Abstract
Recent advances in the in vitro cultivation of Cryptosporidium parvum using hollow fiber bioreactor technology (HFB) have permitted continuous growth of parasites that complete all life cycle stages. The method provides access to all stages of the parasite and provides a method for non-animal production of oocysts for use in clinical trials. Here we examined the effect of long-term (>20 months) in vitro culture on virulence-factors, genome conservation, and in vivo pathogenicity of the host by in vitro cultured parasites. We find low-level sequence variation that is consistent with that observed in calf-passaged parasites. Further using a calf model infection, oocysts obtained from the HFB caused diarrhea of the same volume, duration and oocyst shedding intensity as in vivo passaged parasites.
Author summary
Cryptosporidium parvum and C. hominis are waterborne parasites that are the second to third leading cause of diarrheal disease, and a major contributor to childhood deaths worldwide. Traditionally these intestinal parasites have proven difficult to culture for more than 2–3 days, which hampers long term in vitro studies. We reasoned that cultures of intestinal epithelial cells as monolayers in static plates results in the production of unpolarized epithelial cells. Utilizing hollow fiber technology, we have developed a method for producing intestinal epithelial cell growth that simulates the body resulting in polarized intestinal epithelial cells that have basal and apical surfaces, tight junctions, and develop functional villi. Using this system, we have maintained in vitro cultures of C. parvum that produce all life cycle stages for 20 months. Long-term in vitro culture of parasites often results in the development of a phenotype that is no longer pathogenic to the host; In this publication we show that using a calf model C. parvum BGF-T20HF after 20 months of in vitro culture was unchanged with respect to diarrhea output, parasite load, and clinical scores from the isolate used to initiate the culture (BGF-T0). In addition, we can show that the genome of the cultured parasites (BGF-T20HF) has undergone a similar genomic drift as the parent isolate (BGF-T0) used to start the inoculum that had been maintained by passage through fetal calves. Collectively the data supports the use of the in vitro cultured isolate, BGF-T20HF, for human trials, and provides a long-term model for the development of novel chemotherapeutic drugs to treat this disease.
Introduction
Diarrheal disease is a major cause of worldwide morbidity and mortality, particularly amongst children and immunocompromised individuals. Childhood diarrhea alone is estimated to be responsible for 800,000 deaths annually [1]. Data obtained by the global enteric multicenter study [2] demonstrate that most cases can be attributed to rotavirus, Cryptosporidium sp, Enterotoxigenic Escherichia coli, or Shigella. Cryptosporidium parvum and C. hominis are the causative agents of human cryptosporidiosis, a moderate-to-severe diarrheal disease (MSD), that was reported to be the second to third leading cause of diarrheal disease in under 23-month-old pediatric cases in low socio-economic areas, where it is estimated to result in 7.6 million cases and 202,000 deaths annually [3]. In contrast to higher socio-economic areas, those with low-economic resources had a high incidence of pediatric deaths due to contaminated drinking water supplies resulting from unsanitary conditions and lack of suitable water purification systems [1]. Whereas in higher socio-economic areas, outbreaks of cryptosporidiosis are commonly associated with recreational water supplies such as swimming pools, water parks, hot tubs, and spas. Death results from severe dehydration, and survivors often have long term effects resulting in malnutrition, stunted growth, and cognitive impairment [4]. Current recommended therapy for the disease is nitazoxanide, which is not approved for use in children under 12 months of age, which is the population most vulnerable to the effects of cryptosporidiosis [5]. Advances in the molecular biology and in vitro culture of the parasite [6–11] have provided an impetus to the drug discovery platform resulting in several promising leads [12–15]. The inability to obtain a complete sexual life cycle of the parasite during in vitro growth has hindered biochemical and molecular studies. Conventional 2D cultures do not provide the structural or environmental requirements to permit continuous growth of the parasite and results in asexual reproduction that fails to perpetuate the infection beyond 2–3 days [11]. The use of the hollow fiber bioreactor (HFB) facilitates the creation of a three-dimensional culture system, where the basal surface of the intestinal epithelial cells are attached to the outer surface of the porous hollow fibers thus permitting access to nutrients and oxygen flowing inside the fibers or the intracapillary space (ICS), and the apical surface of the epithelial cells differentiate to form a multilayered polarized surface on the outside of the fibers or the extra-capillary space (ECS). Using this method, we have developed a nutrient rich, low redox ECS medium that replicates the lumen of the gut allowing C. parvum to be maintained in continuous in vitro culture for >20 months, during which time oocysts were transferred to secondary and tertiary HFB where they continued to grow. We have previously shown that oocysts from the HFB were infective in vivo using several mouse models [16]. However, the C. parvum mouse model provides limited information on the pathology of the infection, as the mouse model fails to develop diarrhea, hence the severity of the mouse model infection is based upon reduced weight gain compared to control mice, and numbers of oocysts shed. The ability of the HFB to generate large numbers (5 x 107/mL) of axenic oocysts that do not require chlorination or treatment with strong oxidizing agents is an advantage for biochemical and medical studies using oocysts. However, in vitro cultivation of cells involves an adaptation process that can result in a population of parasites exhibiting significant differences to the original wild-type inoculum [17]. These adaptive responses can negatively impact virulence factors and result in significantly reduced infections in animal models particularly after long term in vitro culture [18–20]. Additionally, many studies have observed that in vitro cultured parasites lose virulence factors and have limited use as models of infection [17]. Hence it was the goal of this study to evaluate C. parvum oocysts generated by the HFB in vitro culture method for potential changes in their repertoire of genes, the presence of insertions and deletions (InDels), and changes in Single Nucleotide Variants (SNV’s). We also compared the clinical outcome of HFB in vitro cultured oocysts with those obtained from calf-passaged oocysts in the calf model [21], which together with the gnotobiotic piglet model [22] are the primary models demonstrating clinical symptoms including significant and sustained diarrhea due to cryptosporidiosis.
Materials and methods
Ethics statement
All in vivo studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health [23]. The calf model protocol (09–120) was approved by the Institutional Animal Care and Use Committee of the University of Arizona, Tucson, AZ (Animal Welfare Assurance number A-3248-01). Calf studies were performed in compliance with guidelines in the Animal Welfare Act and Guide for the Care and Use of Agricultural Animals in Research and Teaching [24]. The animal biosafety level 2 (ABSL-2) facilities used were fully accredited by the American Association for Laboratory Animal Care. All efforts were made to minimize suffering of animals employed in these studies.
Origin of C. parvum IOWA isolates used in this study
BGF-T0 was purchased from Bunch Grass Farms in March 2016 and sequenced at the Beijing Genomics Institute (BGI). BGF- 2017 was purchased from Bunch Grass Farms and sequenced by the GGBC at the University of Georgia. BGF-T20HF–Originated from BGF-T0 following 20 months of continuous culture in the HFB and was sequenced by BGI. IOWA-ATCC–Genomic DNA was ordered from ATCC (catalog number ATCCPRA-67DQ) and sequenced at the Wellcome Sanger Institute (WSI) as in [25]. IOWA-2017 was produced at the Cryptosporidium Production Lab (University of Arizona).
Hollow fiber culture of C. parvum
C. parvum was cultured using a hollow fiber bioreactor [8] containing a 20 kD MW cut off polysulfone fiber cartridge (FiberCell Systems, Frederick, MD). HCT-8 cells were grown on the extra capillary surface of the fibers, and nutrients provided to the basal cell surface from the intracapillary space which contained minimum essential media plus 10% horse serum (MEM + 10% HS) and the following supplements: 0.058 g/L heparin, 0.29g/L L-glutamine, 23.8 g/L HEPES pH 7.8, 4.5 g/L D-glucose, 0.035 g/L ascorbic acid, 0.04 g/L p-aminobenzoic acid, 0.02 Ca pantothenate, 0.001 folic acid. The intracapillary space medium was pumped at 2.5 mL/min from a 1L reservoir. The extra capillary space contained the following supplements dissolved in MEM + 10% HS: 135 mg/L taurodeoxycholate, 4.5 mg/L thioglycolic acid, 49.5 mg/L mannitol, 3.0 mg/L each of glutathione, taurine, betaine, and cysteine, plus 1.34 mg/mL oleic acid, and 3.6 mg/L cholesterol. When the glucose concentration of the intracapillary space dropped to 50% (2.25 g/L) in 24 h, 106 C. parvum oocysts were inoculated into the extra capillary space as previously described [16].
C. parvum Enumeration
Samples (2 mL) were removed from the HFB and total RNA was isolated from pellets obtained by centrifugation at 6449 x g for 5 min (Beckman-Coulter, Indianapolis, IN, USA) using iScript RT-qPCR sample preparation kit (Bio-Rad Labs, Hercules, CA, USA) as previously described [8]. Total RNA was obtained using RNeasy (Qiagen Inc, Valencia, CA, USA) and quantitated using a Qubit 3.0 fluorometer (Life Technologies, Thermo-Fisher Scientific Inc., Waltham, MA, USA). C. parvum 18S rRNA was amplified by qRT-PCR using an iScript One-Step qRT-PCR kit with SYBR green (Bio-Rad Labs) containing specific primers for C. parvum 18S-rRNA (Cp18S-995F: 5′-TAGAGATTGGAGGTTCCT-3′ and Cp18S-1206R: 5′-CTCCACCAACTAAGAACGCC-3′). Total RNA, reagents and primers were incubated at 48°C for 30 min, followed by 95°C for 10 min, and subjected to 40 cycles of 95°C for 15 s and 60°C for 1 min. A melting curve was performed by heating to 95°C for 15 s, followed by 60°C for 15 s and 95°C for 15 s, using a Quant Studio 6 flex Real-Time PCR system (Life Technologies, Thermo-Fisher Scientific Inc, Waltham, MA, USA). Oocyst standards of 105, 106, and 107 oocysts were included, and parasite numbers were evaluated from a graph of the log C. parvum oocysts versus CT for the parasite SSU rRNA [25]. This method was selected since RNA has a turnover time of hours compared to DNA and hence is a better indication that we are enumerating live parasites. For genomic and calf-model infections oocysts were purified using Dynabeads anti-Cryptosporidium (Thermo Fisher Scientific, Waltham, MA) as described using the manufacturers’ instructions.
Fluorescent labelled antibody staining
Oocysts and motile stages from the HFB were observed using antibody-specific fluorescent dye conjugates. Briefly, 1 mL sample from the HFB was centrifuged at 16,162 x g for 1 min (Sorvall Biofuge Fresco, Thermo Fisher Scientific) and the pellet resuspended in 100 μL PBS. Oocysts were stained using 25 μL each of an FITC labelled mouse monoclonal antibody to C. parvum oocyst surface proteins (Crypt-a-Glo, Waterborne Inc., New Orleans LA, USA) and a fluoresceien-labelled polyclonal antibody (Sporo-Glo, Waterborne Inc., New Orleans, LA, USA) to motile stages, for 30 min in the dark, washed twice with PBS and examined using a fluorescence microscope (Nikon Eclipse Ts2R inverted fluorescence microscope) with an excitation wavelength of 410–485 nm and an emission wavelength 515 nm (Crypt-a-Glo), or an excitation wavelength of 535–550 nm, and an emission wavelength 580 nm (Sporo-Glo).
Sequencing protocol
BGF-T0 was acquired from Bunch Grass Farms in March 2016 and underwent sequencing at the Beijing Genomics Institute (BGI) using their BGISEQ DNBSEQ-G400 short-read paired-end sequencing platform. BGF-2017, obtained from Bunch Grass Farms, was subjected to pair-end short-read sequencing by the GGBC at the University of Georgia, employing the Illumina MiSeq sequencer. BGF-T20HF, originating from BGF-T0 through 20 months of continuous cultivation in the HFB, was also sequenced at BGI using the same short-read platform as BGF-T0. For IOWA-ATCC, genomic DNA was procured from ATCC (catalog number ATCCPRA-67DQ) and subsequently sequenced at the Wellcome Sanger Institute (WSI) as described in reference [22]. The production of IOWA-2017 took place at the Cryptosporidium Production Lab (University of Arizona). The mean coverage levels for BGF-T0, BGF-T20F, and BGF-2017 were determined to be 1421x, 60x, and 237x, respectively.
Variant call analysis
Illumina short-read sequences from (BGF-T0, BGF-T20HF and BGF-2017) were aligned to C. parvum IOWA-ATCC v50 [25] available at CryptoDB.org [26] with the addition of three extra sub-telomeric regions for chromosomes 7 and 8 (GenBank Accessions:MZ892386-8) using BWA-mem v0.7.17 [27]. The alignments were then submitted to Picard Toolkit v2.16.0 [28] to parse these alignments and remove duplicates. The genome analysis Tool Kit v4 (GATK4) [29] Haplotype Caller was used to call variants. All variants were filtered using GATK Variant Filtration with the following parameters: phred quality > 30, depth > 10, mapping quality > 40 and fisher statistics < 60.0. The final filtered variant call file (vcf) was then annotated using SNPeff v5.1 [30] using the C. parvum IOWA-ATCC v50 genome annotation as a custom database.
Comparative sequence analysis
All annotated vcf files were submitted to SnpSift extractFields v5.1 [31] to generate variant tables. They were then compared using BedTools v2.29.2 [32] intersect and subtract to identify the shared and unique variants for each sample. Plots were generated using Venny v2.1 [33].
Specific Gene alignments
To generate gene sequences for each isolate, we used a de novo assembly approach using SPAdes v3.15 [34], followed by a reference guided scaffolding approach using Ragtag v2.1 [35], and annotation transfer using Liftoff v1.6.1 [36]. Protein sequences were generated using AGAT v0.4.0 [37] and aligned using MAFFT v7 [38]. To determine if the called variants were affecting protein domains, all target genes were submitted to InterProScan v5 [39].
Calf model infection
A total of 9 calves (3 per group) were obtained from the same United States Department of Agriculture (USDA)-licensed closed-herd dairy vendor [21]. Calves were fed commercial colostrum replacer within 2 h after birth (bovine IgG colostrum replacement; Land O’Lakes, Shoreview, MN) per label instructions. Triplicates were randomly assigned using the Microsoft Excel random number generation tool (Redmond, WA) to be infected with BGF-T20HF (oocysts collected from the HFB culture), BGF-2017 (a recent sample obtained from Bunch Grass Farms), and IOWA-2017 (the parent isolate for BGF routinely calf passaged at the University of Arizona (Fig 1B). Experimental personnel were blind to isolate assignments during the study. All calves were housed in an ABSL-2 facility in separate containment rooms. Precautions and disinfection measures were taken for the deliveries and housing of these calves to prevent unintended Cryptosporidium or other enteropathogen infection. The calves were fed antibiotic-free milk replacer (Nutrena Snowflakes calf milk II-Utiliz milk replacer; Cargill Animal Nutrition, Minneapolis, MN) twice daily from 12 h of age until termination of the experiment at day 10 postinfection (PI). An oral electrolyte solution (Re-Sorb; Pfizer) was supplemented once diarrhea developed in an animal. At 36 to 48 h of age (study day 0), each calf was infected by oral inoculation of 5 × 107 purified disinfected C. parvum oocysts (BGF-T20HF, BGF-2017, and IOWA-2017). Stool samples were collected every 24 h starting on study day 3 PI. The total volume of feces excreted for successive 24-h collections was recorded. Total daily oocyst counts for each calf were determined as previously described [40]. Briefly, qPCR was used to quantify C. parvum oocysts from feces collected over successive 24-h periods which had been well mixed using a commercial blender to ensure sample uniformity. Calves were also assigned numerical scores for the following variables twice daily: clinical symptoms, general health (willingness to rise, stance, rectal temperature, appetite and food intake, attitude, and hydration status), presence or absence of diarrhea, and fecal consistency [41]. All calves were euthanized on study day 10 PI.
Fig 1. Diagramatic section through the hollow fiber bioreactor.
The cartridge contains a series of hollow fibers through which the host cell growth media, MEM plus supplements and 10% horse serum is pumped through the intracapillary space (ICS). The extracapillary space (ECS) contains the host epithelial cells (HCT-8) that attach to and grow around the outside of the fibers forming a 3D matrix that receives nutrients from the basal surface. C. parvum is inoculated into the ECS after the host cell 3D layer has developed as determined by a drop in the ICS glucose concentration of 50% or more in 24h. C. parvum sporozoites attach to the apical surface of the host epithelial cells as they do in the intestinal tract. (A) Diagrammatic section through the HFB showing the extracapillary space (ECS) and host cells growing around the intracapillary space (ICS). (B) C. parvum oocysts from the HFB stained with Crypt-a-Glo. (C) Sporo-Glo stained merozoites and sporozoites from the HFB. (D) Electron microscope image of the HCT-8 cells grown on the fibers showing the presence of microvilli; obtained by sectioning a 3-month cartridge. (E) Growth of C. parvum based upon qRT-PCR of samples collected from the HFB during the 20-month culture period as described in the methods. (F) Typical CT plot used to quantitate C. parvum growth.
Results
Cryptosporidium parvum culture
In vitro cultures of C. parvum (IOWA isolate) were generated using an inoculum of 106 oocysts supplied by Bunch Grass Farms (BGF-T0) and maintained for 20 months using the HFB culture method [8,16] (Fig 1). The culture produced approximately 107−108 oocysts/mL when sampled every 7–10 days (HFB produced oocysts are referred to as BGF-T20HF; Fig 2).
Fig 2. Strain passage and designation diagram.
Cryptosporidium parvum IOWA oocysts, designated, BGF-T0 were purchased from Bunch Grass Farms in March 2016 and sequenced by BGI. C. parvum IOWA oocysts designated BGF- 2017 were purchased from Bunch Grass Farms and sequenced by the GGBC at the University of Georgia. BGF-T20HF–Originated from BGF-T0 following 20 months of continuous culture in the HFB and sequenced at BGI. IOWA-ATCC–Genomic DNA was ordered from ATCC (catalog number ATCCPRA-67DQ) and sequenced at the WSI as in (20). IOWA-2017 was produced at the Cryptosporidium Production Lab (University of Arizona).
Calf infections
Infectivity and clinical scores from the calf clinical model for cryptosporidiosis were obtained for the C. parvum isolate after 20 months of continuous culture in the HFB (BGF-T20HF) and compared with calf-passaged isolates from the same supply BGF-2017 in addition to C. parvum IOWA-2017, which was maintained at the University of Arizona (Table A in S1 Table). Infectivity and clinical scores were obtained from days 3–10 PI (Fig 3A–3F) in triplicate calf infections. One of the BGF-T20HF oocyst samples used resulted in delayed onset of diarrhea which is responsible for the large SD bars for the average daily fecal volume graph (Fig 3A); however, the sum of the total fecal volumes for each group showed no significant differences (Fig 3B). The daily oocyst shedding by calves infected with BGF-T20HF, BGF-2017 and IOWA-2017 were determined daily from 3–10 days PI (Fig 3C). The statistical difference in the number of oocysts shed in the different infected groups was subjected to Kruskal Wallis analysis which indicated the p-value (0.13) was greater than the significance level (α = 0.05), hence the difference between the mean ranks of all groups was not big enough to be statistically different. This conclusion was supported by the test statistic (H = 4.0808) which is in the 95% region of acceptance [0, 5.9915] supporting the conclusion that there is no significant difference between the mean ranks of any pair (Fig 3C). This conclusion was supported using Mann Whitney analysis of the average total oocysts shed for BGF-T20HF compared to BGF-2017 using a 1 tail test with a p of 0.01 resulted in a U value of 18 (critical value of U at p <0.01 is 9), and a z-score of -1.41778 (p value is 0.0778) indicating there was no significant difference between the data obtained for BGF-T20HF and BGF-2017. Mann Whitney analysis of BGF-T20HF compared to IOWA-2017 confirmed the lack of statistical difference between the hollow fiber cultured oocysts and animal passaged oocysts, resulting in a U value of 19 and a z-score of -1.31276. Fecal consistency was scored from 1–4 (Fig 3D) with higher scores representative of more fluid feces (four being the highest score and one the lowest). One calf infected with the BGF-T20HF oocysts had delayed onset diarrhea causing the dip at day 4 PI which resulted in the differences observed between BGF-T20HF and IOWA-2017 at days 6 and 7 PI.
Fig 3. Infectivity and clinical scores for C. parvum isolates BGF-T20HF, compared with calf-passaged isolates BGF-2017 and IOWA-2017.
(A) Average daily fecal volume per calf ± SD of the number of calves in parenthesis BGF-T20HF (4), BGF-2017 (4), IOWA-2017 (5). One HFB (hollow fiber bioreactor derived oocysts) calf was delayed in onset of diarrhea, hence reason for high SDs. (B) Average total fecal volume per calf ± SD of the number of calves in parenthesis BGF-T20HF (4), BGF-2017 (4), IOWA-2017 (5). (C) Daily oocyst numbers shed by BGF-T20HF, BGF-2017, and IOWA-2017. The hollow fiber cultured isolate BGF-T20HF had a similar shedding profile to the parent isolate, BGF-2017 from days 3–10. Non-parametric statistical analysis by the Kruskal-Wallis H-test revealed that there were no significant differences between the groups. (D) Fecal consistency BGF-2017, BGF-T20HF, and IOWA-2017. (E) Daily clinical evaluation. Lower score values indicate healthier calves. Onset of diarrhea was delayed in one BGF-T20HF infected calf. (F) Clinical evaluation score means.
Daily clinical evaluation scores (lower score values indicate healthier calves) of infected versus control calves was performed and the means for each group of 3 calves determined which revealed that BGF-T20HF and the parent isolate BGF-2017 did not statistically differ (Fig 3E). Further, the average of the total clinical evaluation scores for days 3–10 PI indicate no statistical differences between all isolates used in the study (Fig 3F). Cumulatively, the results obtained from the in vivo data indicate there is no changes in the infection sequelae of the HFB cultured parasites, BGF-T20HF, in the calf model after 20 months of continuous in vitro growth.
Genome sequence data
Genome sequences were generated at BGI from ~1 μg of DNA using the 20-month HFB in vitro cultured parasites (BGF-T20HF) and 1μg of DNA from the parental BGF-T0 used to initiate the HFB culture. The generated sequences were then compared to a new genome sequence assembly designated C. parvum IOWA-ATCC [23] in conjunction with a genome sequence for BGF-2017 (supplied by Boris Striepen). Genome-wide, comparison of variants revealed that BGF-T20HF contains less variation overall, except for synonymous substitutions (Table 1 and Fig 4A). Analysis of deletion (D) and insertion (I) events revealed (44 D, 38 I) for the parental BGF-T0, (25 D, 21 I) for BGF-T20HF, and (62 D, 29 I) for BGF-2017 relative to the C. parvum IOWA-ATCC genome sequence (Table B in S1 Table). BGF-T20HF did not show copy number variation of genes or genome segments when compared to BGF-T0 (S1 Fig). Comparative analyses of the coding regions (CDSs) from these 4 genome sequences revealed that BGF-T0, BGF-T20HF and BGF-2017 contain 7, 10 and 1 synonymous (S) and 20, 12 and 5 non-synonymous (N) substitutions respectively, relative to C. parvum IOWA-ATCC (Table 1). Surprisingly, more non-synonymous substitutions are observed in all strains. Overall, sequence analysis indicates a less diverse population and only minor differences in variation between BGF-T20HF relative to parasites passaged in calves (BGF-T0, BGF-2017, IOWA-ATCC) (Tables 1 and B-E in S1 Table).
Table 1. Classification of the variant effects observed* in all BGF samples when compared to C. parvum IOWA-ATCC.
| Variant effect | BGF-T0 | BGF-T20HF | BGF-2017 |
|---|---|---|---|
| 3′ UTR variant | 1 | 0 | 0 |
| 5′ UTR variant | 1 | 1 | 2 |
| Conservative in-frame deletion | 2 | 1 | 2 |
| Conservative in-frame insertion | 5 | 1 | 2 |
| Disruptive in-frame deletion | 6 | 3 | 11 |
| Disruptive in-frame insertion | 1 | 2 | 0 |
| Downstream gene variant | 15 | 7 | 10 |
| Frameshift variant | 10 | 2 | 10 |
| Frameshift variant & stop gained | 1 | 1 | 1 |
| Non-synonymous variant | 20 | 12 | 5 |
| Splice donor variant & intron variant | 1 | 1 | 1 |
| Splice region variant & intron variant | 2 | 0 | 2 |
| Synonymous variant | 7 | 10 | 1 |
| Upstream gene variant | 50 | 43 | 64 |
| Total number of variants | 122 | 84 | 111 |
*Tables B-D in S1 Table
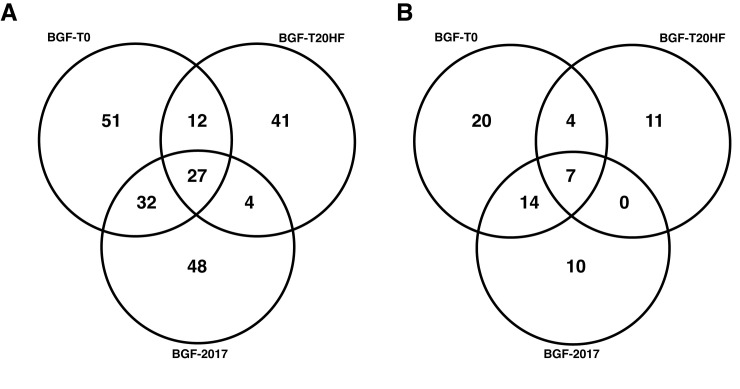
Fig 4. Venn diagram of observed Genome-wide variation found.
(A) All called variants; and (B) Moderate to high-impact variants found in protein coding regions. Data are located in Tables E and F in S1 Table.
When examining CDSs, low levels of variation are detected in all strains and some variants are shared. Variants that change the CDS are enumerated in Fig 4B and Table F in S1 Table. Most of the BGF- T20HF CDS variants were found to be outside of predicted active site or major domain areas (Table C in S1 Table) and are most often associated with non-cytoplasmic regions and coils. However, two variants unique to BGF-T20HF were found to have a missense or disruptive variant that resulted in CDS changes in a predicted InterProScan protein domain. The gene CPATCC_0021250 which encodes a protein phosphatase inhibitor has a mutation (p.Thr32 Asn34del) and CPATCC_0012170 which encodes a P-type ATPase like protein has a missense variant (p.Val12Leu) in a predicted cation-transporting domain. Analysis of the parental BGF-T0 also revealed three variants of putative lesser impact that are not detected in BGF-T20HF or the other strains. These variants are located in CPATCC_0037840, CPATCC_0032770 and CPATCC_0023800 (see Tables B and E in S1 Table).
Variants of lesser significance detected in BGF-T0 only
Analysis of the parental BGF-T0 also revealed three variants of putative lesser impact that are not detected in BGF-T20HF or the other strains. These variants are in CPATCC_0037840 which encodes an extracellular membrane protein with a signal peptide which had Ser336, a polar uncharged amino acid changed to the hydrophobic residue Leu336 in the third loop (amino acids 323–343) of the 9-loop transmembrane domain. It is unlikely however, that this change will have a significant effect on the function of the transmembrane loop. CPATCC_0032770 which encodes casein kinase had a single amino acid change at position Thr219 where the polar uncharged amino acid threonine was replaced by a hydrophobic residue, Isoleucine in the protein kinase domain (amino acids 9–278). Finally the polar, uncharged residue, Ser4 was replaced by the hydrophobic residue Phe4 in the signal peptide domain MRNSVILKIILFSFLDLIYS of the cysteine-rich secretory protein, CPATCC_0023800, which does not affect the signal peptide and is outside of the SCP domain. Overall it is our conclusion that the observed changes are not in critical domains and are unlikely to affect peptide function.
Discussion
The use of hollow fiber biotechnology has been successfully employed for the in vitro growth of all parasite life cycle stages of Plasmodium falciparum [42,43], and C. parvum [8]; it has also been shown to have great potential for the mass production of specific parasite stages, such as Plasmodium falciparum sporozoites for use in vaccine production [44]. The technique enables the development of a 3D culture environment allowing the formation of a polarized apical cell surface and a basal cell surface for transport of nutrients and oxygen to the host cells, reproducing the cellular environment found in host tissues. There have been many recent advances in the in vitro cultivation of C. parvum which permit access to laboratory cultured parasite stages such as the organoid culture method [45,46], that permits access to parasite stages for morphological and molecular analysis. The advantage of the hollow fiber bioreactor is that it provides a method to produce large quantities of in vitro cultured C. parvum oocysts that are free from the harsh chlorination treatment necessary for the sterilization of animal generated oocysts. Parasites generated using this method have several advantages over animal generated parasites notably they provide a novel method for the generation of parasite stages needed for vaccine development, which has shown to have great promise for a malaria vaccine [44]. In vitro cultured parasites permit extended (beyond 48 h) drug testing protocols to be employed and provide a method for preliminary pharmacokinetic/pharmacodynamic data to be obtained for candidate chemotherapeutic agents [47,48]. Currently no other in vitro culture method provides access to sufficient oocyst numbers (107 to 108 oocysts/mL) that can be generated in a GLP facility for use in human clinical trials. However, long term in vitro culture of parasites can result in loss of virulence factors and subtle genetic variation that impact their ability to be useful models for chemotherapeutic trials [17–20]. For these reasons we evaluated in vitro cultured C. parvum after 20 months of continuous culture in the HFB for virulence and genomic changes. We found minor differences in the virulence of C. parvum BGF-T20HF compared to either the parent IOWA isolate BGF-2017 or the University of Arizona isolate IOWA-ATCC.
C. parvum IOWA oocysts, as a population and not a cloned strain, have been propagated in bovine and murine models in multiple locations since the late 1970’s. Molecular divergence was observed when examining 4 loci from 19 samples collected in 2006 from different locations or at different times with some samples being more similar than others [49]. Thus, the number of whole genome differences detected here among the 4 isolates examined (regardless of mode of propagation) is not surprising. However, it serves as a reminder to the community of the need for careful sample naming, lineage tracking, and sequence naming given changes that occur with time via drift and the generation of variants during replication and mutational events.
In conclusion, in vitro cultures of C. parvum using the hollow fiber bioreactor for a period of 20 months produced parasites that had no decrease in virulence properties, had similar clinical scores in a calf model, and demonstrated similar patterns of genomic variation as found within animal-cultured parasites. Hence the HFB permits extended in vitro (beyond 48 h) drug testing protocols to be employed, in addition to preliminary pharmacokinetic/pharmacodynamic data to be obtained which is currently only available using animal models. In addition, because in vitro cultured parasites do not require treatment with chlorine-based reagents to remove bacteria and other gut flora they provide a more controlled and GLP amenable source of oocysts for use in clinical trials.
Supporting information
Sequence reads were aligned to C. parvum IOWA-ATCC genome assembly to assess deletions, insertions, and duplications. Both sample read depths were normalized by the whole genome average depth to get the estimated copy number across all chromosomes.
(JPG)
Table A. Sample Summary. Table B. BGF-T0 variants. Table C. BGF-T20HF variants. Table D. BGF-2017 variants. Table E. Shared and unique variants. Table F. Variants of putative effect.
(XLSX)
Acknowledgments
The authors acknowledge Dr. Stephen Ward (Bill and Melinda Gates Foundation) for helpful discussions in the planning and execution of this research. Beijing Genomics Institute at Shenzhen, China for Next-Gen sequencing of the C. parvum genome (BGF-T0 and BGF-T20HF).
Data Availability
All data needed to evaluate the conclusions of this paper are presented in the paper, its supplementary materials or deposited in an online database. Nucleotide sequences have been deposited in the GenBank under BioProject PRJNA877237 (SRA accessions: SRR24474866, SRR24474867). BGF-2017 raw Illumina reads are available under SRA accession SRR11516703.
Funding Statement
Financial support was provided by The Bill and Melinda Gates Foundation awards OPP1117675 (N.Y.), OPP1151701 (J.C.K), Investment 44418 from the Bill and Melinda Gates Foundation (M.W.R.). Investment GH VAP NG-ID20 from Bill and Melinda Gates Foundation (NY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M.M., R.P.B., and D.S., received salary support from the BMGF.
References
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–222. doi: 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 2.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15(1):85–94. doi: 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, et al. The Burden of Cryptosporidium Diarrheal Disease among Children < 24 Months of Age in Moderate/High Mortality Regions of Sub-Saharan Africa and South Asia, utilizing Data from the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis. 2016;10(5):e0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrant DI, Moore SR, Lima AA, Patrick PD, Schorling JB, Guerrant RL. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am J Trop Med Hyg. 1999;61(5):707–713. doi: 10.4269/ajtmh.1999.61.707 [DOI] [PubMed] [Google Scholar]
- 5.Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet. 2002;360(9343):1375–1380. doi: 10.1016/S0140-6736(02)11401-2 [DOI] [PubMed] [Google Scholar]
- 6.Costa LB, Noronha FJ, Roche JK, Sevilleja JE, Warren CA, Oria R, et al. Novel in vitro and in vivo models and potential new therapeutics to break the vicious cycle of Cryptosporidium infection and malnutrition. J Infect Dis. 2012;205(9):1464–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523(7561):477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morada M, Lee S, Gunther-Cummins L, Weiss LM, Widmer G, Tzipori S, et al. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol. 2016; 46(1):21–29. [DOI] [PubMed] [Google Scholar]
- 9.DeCicco RePass MA, Chen Y, Lin Y, Zhou W, Kaplan DL, Ward HD. Novel Bioengineered Three-Dimensional Human Intestinal Model for Long-Term Infection of Cryptosporidium parvum. Infect Immun. 2017;85(3):e00731–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baydoun M, Benamrouz Vanneste S, Creusy C, Guyot K, Gantois N, Chabe M, et al. Three-dimensional (3D) culture of adult murine colon as an in vitro model of cryptosporidiosis: Proof of concept. Sci Rep. 2017;7(1):17288. doi: 10.1038/s41598-017-17304-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sateriale A, Slapeta J, Baptista R, Engiles JB, Gullicksrud JA, Herbert GT, et al. A Genetically Tractable, Natural Mouse Model of Cryptosporidiosis Offers Insights into Host Protective Immunity. Cell Host Microbe. 2019;26(1):135–146. doi: 10.1016/j.chom.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manjunatha UH, Vinayak S, Zambriski JA, Chao AT, Sy T, Noble CG, et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017;546(7658):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi R, Hulverson MA, Huang W, Vidadala RSR, Whitman GR, Barrett LK, et al. Bumped kinase inhibitors as therapy for apicomplexan parasitic diseases: lessons learned. Int J Parasitol 2020;50(5):413–422. doi: 10.1016/j.ijpara.2020.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulverson MA, Choi R, McCloskey MC, Whitman GR, Ojo KK, Michaels SA, et al. Repurposing infectious disease hits as anti-cryptosporidium leads. ACS Infect Dis 2021;7(5):1275–1282. doi: 10.1021/acsinfecdis.1c00076 [DOI] [PubMed] [Google Scholar]
- 15.Zhang CX, Love MS, McNamara CW, Chi V, Woods AK, Joseph S, et al. Pharmacokinetics and pharmacodynamics of clofazimine for the treatment of cryptosporidiosis. Antimicrob Ag Chemother 2022;66(1):e0156021.doi:10.1128/AAC.01560.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarlett N, Morada M. Long-term in vitro Culture of Cryptosporidium parvum. Bio Protoc. 2018;8:e2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford SE, Chintala MM, Bushek D. Comparison of in vitro-cultured and wild-type Perkinsus marinus. I. Pathogen virulence. Dis Aquat Organ. 2002;51(3):187–201. [DOI] [PubMed] [Google Scholar]
- 18.Brown AC, Guler JL. From Circulation to Cultivation: Plasmodium in vivo versus in vitro. Trends Parasitol. 2020;36(11):914–926. doi: 10.1016/j.pt.2020.08.008 [DOI] [PubMed] [Google Scholar]
- 19.Ali KS, Rees RC, Terrell-Nield C, Ali SA. Virulence loss and amastigote transformation failure determine host cell responses to Leishmania mexicana. Parasite Immunol. 2013;35(12):441–456. [DOI] [PubMed] [Google Scholar]
- 20.Moreira D, Santarem N, Loureiro I, Tavares J, Siva AM, Amorim AM, et al. Impact of continuous axenic cultivation in Leishmania infantum virulence. PLoS Negl Trop Dis. 2012; 6(1):e1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riggs MW, Schaefer DA. Calf Clinical Model of Cryptosporidiosis for Efficacy Evaluation of Therapeutics. Methods Mol Biol. 2020;2052:253–282. doi: 10.1007/978-1-4939-9748-0_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Beamer G, Tzipori S. The piglet acute diarrhea model for evaluating efficacy of treatment and control of cryptosporidiosis. Human Vaccines Immunotherapeutics. 2019;15(6):1445–1452. doi: 10.1080/21645515.2018.1498436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Research Council. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. 2011. [Google Scholar]
- 24.Federation of Animal Science Societies. Guide for the care and use of agricultural animals in research and teaching, 3rd ed. Federation of Animal Science Societies, Champaign, IL. 2020. [Google Scholar]
- 25.Baptista RP, Sateriale A, Sanders MJ, Brooks KL, Tracey A, Ansell BRE, et al. Long-read assembly and comparative evidence-based reanalysis of Cryptosporidium genome sequences reveal expanded transporter repertoire and duplication of entire chromosome ends including subtelomeric regions. Genome Res. 2022;32(1):203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34(Database issue):D419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Institute Broad, Tools Picard [Google Scholar]
- 29.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cingolani P, Patel VM, Coon M, Nguyen T, Land SL, Ruden DM, et al. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front Genet. 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England). 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveros JC. Venny: An interactive tool for comparing lists with Venn’s diagrams. (2007–2015). [Google Scholar]
- 34.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Computational Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, et al. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019;20(1):224. doi: 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shumate A, Salzberg SL. Liftoff: accurate mapping of gene annotations. Bioinformatics. 2020;37(12):1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dainat J. AGAT: Another Gff Analysis Toolkit to handle annotations in any GTF/GFF format. [Google Scholar]
- 38.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer DA, Betzer DP, Smith KD, Millman ZG, Michalski HC, Menchaca SE, et al. Novel bumped kinase Inhibitors are safe and effective therapeutics in the calf clinical model for cryptosporidiosis. J Infect Dis. 2016;214(12):1856–1864. doi: 10.1093/infdis/jiw488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imboden M., Schaefer D. A., Bremel R. D., Homan E. J., Riggs M. W. Antibody fusions reduce onset of experimental Cryptosporidium parvum infection in calves. Vet Parasitol 2012;188:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T, Glushakova S, Zimmerberg J. A new method for culturing Plasmodium falciparum shows replication at the highest erythrocyte densities. J Infect Dis 2003;187(1):159–162. [DOI] [PubMed] [Google Scholar]
- 43.Preechapornkul P, Chotivanich K, Imwong M, Dondorp AM, Lee SJ, Day NP, et al. Optimizing the culture of Plasmodium falciparum in hollow fiber bioreactors. Southeast Asian J Trop Med Public Health 2010;41(4):761–769. [PMC free article] [PubMed] [Google Scholar]
- 44.Eappen AG, Li T, Marquette M, Chakravarty S, Kc N, Zanghi G, et al. In vitro production of infectious Plasmodium falciparum sporozoites. Nature 2022;612(7940):534–539. [DOI] [PubMed] [Google Scholar]
- 45.Heo I, Dutta D, Schaefer DA, Lakobachvilli N, Artegiani B, Sachs N, et al. Modelling Cryptosporidium infection in human small intestine and lung organoids. Nat Microbiol 2018;3(7):814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhalchandra S, Lamisere H, Ward H. Intestinal organoid/enteroid-based models for Cryptosporidium. Curr Opin Microbiol 2020;58:124–129. doi: 10.1016/j.mib.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold SLM, Choi R, Hulverson MA, Whitman GR, Mccloskey MC, Dorr CS, et al. P-Glycoprotein-mediated efflux reduces the in vivo efficacy of a therapeutic targeting the gastrointestinal parasite Cryptosporidium. J Infect Dis 2019;220(7):2288–2298. doi: 10.1093/infdis/jiz269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarlett N, Morada M, Gobin M, Van Voorhis W, Arnold S. In vitro Culture of Cryptosporidium parvum Using Hollow Fiber Bioreactor: Applications for Simultaneous Pharmacokinetic and Pharmacodynamic Evaluation of Test Compounds. Methods Mol Biol. 2020;2052:335–350. [DOI] [PubMed] [Google Scholar]
- 49.Cama VA, Arrowood MJ, Ortega YR, Xiao L. Molecular Characterization of the Cryptosporidium parvum IOWA Isolate Kept in Different Laboratories. J Eukaryot Microbiol. 2006;53(Suppl 1), S40–42. [DOI] [PubMed] [Google Scholar]