Abstract
Sen1p from Saccharomyces cerevisiae is a nucleic acid helicase related to DEAD box RNA helicases and type I DNA helicases. The temperature-sensitive sen1-1 mutation located in the helicase motif alters the accumulation of pre-tRNAs, pre-rRNAs, and some small nuclear RNAs. In this report, we show that cells carrying sen1-1 exhibit altered accumulation of several small nucleolar RNAs (snoRNAs) immediately upon temperature shift. Using Northern blotting, RNase H cleavage, primer extension, and base compositional analysis, we detected three forms of the snoRNA snR13 in wild-type cells: an abundant TMG-capped 124-nucleotide (nt) mature form (snR13F) and two less abundant RNAs, including a heterogeneous population of ∼1,400-nt 3′-extended forms (snR13R) and a 108-nt 5′-truncated form (snR13T) that is missing 16 nt at the 5′ end. A subpopulation of snR13R contains the same 5′ truncation. Newly synthesized snR13R RNA accumulates with time at the expense of snR13F following temperature shift of sen1-1 cells, suggesting a possible precursor-product relationship. snR13R and snR13T both increase in abundance at the restrictive temperature, indicating that Sen1p stabilizes the 5′ end and promotes maturation of the 3′ end. snR13F contains canonical C and D boxes common to many snoRNAs. The 5′ end of snR13T and the 3′ end of snR13F reside within C2U4 sequences that immediately flank the C and D boxes. A mutation in the 5′ C2U4 repeat causes underaccumulation of snR13F, whereas mutations in the 3′ C2U4 repeat cause the accumulation of two novel RNAs that migrate in the 500-nt range. At the restrictive temperature, double mutants carrying sen1-1 and mutations in the 3′ C2U4 repeat show reduced accumulation of the novel RNAs and increased accumulation of snR13R RNA, indicating that Sen1p and the 3′ C2U4 sequence act in a common pathway to facilitate 3′ end formation. Based on these findings, we propose that Sen1p and the C2U4 repeats that flank the C and D boxes promote maturation of the 3′ terminus and stability of the 5′ terminus and are required for maximal rates of synthesis and levels of accumulation of mature snR13F.
snoRNAs (small nucleolar RNAs) are required for processing and posttranscriptional modification of rRNA (22, 23, 36). Several snoRNAs facilitate endonucleolytic cleavages in pre-rRNA. In addition, numerous snoRNAs that contain C and D boxes, short conserved sequences found near snoRNA termini, are involved in the formation of 2′-O-methylribose residues in rRNA (7, 8, 17, 35, 37). Other snoRNAs that contain the sequence ACA near their 3′ ends facilitate the formation of pseudouridine in rRNA (25). Many yeast snoRNAs contain trimethylguanosine (TMG) caps at their 5′ ends (23). TMG caps are hypermethylated forms of monomethylguanosine caps that are added cotranscriptionally to the 5′ end of RNA polymerase II primary transcripts. The presence of a TMG cap may function to protect the 5′ ends of snoRNAs from exonucleolytic decay.
Most snoRNAs are synthesized via RNA maturation pathways that convert pre-snoRNAs to mature snoRNAs. Much is now known about the cis-acting requirements of snoRNA processing. C/D box snoRNAs contain C boxes near their 5′ termini and D boxes near their 3′ termini. These RNAs bind to fibrillarin, an abundant nucleolar protein, and fold into secondary structures containing a base-paired stem that forms between sequences immediately 5′ to C boxes and 3′ to D boxes. The C/D box-fibrillarin snoRNP complex is required for the formation of 5′ and 3′ termini of mature snoRNAs. Mutations in the yeast and vertebrate U14 C and D boxes that perturb base pairing cause underaccumulation of U14 (16, 41, 43). Compensatory mutations that restore base pairing also restore normal accumulation. These results indicate that the C/D box-fibrillarin snoRNP complex is important for the formation and stability of C/D box snoRNAs.
In addition to fibrillarin, other trans-acting factors are required for the maturation and stability of snoRNAs. The C/D box snoRNAs U14, U16, and U18 undergo endonucleolytic cleavages and/or end-trimming steps that convert the primary transcripts into mature snoRNAs (4, 5, 41). Competition experiments suggest the existence of common trans-acting factors that facilitate the processing of these snoRNAs (6, 41). The D box of Xenopus U3, U8, and U14 snoRNAs is required for nuclear retention, cap hypermethylation, and stability (34). Nuclear retention of U3, U8, and U14 is saturable, suggesting the existence of shared trans-acting factors that bind D boxes. Furthermore, maturation of yeast U14 and snR190, which are derived from a shared polycistronic precursor, requires the 5′→3′ exonucleases Rat1p and Xrn1p (19, 27). Some snoRNAs contain near their 3′ ends the sequence ACA which facilitates 3′ end formation. When the ACA box of yeast snR11 is mutated, underaccumulation of mature snR11 results. ACA snoRNAs associate with the protein Gar1p, which may be involved in stability or 3′ end formation (2). Yeast ACA pre-snoRNAs are processed in human cell extracts, suggesting that snoRNA processing enzymes are conserved between yeast and human genomes (14).
In this report, we show that the putative nucleic acid helicase Sen1p is required for the maturation and stability of snoRNAs in yeast. The SEN1 gene was cloned by complementation of the temperature-sensitive mutation sen1-1, which causes a 10-fold increase in the accumulation of pre-tRNAs at both permissive and restrictive temperatures and a 90% reduction in the in vitro activity of tRNA-splicing endonuclease (42). 252 kDa Sen1p localizes to the nucleus and contains a consensus P-loop nucleoside triphosphate binding motif and an RNA helicase motif similar to that of the mouse protein Mov10 and yeast Upf1p (10, 18, 20, 21, 38). Upf1p has ATP-dependent nucleic acid helicase activity in vitro (9). The sen1-1 mutation causes a G→A change at a position that is conserved between the Sen1p and Upf1p helicase motifs (10).
The effects of loss of Sen1p function are not limited to tRNA splicing. The sen1-1 mutation causes mislocalization of nucleolar proteins and defects in several steps in pre-rRNA processing (38, 39). Recently, it was shown that Sen1p forms RNP complexes with snoRNAs since it coimmunoprecipitates with more than 20 snoRNA species (39). In addition, chimeric mRNAs containing a portion of antisense U6 snRNA accumulate in excess in cells carrying a mutation in the SEN1 gene (33). It therefore appears likely that Sen1p performs a function that affects multiple RNA biosynthetic pathways. In this report, we show that yeast snR13, a TMG-capped snoRNA that contains C and D boxes (23), exists in wild-type cells in three forms, including an abundant RNA that is presumed to be the functional snR13 snoRNA as well as much less abundant 3′ extended and 5′ truncated forms. Our results suggest that Sen1p is required for the maturation and stability of snR13.
MATERIALS AND METHODS
Strains and media.
The strains used in this study were as follows: 1971 (MATα ura3-52 leu2-3,112 pep4-3), FWY1 (MATα ura3-52 leu2-3,112 sen1-1 pep4-3), TR110 (MATa ura3 leu2 snR13-Δ1), and TR116 (MATα ura3 leu2 his3 snR13-Δ1 sen1-1). Strain FWY1 is an isogenic derivative of strain 1971 that was created by gene replacement (30). snR13-Δ1 was substituted for the wild-type chromosomal locus by gene replacement. Strain TR116 was derived from a cross between an snR13-Δ1 strain and strain FWY1. Standard yeast media were used for cell culture except for in vivo labeling with orthophosphate, where low-phosphate YEPD was used (29). Temperature shift experiments were performed by adding an equal volume of medium prewarmed to 49°C to cultures grown at 25°C. In strains carrying sen1-1, 25°C is a permissive temperature and 37°C is a restrictive temperature for growth (42).
Plasmids.
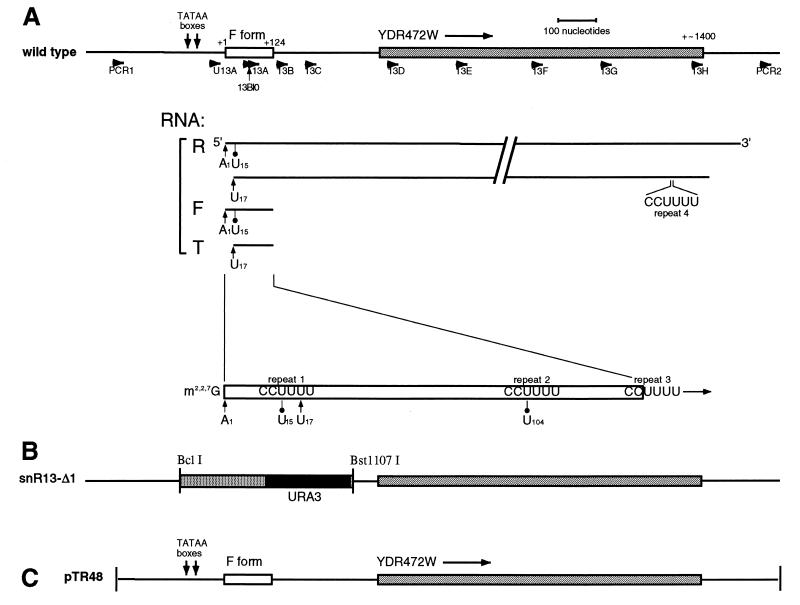
pTR48 (snR13 LEU2 CEN) was constructed by PCR amplification of a region of wild-type genomic DNA with oligonucleotide primers PCR1 and PCR2 (Table 1; Fig. 1), using Pfu DNA polymerase (Stratagene Corp.). The resulting PCR product was ligated into the SmaI site of pRS315, which carries a centromere and the LEU2 gene. pTR48 contains the entire snR13 transcriptional unit. The 5′ end of cloned DNA present in pTR48 begins 260 bases of DNA upstream from the snR13 +1 position and extends approximately 450 bases past the 3′ end of the reduced-mobility form. pTR43 was constructed by PCR amplification of wild-type DNA, using the primers PCR1 and 13D (Table 1; Fig. 1). The product was subcloned into the SmaI site of the yeast 2μm vector Yep351 (15). The gene disruption snR13-Δ1 was constructed by replacing a Bst1107I/BclI restriction fragment containing snR13F DNA with a restriction fragment containing the URA3 gene. snR13-Δ1 was substituted for the wild-type chromosomal locus by standard one-step gene replacement (30). Strain TR116 was derived from a cross between an snR13-Δ1 strain and strain FWY1. DNA fragments carrying mutant alleles of snR13 were inserted into a CEN LEU2 vector as follows: pTR51 (snR13-R3AA), pTR52 (snR13-R1AA), pTR53 (snR13-R3SUB), and pTR56 (snR13-RNEW).
TABLE 1.
Oligonucleotides used
| Name | Nucleotide sequence |
|---|---|
| U13A | 5′-AATCCAATTAACAAAAG-3′ |
| 13A | 5′-AGTAAAAAAAGGTAGCTTGAG-3′ |
| 13B | 5′-ACTAAGATTTTCTACGG-3′ |
| 13C | 5′-CCCAACGTACTAACATC-3′ |
| 13D | 5′-TGTTGGTCAGATGCGCTTGG-3′ |
| 13E | 5′-TTCAAAATCGCCTGCAG-3′ |
| 13F | 5′-CGTTCATTGGCAGATG-3′ |
| 13G | 5′-GGGTTAAAGTGGGGAAG-3′ |
| 13H | 5′-AAATCTCAAACCTTCCC-3′ |
| 13BIO | 5′-CCCCCCCCCCAAGTAAAAAAAGGTAGCTTGAGT-3′ |
| R3AA | 5′-CTTTTATCTGACAATTTAACTTCCCCG-3′ |
| R1AA | 5′-CAGGAAGTTTTTTCAATTTTATATGATG-3′ |
| RNEW | 5′-GTTGCTGTTTGGCCTTTTGCCAAATCAG-3′ |
| R3SUB | 5′-CCTTTTTTTACTTTTATCTGAAAAAAAAACTTCCCCGTAGAAAATC-3′ |
| SUF4A | 5′-CGTGTCCCCACCTTGGAAG-3′ |
| snR11 | 5′-AAACGATTAAGTAAACTATA-3′ |
| snR31 | 5′-ATCAGGTGTTTAAAAAGGCGAGG-3′ |
| PCR1 | 5′-GATTCAAGTTGAAAATGAATAG-3′ |
| PCR2 | 5′-TAAACTAAAACAAACACTACCAC-3′ |
| CMUT | 5′-CCACCGCGGTGGCATGCCAATTCGCCC-3′ |
FIG. 1.
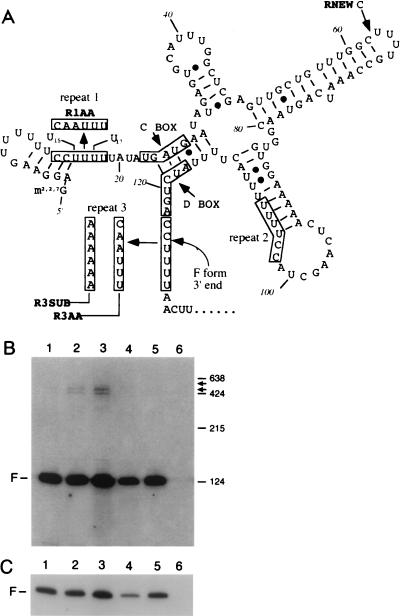
(A) Schematic representation of the SNR13 chromosomal region on chromosome IV showing the linear relationships between snR13R, snR13F, and snR13T. The upper line represents SNR13 DNA. YDR472w (shown 5′ to 3′) is an ORF contained within the snR13R 3′ tail. The positions where oligonucleotides used in this study anneal are shown below the chromosome (PCR1, U13A, 13BIO, 13A through 13G, and PCR2). snR13R, -F, and -T are drawn to scale in the 5′-to-3′ direction and indicated by bold horizontal lines. The positions of primer extension stops and the positions of four C2U4 repeats are shown. (B) Structure of the null allele snR13-Δ1. (C) DNA from the SNR13 region contained in centromeric plasmid pTR48 (see Materials and Methods).
General RNA methods.
RNA was prepared by hot phenol extraction (20). Midwestern blotting was performed as described previously (28). Oligonucleotide-directed RNase H digests were performed by adding 1 pmol of oligonucleotide to samples containing 10 μg of total yeast RNA in 40 mM Tris HCl (pH 7.7)–4 mM MgCl2–1 mM dithiothreitol–30 μg of bovine serum albumin per ml. Samples were denatured for 5 min at 65°C and allowed to cool slowly to 25°C; then 0.1 U of RNase H was added, and samples were digested for 30 min. Reactions were stopped by adding an equal volume of 95% formamide–20 mM EDTA–0.05% bromophenol blue–0.05% xylene cyanol. Primer extension was performed using with avian myeloblastosis reverse transcriptase (Promega Corporation) at 41°C. Secondary structure predictions were performed with the Genetics Computer Group Fold program, using default parameters (11). Oligonucleotides used in this study are described in Table 1; the locations in snR13 to which oligonucleotides anneal are shown in Fig. 1.
Northern blotting was performed as described by van Tol et al. (40) except that 1% sodium dodecyl sulfate (SDS) was substituted for 0.1% SDS in the hybridization solution. Size standards were visualized by probing Northern blots as Midwestern blots to detect TMG-capped RNAs of known size, except for Fig. 5, where end-labeled HinfI-digested φX174 DNA was used (Promega). Data were quantitated with a Molecular Dynamics PhosphorImager, using object average background correction. Experiments were performed with duplicate blots of RNA from independent transformants. SUF4 glycine tRNA was used as a loading control and was detected with complementary oligonucleotide SUF4A (Table 1). SUF4 tRNA is encoded by an intronless tRNA gene (24). The abundance of SUF4 tRNA is unaffected by loss of SEN1 function. The signal intensity of bands was divided by the signal generated by the SUF4 tRNA to correct for loading errors. Means and standard deviations were calculated from data collected in duplicate and standardized for loading.
FIG. 5.
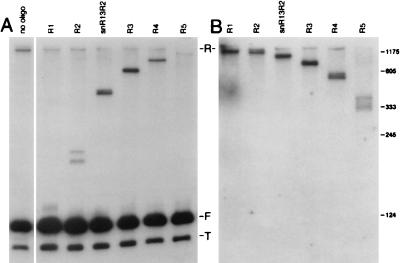
RNase H mapping of snR13R RNA. RNA was extracted from strain FWY1 (sen1-1) 4 h after temperature shift. Lanes are labeled with the names of the oligonucleotides used for each RNase H cleavage reaction. The positions to which they anneal are shown in Fig. 1. Samples were gel fractionated and transferred to a nylon membrane. Numbers at the right indicate the nucleotide positions of RNA size standards. (A) RNase H cleavage products were probed with oligonucleotide 13A, which detects RNA fragments 5′ of the cleavage site. The reaction in the left lane (no oligonucleotide [oligo]) shows the positions of full-length RNAs resulting when the cleavage reaction was performed in the absence of any oligonucleotide. (B) The blot shown in (A) was stripped and reprobed with oligonucleotide 13H (Fig. 1), which detects RNA fragments 3′ of the cleavage site.
Analysis of in vivo-labeled RNAs.
snR13F RNA was prepared for thin-layer chromatography (TLC) by labeling for 7 h at 25°C with 1 mCi of [32P]orthophosphate. snR13 RNAs were purified from hot phenol-extracted total RNA by hybridization to the 5′-biotinylated oligonucleotide 13BIO (Table 1; Fig. 1). 13BIO-snR13 hybrids were bound to streptavidin-conjugated magnetic beads (Promega) in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and washed four times in 0.1× SSC, using a magnet for retention of the DNA-RNA hybrids. Elution was accomplished by resuspension in water. snR13F RNA was purified from the hybrid-selected RNAs by elution from a preparatory polyacrylamide gel in 0.3 ml of 0.5 M ammonium acetate–1 mM EDTA–0.2% SDS. Conditions for RNase T1 and T2 digestion were described previously (3). Digestion products were separated by two-dimensional cellulose TLC. The solvent system for the first dimension was isobutyric acid–0.5 M NH4OH in a 5:3 (vol/vol) ratio. After drying in a cool stream of air, chromatography in the second dimension was accomplished with isopropanol-concentrated HCl-water in the ratio 14:3:3 (vol/vol/vol) as the solvent system (26).
To analyze RNAs labeled by an in vivo pulse, cells of strain 1971 and FWY1 were grown in low-phosphate YEPD to mid-log phase prior to in vivo labeling. Labeling was performed by adding 1 mCi of [32P]orthophosphate and allowing growth to proceed for 1 h. Cells pellets were frozen for later RNA extraction. snR13 RNAs were purified by hybrid selection as described above, using oligonucleotide 13BIO. Samples containing hybrid-selected RNAs were quantitated by scintillation counting, and volumes containing equal counts were fractionated by polyacrylamide gel electrophoresis under denaturing conditions.
Mutagenesis.
Site-directed mutagenesis was performed on pTR48 (Fig. 1), using an oligonucleotide-directed method that extends 5′-phosphorylated primers containing desired mutations with Klenow enzyme followed by ligation (Chameleon site-directed mutagenesis system; Stratagene). Phosphorylated oligonucleotides R3AA, R1AA, RNEW, and R3SUB were used as primary primers, and CMUT was used as the secondary primer (Table 1; Fig. 1). CMUT is complementary to a polylinker and changes a unique SacI restriction site present on pTR48 to an SphI site. The primers were annealed, extended, and ligated, and the resulting products were transformed into a mutS mismatch repair-deficient Escherichia coli strain. Plasmids were isolated and transformed into E. coli DH5α. Plasmids prepared from DH5α that contained a new SphI site and displayed correct restriction digest patterns were sequenced through the entire snR13F region to identify plasmids carrying a mutant snR13 gene.
RESULTS
Inactivation of Sen1p affects the accumulation of TMG-capped snoRNAs.
Midwestern blotting was used to assess whether specific TMG-capped RNAs change in abundance when Sen1p is inactivated. RNA was extracted from two strains with isogenic genetic backgrounds, FWY1 (sen1-1) and 1971 (SEN1) (Materials and Methods). Samples were taken at 4-h time intervals following a shift of each strain from 25 to 37°C. Antibodies against TMG caps were used to probe total RNA transferred to a nylon membrane (28). The previously described array of TMG-capped RNAs detected by Midwestern blotting include four of the five spliceosomal snRNAs and a substantial number of snoRNAs.
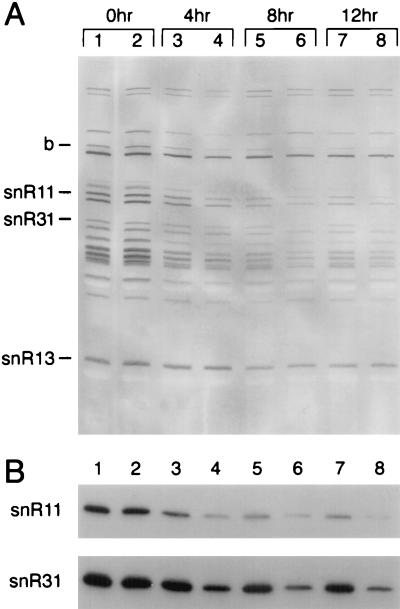
Several TMG-capped RNAs undergo a temporal decrease in abundance in the temperature-shifted sen1-1 strain (Fig. 2A), the most prominent of which are snR11 (2) and snR31 (1). The identities of these RNAs in the Midwestern blot mobility pattern were established previously (28). Another TMG-capped RNA designated b also decreased in abundance. This RNA comigrates with snR42 (2). We reprobed the Midwestern blot in Fig. 2A with 32P-end-labeled oligonucleotides complementary to a select subset of RNAs (Fig. 2B). The results confirmed that the snoRNAs snR11 and snR31 undergo a temporal decrease in abundance as a result of inactivation of sen1-1. At 4 h following temperature shift, snR11 and snR31 were reduced in the sen1-1 mutant to 30% of the amount of the corresponding RNA found in the isogenic wild-type strain 1971.
FIG. 2.
Detection of TMG-capped RNAs by Midwestern and Northern blotting. (A) Midwestern blot. Total RNA was extracted from strain 1971 (SEN1) (lanes 1, 3, 5, and 7) and FWY1 (sen1-1) (lanes 2, 4, 6, and 8) at the times indicated following a temperature shift from 25 to 37°C. RNA was fractionated on a 9% polyacrylamide gel, transferred to nylon, and detected with anti-TMG antibodies (28). snR11 and snR31 decrease in abundance as a consequence of thermal inactivation of Sen1p. RNA species b shows a similar sen1-1-dependent decrease in abundance but has not yet been positively identified (28). (B) The blot shown in panel A was reprobed with 32P-end-labeled oligonucleotides complementary to snR11 and snR31. A probe complementary to SUF4, a glycine tRNA derived from an intronless precursor (24), was used for a loading control (not shown).
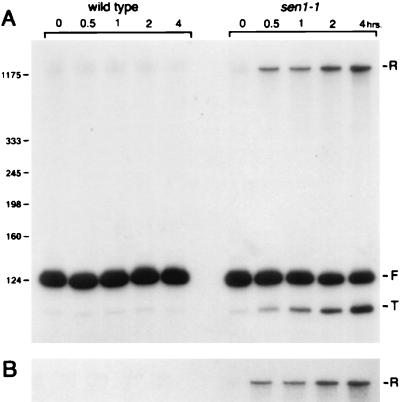
Using conventional Northern blotting, we also found that unexpected forms of snR13 accumulate as a result of inactivation of sen1-1 (Fig. 3A). Two new forms of snR13 accumulated rapidly in temperature-shifted sen1-1 cells. One had lower mobility than the 124-nucleotide-long mature snR13, and one had a greater mobility. We designated the three forms of snR13 detected in temperature-shifted sen1-1 cells R (for reduced mobility), F (for final form), and T (for truncated form) (Fig. 3A). Form F (snR13F) remained relatively constant in abundance over time in both wild-type and sen1-1 cells. In sen1-1 cells, an increase in the abundance of forms R and T was detected as early as 30 min following temperature shift. Forms R and T were both detected at low levels in wild-type cells. Both RNAs increased in abundance following temperature shift of the sen1-1 strain with similar kinetics.
FIG. 3.
Accumulation of snR13-related RNAs in a strain carrying sen1-1. (A) Strains 1971 (wild type) and FWY1 (sen1-1) were grown at 25°C and then shifted to 37°C. Total RNA was extracted at the time intervals indicated and analyzed by Northern blotting using oligonucleotide 13A (Fig. 1) as the probe. SUF4 tRNA was used as a loading control (not shown). Three RNAs were detected: snR13R (retarded mobility), snR13F (final form), and snR13T (truncated form). Size standards are indicated in nucleotides on the left. (B) The blot shown in panel A was stripped and reprobed with oligonucleotide 13D, which anneals downstream from the 3′ end of snR13F (Fig. 1). This probe detects only snR13R RNA.
Because SEN1 is an essential gene (10), we assessed whether the onset of altered RNA accumulation precedes loss of cell viability following a shift to the restrictive temperature. There was no loss of cell viability in strain FWY1 during the first 4 h following temperature shift. During this period, cell doubling times for strains FWY1 and 1971 were similar. The plating efficiency of FWY1 leveled off and then began to decline 6 h after temperature shift.
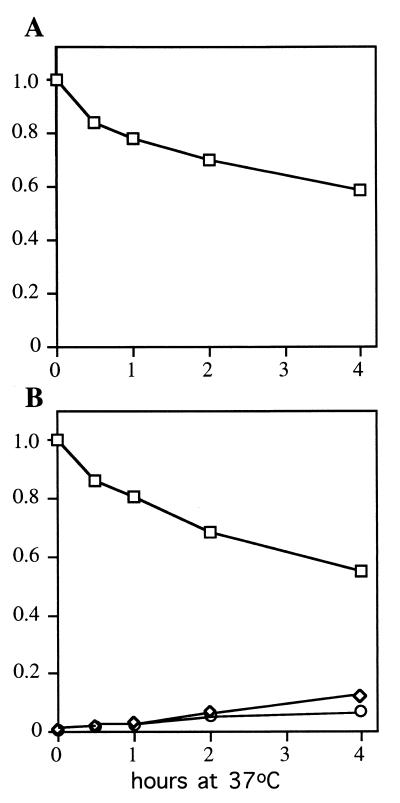
The changes in RNA accumulation shown in Fig. 3 commence well before the onset of cell death. Using a PhosphorImager, we quantitated in duplicate the levels of snR13R, -F, and -T at each time point following temperature shift (Fig. 4). In strain 1971 (SEN1), snR13F RNA shows a temporal decline caused by shifting temperature. The levels of snR13R and snR13T were too low for reliable quantitation in wild-type cells. In strain FWY1 (sen1-1), snR13F shows a temporal decline slightly steeper than that observed for strain 1971. snR13R and snR13T begin to accumulate immediately following temperature shift of sen1-1 cells. The relative abundance of snR13R may be underestimated because transfer of larger RNAs from the gel to the membrane is less efficient than transfer of smaller RNAs. The changes in accumulation of snR13 RNAs in temperature-shifted FWY1 (sen1-1) are fully complemented by plasmids containing wild-type SEN1.
FIG. 4.
Kinetics of accumulation of snR13 RNAs following inactivation of Sen1p. Data from the Northern blot shown in Fig. 3A and from a duplicate experiment were quantitated with a PhosphorImager, averaged, and plotted. RNA accumulation levels were normalized by using SUF4 tRNA as a loading control. The data are expressed as a proportion of snR13F accumulation at time zero. (A) Accumulation of snR13F (□) at time intervals following a shift from 25 to 37°C in strain 1971 (SEN1). The abundances of snR13R and snR13T were too low for reliable quantitation. (B) Accumulation of snR13R (○), snR13F (□), and snR13T (◊) at time intervals following a shift from 25 to 37°C in strain FWY1 (sen1-1).
It has been reported that snR13 is an essential gene (2). To confirm this and to test whether perturbations in the abundance of snR13 RNAs can themselves affect growth, we constructed the haploid strain TR110, which carries the null allele snR13-Δ1 (Materials and Methods) (Fig. 1B). This strain was found to be viable and failed to produce any detectable snR13F RNA as judged by Northern blotting, indicating that the RNA is not essential for growth. Loss of snR13 function does not contribute to growth inhibition in strains that carry sen1-1. Plasmid pTR48 (Fig. 1C) contains the entire snR13 transcriptional unit cloned into the yeast centromeric vector pRS315 (32). A transformants of strains TR110 carrying plasmid pTR48 produced mature snR13F, indicating that all of the sequences necessary for snR13 synthesis are present on the plasmid. This transformant had the same growth rate as the parental TR110 strain.
Novel forms of snR13 RNA consist of 5′ truncations and 3′ extensions of snR13F.
Since forms R and T appeared to be related to snR13F, we considered several possible causes for the slow mobility observed for band R, including a 5′ extension, a 3′ extension, or possibly an unusual topological structure. We reprobed the blot shown in Fig. 3A with oligonucleotide 13D (Fig. 1A; Table 1), which anneals downstream from the normal 3′ end of snR13F and which should not anneal with the F or the T form. Since this probe detected only form R (Fig. 3B), we performed further experiments to delineate the extent of the 3′ extension.
The low mobility of snR13R is due to a ∼1,300-nucleotide 3′ extension, as shown by a series of RNase H cleavage reactions (12, 31). Total RNA was extracted from temperature-shifted strain 1971 (sen1-1) and annealed with oligonucleotides 13B, 13C, 13D, 13E, 13F, and 13G, which are complementary to sequences downstream of the 3′ end of snR13F (Fig. 1; Table 1). Following RNase H treatment, samples were subjected to denaturing gel electrophoresis and transferred to a nylon membrane that was probed with 32P-end-labeled oligonucleotide 13A, which anneals within snR13F (Fig. 5A). Each site-directed digest generated two 5′ fragments derived from cleaved snR13R RNA that migrated as a doublet. This finding indicates that snR13R RNA consists of two subpopulations with two discrete 5′ ends. The two 5′ ends of snR13R RNA appeared to be coincident with the 5′ ends observed in forms F and T because the doublets approached the mobilities observed for forms F and T as the RNase H digests were targeted successively closer to the 3′ ends. This result suggests that snR13T is truncated at the 5′ end.
To analyze the 3′ end of snR13R RNA, the blot shown in Fig. 5A was stripped and reprobed with end-labeled oligonucleotide 13H, which anneals ∼1,300 nucleotides downstream of the 3′ end of snR13F (Fig. 1). The detection of doublets in each lane using this probe indicated that snR13R consists of two subpopulations with two discrete 3′ ends (Fig. 5B). Overall, the RNase H cleavage experiments using oligonucleotide 13A and 13H to probe the Northern blots show that there are at least two and possibly as many as four forms of snR13R, depending on how the 5′ and 3′ ends are distributed relative to each other in the population.
Detection of snR13R RNAs with oligonucleotide 13H indicates that snR13R RNAs are long enough to contain the entire open reading frame (ORF) YRD472w which was discovered in the S. cerevisiae genomic sequencing project. YDR472w is located 3′ to snR13 (Fig. 1). If translated, this ORF would give rise to a protein consisting of 283 amino acid residues. Probe 13H failed to detect bands other than the snR13R RNAs (data not shown), suggesting that an independent mRNA corresponding to ORF YRD472w either does not exist or is present below the level of detection with end-labeled 13H as the probe.
Primer extension analysis was used to map the 5′ ends and the locations of internal stops in the snR13 RNAs. Total RNA was extracted from strains 1971 (SEN1) and FWY1 (sen1-1) shifted from 25 to 37°C for 4 h. Primer extension reactions were performed with two 32P-end-labeled oligonucleotides, 13A, which anneals within snR13F RNA, and 13B, which anneals downstream of the 3′ end of snR13F RNA. The results of these studies are summarized in Fig. 1A.
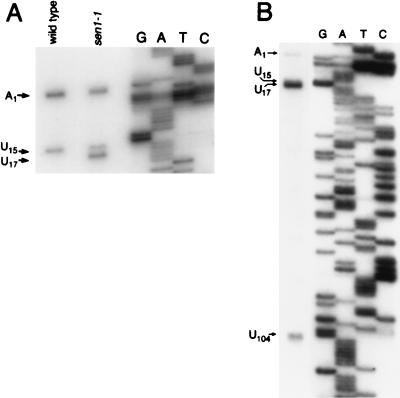
With oligonucleotide 13A as the primer, total RNA extracted from temperature-shifted strain 1971 (SEN1) gave rise to two primer extension cDNA products at 41°C (Fig. 6A). One cDNA resulted from termination of reverse transcription at the 5′ most nucleotide (other than the TMG cap) in snR13F RNA. This product is designated A1 because it terminated opposite the first residue in snR13F RNA, which is an adenosine (22). The second cDNA mapped to position U15. PhosphorImager quantitation of several primer extension reactions consistently showed that stop U15 occurred 40% as frequently as stop A1. The same primer extension reactions were repeated at 50°C to see if increasing the temperature would allow elongation through stop U15, but the U15 product persisted with no decrease in abundance relative to stop A1.
FIG. 6.
Primer extension of snR13 RNAs. (A) Primer extension of total RNA from strains 1971 (SEN1) and FWY1 (sen1-1) grown for 4 h at 37°C. Oligonucleotide 13A (Fig. 1) was end labeled and used for both primer extension mapping and dideoxy sequencing from plasmid pTR43 (Materials and Methods). cDNA band U15 is due to internal termination within snR13F. U17 arises from the 5′ end found in snR13T and a subpopulation of snR13R RNA. (B) Primer extension of total RNA from strain FWY1 (sen1-1) grown for 4 h at 37°C, using an end-labeled oligonucleotide 13B (Fig. 1). Stops A1 and U17 correspond to two distinct 5′ ends of snR13R RNA. Stops U15 and U104 probably arise due to internal termination of reverse transcriptase within snR13R RNA.
When total RNA from temperature-shifted strain FWY1 (sen1-1) was extended by using primer 13A, the resulting cDNAs terminated at A1, U15, and U17 (Fig. 6A). To establish which RNAs contributed to each stop, we analyzed each population of snR13 RNAs by gel purifying the R, F, and T forms for use as separate templates for primer extension. Template RNAs primed with oligonucleotide 13A that gave rise to cDNAs ending at A1 also gave rise to cDNAs ending at U15. The U17 cDNA arises from two sources of template RNA that share a common 5′ truncation of 16 nucleotides. One source is snR13T RNA, which accumulates to higher levels following temperature shift. The second source is the same subpopulation of R form RNA that gave rise to the lower bands in the doublets generated by the RNase H digestions (Fig. 5A).
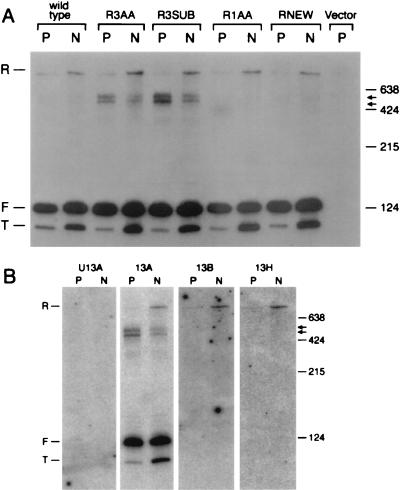
Using as the primer oligonucleotide 13B (Fig. 1A), which extends only those RNAs containing a 3′ terminus downstream of the snR13F 3′ terminus, we analyzed cDNA stops using total RNA from the temperature-shifted strain FWY1 (sen1-1) (Fig. 6B). A primer extension stop mapping to position U104 was detected in addition to stops at A1, U15, and U17. Inspection of the primary sequence of snR13F RNA revealed that primer extension stops U15 and U104 mapped within a repeated C2U4 sequence (Fig. 1A). The normal 3′ end reported for snR13 (23) maps within a third C2U4 repeat, and a fourth repeat occurs within ORF YDR472w. We used suitably positioned primers to test the ability of repeats 3 and 4 to give rise to premature primer extension stops but detected none (data not shown). We also performed primer extension on RNA extracted from the wild-type strain by using oligonucleotide 13B. Although the steady-state levels of snR13R RNA were very low in the wild type, we were able to detect stops A1 and U15 in such reactions (data not shown). This finding confirms that small amounts of 5′-truncated and 3′-extended forms of snR13 are present in wild-type cells.
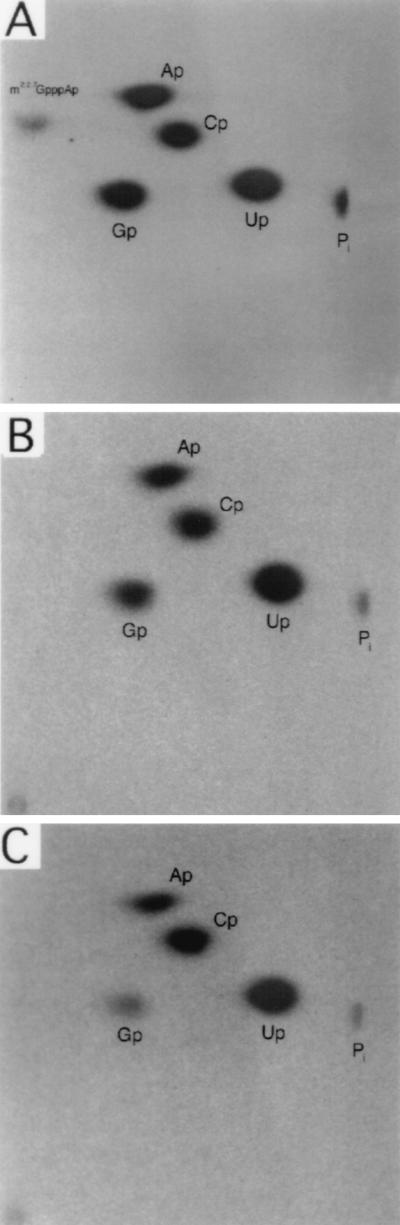
To determine whether primer extension stops U15 and U104 are due to the presence of base modifications, we examined the nucleotide composition of wild-type 32P-labeled snR13F RNA purified by hybrid selection using the biotinylated oligonucleotide 13BIO (Fig. 1A) followed by gel fractionation and elution of bands from the gel (Materials and Methods). snR13F was digested to nucleoside 3′-monophosphates with RNase T2 and subjected to two-dimensional analysis on cellulose thin layers (Fig. 7A). The nucleoside monophosphate molar ratios agreed closely with the molar ratios predicted from the snR13 sequence (Table 2). A spot that failed to migrate significantly in the second dimension was identified. This spot most likely corresponds to the TMG cap dinucleotide expected to be liberated upon digestion with RNase T2. No other modifications were detected.
FIG. 7.
TLC of snR13F purified from strain 1971 (SEN1) (see Materials and Methods). In each chromatogram, the loading origin is in the lower left corner, with the first dimension developed from bottom to top and the second dimension developed from left to right. Quantitative analysis of each chromatogram is provided in Table 2. (A) Chromatogram of the products of RNase T2 digestion of snR13F RNA. Spots correspond to nucleoside 3′ monophosphates and the presumed cap dinucleotide. (B) Chromatogram of the products of RNase T2 digestion of RNA fragment T1a fragment (U7 to G24). (C) Chromatogram of the products of RNase T2 digestion of RNA fragment T1b (A88 to G121).
TABLE 2.
Base composition of snR13F
| snR13F | RNase T2 digestion product | Molar ratio
|
|
|---|---|---|---|
| Predicteda | Observedb | ||
| Full length | Ap | 0.22 | 0.20 |
| Cp | 0.16 | 0.16 | |
| Gp | 0.20 | 0.21 | |
| Up | 0.39 | 0.41 | |
| m2,2,7GpppAp | 0.03 | 0.02 | |
| Fragment T1a (U7–G24) | Ap | 0.13 | 0.12 |
| Cp | 0.22 | 0.20 | |
| Gp | 0.04 | 0.04 | |
| Up | 0.61 | 0.64 | |
| Fragment T1b (A88–G121) | Ap | 0.11 | 0.14 |
| Cp | 0.11 | 0.14 | |
| Gp | 0.06 | 0.12 | |
| Up | 0.72 | 0.60 | |
Derived from predicted snR13 primary sequence.
Average of two independent experiments.
We confirmed that the spot thought to represent the cap dinucleotide was not in fact due to a modification at position C14 or C103. Modification of C14 or C103 could cause premature termination of reverse transcriptase at cDNA positions U15 and U104. Purified snR13F RNA was digested with RNase T1 and fractionated on a one-dimensional polyacrylamide gel. Based on the sequence, the two largest T1 fragments, T1a (U7 to G24) and T1b (A88 to G121), contain the regions surrounding the U15 and U104 primer extension stops. The T1a and T1b RNA fragments were purified by elution from the gel and digested with RNase T2. When the digestion products were separated by TLC, the only spots identified from T1a and T1b were those corresponding to the four major nucleoside monophosphates (Fig. 7B and C). When the molar ratios of nucleoside monophosphates derived from snR13F, T1a, and T1b were calculated (Table 2), the experimentally determined ratios for snR13F and T1a were very close to theoretical expectation. The results for T1b deviated somewhat from theoretical expectation, possibly due to contamination with another fragment. Overall, these experiments indicate that primer extension stops arising at U15 and U104 are not caused by nucleoside modifications.
Newly synthesized snR13 RNAs.
To visualize newly synthesized RNA, we examined in vivo pulse-labeled RNA from strains 1971 (SEN1) and FWY1 (sen1-1). Pulse-labeling at 25°C was accomplished by adding 1 mCi of [32P]orthophosphate for 1 h to SEN1 or sen1-1 cells growing in low-phosphate YEPD at 25°C. Pulse-labeling at 37°C was accomplished by shifting the cells from 25 to 37°C for 1 h followed by an additional hour of labeling at 37°C with 1 mCi of [32P]orthophosphate. Total labeled RNA was extracted, and the snR13 RNAs were purified by hybrid selection using biotinylated oligonucleotide 13BIO (Fig. 1A).
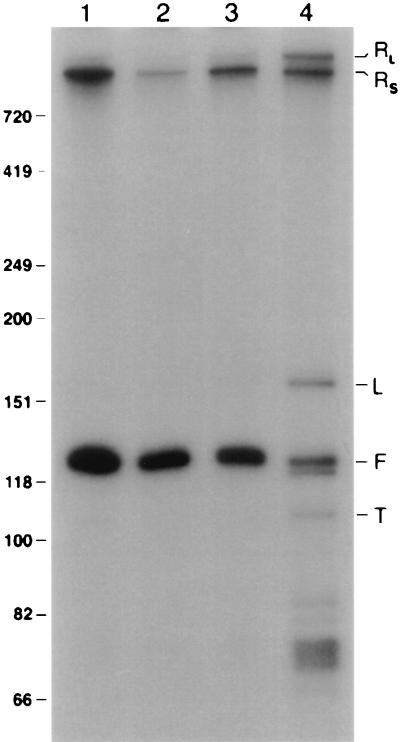
The purified RNAs were fractionated on a denaturing polyacrylamide gel (Fig. 8). Two slowly migrating forms of snR13, RL, and RS, were detected in wild-type cells at positions on the gel corresponding to approximately 1,400 and 1,200 nucleotides, respectively. RL was barely detected in wild-type cells pulse-labeled at 25 or 37°C (Fig. 8, lanes 1 and 2). RS was readily detected in wild-type cells pulse-labeled at 25°C (Fig. 8, lane 1). The abundance of RS was decreased in wild-type cells labeled at 37°C (Fig. 8, lane 2). Newly synthesized snR13F was readily detected in wild-type cells labeled at 25 or 37°C (Fig. 8, lanes 1 and 2).
FIG. 8.
In vivo pulse-labeling of snR13 RNAs. Following labeling, RNAs were purified by using the biotinylated oligonucleotide 13BIO (Fig. 1; Materials and Methods). Lane 1, RNA from strain 1971 (SEN1) labeled for 1 h at 25°C; lane 2, RNA from strain 1971 shifted to 37°C for 1 h and then 32P labeled for 1 h at 37°C; lane 3, RNA from strain FWY1 (sen1-1) labeled for 1 h at 25°C; lane 4, RNA from strain FWY1 shifted to 37°C for 1 h and then 32P labeled for 1 h at 37°C. The identities of the RNAs are indicated on the right; RNA size standards are indicated in nucleotides on the left.
sen1-1 mutant cells contained RL and RS in proportions similar to those in wild-type cells at the permissive temperature of 25°C (Fig. 8, lane 3). RL accumulated in these cells as a result of temperature shift (Fig. 8, lane 4). Contrary to the case for wild-type cells, the abundance of RS did not change significantly in temperature-shifted sen1-1 cells. snR13F was readily synthesized in sen1-1 cells at 25°C, but only small amounts of snR13F and snR13T were detected in the mutant when pulse-labeling was performed at the nonpermissive temperature (Fig. 8, lanes 3 and 4). This result indicates that snR13F RNA detected by Northern blotting of total RNA extracted from temperature-shifted sen1-1 cells is composed of stable RNA that was synthesized prior to inactivation of Sen1p. Although snR13T was synthesized at a relatively low rate in sen1-1 cells at 37°C as judged by this experiment, it accumulated to levels detectable on Northern blots. Several additional bands were detected in sen1-1 cells pulse-labeled at 37°C (Fig. 8, lane 4) that were not detected by conventional Northern blotting. These include species L, which is approximately 165 nucleotides long, as judged by its mobility, and a number of bands migrating faster than snR13F, including snR13T. The significance of these labile RNAs is not yet known.
Identification of a sequence important in snR13 RNA synthesis.
Inspection of the snR13 primary sequence revealed that the sequence C2U4 is reiterated three times in snR13F (Fig. 9A). The normal 3′ end reported for snR13 (23) (GenBank entry SCU16692) is embedded within C2U4 repeat 3. The premature primer extension stop at position U15 and the 5′ truncation at U17 map within C2U4 repeat 1.
FIG. 9.
Effects of mutations in snR13 on RNA accumulation. (A) Predicted secondary structure of wild-type snR13F RNA, generated by using the Genetics Computer Group Fold program (11). The locations of the C and D boxes, the primer extension stops, and the mutations R1AA, R3SUB, R3AA, and RNEW are shown (see text). The 3′ end of wild-type snR13F is formed between residues C124 and U125. (B) Effects of snR13 mutations. Strain TR110, which carries snR13-Δ1, was transformed with plasmids as indicated below. Transformants were grown at 30°C followed by RNA extraction and Northern blotting using oligonucleotide 13A (Fig. 1) as the probe. Lane 1, pTR48 (wild-type SNR13); lane 2, pTR51 (snR13-R3AA); lane 3, pTR53 (snR13-R3SUB); lane 4, pTR52 (snR13-R1AA); lane 5, pTR56 (snR13-RNEW); lane 6, pRS315 (vector only). Arrows to the right indicate the mobilities of the novel RNAs. Size standards are indicated in nucleotides to the right. (C) A shorter exposure of the Northern blot shown in panel B shows that snR13F is decreased in abundance in lane 4 (see text).
To determine whether the C2U4 repeats are important in snR13 synthesis, four site-directed mutations were constructed in an snR13 gene contained in plasmid pTR48 (Fig. 1C and 9A). Two mutations perturb the sequence of C2U4 repeat 3 in which the site of 3′ end formation is embedded. The snR13-R3AA allele codes for RNA in which C2U4 repeat 3 is changed from C2U4 to CA2U3. The snR13-R3SUB allele codes for an RNA containing a substitution of the entire repeat 3 with the sequence A6. The snR13-R1AA allele codes for an RNA in which the 5′-proximal C2U4 repeat 1 is changed from C2U4 to CA2U3. The snR13-RNEW allele codes for an RNA containing a new C2U4 repeat in the third loop of the predicted snR13 secondary structure. snR13-RNEW was created by changing a naturally occurring CU4 sequence to C2U4.
Plasmids carrying each mutant allele, the wild-type SNR13 gene, and the vector alone were introduced into strain TR110, which carries snR13-Δ1 (Materials and Methods). Transformants were grown in selective medium at 25°C, followed by RNA extraction and Northern blotting with probe 13A. The mutations snR13-R3AA and snR13-R3SUB gave rise to a novel doublet with an apparent mobility in the range of 500 nucleotides (Fig. 9B, lanes 2 and 3). The snR13-R3SUB mutation caused a fourfold-greater accumulation of the novel doublet than did the snR13-R3AA mutation. The snR13-R1AA and snR13-RNEW mutations did not cause accumulation of this doublet, showing that formation of the novel doublet is a specific result of perturbation of C2U4 repeat 3 at the 3′ end of snR13. The snR13-R1AA mutation in repeat 1 caused a twofold reduction in snR13F accumulation compared to the wild type (Fig. 9C, lane 4). snR13-RNEW failed to cause any changes in snR13 RNA accumulation (Fig. 9B, lane 5), showing that C2U4 is insufficient by itself to specify a site for 3′ end formation in the absence of a proper context.
Epistatic relationship between sen1-1 and mutations in snR13.
We assessed the consequences of combining mutations in C2U4 repeats with sen1-1. To accomplish this, plasmids carrying each of the snR13 mutant alleles were transformed into the haploid strain TR116, which carries the chromosomal mutations sen1-1 and snR13-Δ1 (Materials and Methods). Strain TR116 was also transformed with plasmid pTR48, which carries DNA coding for the entire wild-type snR13 transcriptional unit (Fig. 1C). Transformants were grown at the permissive temperature of 25°C in selective medium to maintain the plasmids. In addition, transformants were grown to mid-log phase in selective medium at 25°C and then shifted to the nonpermissive temperature of 37°C for 4 h.
RNA was extracted from the transformants of strain TR116 at each growth temperature and analyzed by Northern blotting using probe 13A (Fig. 4A), which hybridizes to forms R, F, and T and the novel doublet RNAs. A transformant carrying plasmid pTR48 gave rise to snR13F and increased amounts of snR13R and snR13T at 37°C (Fig. 10A). As expected, transformants carrying snR13-R3AA and snR13-R3SUB gave rise to the novel doublet at the permissive temperature. When cells containing snR13-R3AA or snR13-R3SUB were shifted to 37°C, the accumulation of snR13R RNA increased threefold while the accumulation of the novel doublet was decreased by a corresponding amount. Therefore, double mutants containing sen1-1 and snR13-R3AA or snR13-R3SUB resemble sen1-1 single mutants when placed at a nonpermissive temperature for sen1-1. This epistatic relationship suggests that Sen1p and C2U4 repeat 3 affect a common pathway. Furthermore, these results indicate that Sen1p functions prior to the action of C2U4 repeat 3, allowing division of snR13 synthesis into two discrete steps, the first requiring Sen1p and the second requiring C2U4 repeat 3. Transformants carrying snR13-R1AA or snR13-RNEW produced a banding pattern identical to a transformant carrying pTR48, indicating no epistatic interaction between sen1-1 and these snR13 alleles.
FIG. 10.
(A) Epistatic interactions between sen1-1 and mutations in SNR13. RNA was extracted from strain TR116 (sen1-1 snR13-Δ1) transformed with pTR48 (wild type) and with plasmids containing the snR13 mutations R3AA, R3SUB, R1AA, and RNEW RNA was (see Materials and Methods). Probe 13A (Fig. 1) was used to probe a Northern blot. RNA was extracted from cells grown at 25°C, a permissive temperature for sen1-1 (P), and from cells grown at 25°C followed by a shift to 37°C, a nonpermissive temperature for sen1-1, for 4 h (N). Arrows to right indicate positions of the novel RNAs. (B) Analysis of novel doublet RNA mobility by Northern blotting of RNA extracted from TR116 transformed with pTR51 (sen1-1 snR13-R3SUB) grown at 25°C (P) and then shifted to 37°C (N). The blots were probed with oligonucleotides U13A, 13A, 13B, and 13H (Fig. 1). Sizes are indicated in nucleotides on the right.
The novel doublet RNAs that arise due to the R3SUB mutation in C2U4 repeat 3 were analyzed further to assess whether they migrate on gels according to size (Fig. 10B). The doublet RNAs have an apparent mobility in the range of 500 nucleotides on Northern blots probed with oligonucleotide 13A. Oligonucleotide 13B, which anneals to snR13R just downstream from the site of 3′ end formation, hybridized to snR13R but failed to detect the novel doublet. Probe 13H, which anneals near the 3′ end of snR13R, hybridized to snR13R but not the novel doublet. These probes might be excluded from annealing to novel double RNAs for some structural reason. Alternatively, the novel doublet RNAs may lack 3′ extensions long enough to account for their apparent mobility on gels. We also used as a probe oligonucleotide U13A, which is expected to hybridize to a sequence just upstream of nucleotide +1 of snR13F. This oligonucleotide failed to detect any of the snR13 RNAs. Since probes U13A and 13B are separated by a distance of 164 nucleotides, the positions of migration of the novel doublet RNAs in the 500-nucleotide range may be caused by topological constraints that retard their migration on gels. Similar results were observed using a strain that carries the R3AA mutation (data not shown).
DISCUSSION
In this report, we describe a family of RNAs that are derived from the snR13 locus of S. cerevisiae. The most abundant of these, snR13F, is a TMG-capped, C/D box-containing snoRNA that most likely functions in rRNA maturation in the nucleolus (23). Three overlapping RNAs, snR13R, snR13F, and snR13T, were detected in wild-type cells by Northern blotting. The steady-state levels of these RNAs changed when strains carrying the temperature-sensitive mutation sen1-1 were shifted to the restrictive temperature. snR13R and snR13T increased in abundance with similar kinetics. The level of snR13F declined in both wild-type and sen1-1 cells at the restrictive temperature, obscuring any potential change due to the sen1-1 mutation itself. The reason for this decline at an elevated temperature is unknown.
When in vivo-labeled, nascent snR13 RNA was analyzed in both wild-type and temperature-shifted sen1-1 cells, similar changes were observed except that the amount of snR13F RNA synthesized was severely reduced in sen1-1 cells shifted to the restrictive temperature. This result indicates that the rate of synthesis is significantly reduced. We identified in wild-type and temperature-shifted sen1-1 cells several additional nascent RNAs that were not detected on Northern blots. Two long forms of snR13, called RL and RS, were detected. RL is probably equivalent to the snR13R RNAs detected on Northern blots, because it responds to inactivation of Sen1p in the same way as snR13R. Similar amounts of RS were detected at the permissive and restrictive temperature. The relationship of RS to RL is not yet known. Our results indicate that a function provided by Sen1p, which is most likely an RNA unwinding activity, is required for the synthesis of snR13F. This is consistent with an interpretation in which snR13F is derived from RL by a posttranscriptional pathway leading to 3′ end formation.
We used RNase H mapping and primer extension to characterize the three stable RNAs that accumulate on Northern blots. These RNAs are distinguished by a 5′ truncation, a 3′ extension, or both. snR13R is a mixed population of RNAs that share in common a ∼1,300-nucleotide extension of the 3′ end of snR13F. RNase H mapping indicates that R form RNAs contain two discrete 5′ ends and two discrete 3′ ends. Depending on the distribution of 5′ and 3′ ends, there are two to four species of snR13R. Using primer extension, we showed that one of the 5′ ends of snR13R is identical to the 5′ end of snR13F (residue A1 in Fig. 1 and 6). Since Midwestern blotting and TLC analysis indicate that snR13F contains a TMG cap, it seems likely that a subpopulation of snR13R may also contain a TMG cap. We could not detect a TMG-capped R form on Midwestern blots, but this could be because of low abundance, poor transfer to nylon membranes, or comigration with other TMG-capped RNAs.
The changes in synthetic rates and levels of accumulation of the snR13 RNAs result from a requirement of Sen1p to both stabilize the 5′ terminus and to promote formation of the 3′ terminus from a 3′-extended RNA. The truncated 5′ end found among snR13R RNAs begins at nucleotide U17 and is therefore missing the first 16 nucleotides found in snR13F RNA. snR13T also begins at U17. snR13T RNA was detected on Northern blots but not on Midwestern blots and is therefore not likely to contain a TMG cap. Based on these findings, we propose that snR13F, snR13T, and the 5′-truncated version of snR13R are all derived from a common monomethyl-G-capped primary transcript that becomes hypermethylated to form a single TMG-capped RNA.
The 5′ truncation of snR13R and snR13T may result from exonucleolytic degradation or endonucleolytic cleavage of a single primary transcript that begins at nucleotide A1, producing a capless, 5′-truncated RNA. If the two 5′ ends were derived from two primary transcripts initiating at two different transcriptional start sites, all of the RNAs might be expected to have a TMG cap which is synthesized in conjunction with transcription by RNA polymerase II. The lack of a TMG cap in snR13T is inconsistent with this model. Our interpretation of the data implies that Sen1p is required for maintenance of the 5′ end. In absence of Sen1p function, the 5′ end is removed up to a position approaching the location of the C/D base-paired stem.
Primer extension stops were detected at positions U15 and U104. Both stops map within the sequence C2U4 that is repeated three times in snR13F and snR13T and once more in snR13R. The truncated 5′ end beginning at U17 in a subpopulation of snR13R and in snR13T maps within C2U4 repeat 1. The 3′ ends of snR13F and snR13T are located within C2U4 repeat 3. Because of the apparent significance of C2U4 repeats, we analyzed a substitution of CA2U3 for C2U4 repeat 1, which contains the U15 and U17 primer extension stops, and substitutions of CA2U3 and A6 for C2U4 repeat 3, which contains the 3′ terminus of the snR13F and snR13T RNAs (Fig. 1 shows the locations of these repeats).
If C2U4 sequence repeats 1 and 3 were to function as signals for transcription initiation and termination, respectively, we would expect that mutations that impair the function of these sequences might cause increased accumulation of RNAs containing extended 5′ and/or 3′ ends. Our results show that this is not the case. The mutation in repeat 1 caused decreased accumulation of snR13F RNA without a concomitant increase in 5′-truncated snR13T and 3′-extended snR13RR. Both of the mutations in repeat 3 failed to cause detectable increases in the accumulation of snR13T and snR13R RNAs. Instead, these mutations resulted in the accumulation of a pair of novel RNAs that differ markedly from R forms. The novel doublet RNAs failed to migrate according to size and may not contain a significant 3′ extension, indicating a potential level of structural complexity not expected for simple 3′-extended transcripts. These results make it unlikely that C2U4 repeats function in transcription initiation or termination. Instead, they are likely to play a role in maturation.
The predicted secondary structure of snR13F (Fig. 9) suggests that C2U4 repeats 1 and 3 may reside near each other in three-dimensional space. The two repeats flank a base-paired region that overlaps with the C and D boxes. In our studies we detected 5′ and 3′ ends within repeats 1 and 3, respectively, but failed to detect RNAs with an endpoint in repeat 2 or in a new repeat created by the snR13-RNEW mutation. These results indicate that the presence of a C2U4 repeat is not sufficient to specify a site for end formation. According to this interpretation, C2U4 repeats may play a role in maturation by providing a preferred context to the neighboring base-paired C/D box. Additional evidence supporting this view comes from a comparison of sequences surrounding C and D boxes in other snoRNAs. When the sequences of other snoRNAs were examined, we found that several contained either an exact C2U4 match or a close match differing by a single nucleotide (Table 3). However, some snoRNAs such as snR10 and snR31 do not possess any sequences resembling C2U4 (2, 36). These observations indicate that C2U4 is most likely a preferred sequence but not one that is globally required in all snoRNAs.
TABLE 3.
Yeast RNAs with snR13-like 3′ sequences
| RNA | Sequencea |
|---|---|
| snR13 | UUAUCUGACCuuuuaacuu |
| U14 | GAUCUGAGCCuuuauuuuu |
| snR11 | CGCACAUACcuuuuuuuuu |
| snR40 | AUUCUGAUCcauuuuccuu |
| snR45 | UUCUGAUUCUUuuuucuuu |
| U4 | UUGGAAUACCUUuuuucau |
| U5S | UUUUGGAACUuuucugccc |
Sequences found in mature RNAs are in uppercase; 3′ flanking sequences are in lowercase; D boxes are underlined. Loss of function of Sen1p affects the accumulation of all listed RNAs except U4 (36).
The phenotypes conferred by sen1-1 and by mutations in C2U4 repeats 1 and 3 resemble the phenotypes of C/D box mutations described by others (6, 13, 41). For example, deletion of a short stem that immediately flanks the C and D boxes in human U14 snoRNA results in maturation defects. Also, Xenopus U16 and U18 RNAs require intact C and D boxes for end maturation. The 3′ end of Xenopus U16 is formed within a sequence context identical to that found for snR13 and proceeds by a reaction requiring protein factors that have been partially purified. We speculate that one of the factors could be Sen1p in yeast and its homologue in higher eukaryotes.
We tested these ideas further by analyzing double mutants carrying sen1-1 and each of the mutations in C2U4 repeats 1 and 3. When mutations in repeat 1 were combined in the same haploid strain with the sen1-1 mutation, the abundance of snR13F was significantly reduced at the restrictive temperature. We have not yet determined whether this is due to a reduced rate of formation of snR13F or to an increased rate of decay of the mature RNA. When mutations in repeat 3 were combined with the sen1-1 mutation, R forms accumulated at the expense of the novel doublet RNAs upon a shift from permissive to restrictive temperature. This finding indicates that an epistatic relationship exists between Sen1p and C2U4 repeat 3. Based on this finding, we propose that Sen1p is required prior to repeat 3. Furthermore, the epistatic relationship suggests a connection between Sen1p, C2U4 repeat 3, and the C/D box-fibrillarin snoRNP complex. Repeat 3 may provide a good sequence context that influences the binding of C/D box proteins that both protect snR13F from degradation and promote 3′ end maturation. Sen1p might be recruited into the snoRNP complex to facilitate 3′ end formation. Alternatively, Sen1p might promote the assembly of a snoRNP complex.
There are two reasons for suspecting that Sen1p may play a direct role in the formation of RNA termini and may be recruited into or assist in the assembly of snoRNP complexes. The changes in synthetic rates and levels of accumulation caused by loss of Sen1p function commence immediately upon temperature shift, which would be expected if Sen1p plays a direct role in snoRNA maturation. By contrast, the accumulation of tRNA precursors observed in strains carrying sen1-1 does not correlate with growth temperature, indicating that an indirect effect of Sen1p on tRNA splicing is likely (42). Most importantly, snR13F coimmunoprecipitates with wild-type Sen1p, suggesting that an RNA-protein physical interaction is likely (39). It remains to be determined whether snR13R and snR13T RNAs are also present in precipitates because their abundance in wild-type strains is very low.
Direct evidence that the novel doublet RNAs represent trapped intermediates in a maturation pathway is still lacking. We were unable to detect newly synthesized RNAs corresponding to the novel doublet RNAs in wild-type cells, but they may still exist as extremely labile RNAs. If they correspond to intermediates in 3′ end formation, their existence suggests that 3′ end formation involves more than one step. We are currently investigating the structures of these RNAs to determine why they exhibit anomalous migration on gels and whether they differ in length at the 5′ end in a manner similar to snR13F, snR13T, and the subpopulations of the snR13R RNAs.
Inactivation of Sen1p caused time-dependent changes in the accumulation of the snoRNAs snR10, snR11, snR13, snR31, snR40, and snR42, which include both C/D box and ACA snoRNAs, and the snRNA U5 (28, 39). In addition, other RNAs have recently been shown to exhibit altered accumulation upon inactivation of Sen1p, including RNAs of unexpected size containing U5, snR40, and snR45 sequences (39). Since loss of Sen1p function affects multiple RNAs and since it is recruited into RNP complexes containing as many as 20 snoRNAs (39), we suspect that Sen1p plays a role in the maturation of numerous snoRNAs.
The sen1-1 mutation causes pleiotropic defects in RNA metabolism, including effects on tRNA and rRNA processing (39, 42). Also, temperature-shifted sen1-1 cells exhibit altered localization of nucleolar proteins such as fibrillarin (38). Since changes in rRNA processing and nucleolar organization do not appear until several hours after temperature shift of sen1-1 cells but perturbations of snR13 synthesis commence immediately, it seems likely that global perturbations in snoRNA synthesis are primary defects in sen1-1 cells. The nucleolar and rRNA defects probably develop as a secondary consequence because rRNA maturation requires snoRNAs. The primary defect responsible for the accumulation of intron-containing pre-tRNAs in sen1-1 cells is still unknown. Our studies raise the possibility that a small RNA whose synthesis depends on Sen1p could be required for tRNA synthesis.
snR13R RNAs extend into and include all of the downstream ORF YDR472w. At present, we are unable to interpret the significance of this finding. Recent evidence indicates that YDR472w produces a polyadenylated mRNA that is likely to be translated into a protein product (21a). Studies are in progress to determine whether the snR13R RNAs encode a protein in addition to producing a snoRNA.
ACKNOWLEDGMENTS
We thank Francis Webb for construction of strain FWY1 and Jeff Dahlseid for insightful discussions during the course of this project. We also thank David Brow, Michael Lelivelt, Renee Shirley, and John Shannonhouse for critical readings of the manuscript.
This work was supported by the College of Agricultural and Life Sciences, University of Wisconsin, Madison, PHS grant GM40310 (M.R.C.), and PHS training grant in genetics GM07133 (T.P.R.).
Footnotes
University of Wisconsin Laboratory of Genetics paper 3486.
REFERENCES
- 1.Balakin A G, Schneider G S, Corbett M S, Ni J, Fournier M J. SnR31, snR32, and snR33: three novel, non-essential snRNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5391–5397. doi: 10.1093/nar/21.23.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 3.Brownlee G G. Determination of Sequences in RNA. New York, N.Y: American Elsevier Publishing; 1972. [Google Scholar]
- 4.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 6.Caffarelli E, Maggi L, Fatica A, Jiricny J, Bozzoni I. A novel Mn++-dependent ribonuclease that functions in U16 snoRNA processing in X. laevis. Biochem Biophys Res Commun. 1997;233:514–517. doi: 10.1006/bbrc.1997.6487. [DOI] [PubMed] [Google Scholar]
- 7.Cavaille J, Bachellerie J P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 8.Cavaille J, Nicoloso M, Bachellerie J P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 9.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 10.DeMarini D J, Winey M, Ursic D, Webb F, Culbertson M R. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Jaeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979;7:179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. A novel small nucleolar RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 15.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 16.Huang G M, Jarmolowski A, Struck J C, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′,3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 18.Koonin E V. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 19.Lafontaine D, Tollervey D. Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem Cell Biol. 1995;73:803–812. doi: 10.1139/o95-088. [DOI] [PubMed] [Google Scholar]
- 20.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 21.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Lelivelt, M., and M. Culbertson. Unpublished data.
- 22.Mattaj I W, Tollervey D, Seraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993;7:47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 24.Mendenhall M D, Leeds P, Fen H, Mathison L, Zwick M, Sleiziz C, Culbertson M R. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Saccharomyces cerevisiae. J Mol Biol. 1987;194:41–58. doi: 10.1016/0022-2836(87)90714-5. [DOI] [PubMed] [Google Scholar]
- 25.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura S. Modified Nucleosides in tRNA. In: Schimmel P R, Soll D, Abelson J N, editors. Transfer RNA: structure, properties, and recognition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1979. pp. 59–79. [Google Scholar]
- 27.Petfalski E, Dandekar T, Henry V, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen T P, Culbertson M R. Analysis of yeast trimethylguanosine-capped RNAs by midwestern blotting. Gene. 1996;182:89–96. doi: 10.1016/s0378-1119(96)00519-7. [DOI] [PubMed] [Google Scholar]
- 29.Rubin G M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973;248:3860–3875. [PubMed] [Google Scholar]
- 30.Scherer S, Davis R W. Replacement of chromosomal segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibahara S, Mukai S, Nishihara T, Inoue H, Ohtsuka E, Morisawa H. Site-directed cleavage of RNA. Nucleic Acids Res. 1987;15:4403–4415. doi: 10.1093/nar/15.11.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinmetz E J, Brow D A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terns M P, Grimm C, Lund E, Dahlberg J E. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tollervey D. Small nucleolar RNAs guide ribosomal RNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- 36.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 37.Tycowski K T, Smith C M, Shu M D, Steitz J A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ursic D, DeMarini D J, Culbertson M R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- 39.Ursic D, Himmel K L, Gurley K A, Webb F, Culbertson M R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Tol H, Stange N, Gross J, Beier H. A human and a plant intron-containing tRNATyr gene are both transcribed in a HeLa cell extract but spliced along different pathways. EMBO J. 1987;6:35–41. doi: 10.1002/j.1460-2075.1987.tb04715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 42.Winey M, Culbertson M R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]