Abstract
The deadenylation of maternal mRNAs in the Xenopus embryo is a sequence-specific process. One cis element that targets maternal mRNAs for deadenylation after fertilization is the embryo deadenylation element (EDEN). This element, composed of U/R repeats, is specifically bound by a protein, EDEN-BP. In the present study we show that the rate at which an RNA containing an EDEN is deadenylated can be increased by the presence of an additional cis element composed of three AUU repeats. This effect was observed for a natural EDEN (c-mos) and two synthetic EDENs. Hence, the enhancement of EDEN-dependent deadenylation conferred by the (AUU)3 motif is not due to an interaction with a particular EDEN sequence. Mutation of the (AUU)3 motif abrogated the enhancement of EDEN-dependent deadenylation. These data indicate that the rate at which a specific maternal mRNA is deadenylated in Xenopus embryos is probably defined by a cross talk between multiple cis elements.
During the first hours of embryonic development in most if not all animal species, transcription is effectively silent. Accordingly, the developing embryos are dependent on the stock of maternal gene products for control of their growth and subsequent differentiation. In Xenopus, Drosophila, and mouse oocytes and embryos the recruitment or release of maternal mRNAs into or from polysomes is correlated, respectively, with the lengthening or shortening of the poly(A) tail (see reference 22 for a review).
Several cis-acting RNA elements that control poly(A) tail length have been identified. The first such element to be characterized was the cytoplasmic polyadenylation element (CPE). This is a uridine-rich element present in the 3′ untranslated regions (UTRs) of maternal Xenopus mRNAs that are polyadenylated during oocyte maturation (5, 12). mRNAs devoid of this element are deadenylated by a process called default deadenylation (6, 23).
Sequence elements similar to the Xenopus CPE have been identified in the 3′ UTRs of maternal mouse mRNAs (8). In the case of mice it was shown that this CPE-like element directed not only the cytoplasmic polyadenylation during oocyte maturation but also the deadenylation of these same mRNAs in primary oocytes. Hence, this element was named an adenylation control element (ACE). The cytoplasmic polyadenylation directed by both CPEs and ACEs requires the AAUAAA nuclear polyadenylation signal (5, 8, 12). In addition, CPE-driven polyadenylation can be affected by auxiliary cis elements. For instance, the 3′ UTRs of Cl1 and Cl2 Xenopus mRNAs contain silencing elements that repress the function of the CPE during oocyte maturation but not after fertilization (19, 20).
The Eg family of maternal mRNAs were originally identified as mRNAs that are poly(A)+ in the egg and deadenylated after fertilization (16). c-mos mRNA was later shown to present the same adenylation-deadenylation behavior (17). This postfertilization deadenylation of Xenopus Eg mRNAs is a transcript-specific process that requires sequence information present in the 3′ UTRs of these mRNAs (3). The sequence elements that direct the postfertilization deadenylation of Xenopus Eg5 and c-mos mRNAs (embryo deadenylation elements, [EDENs]) have been identified (14). In addition, the 53-kDa protein (EDEN-BP) that binds to these elements has been shown to be a necessary component of the EDEN-dependent deadenylation process in embryos (14).
Independently, Stebbins-Boaz and Richter (21) have studied cis elements implicated in the control of Eg1-cdk2 mRNA poly(A) tail length in Xenopus oocytes and embryos. They localized two cis elements (5′cdk2 and 3′cdk2) in the 3′ UTR of this mRNA that appear to act synergistically to cause the deadenylation of this mRNA in embryos but not in maturing oocytes. Although binding of EDEN-BP was detected for the full-length Eg1-cdk2 3′ UTR (10, 21), no binding of EDEN-BP was detected for either the 5′cdk2 or the 3′cdk2 element (12a, 21). This implies that the deadenylation activity targeted by these elements is independent of EDEN-BP. However, previous unpublished data from this laboratory (12a) suggested that the (AUU)3 motif present in the 3′cdk2 element could modulate the deadenylation kinetics of EDEN-containing RNAs. The present study was therefore performed to characterize this possible interplay between the two cis elements. We show here that this (AUU)3 motif acts as an enhancer of EDEN-dependent deadenylation. This enhancer activity was observed for two distinct EDEN sequences, implying that it is a general effect.
MATERIALS AND METHODS
Plasmid constructions.
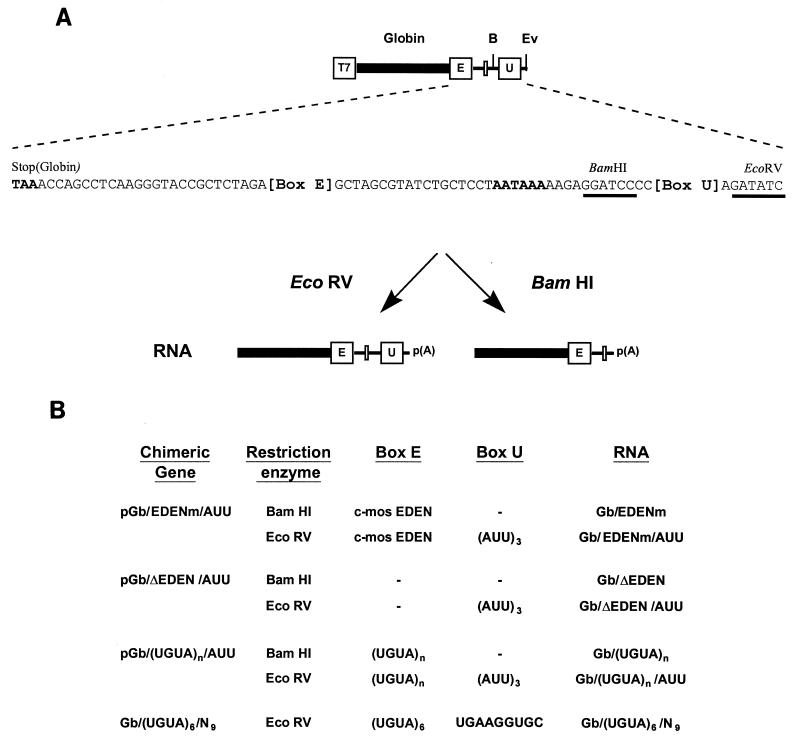
The generic structure of the different chimeric genes used is given in Fig. 1A, as is the sequence of the 3′ UTR common to all of these chimeric genes.
FIG. 1.
Structure of chimeric genes and composition of RNAs. (A) Generic structure and sequence of the 3′ UTR of the chimeric genes used in this study as transcription templates. Transcription was driven by the T7 promoter. The reporter sequence containing the globin open reading frame is indicated by a thick line; the position of the c-mos EDEN or of the n-fold UGUA repeats [(UGUA)n] is indicated by a boxed “E.” The position of the (AUU)3 motif or mutations of this motif is represented by a boxed “U.” The position of the AAUAAA nuclear polyadenylation signal is indicated by a narrow open box. In the 3′ UTR sequence the globin translational stop codon and the nuclear polyadenylation signals are in bold type. The positions of the EcoRV (Ev) and BamHI (B) restriction sites used to make the transcription templates for RNAs with or without the (AUU)3 motif are also shown and underlined in the sequence. p(A), poly(A) tail added to the RNA by E. coli PAP after transcription. (B) For each chimeric gene, the restriction enzymes used and the corresponding RNAs are indicated. The nature of the sequence at positions E and U is also indicated. The sequence of the c-mos EDEN is 5′TATATGTATGTGTTGTTTTATGTGTGTGTGTGTGCT3′.
pGb/EDENm/AUU was derived from pGbORF/mosEDEN (14). In this plasmid the BamHI restriction site in the globin reading frame has been mutated to a SalI site (1). The portion corresponding to the 3′ UTR of the chimeric gene containing the 36-nucleotide c-mos EDEN (EDENm) and the (AUU)3 motif was amplified by PCR with the following oligonucleotides as 5′ and 3′ primers: 5′CCTCAAGGGTACCGCTCTAG3′ and 5′GTAGCCGATATCTAATAATAATGGGGATCCTCT3′. The 3′ primer introduced an EcoRV site immediately 3′ to the (ATT)3 motif. The BamHI site 5′ to this motif was retained. The PCR product was restricted by XbaI and EcoRV and cloned into pGbORF/mosEDEN which had been cleaved by the same restriction enzymes.
To construct pGb/ΔEDEN/AUU, the pGb/EDENm/AUU plasmid was digested with XbaI and NheI, which excise the c-mos EDEN, and then was blunt ended and religated.
pGb/(UGUA)9/AUU was constructed by PCR-mediated mutagenesis with pGb/EDENm/AUU as the PCR template. The 3′ primer (5′CAGATACGCTAGCTACATACATACATACATACATACATACATACATACATCTAGAGC GGTA3′) introduced a sequence consisting of nine repetitions of TGTA that is the same length as the c-mos EDEN. The T7 sequencing primer was used as the 5′ PCR primer. The amplified product was cloned into pGEM-T (Promega). Restriction by EcoRI and NheI released the region containing the globin reading frame and the modified EDEN, which was cloned into pGb/EDENm/AUU which had been restricted by the same enzymes.
To construct pGb/(UGUA)2/AUU, pGb/(UGUA)4/AUU, and pGb/(UGUA)6/AUU by PCR-mediated mutagenesis, pGb/(UGUA)9/AUU was used as a template with the following oligonucleotides as 3′ PCR primers, respectively: 5′CAGATACGCTAGCTACATACATCTAGAGCGGTAC3′, 5′GCTAGCTACATACATACATACATCTAGAGCGGTACC3′, and 5′GCTAGCTACATACATACATACATACATACATCTAGAGCGGTACC3′. The 5′ PCR primer was 5′GAATTCGAGCTCGGTACCCAG3′. The amplified PCR products were cloned into pGEM-T to create pGEM-L2, pGEM-L4, and pGEM-L6, respectively. These plasmids were restricted by NheI and SalI, and the fragments containing the portion of the globin coding sequence corresponding to the C terminus and the 3′ UTRs were cloned into pGb/EDENm/AUU which had also been restricted by NheI and SalI.
The Gb/(UGUA)6/N9 RNAs were synthesized directly from DNA templates amplified by PCR. pGb/(UGUA)6/AUU was used as the PCR template with a 5′ primer (5′GTAAAACGACGGCAGTAATTG3′) situated 5′ to the T7 promoter and a 3′ primer (5′GCTTGATATCTGCACCTTCAGGGGATCCTCTTTTTATTAGGAG3′) in which the (AUU)3 motif was replaced by the randomly chosen sequence underlined.
All constructions as well as the PCR-transcription templates were verified by sequencing.
RNA synthesis.
32P-labeled and capped transcripts were made in vitro with the Promega Riboprobe transcription kit as previously described (10). The transcription templates were linearized by restriction with the enzymes indicated in Fig. 1B. In vitro polyadenylation was achieved by incubating (5 min at 37°C) 1 to 5 μg of in vitro-synthesized RNA with 3 U of Escherichia coli poly(A) polymerase (PAP; Pharmacia) in a 50-μl reaction volume (50 mM Tris-HCl, pH 8.0; 250 mM NaCl; 10 mM MgCl2; 1 mM MnCl2; 2 mM dithiothreitol; 50 ng of bovine serum albumin per μl; 0.5 mM ATP; and 1.6 U of RNasin per μl). The reaction was stopped by adding 50 μl of 100 mM Tris-HCl (pH 7.5)–300 mM NaCl–10 mM EDTA–2% sodium dodecyl sulfate (SDS). The RNA was phenol-chloroform extracted twice and ethanol precipitated. The polyadenylated radiolabeled RNA was purified by electrophoresis on 5% denaturing polyacrylamide gels as described previously (1). RNA size markers were synthesized as described by Audic et al. (2).
Biological and analytical methods.
Embryos were obtained by standard procedures and incubated in F1 solution (15). Embryos at the two-cell stage were injected with 18.4 nl of in vitro transcript (about 1,000 cpm; 0.25 to 1 fmol of RNA). The embryos were allowed to develop at 22°C. At various times, five-embryo lots were taken, and the total RNA was extracted as described by Harland and Misher (7). When required for digestion with RNase H, the RNA samples were resuspended in water and treated as described by Osborne et al. (13). The samples of total RNA or the RNase H-treated samples were dissolved in 50% formamide and analyzed by electrophoresis on 4% polyacrylamide-urea gels as described by Audic et al. (1). The proportion of deadenylated RNA was quantified by the ImageQuant program (Molecular Dynamics) to treat numerical data produced by scanning multiple autogradiograms. The amount of poly(A)− RNA, the final deadenylation product, was taken as the signal within a window between A0 and A20. The total amount of RNA was considered the summation, in each lane, between A0 and the top of the signal in the 0-h lane.
For UV cross-linking, 32P-labeled RNAs were incubated in activated Xenopus egg extracts, irradiated with UV light, and treated with RNase A as described by Legagneux et al. (10). The radiolabeled proteins were analyzed by SDS-polyacrylamide gel electrophoresis and revealed by autoradiography. Immunodepletion of egg extracts with anti-EDEN-BP antibodies fixed to protein A-Sepharose 4B beads (Pharmacia) was done according to the method of Paillard et al. (14).
RESULTS
To analyze the effect of the (AUU)3 motif on EDEN-dependent deadenylation a chimeric gene was constructed with the Xenopus laevis β-globin coding sequence and a 3′ UTR that contained the c-mos EDEN followed by the nuclear polyadenylation signal (AATAAA) and the (ATT)3 motif. An EcoRV restriction site was placed just 3′ to the (ATT)3 motif, and a BamHI site was placed 5′ to this motif. Templates for in vitro transcription that did or did not contain this motif were generated by restriction of this plasmid with either EcoRV or BamHI, respectively (Fig. 1B).
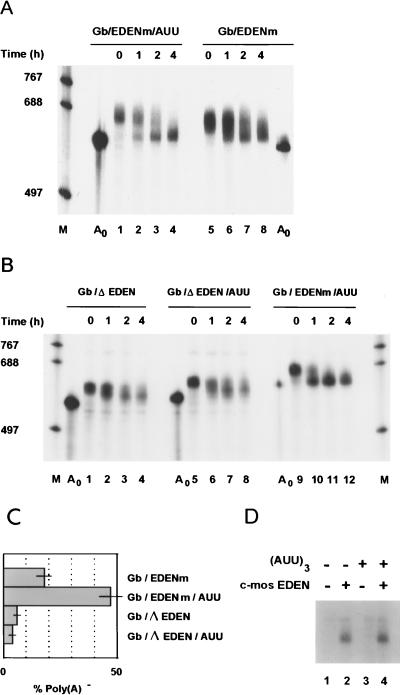
The capped 32P-labeled RNAs transcribed in vitro from these templates were polyadenylated by E. coli PAP and injected into two-cell embryos. Samples of five embryos were taken immediately after injection (0 h) and then at 1, 2, and 4 h after injection. Total RNA was extracted from these samples and analyzed by electrophoresis on polyacrylamide-urea gels followed by autoradiography. The results of such an experiment are shown in Fig. 2. The Gb/EDENm/AUU mRNA was deadenylated after injection (Fig. 2A, lanes 1 to 4) with kinetics very similar to that previously observed for the GbORF/mosEDEN chimeric mRNA that contains the same 3′ UTR (14). The injected RNA molecules were deadenylated in an asynchronous manner, with a coexistence at the 1-h point of the final deadenylated product and of molecules that migrated at the position of the injected poly(A)+ RNAs. For the RNAs devoid of the (AUU)3 motif (Fig. 2A, lanes 5 to 8) the deadenylation appeared to be less efficient. At the 1-h point a smaller proportion of these injected RNAs had been shortened to the size of the final deadenylation product (Fig. 2A; compare lanes 2 and 6), and at the 2-h point the signal from RNAs of intermediate size (partially deadenylated molecules) was greater for the RNAs devoid of the (AUU)3 motif (compare lanes 3 and 7). This difference between the rates of deadenylation of the RNAs containing and devoid of the (AUU)3 motif was confirmed by quantification of these data (Fig. 2C). The presence of the (AUU)3 motif increased the rate at which an RNA containing the c-mos EDEN was deadenylated about 2.5-fold.
FIG. 2.
Effect of the (AUU)3 motif on c-mos EDEN-dependent deadenylation. (A) Between 0.15 and 0.3 fmol of the 32P-labeled and capped Gb/EDENm/AUU and Gb/EDENm RNAs were injected as polyadenylated transcripts into two-cell embryos as described in Materials and Methods. Samples of five embryos were taken for RNA extraction at the indicated times after injection, and aliquots of 2.5-embryo equivalents were analyzed in a 5% polyacrylamide-urea gel. After being dried the gel was subjected to autoradiography. Lane A0, nonadenylated RNAs before injection; lane M, RNA molecular weight markers; sizes are indicated on the left. (B) The chimeric RNAs devoid of an EDEN sequence (Gb/ΔEDEN and Gb/ΔEDEN/AUU) were synthesized as 32P-labeled, capped transcripts and polyadenylated in vitro prior to injection into two-cell embryos. As a control the polyadenylated Gb/EDENm/AUU RNA was similarly prepared and injected into two-cell embryos. At the indicated times after injection aliquots of five embryos were taken and further processed as described for panel A. Lanes A0 and M, same as for panel A. (C) Quantification of the proportion of poly(A)− RNA produced 4 h after injection into two-cell embryos. For the chimeric transcripts used for panels A and B, the amount of poly(A)− RNA, the final deadenylation product, relative to total RNA in each lane was calculated as described in Materials and Methods. (D) Effect of the (AUU)3 motif on the UV cross-linking of EDEN-BP to the c-mos EDEN. RNAs containing (lanes 2 and 4) or devoid of (lanes 1 and 3) the c-mos EDEN and with (lanes 3 and 4) or without (lanes 1 and 2) the (AUU)3 motif were incubated in activated egg extracts. After irradiation with UV light and treatment with RNase A, the proteins were resolved on a 10% polyacrylamide gel in the presence of SDS. Radiolabeled proteins were revealed by autoradiography of the dried gel.
A close examination of the data in Fig. 2A indicates that the size of the in vivo-deadenylated RNAs is slightly greater than that of the in vitro-synthesized A0 transcripts, which implies that in vivo the final deadenylated product retains a short oligo(A) tail. This is similar to the situation described for several yeast mRNAs (see reference 9 for a review) and for mRNAs containing c-fos destabilizing elements expressed in NIH 3T3 cells (18). In these cases the poly(A) tail is shortened to an oligo(A) tail before the body of the mRNA is degraded. In primary mouse oocytes the deadenylation of maternal mRNAs driven by the ACE also leaves an oligo(A) tail (8). With the RNA size markers and the nonadenylated in vitro transcript, it was estimated that, for the RNAs deadenylated via the EDEN-dependent pathway in Xenopus embryos, the oligo(A) tail contained 15 to 20 A’s (an average of five independent experiments). Hereafter we refer to the final product of in vivo deadenylation as poly(A)− RNA and the nonadenylated in vitro transcript as A0 RNA.
To ensure that the (AUU)3 motif stimulated EDEN-dependent deadenylation, RNAs with or without the (AUU)3 motif and from which the EDENm element had been deleted (Gb/ΔEDEN/AUU and Gb/ΔEDEN, respectively) were injected as 32P-labeled, capped transcripts into two-cell embryos after in vitro polyadenylation. These RNAs did not bind EDEN-BP (Fig. 2D, lanes 1 and 3). As a positive control of EDEN-dependent deadenylation in this experiment the in vitro-polyadenylated Gb/EDENm/AUU RNA was also injected. Total RNA was extracted from these injected embryos and analyzed (Fig. 2B) as described above.
The Gb/ΔEDEN RNA (Fig. 2B, lanes 1 to 4) and the Gb/ΔEDEN/AUU RNA (lanes 5 to 8) were deadenylated synchronously, which is indicative of default deadenylation (14). In comparison, the Gb/EDENm/AUU RNA (Fig. 2B, lanes 9 to 12) showed the expected asynchronous deadenylation with the coexistence of the initial poly(A)+ RNA with the final deadenylated product. Quantification of the amounts of poly(A)− Gb/ΔEDEN and Gb/ΔEDEN/AUU RNAs at the 4-h point, relative to the total amount of these RNAs at this time (Fig. 2C), showed that the (AUU)3 motif did not increase the rate of deadenylation in the absence of an EDEN.
One explanation for the observed increase in EDEN-dependent deadenylation when the (AUU)3 motif was present in the RNA is that this motif causes an increase in EDEN-BP binding. To test this possibility, UV-cross-linking experiments were performed with the RNAs containing or devoid of an EDEN and with or without the (AUU)3 motif. The data (Fig. 2D) show that the EDEN-BP cross-linking signal, observed only with the EDEN-containing RNAs (lanes 2 and 4), was not significantly affected by the presence of the (AUU)3 motif.
The natural EDENs characterized to date are not strictly conserved sequence motifs; the only identified consensus is a repetition of UR dinucleotides. Hence, the effect of the (AUU)3 motif described above could eventually be attributed to a particular interaction or synergy between the c-mos EDEN and the (AUU)3 motif. Therefore, to further characterize the effect of the (AUU)3 motif on EDEN-dependent deadenylation we decided to make a synthetic EDEN based on a repetition of the simple UGUA motif. These synthetic EDENs were embedded into the 3′ UTR of β-globin mRNA.
Cross-linking of EDEN-BP to UGUA repeats.
EDEN-BP is a necessary component of EDEN-dependent deadenylation (14). Therefore, binding of EDEN-BP is a primary prerequisite of a synthetic EDEN. Initial UV-cross-linking experiments using radiolabeled RNAs [Gb/(UGUA)n], whose 3′ UTRs contained two, four, six, or nine repetitions of the UGUA motif, showed that a 32P-labeled protein of about 53 kDa was detected only with RNAs containing four or more UGUA repeats (data not shown).
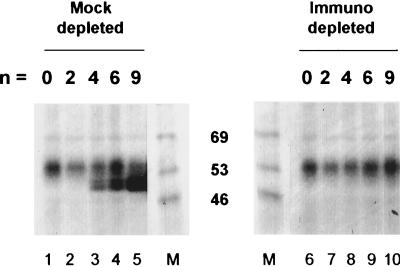
To verify that the 53-kDa 32P-labeled protein produced in these UV-cross-linking experiments corresponded to EDEN-BP, similar experiments were performed with EDEN-BP-depleted and mock-depleted egg extracts. In these experiments the RNA with no UGUA repeats corresponds to the Gb/ΔEDEN RNA. Mock depletion with preimmune serum does not affect the UV-cross-linking pattern of EDEN-containing RNAs (14). The proteins that are UV cross-linked to an EDEN generally migrate as a doublet (p53-p55) (10). In the experiment whose results are shown here (Fig. 3) all the RNAs (n = 0 to 9) bound a protein in the mock-depleted extract of about 60 kDa (lanes 1 to 5). Those with four or more UGUA repeats (Fig. 3, lanes 3 to 5) produced an additional signal of about 53 kDa that was clearly resolved as a doublet. The UV-cross-linking signal for the doublet at 53 kDa increased with the number of UGUA repeats, whereas that for the protein at 60 kDa was constant. After immunodepletion (Fig. 3, lanes 6 to 10) no signal corresponding to the p53-p55 32P-labeled protein was detected; only the signal for the nonspecific 60-kDa protein was observed. We have also observed that the p53-p55 32P-labeled proteins detected after UV cross-linking with the untreated extracts are immunoprecipitated by anti-EDEN-BP antiserum (data not shown). Therefore, four or more UGUA repeats can bind EDEN-BP and thus constitute potential synthetic EDENs.
FIG. 3.
Detection by UV cross-linking of EDEN-BP binding to RNAs containing UGUA repeats. 32P-labeled Gb/(UGUA)n RNAs were incubated in activated egg extracts that had been treated with preimmune serum (mock depleted) or anti-EDEN-BP serum (immunodepleted). After irradiation with UV light the samples were processed as described in the legend to Fig. 2D. The numbers (n) of UGUA repeats in the different chimeric RNAs are indicated above the lanes. Lane M, molecular mass markers; sizes (in kilodaltons) are indicated between the two panels.
UGUA repeats constitute a functional EDEN whose efficiency is enhanced by the (AUU)3-containing element.
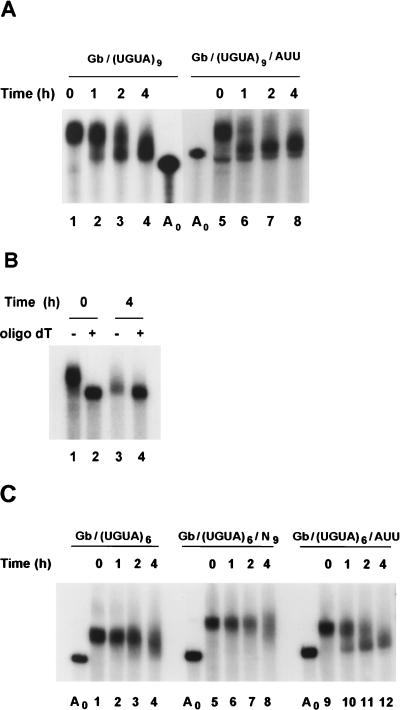
The putative EDEN (UGUA)9 is the same length as the c-mos EDEN and binds EDEN-BP proficiently. Therefore, RNAs containing this motif in the 3′ UTR and with [Gb/(UGUA)9/AUU] or without [Gb/(UGUA)9] the (AUU)3 motif (Fig. 1) were synthesized as 32P-labeled, capped molecules. A poly(A) tail was added to these RNAs by in vitro polyadenylation with PAP prior to injection into two-cell embryos. Total RNA was extracted from these embryos at 0, 1, 2, and 4 h postinjection and analyzed by electrophoresis on polyacrylamide-urea gels followed by autoradiography. In the experiment whose results are shown in Fig. 4A, the Gb/(UGUA)9 RNA (lanes 1 to 4) was deadenylated efficiently; poly(A)− RNA was already detected at the 1-h point (lane 2). In the presence of the (AUU)3 motif (Fig. 4A, lanes 5 to 8) the initial rate of deadenylation was increased; however, at the 4-h point the amounts of deadenylated product for these two RNAs appeared to be similar. Quantification of these data showed that for both of these RNAs the proportion of the deadenylated product, relative to the total amount of RNA, at the 4-h point was 35% ± 5% (mean ± standard deviation).
FIG. 4.
The (AUU)3 motif can stimulate EDEN-dependent deadenylation. (A) The Gb/(UGUA)9 and Gb/(UGUA)9/AUU RNAs were synthesized as 32P-labeled, capped transcripts and then polyadenylated in vitro with E. coli PAP. Between 0.15 and 0.3 fmol of these RNAs was injected into two-cell embryos. At the indicated times after injection, aliquots of five embryos were taken for RNA extraction and analysis as described in the legend to Fig. 2. Lanes A0, nonadenylated RNAs before injection. (B) Samples of five embryos injected with the radiolabeled, capped polyadenylated Gb/(UGUA)9 RNA were taken for RNA extraction at the indicated times after injection. Half of each sample was treated with RNase H in the presence (+) or absence (−) of oligo(dT). The samples were then analyzed as described in the legend to Fig. 2. (C) The three different chimeric RNAs, Gb/(UGUA)6, Gb/(UGUA)6/AUU, and Gb/(UGUA)6/N9, that all contained six UGUA repeats in the 3′ UTR were synthesized and polyadenylated in vitro. Two-cell embryos injected with these 32P-labeled RNAs were further processed as described for panel A. In Gb/(UGUA)6/AUU the (AUU)3 motif was present between the AAUAAA signal and the poly(A) tail. In Gb/(UGUA)6/N9 the (AUU)3 motif was replaced by an unrelated sequence, and in Gb/(UGUA)6 the motif was absent. Lanes A0, nonadenylated RNAs before injection.
An oligo(dT)-directed RNase H analysis was performed to confirm that the observed shortening of the RNA molecules was due to deadenylation. The results of this analysis for the RNA samples prepared from embryos taken 0 and 4 h after injection are shown in Fig. 4B. The in vitro deadenylation of the RNAs in the 0- and 4-h samples produced molecules of identical sizes (Fig. 4B; compare lanes 2 and 4). For the 4-h sample the small decrease in size caused by the in vitro deadenylation (Fig. 4B; compare lanes 3 and 4) corresponds to the removal of the residual oligo(A) tail from the in vivo-deadenylated RNAs.
In the UV-cross-linking experiments (Fig. 3) the (UGUA)6 element produced a less intense signal for EDEN-BP than the (UGUA)9 element; it could, therefore, be a less efficient EDEN. To determine the effect of the (AUU)3 motif on the kinetics of deadenylation conferred by the (UGUA)6 element, the RNAs containing this element with [Gb/(UGUA)6/AUU] or without [Gb/(UGUA)6] the (AUU)3 motif were synthesized as 32P-labeled, capped molecules. In addition, a third radiolabeled RNA [Gb/(UGUA)6/N9], in which the (AUU)3 motif was replaced by a randomly chosen sequence was synthesized (Fig. 1). These three RNAs were polyadenylated in vitro with PAP and injected into two-cell embryos. Total RNA was extracted from these embryos at 0, 1, 2, and 4 h postinjection and analyzed by electrophoresis on polyacrylamide-urea gels followed by autoradiography (Fig. 4C). The Gb/(UGUA)6 RNA (Fig. 4C, lanes 1 to 4) was deadenylated, but the rate of this deadenylation was lower than that observed for the Gb/(UGUA)9 RNA (compare lanes 1 to 4 in Fig. 4A and C). The presence of the (AUU)3 motif in the Gb/(UGUA)6/AUU RNA clearly increased the rate of deadenylation (Fig. 4C, lanes 9 to 12). At 1 h postinjection (Fig. 4C, lane 10), RNA had already accumulated at the poly(A)− position. Quantification of these data showed that at 4 h postinjection 40% ± 5% of the Gb/(UGUA)6/AUU RNA had been deadenylated. In the absence of the (AUU)3 motif, only 17% ± 2% of Gb/(UGUA)6 RNA had been deadenylated at this time. The stimulation of EDEN-dependent deadenylation by the (AUU)3 motif is sequence specific, because mutating the nine nucleotides of this motif (Fig. 4C, lanes 5 to 8) caused the deadenylation kinetics to revert to the original pattern observed for the Gb/(UGUA)6 RNA.
DISCUSSION
In the work presented here we have shown that an (AUU)3 motif can increase the rate of deadenylation [formation of poly(A)− RNA] targeted by an EDEN. This effect was observed for both a natural EDEN (c-mos EDEN) and two synthetic EDENs, composed of UGUA repeats, which have graded efficiencies.
The (AUU)3 motif was originally part of the 3′cdk2 element described by Stebbins-Boaz and Richter (21). In the context of the chimeric RNA used by these authors (5′cdk2/B4/3′ckd2) the 5′cdk2 and 3′cdk2 elements act synergistically to confer deadenylation. These elements are not related to the EDEN either in sequence or in protein binding capacity. This suggests that the (AUU)3 motif may act as a general enhancer of sequence-specific deadenylation in embryos.
Although the 5′cdk2 and 3′cdk2 elements do not bind EDEN-BP, the complete Eg1-cdk2 3′ UTR does contain an EDEN-BP binding site (see the introduction). Hence, this mRNA is a potential substrate for EDEN-dependent deadenylation. The data presented here show that if this were the case then the 3′cdk2 element would work in synergy not only with the 5′cdk2 element but also with the EDEN. In essence this implies that the Eg1-cdk2 mRNA would be a substrate for two distinct deadenylation pathways. Since the Eg1-cdk2 mRNA encodes an important cell cycle-regulatory protein (for a review, see reference 4), multiple deadenylation pathways may have evolved to ensure its translational silencing after fertilization.
Maternal mRNAs that are deadenylated in embryos contain a CPE that ensures their poly(A)+ status in the laid egg. Therefore, one possible way to enhance the rate of deadenylation in embryos would be to inhibit CPE-dependent polyadenylation. However, when a chimeric RNA that contained the Eg5 3′ UTR and the cloned 65-adenosine tail with the associated (AUU)3 motif but with its EDEN deleted, was injected into two-cell Xenopus embryos it was further polyadenylated (14). Hence, in this context at least, the (AUU)3 motif does not prevent polyadenylation. It is not clear at present how the (AUU)3 motif exerts its effect on EDEN-dependent deadenylation. There is no complementary sequence in the β-globin 3′ UTR that could base-pair with the (AUU)3 motif and thereby form a secondary structure. In addition, no factor that associates with this motif could be detected by UV cross-linking (unpublished data). Therefore, the mechanism by which the (AUU)3 motif enhances deadenylation remains to be determined.
Of the synthetic EDENs used during this work, the one containing nine UGUA repeats was the most efficient in directing deadenylation. The element with six UGUA repeats had an intermediate deadenylation activity. This effect of the number of UGUA repeats on EDEN-dependent deadenylation activity is suggestive of a correlation between the affinity of EDEN-BP binding and the size (length) of the target sequence. Increased affinity would probably lead to an increased temporal stability of the deadenylation complex, which in turn could cause an increase in deadenylation activity.
No binding of EDEN-BP was detected when only two UGUA repeats (four contiguous UR dinucleotides) were present (Fig. 3, lane 2). For the Eg5 EDEN, mutants that retained six contiguous UR dinucleotides bound EDEN-BP (14). From this it could be implied that an EDEN-BP binding site should contain at least five contiguous UR dinucleotides. However, in at least one natural EDEN this does not appear to be the case. The 3′ UTR of Eg2 mRNA does not contain more than four contiguous UR dinucleotides. Hence, it may be the density of UR dinucleotides rather than the number of contiguous repeats that is the important parameter in defining an EDEN. The recent cloning of EDEN-BP will allow us to use a SELEX approach to better define the constraints for EDEN-BP binding.
Finally, the data presented here show that the deadenylation of maternal mRNAs in Xenopus embryos can be controlled by multiple cis elements. The Eg mRNAs contain both a CPE and an EDEN, and although the EDEN does not act simply as an inhibitor of CPE function (11), it is conceivable that there could be a competition between these two antagonistic functions. Hence, the sequential changes in poly(A) tail length observed for the maternal Eg mRNAs in vivo will probably be defined by the net result of a cross talk between the various cis elements present in a given mRNA.
ACKNOWLEDGMENTS
We thank Vincent Legagneux and Luc Paillard for constructive discussion during this work and for criticism of the manuscript.
This work was supported by grants from the European Economic Community Biotechnology Program (BIO4-CT95-0045), the Ministère Chargé de la Recherche (ACC-SV4), and the Association pour la Recherche sur le Cancer (contract 6788).
REFERENCES
- 1.Audic Y, Omilli F, Osborne H B. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audic Y, Omilli F, Osborne H B, Landais L. Design and use of easily made RNA size markers. BioTechniques. 1997;23:612–616. doi: 10.2144/97234bm11. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne H B. The deadenylation conferred by the 3′ untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevalier S, Blow J J. Cell cycle control of replication initiation in eukaryotes. Curr Opin Cell Biol. 1996;8:815–821. doi: 10.1016/s0955-0674(96)80082-2. [DOI] [PubMed] [Google Scholar]
- 5.Fox C A, Sheets M D, Wickens M. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 6.Fox C A, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- 7.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102:837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- 8.Huarte J, Stutz A, O’Connel M L, Gubler P, Belin D, Darrow A L, Strickland S, Vassali J D. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 10.Legagneux V, Bouvet P, Omilli F, Chevalier S, Osborne H B. Identification of RNA-binding proteins specific to Xenopus Eg maternal mRNAs: association with the portion of Eg2 mRNA that promotes deadenylation in embryos. Development. 1992;116:1193–1202. doi: 10.1242/dev.116.4.1193. [DOI] [PubMed] [Google Scholar]
- 11.Legagneux V, Omilli F, Osborne H B. Substrate-specific regulation of RNA deadenylation in Xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- 12.McGrew L L, Dworkin-Rastl E, Dworkin M B, Richter J D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 12a.Omilli, F. Unpublished data.
- 13.Osborne H B, Duval C, Ghoda L, Omilli F, Bassez T, Coffino P. Expression and post-transcriptional regulation of ornithine decarboxylase during early Xenopus development. Eur J Biochem. 1991;202:575–581. doi: 10.1111/j.1432-1033.1991.tb16410.x. [DOI] [PubMed] [Google Scholar]
- 14.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H B. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paris J, Osborne H B, Couturier A, LeGuellec R, Philippe M. Changes in the polyadenylation of specific stable RNAs during the early development of Xenopus laevis. Gene. 1988;72:169–176. doi: 10.1016/0378-1119(88)90139-4. [DOI] [PubMed] [Google Scholar]
- 16.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 17.Sheets M D, Fox C A, Hunt T, Van de Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 18.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 19.Simon R, Richter J D. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol Cell Biol. 1994;14:7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon R, Tassan J P, Richter J D. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev. 1992;6:2580–2591. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- 21.Stebbins-Boaz B, Richter J D. Multiple sequence elements and a maternal mRNA product control cdk2 RNA polyadenylation and translation during early Xenopus development. Mol Cell Biol. 1994;14:5870–5880. doi: 10.1128/mcb.14.9.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stebbins-Boaz B, Richter J D. Translational control during early development. Crit Rev Eukaryo Gene Expr. 1997;7:73–94. doi: 10.1615/critreveukargeneexpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 23.Varnum S M, Wormington W M. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: a default mechanism for translational control. Genes Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]