop did not dramatically alter the packaging of Tf1 mRNA suggests that these mutations caused reduced transposition by inhibiting a process that occurred after particle formation. Since the mutations within the U5-IR stem-loop did inhibit the generation of the self-primer, these sequences appeared to contribute directly to the early steps of reverse transcription. The nucleotide substitutions in the U5-IR sequence likely altered contacts that are recognized by RT during the establishment of a preinitiation complex. The idea that RT is the factor that is unable to recognize the mutagenized U5-IR structures is supported by previous work that showed that the active site of RNase H in RT is required for cleavage of the Tf1 primer (14). Although the mutations in the U5-IR of Tf1 inhibit the priming of Tf1 reverse transcription, the importance of the U5-IR structure in the reverse transcription of RSV suggests that the function of the RNA structure is not specific for the mechanism of self-priming.
The extensive similarity between the structure we have identified in the RNA of Tf1 and the U5-IR stem-loop of RSV (Fig. 1B and C) included the observation that the stems in Tf1 and RSV were both predicted to be 7 bp long. This as well as other similarities compelled us to compare the functions of these sequences. Mutations that disrupted the stem of the RSV structure resulted in a decrease of viral DNA synthesis in infected cells. Upon further investigation, the mutations in the U5-IR stem of RSV also resulted in decreased initiation of reverse transcription as measured in melittin-permeabilized virions by the incorporation of the first two deoxyribonucleotides of the cDNA (8). The contribution of the U5-IR stem of RSV to the initiation of reverse transcription suggests that these sequences in both RSV and Tf1 possess a general function required for the creation of productive preinitiation complexes.
In a separate study, the function of the five ribonucleotides of the U5-IR loop in RSV was examined. A virus that contained substitutions for all 5 nt in the loop possessed significant activity when permeabilized virions were assayed for the ability to incorporate the first 2 nt of the reverse transcript (19). This high level of initiation activity was slightly lower than the activity of wild-type virus. The minor role of the nucleotides in the U5-IR loop of RSV was similar to the wild-type levels of transposition produced by copies of Tf1 that had substitutions throughout the U5-IR loop. These results indicate that the loop nucleotides do not contribute important sequence-specific contacts with other components of the reverse transcription machinery. In addition, the observation that the U5-IR loop of Tf1 could be reduced from 25 to 6 nt and that this version of Tf1 could be rescued by the addition of a GC base pair in the U5-IR stem provided further evidence that the contributions to the initiation of reverse transcription made by the RNA structure of Tf1 were similar to the contributions provided by its counterpart in RSV.
Virions expressed by versions of RSV with altered U5-IR loops were examined for defects in mRNA packaging that could account for inefficient initiation of reverse transcription. In the case of two mutations, I4L and I12Lpk, the addition of nucleotides into the loop caused defects in the initiation of reverse transcription. However, the virions isolated from culture supernatants were shown to contain normal levels of RSV mRNA (19). Extensive insertions of nucleotides that extended the length of the stem did cause defects in mRNA packaging that may have resulted not from a loss of U5-IR function but from the disruption of other interactions required for particle assembly. Nevertheless, the lack of any packaging defects resulting from the I4L and I12Lpk mutations indicates that the U5-IR structure in RSV is similar to that in Tf1 in that it does not contribute directly to mRNA packaging.
The wild-type levels of Tf1 activity resulting from the reduction of the U5-IR loop to 6 nt required that an AU base pair adjacent to the loop be replaced by a GC base pair. The need for a greater number of GC base pairs in the stem of Tf1 than in the stem of RSV may be in part due to the sequence of the RSV loop. We noted that the nucleotide base pair in the stem at the junction of the loop of RSV is CG and the nucleotides in the loop that flank the stem are A and G. Based on thermodynamic considerations, both of these nucleotide combinations have been predicted to provide the most-stable RNA hairpin (24). The original nucleotide combinations in Tf1 were predicted to provide a much less stable hairpin given the same size of stem and loop.
The contribution to the initiation of reverse transcription made by the U5-IR structure may be a common feature of LTR retroelements since predictions of RNA structure suggest that in addition to the mRNA of RSV, similar U5-IR stem-loops may form in the mRNA of human immunodeficiency virus type 1 (3, 10), murine leukemia virus, reticuloendotheliosis virus, human immunodeficiency virus type 2, feline immunodeficiency virus, squirrel monkey retrovirus, and Mason-Pfizer monkey virus (12). Experimental support for the formation of a U5-IR structure in the mRNA of murine leukemia virus consists of a deletion of 14 nt that removed one strand of the predicted stem (22). This deletion created a virus that was defective for reverse transcription.
Further comparison of the U5-IR stem-loop of Tf1 to that of RSV indicates that both are located in similar positions relative to the PBS. In summary, the results from our study support the conclusion that the U5-IR structures of two divergent elements, Tf1 and RSV, are conserved in size, position, and function. This conservation of RNA structure suggests that the development of therapeutic agents that mimic U5-IR function may prove to be a useful antiviral approach.
ACKNOWLEDGMENT
We thank Van-Dinh Dang for providing the plasmid that contained the 8-amino-acid deletion in Tf1 Gag.
REFERENCES
- 1.Aiyar A, Cobrinik D, Ge Z, Kung H-J, Leis J. Interaction between retroviral U5 RNA and the TψC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwood A, Lin J, Levin H. The retrotransposon Tf1 assembles virus-like particles with excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin F, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 5.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 6.Braiterman L T, Boeke J D. Ty1 in vitro integration: effects of mutations in cis and in trans. Mol Cell Biol. 1994;14:5731–5740. doi: 10.1128/mcb.14.9.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman K B, Byström A S, Boeke J D. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci USA. 1992;89:3236–3240. doi: 10.1073/pnas.89.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Dang, V., and H. L. Levin. Unpublished data.
- 9.Gabriel A, Boeke J D, editors. Retrotransposon reverse transcription. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- 10.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA3Lys (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 11.Joyce C M, Grindley N. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leis J, Aiyar A, Cobrinik D. Regulation of initiation of reverse transcription of retroviruses. In: Skalka A, Goff S, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 33–47. [Google Scholar]
- 13.Levin H L. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin H L. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin H L, Boeke J D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992;11:1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin H L, Weaver D C, Boeke J D. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. . (Erratum, 13: 1494, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Lin, J., and H. Levin. Unpublished data.
- 17.Lin J H, Levin H L. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- 18.Lin J H, Levin H L. Self-primed reverse transcription is a mechanism shared by several LTR-containing retrotransposons. RNA. 1997;3:952–953. . (Letter.) [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J T, Ge Z, Morris S, Das K, Leis J. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J Virol. 1997;71:7648–7656. doi: 10.1128/jvi.71.10.7648-7656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore S P, Powers M, Garfinkel D J. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 22.Murphy J E, Goff S P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J Virol. 1989;63:319–327. doi: 10.1128/jvi.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 24.Serra M J, Barnes T W, Betschart K, Gutierrez M J, Sprouse K J, Riley C K, Stewart L, Temel R E. Improved parameters for the prediction of RNA hairpin stability. Biochemistry. 1997;36:4844–4851. doi: 10.1021/bi962608j. [DOI] [PubMed] [Google Scholar]
- 25.Telesnitsky A, Goff S P. Reverse transcription and the generation of retroviral DNA. Plainview, N.Y: Cold Spring Harbor Press; 1997. [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield J K, Kang S-M, Morrow C D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]