Abstract
We have identified gas5 (growth arrest-specific transcript 5) as a non-protein-coding multiple small nucleolar RNA (snoRNA) host gene similar to UHG (U22 host gene). Encoded within the 11 introns of the mouse gas5 gene are nine (10 in human) box C/D snoRNAs predicted to function in the 2′-O-methylation of rRNA. The only regions of conservation between mouse and human gas5 genes are their snoRNAs and 5′-end sequences. Mapping the 5′ end of the mouse gas5 transcript demonstrates that it possesses an oligopyrimidine tract characteristic of the 5′-terminal oligopyrimidine (5′TOP) class of genes. Arrest of cell growth or inhibition of translation by cycloheximide, pactamycin, or rapamycin—which specifically inhibits the translation of 5′TOP mRNAs—results in accumulation of the gas5 spliced RNA. Classification of gas5 as a 5′TOP gene provides an explanation for why it is a growth arrest specific transcript: while the spliced gas5 RNA is normally associated with ribosomes and rapidly degraded, during arrested cell growth it accumulates in mRNP particles, as has been reported for other 5′TOP messages. Strikingly, inspection of the 5′-end sequences of currently known snoRNA host gene transcripts reveals that they all exhibit features of the 5′TOP gene family.
In the nucleolus of eukaryotic cells, ribosomal DNA is transcribed by RNA polymerase I into long precursor (pre-rRNA) transcripts, which are modified by methylation and pseudo-uridylation, cleaved to yield 18S, 5.8S, and 28S rRNAs, and then assembled into the mature large and small ribosomal subunits prior to export to the cytoplasm (for reviews see references 22, 26, and 77). A large number of small nucleolar ribonucleoprotein (snoRNP) particles have emerged as key players in this biosynthetic process. Currently more than 70 snoRNA species have been identified (for reviews see references 51, 75, and 82). All snoRNAs, with the exception of MRP RNA, can be divided into two classes: those that possess boxes C (RUGAUGA) and D (CUGA), which are required for association with the abundant nucleolar autoantigen fibrillarin (see reference 51), and those that possess boxes H (ANANNA) and ACA, which mediate the binding of Gar1 protein (4, 8, 24, 37). Only a few snoRNAs have been found to be required for growth in yeast (U3 [6, 29], U14 [43], MRP [14, 73], snR10 [80, 81], and snR30 [5, 54]) or for specific pre-rRNA cleavage events in Xenopus oocytes (U3 [34, 72], U8 [63], and U22 [85]).
Recently, box C/D snoRNAs and box H/ACA snoRNAs were found to target specific sites in pre-rRNA for 2′-O-methylation and pseudouridylation, respectively (for reviews see references 46, 48, 62, 75, and 82). These modification reactions are mediated by extensive regions (10 to 21 nt) of complementarity between the so-called “antisense” snoRNAs and sequences flanking the rRNA sites to be modified. Specifically, U24, U20, and U25 were shown to direct site-specific ribose methylation of pre-rRNA in HeLa cells (39), yeast (13), and Xenopus oocytes (87), respectively, and snR8, snR3, snR33, and snR5 (among others) were demonstrated to target pre-rRNA for pseudouridylation in yeast (23, 57). The presence of ∼200 modified nucleotides in vertebrate rRNA (47) suggests that more than half of the antisense snoRNAs remain to be identified.
A unique feature of snoRNAs is that most are encoded within the introns of protein-coding genes (reviewed in reference 51). This economic use of introns is commonplace among intron-rich organisms, such as vertebrates, where antisense snoRNAs have been found to be exclusively intron encoded. By contrast, in Saccharomyces cerevisiae many snoRNAs are produced from independent transcription units. A vertebrate host gene intron encodes only a single snoRNA, whereas in yeast and plants, some snoRNA genes are located in polycistronic arrays without exons separating the snoRNA sequences (41). In all species investigated, the intron-encoded snoRNAs are transcribed from their host genes by RNA polymerase II as portions of the pre-mRNA. The functional snoRNAs are then produced by exonucleolytic trimming that follows either splicing (12, 38, 86) or endonucleolytic cleavage of intron sequences (9, 10).
The mode by which snoRNA sequences became inserted into the introns of their host genes is not known. Interestingly, the host gene for a particular snoRNA can differ even among closely related vertebrates, suggesting that intron-encoded snoRNAs may be highly mobile genetic elements (see reference 51). Likewise, the reason particular genes have been chosen as hosts for intron-encoded snoRNAs has been unclear. Initially, it appeared that all snoRNA host genes generate protein products that function in ribosome biogenesis or in translation; ribosomal proteins (rp) L1, L5, L7a, S8, nucleolin, and eIF4AI are a few examples (see references 66, 68, 67, 59, 58, and 24, respectively). Such genetic organization could provide coregulation of protein components of the translational machinery and snoRNAs, which contribute to rRNA maturation (76). However, the discovery of other snoRNA host genes lacking obvious ribosome-related functions (for example, ATP synthase β [39]) suggested that host genes may have been chosen merely to meet the need for transcription rates high enough to produce a sufficient level of snoRNAs (∼104 copies/cell) to base pair with the cell’s nascent pre-rRNA molecules.
UHG (U22 host gene) is an unusual snoRNA host gene because it does not appear to specify a protein product. It generates, in addition to U22, seven different box C/D antisense snoRNAs (U25 to U31) (84). Comparison of mouse and human UHG sequences revealed that its introns are more conserved than its exons, suggesting that the snoRNAs may be the only functional portions of the transcript. Both human and mouse UHG messages are riddled with stop codons in all three reading frames; the longest open reading frames (ORFs) would produce peptides of 51 and 40 amino acids for human and mouse UHG, respectively. Nonetheless, the UHG transcript resembles a typical rp mRNA in that it begins with a C residue, is spliced and polyadenylated, and is associated with ribosomes. However, unlike rp mRNAs, the spliced UHG RNA is almost undetectable in HeLa cells. Inhibition of translation in HeLa cells with the initiation inhibitor pactamycin or elongation inhibitors cycloheximide or puromycin results in a 15-fold increase in the level of spliced UHG transcript (84). This link between the levels of UHG RNA and active translation, in conjunction with its numerous stop codons, suggests that it may be a candidate for the nonsense-mediated decay pathway (49, 84).
Here we report the identification of a second member of the UHG class of snoRNA host genes. Growth arrest-specific transcript 5 (gas5) was initially discovered in a screen for potential tumor suppressor genes expressed at high levels during growth arrest (74). The murine gas5 gene produces a ubiquitous, polyadenylated, alternatively spliced message which is almost undetectable in actively growing cells yet is highly expressed in cells undergoing serum starvation or density arrest (15, 16). We demonstrate that gas5 is a multi-snoRNA host gene which encodes 9 (in mouse) or 10 (in human) antisense snoRNAs. By mapping the 5′ end of the gas5 transcript and comparing it with other known snoRNA host genes, we observe that all known snoRNA host genes exhibit characteristics which define the 5′TOP (terminal oligopyrimidine) class of genes. We provide evidence that membership in the 5′TOP family explains why the abundance of the gas5 spliced product is growth dependent. Furthermore, the discovery that all snoRNA host genes contain 5′TOP sequences may illuminate why certain genes have been selected to serve as snoRNA host genes.
MATERIALS AND METHODS
Cloning mouse and human gas5 genes.
The partial sequence of U80 was obtained by a reverse transcriptase PCR cloning approach (61) in which cDNAs were generated by first ligating an antisense oligonucleotide complementary to the T3 promoter (5′-TTTAGTGAGGGTTAAT-3′dA) onto the 3′ ends of RNAs isolated from HeLa cells by immunoprecipitation with anti-fibrillarin (anti-fb) antibodies (see below) followed by primer extension using a sense T3 primer (5′-ATTAACCCTCACTAAA). cDNAs between 70 and 90 nucleotides in length were gel purified and subjected to PCR using the T3 primer and oligonucleotide 124 (5′-GTGAACAATCCAACGCTGA) which corresponds to residues 3607 to 3631 of 28S rRNA. The resulting PCR product(s) was cloned into pGEM-3Z and sequenced.
To obtain a complete sequence for the mouse gas5 gene, PCR was performed between intron 1 (158, 5′-GACGTAGGATCCTGCTGGATATGTGCAACT) and intron 4 (159, 5′-TCATCGAAGCTTGTTAACGACCACTAGCTC) and between intron 1 (160, 5′-GACGTAGGATCCGTATGCAATTTCCTGAGT) and intron 3 (161, 5′-TCATCGAAGCTTAGCAAATATGATGTCATC) using mouse genomic DNA (Clontech Laboratories). The PCR products were cloned into BamHI-HindIII-digested pGEM-3Z. Cloning of these segments provided complete sequences for U74 and U75 and confirmed that intron 4 does not contain a consensus box C sequence. The mouse gas5 promoter was also recloned by PCR from mouse genomic DNA (Clontech Laboratories) by using primers upstream of the TATA box (173, 5′-CTTGAGGAGGAGTCTGAG) and complementary to intron 1 (166, 5′-TCAGTTGTCCCTACCAACATAGCCT). PCR products were ligated into the pCR2.1 TA vector (Invitrogen). The promoter was found to differ from the reported gas5 sequence (G instead of A) at position −2 upstream of the gas5 transcription start site (see Fig. 3b).
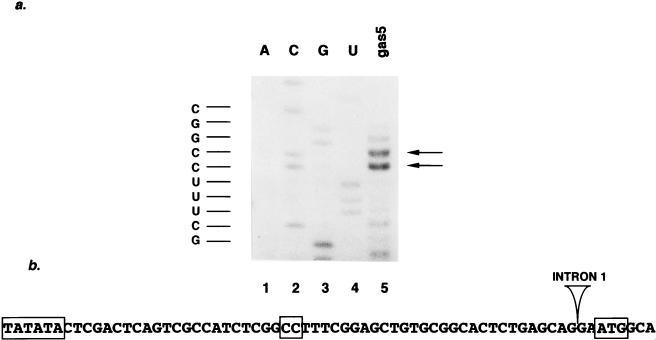
FIG. 3.
gas5 is member of the 5′TOP gene family. (a) Extension of a deoxyoligonucleotide primer complementary to mouse gas5 (lane 5) was performed using mRNA isolated from NIH 3T3 cells treated with cycloheximide. The products were run adjacent to a sequencing ladder of DNA containing the mouse gas5 exon 1 and promoter (lanes 1 to 4). (b) The transcription start site maps to two adjacent cytidine residues located 29 and 30 nucleotides downstream of a TATA box and 32 and 33 nucleotides upstream of a consensus translation start site. The TATA element, transcription start sites, and translation start site are boxed; the position of the first intron of the mouse gas5 gene is indicated.
Human gas5 was cloned in multiple steps. First, PCR between U44 and U81 snoRNA sequences (using primers 151, 5′-GATGATAGCAAATGCTGAC, and 150, 5′-AGTAATCAGTGAGAGAGTTCAAG) was performed on HeLa genomic DNA, and the product was cloned into the pCR2.1 TA vector (Invitrogen). Second, primer extension using a primer complementary to human exon 11 (184, 5′-TTTCAAGCAGTAAGCTGCATGC) was used to generate a cDNA from oligo(dT) (Boehringer Mannheim)-selected RNA from HeLa cells treated with 20 μg of cycloheximide/ml for 12 h. The cDNA was extended at its 3′ end with dATP (200 μM) by using terminal deoxynucleotidyltransferase (Gibco BRL); PCR was performed using an oligo(dT) primer and the 184 oligonucleotide. PCR was then performed between exon 2 and U44 using primer 190 (5′-CCTGTGAGGTATGGTGCTGG) and primer 152 (5′-GTCAGCATTTGCTATCATC). To obtain the 5′ and 3′ sequences of the human gas5 gene, cDNAs were generated with either primer 184 (and subsequently tailed with dTTP) or an oligo(dT) primer, respectively, and PCR was then performed with an oligo(dA) primer and primer 184 (for the 5′ end) or oligo(dT) primer and primer 190 (for the 3′ end). (The presence of the 5′TOP sequence necessitated tailing the cDNA with dT instead of dA since the oligo(dT) primer was found to base pair within the 5′TOP cDNA sequence during the PCR.) Finally, PCR was performed on HeLa genomic DNA between exons 1 (200, 5′-GCTTTTTTCGAGGTAGGAGTCG) and 2 (201, 5′-CTGTCCATAAGGTGCTATCC) and exons 11 (202, 5′-GCATGCAGCTTACTGCTTG) and 12 (199, 5′-CTAGCTTGGGTGAGGCAAGAC) to obtain complete intron 1 (U74) and intron 11 (U81) sequences, respectively.
RNA isolation and Northern analysis.
Nuclear RNA for Northern analysis of snoRNAs was isolated from NIH 3T3 cells which were washed twice with cold phosphate-buffered saline (PBS) and sonicated three times with a Branson sonicator on setting 3 for 30 sec in NET-2 buffer containing 400 mM NaCl. Extracts were centrifuged in a Beckman SS-34 rotor for 10 min at 10,000 rpm. The supernatant was subjected to immunoprecipitation using anti-fb or anti-Sm antibodies (83) after removal of aliquots for total RNA samples. RNA was isolated by treatment with 7 mM Tris (pH 7.5), 0.7 mM EDTA, 20 mM NaCl, 0.7% sodium dodecyl sulfate and 30 μg of proteinase K (Beckman)/ml for 30 min at 37°C followed by PCA (phenol-chloroform-isoamylalcohol, 50:48:2) extraction (twice) and EtOH precipitation. RNA was electrophoresed on a 6% polyacrylamide gel (total RNA lanes contained ∼1 × 104 cells/lane; anti-fb and anti-Sm RNA lanes contained ∼2 × 105 cells/lane), transferred to a Zeta-probe (Bio-Rad) membrane, and hybridized using oligonucleotides complementary to predicted snoRNA sequences (U74, 5′-TCAGTTGTCCCTACCAACATAGCCT; U75, 5′-TCTGTCCACTACTCTCATACCATCA; U76, 5′-TCAAGAGTAGCAAATATGATGTCATC; intron 4, 5′-TCCTCAGATACGCAGAAACAATG and 5′-GACTTCAGATCTCCCACCCACTCCT; U44, 5′-TCAGATAGAGCTAATAAGAT; U78, 5′-TCAGCTCAGACATTTGATCAACATC; U79, 5′-TCTTATTCAGAGAGATTCCCA; U80, 5′-GATACATCAGATAGGAGCGAAAGAC; U47, 5′-TCATTTGGCAGAATCATCTACATC; and U81, 5′-AGTAATCAGTGAGAGAGTTCAAG). Northern blots of HeLa and mouse total RNA and anti-fb RNA were probed by using an oligonucleotide complementary to U77, 5′-AATCTGCTGAACTATGCAACCATCA. All probes were 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Pharmacia).
Cellular RNA for Northern analyses of spliced gas5 RNA was obtained from NIH 3T3 cells washed in PBS and resuspended in a solution containing 20 mM HEPES (pH 7.9), 4 mM MgCl2, 500 mM KCl, 0.5% Nonidet P-40, 5 mM dithiothreitol, and 400 U RNasin (Boehringer Mannheim). Samples were then centrifuged in a Beckman SS-34 rotor for 6 min at 12,000 rpm. For cycloheximide, serum starvation, and density arrest time courses, total RNA was treated with proteinase K and extracted with PCA (as described above) and then fractionated on 1% agarose gels containing 2.2 M formaldehyde (17 μg of RNA/lane for cycloheximide and serum starvation time courses, 4 μg of RNA/lane for pactamycin and rapamycin time courses, and 10 μg of RNA/lane for density arrest time course), transferred to a Zeta-probe membrane (Bio-Rad), and probed for gas5 and UHG spliced RNAs, rpS16 and β-actin mRNAs, and U75 snoRNA. gas5, UHG, and rpS16 probes were made by Klenow (Promega) filling of annealed oligonucleotides (mouse gas5, 5′-CCTTTCGGAGCTGTGCGGCATTCTGAGCAGGAATGGCAGTGTGGACCTCTGTGATGGGACATCTTGTGGGAT and 5′-GCTTCCTGACGAGTCCTCGTAAGCCTTCATCCTCCTTTGCCACAGAACTGGCTGTGAGATCCCACAAGATGTCC; mouse UHG, 5′-TTTCCTTGTTCGGGGTTTGAGGTGCCACCTTACAAAAGGATGGGTGTACGCTCTCTTTTCAG AATGTGGTTC and 5′-CGTGTTATTTGTAAAATTGAACAGGCCTGGCTCCAAAGTGTAAAGGGCTTTGAGATGAACCACATTCTGAAA; and mouse rpS16, 5′-CGCGGCGCTGCGGTGTGGAGCTCGTGCTTGTGCTCGGAGCTATGCCGTCCAAGGGTCCGCTGCAGTCCGTGCAGGTCTTCGGACGC AAGAAAACTCTCGCTGTGGCCCACTGCA and 5′-AGCAAAACAGGCTCCAGTAAGTTGTACTGCAGCGCGCGCGGCTCGATCATCTCCAGGGGACGTCCGTTCACCTTGATGAGCCCATTTCCCCGTTTCCAGTCCGCCA CAGCGAG) in 10 μl of 50 mM Tris (pH 7.5) and 10 mM MgCl2, 0.5 U of Klenow fragment (Promega) and 50 μCi of [α-32P]dCTP, and 50 μCi of [α-32P]TTP. β-Actin (human), 18S (Xenopus), and 28S (Xenopus) probes were made by random priming of BamHI-NcoI-digested pCITE-pβAct (a generous gift from George Farr), PstI-BamHI-digested pxlrDNA (a generous gift from Barbara Sollner-Webb), and BamHI-EcoRI-digested pxlrDNA, respectively, using Prim-a-Gene (Promega) and 50 μCi of [α-32P]dATP. The data shown in Fig. 5 are representitive of two independent experiments and were quantitated by PhosphorImager analysis. For all but the density arrest time course, the levels of U75 are approximately equivalent, providing an internal control verifying that equal amounts of RNA were indeed loaded in each lane. The increase in U75 during density arrest is discussed in the text.
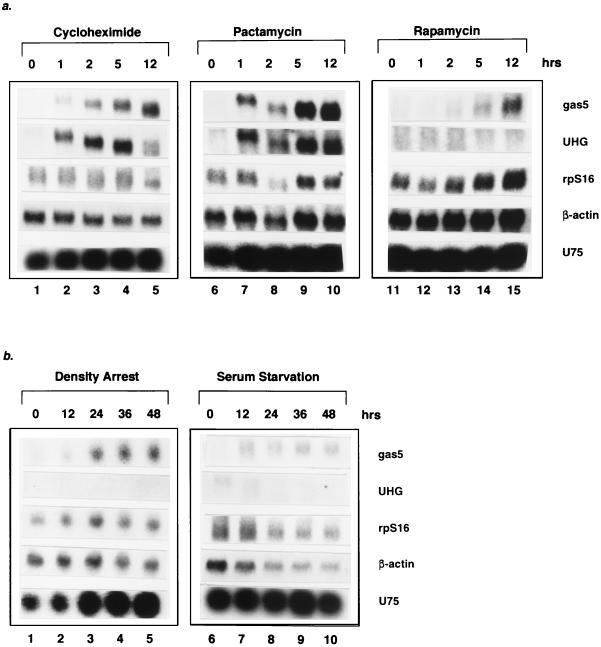
FIG. 5.
Accumulation of spliced gas5 RNA in (a) translation-inhibited or (b) growth-arrested cells. Northern analysis, using probes for gas5 and UHG spliced transcripts as well as U75 snoRNA, β-actin, and rpS16 mRNAs, was performed on total RNA isolated from NIH 3T3 cells treated with 20 μg of cycloheximide/ml (lanes 1 to 5), 280 ng of pactamycin/ml (lanes 6 to 10) or 20 ng of rapamycin/ml (lanes 11 to 15) for 0 (untreated), 1, 2, 5, and 12 h (a) or growth arrested by either contact inhibition (lanes 1 to 5) or serum starvation (lanes 6 to 10) for 0 (growing), 12, 24, 36, and 48 h (b). For each timecourse, equal amounts of RNA were loaded in the lanes (the internal controls show that lane 8 in panel a was underloaded). The progressive increase in mobility of the gas5 and UHG transcripts upon cycloheximide and pactamycin treatment may reflect poly(A) tail shortening.
Mapping the gas5 transcription start site.
NIH 3T3 cells (∼20 × 106) were treated with 20 μg of cycloheximide/ml for 2 h, and cellular RNA was obtained as described above. Following EtOH precipitation, mRNA was isolated using oligo(dT) beads (Boehringer Mannheim) and used in primer extension reactions (89) with a [γ-32P]ATP-5′-end-labeled primer complementary to gas5 exon 1 (172, 5′-CTGCTCAGAATGCCGCAC). The dideoxy sequencing ladder was created by using Sequenase (U.S. Biochemicals) on the plasmid containing the gas5 promoter (see above).
Cell culture, growth arrest, and inhibition of translation.
Mouse embryo NIH 3T3 cells (American Type Culture Collection) were maintained at 37°C in monolayer in Dulbecco’s modified Eagle medium supplemented with 10% calf serum (Gibco BRL), 0.4 mM glutamine, and 1 mg of penicillin-streptomycin/ml. Cells were seeded at low density and split when they were 60 to 80% confluent. For serum starvation experiments, cells were washed twice with PBS, and media containing 0.5% calf serum and 0.4 mM glutamine (but no penicillin-streptomycin) were added. For density arrest experiments, cells were grown to confluency and incubated an additional 12 to 24 h. For inhibition of translation experiments, cells were treated with 20 μg of cycloheximide (Sigma)/ml, 280 ng of pactamycin (National Cancer Institute)/ml, or 20 ng of rapamycin (Sigma)/ml for 0 to 12 h. RNA was isolated as described above.
Sucrose gradients.
Cell extracts were prepared as described above for cellular RNA from NIH 3T3 cells which were growing (∼60 × 106 cells), serum starved (∼45 × 106 cells) for 12 h, treated with cycloheximide for 12 h (∼15 × 106 cells), or incubated at confluency for 24 h (∼15 × 106 cells). Sucrose gradients were made with 10 and 50% sucrose (Sigma) in a solution containing 10 mM HEPES (pH 7.4), 5 mM MgCl2, and 500 mM KCl by using a Biocomp gradient master. Gradients were centrifuged for 10 h at 32,000 rpm in a Beckman SW41 rotor. Gradients were fractionated by hand (into 15 fractions), and RNA was isolated by treatment with proteinase K-sodium dodecyl sulfate, PCA extraction, and EtOH precipitation as described above. Optical density readings (at 260 nm) were made on fractions prior to RNA isolation. Isolated RNA was then electrophoresed on a 1% formaldehyde-agarose gel, as described above, for 6 h at 120 W, transferred to Zeta-blot (Bio-Rad), and probed for gas5 and UHG spliced RNAs, rpS16 and β-actin mRNAs, and 18S and 28S rRNAs, as described above.
Nucleotide sequence accession numbers.
Sequences of the mouse gas5 snoRNAs have been deposited in GenBank (accession no. AJ224029 to AJ224035).
RESULTS
gas5 encodes 10 box C/D snoRNAs.
During an analysis of antisense snoRNAs predicted to direct 2′-O-methylation of rRNA, we obtained the partial sequence of a novel human box C/D snoRNA (U80) with complementarity to nucleotides 1610 to 1624 of 28S rRNA. A GenBank database search produced a 92% match of this sequence to a region within the ninth intron of the murine gas5 gene (16, 74). Inspection of the two database entries for murine gas5 (exons 1 to 3, GenBank accession no. X67267; exons 4 to 12, GenBank accession no. X67268) revealed that six additional introns contained box C, D′, and D sequences along with regions of complementarity to 18S or 28S rRNAs, indicating that gas5 could be a multi-snoRNA host gene similar to UHG.
To characterize the gas5 gene in its entirety, a region extending from the promoter to the fourth intron was cloned from mouse genomic DNA and sequenced; two more snoRNA sequences, located within the first and third introns, then became apparent. The primary structure of the mouse gas5 gene depicting the positions of the nine intron-encoded snoRNAs—named U74, U75, U76, U44, U78, U79, U80, U47, and U81—is presented in Fig. 1a. Transcription followed by alternative splicing (of the seventh exon) and polyadenylation produced two transcripts containing either 11 or 12 exons (16). The fifth and tenth introns of murine gas5 had been previously reported to contain snoRNA sequences homologous to human U44 and U47, respectively (39). The fourth intron of the mouse gas5 gene contains four box D sequences, but no consensus box C sequence.
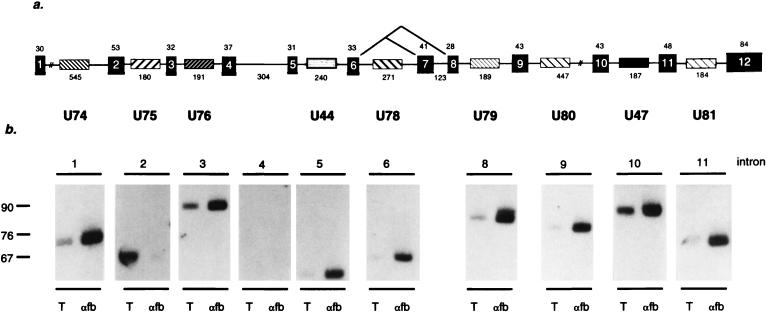
FIG. 1.
gas5 is a multi-snoRNA host gene. (a) Black boxes represent the 12 exons of mouse gas5; cross-hatched boxes represent the snoRNA sequences present within nine of its introns. Numbers above and below the gene are the lengths of exons and introns, respectively. The alternative splicing event that results in the inclusion of the seventh exon is indicated. (b) Northern analyses were performed on mouse total RNA (T) and RNA isolated by immunoprecipitation with anti-fb (αfb) antibodies by using probes derived from intronic sequences of gas5. (The same blot was probed for U75 and U79, indicating that the less-efficient immunoprecipitation of U75 is not due to underloading of the lane.) Each intron except the fourth and seventh produces a stable RNA species between 60 and 90 nucleotides long. The nine mouse gas5 anti-fb immunoprecipitable RNAs are named U74, U75, U76, U44, U78, U79, U80, U47, and U81. The human gas5 gene encodes, in addition to the nine murine snoRNAs, a tenth snoRNA (U77) within intron 4 (data not shown). The sizes of the human exons, in order, are 30, 53, 36, 40, 30, 38, 77 or 38 (alternatively spliced), 54, 30, 23, 48, and 210 nucleotides and introns are 916, 194, 191, 321, 281, 307, 188, 523, 167, 179, and 205 nucleotides.
Genomic and cDNA clones of the human gas5 homologue were obtained by PCR (see Materials and Methods). The genomic organization of human gas5 is very similar to that of the mouse gene: both contain 11 introns and encode the same nine snoRNAs in corresponding introns; however, an additional snoRNA (named U77) is encoded within the fourth intron of the human gene. In addition, each gene possesses a small intron (123 nucleotides in mouse and 167 nucleotides in human) which does not contain a snoRNA-like sequence; in human gas5, the snoRNA-less intron is the ninth intron, while in the mouse gene it is the seventh. By sequencing two human gas5 cDNAs, we found that, like mouse gas5, the human homologue is alternatively spliced. For both genes, the alternative splicing events involve the seventh exon: in the mouse gene there is an alternative 3′ splice acceptor, which serves to include the seventh exon, whereas in the human homologue, an alternative 5′ splice donor is located within the seventh exon (see Fig. 4a).
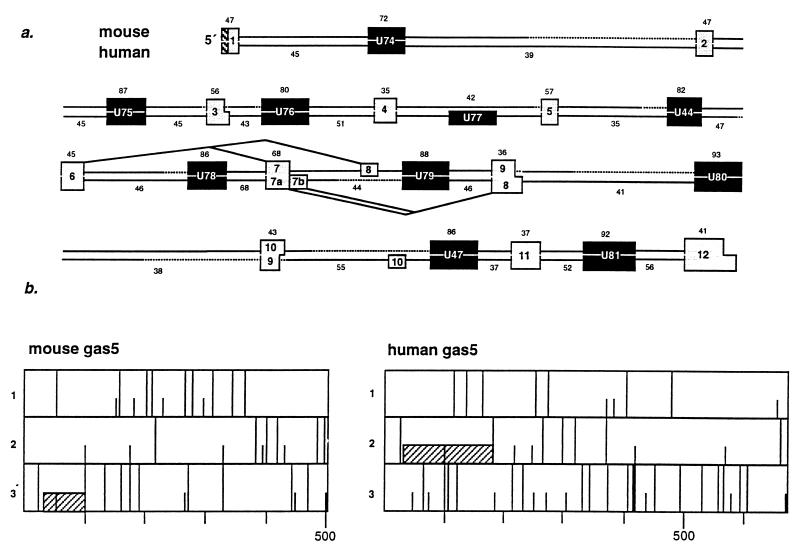
FIG. 4.
gas5 appears to be non-protein coding. (a) Comparison of human and mouse gas5 genes with the Genetics Computer Group software (20) GAP program shows that the only conserved portions are the 5′TOP tract and the intron-encoded snoRNA sequences. Indicated above are the percentages of identity between respective exon (shaded boxes) and snoRNA (filled boxes) sequences; below are the percentages of identity for the portions of intron external to snoRNA sequences (open boxes). The mouse and human gas5 5′TOP sequences (cross-hatched boxes) are 86% identical over seven nucleotides. The dotted line denotes regions in which corresponding introns are not the same length. (b) ORFs of mouse and human spliced gas5 RNAs. Short and long vertical bars represent start and stop codons, respectively, in all three frames. The cross-hatched boxes denote the most likely reading frames. The length in nucleotides of the spliced RNAs is indicated below each diagram.
To demonstrate that stable RNA species are produced from the gas5 introns, Northern analyses were performed. Oligonucleotides complementary to the nine predicted murine snoRNAs were used to probe total RNA and RNA isolated by immunoprecipitation with anti-fb (Fig. 1b) or anti-Sm antibodies (data not shown) from NIH 3T3 cells. As expected, each intron generates a detectable anti-fb immunoprecipitable RNA species. While no stable RNA is generated from the fourth intron of mouse gas5, Northern analysis of HeLa cell RNA confirms that U77 is produced from the fourth intron of the human gene (data not shown). The 10 RNAs range in length from 60 to 85 nucleotides and appear to be of an abundance (∼104 copies/cell) similar to that of other antisense snoRNAs (see reference 51).
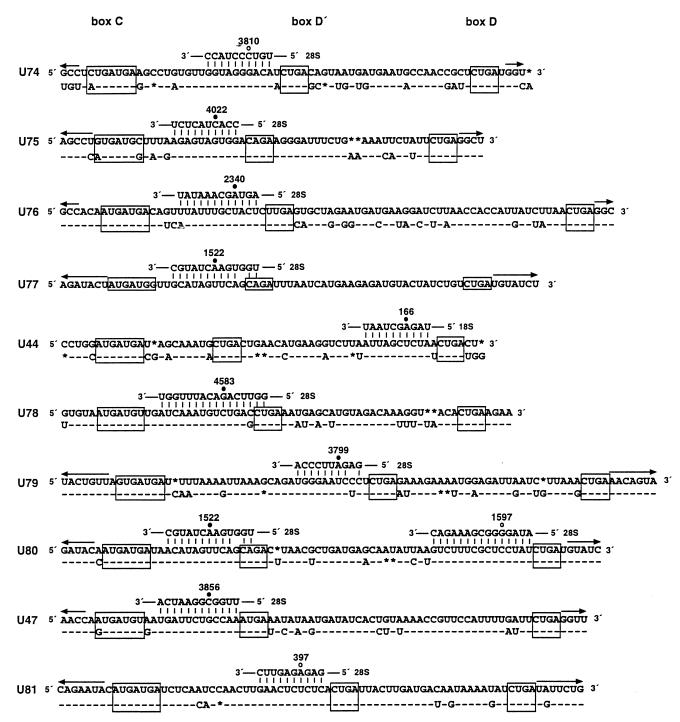
The sequences of human and mouse gas5 snoRNAs are compared in Fig. 2. Each snoRNA possesses boxes C, D′, and D along with extensive complementarity (ranging from 10 to 16 bp) to highly conserved regions of either 18S or 28S rRNA. For 8 of the 10 snoRNAs, reported sites of rRNA ribose methylation (47) reside within these regions of complementarity; we predict that C3810, G1597, and A397 (targeted by U74, U80, and U81, respectively) in 28S rRNA are methylated residues not yet reported. The snoRNA encoded within the ninth intron of gas5, U80, is unusual in that it exhibits two regions of complementarity and therefore has the potential to target two sites in 28S (A1522 and G1597) for methylation; interestingly, the human U77 snoRNA targets the same residue (A1522) for methylation as the upstream site in both human and mouse U80 (Fig. 2). Similar to the other members of this class of snoRNAs, in each known case the methylated rRNA residue is located opposite the snoRNA residue precisely five nucleotides upstream of box D or D′. Since U24 and U25 snoRNAs were previously demonstrated to be required to site-specifically target rRNA for 2′-O-methylation (39, 87), the gas5 snoRNAs are strongly implicated in the 2′-O-methylation of pre-rRNA.
FIG. 2.
Sequences of gas5 intron-encoded snoRNAs. Human sequences above are aligned with mouse homologues below; − and ∗ designate identities and deletions, respectively. Boxes C, D′, and D are outlined. Regions of complementarity to rRNA are presented above the snoRNAs, with the rRNA residues targeted for methylation indicated (47). Closed and open circles indicate reported and predicted sites of rRNA methylation, respectively. The arrows denote terminal base pairing potential.
gas5 contains a 5′TOP sequence.
To precisely map the transcription start site of the gas5 gene, we performed primer extension analysis on oligo(dT)-selected mRNA from NIH 3T3 cells by using a probe to the first exon of mouse gas5. Figure 3a shows that the major transcription start site is located at two adjacent cytidine residues within the sequence TCTCGGCCTTTC; a small fraction of transcripts commences at the guanosine residue preceding the two cytidine residues. The transcription start site(s) is located 29 and 30 nucleotides downstream of the TATA box and 32 and 33 nucleotides upstream of a strong (according to reference 40) translation start site, as shown in Fig. 3b. The first intron is located two nucleotides upstream of this AUG codon. Two human gas5 cDNAs were analyzed by 5′ rapid amplification of cDNA ends and show that the 5′ ends of the human and mouse transcripts are almost identical (CUUUUCG versus CCUUUCG, respectively). The human spliced gas5 RNA also exhibits a short (33 nucleotides) putative 5′ untranslated region (5′UTR).
Recently, a number of genes, including all of those encoding vertebrate ribosomal proteins, were classified as members of the 5′TOP family (for reviews see references 2 and 52). The salient features of this gene class are (i) a tract of 4 to 13 pyrimidines, occasionally interrupted by one or two guanosine residues, surrounding a cytidine transcription start site, and (ii) a short 5′UTR where a translation initiation site conforming to the consensus (40) is located at the first AUG of the message. Since the gas5 transcript in both human and mouse contains a seven-nucleotide 5′ oligopyrimidine tract, a consensus translation start site, and a short 5′UTR, its sequence suggests that it is a member of the 5′TOP gene family.
gas5 appears to be non-protein coding.
Although indirect, the most compelling evidence that UHG does not specify a protein product is that the exons are not conserved between its human and mouse homologues (84). To compare human and mouse gas5 genes, three sequence categories were analyzed: (i) exons, (ii) intron regions in which no snoRNA sequences are located, and (iii) snoRNA-containing sequences. The percentage of identity of each pair of corresponding regions (e.g., human exon 1 with mouse exon 1) is presented in Fig. 4a. Most highly conserved are the first seven nucleotides at the 5′ end of the gas5 transcript (86% identical) and the snoRNA sequences (72 to 93% identical), whereas the average identity between exons and intron regions not encoding snoRNAs is only 49 and 46%, respectively. Interestingly, the second most highly conserved region of gas5 encompasses a site of alternative splicing: the seventh exon and preceding intron are 68% identical (Fig. 4a). Similar to gas5, the 5′ end (first 10 nucleotides) of UHG is conserved in mouse and human (84). Together, these observations suggest that the only functional regions of gas5 and UHG are their 5′TOP sequences and intron-encoded snoRNAs.
While in vitro translation of a gas5 transcript yielded an 8-kDa polypeptide (16), attempts to identify an in vivo protein product of the mouse gas5 gene have been unsuccessful (64a). Presented in Fig. 4b is an analysis of the protein coding potential for mouse and human spliced gas5 RNAs. A strong translation start context (40)—GNNAUGG preceded by a stop codon—surrounds the first AUG codons in both transcripts. Because of the multitude of stop codons, peptides of only 23 amino acids (mouse) or 50 amino acids (human) could be produced from these AUG codons. In the human gas5 transcript, the first ORF is the longest, whereas in the mouse RNA, the longest ORF (39 amino acids) would start at the third AUG codon, which is predicted to be only an adequate translation start site. While the peptides resulting from the first ORFs of the human and mouse transcripts are only 26% homologous, an analysis of a partial rat gas5 clone (64b) reveals that it could produce a 23-amino acid peptide that is 74% identical to the predicted mouse peptide.
Translation inhibition and growth arrest result in accumulation of spliced gas5 RNA.
Despite the unlikelihood that gas5 produces a functional protein product, its spliced RNA is polyadenylated and associated with ribosomes (16). Thus, we suspected that the almost undetectable levels previously reported for the gas5 transcript (15, 16) could be a result of its degradation via a pathway linked to translation, as appears to be the case for UHG (84). Northern analysis was performed on RNA isolated from NIH 3T3 cells treated for up to 12 h with the translation elongation or initiation inhibitors cycloheximide or pactamycin. The results, presented in Fig. 5a, indicate that the low level of gas5 RNA increases dramatically in cells in which translation has been arrested for 12 h (12-fold with cycloheximide, compare lanes 1 and 5; 18-fold with pactamycin, compare lanes 6 and 10). UHG behaves similarly under these conditions (Fig. 5a) (84). In addition, a sevenfold increase in the level of the gas5 spliced RNA was observed when cells were treated with the 5′TOP-specific translation inhibitor rapamycin (compare lanes 11 and 15); in contrast, UHG did not accumulate under these conditions. For each experiment, the levels of β-actin and rpS16 mRNAs, which were examined as controls, did not change dramatically. Since U75 (a snoRNA encoded within gas5; Fig. 5) and U22 (a snoRNA encoded within UHG; data not shown) are abundant, gas5 and UHG are indeed transcribed, but their spliced products are rapidly degraded in a translation-dependent manner.
Increased levels of spliced gas5 RNA have been shown to result from posttranscriptional regulation in growth-arrested cells (15, 16). We performed Northern analyses to assess the gas5 levels in NIH 3T3 cells growth arrested by contact inhibition compared to levels after serum starvation (Fig. 5b, lanes 1 to 5 versus lanes 6 to 10, respectively). As expected, both conditions generate increased levels of spliced gas5 RNA while levels of β-actin and rpS16 mRNAs remain constant or decrease. However, the effects of density arrest and serum starvation are not identical: in density-arrested cells, the level of spliced gas5 RNA increases 5.4-fold and the level of U75 increases 3.0-fold (average of two experiments; compare lanes 1 and 5); when cells are cultured in media containing low levels of serum, the level of spliced gas5 RNA increases 2.5-fold while the level of U75 remains constant (average of two experiments; compare lanes 6 and 10). Under both conditions, the observed increases in spliced gas5 RNA relative to U75 are consistent with previous reports that the gas5 message is regulated posttranscriptionally (15, 16). In contrast to gas5, the level of the UHG message is not increased by either serum starvation or density arrest (Fig. 5b); however, as with U75, the level of U22 (encoded in UHG) increases in density-arrested cells (data not shown).
gas5 spliced RNA shifts into mRNP particles during growth arrest.
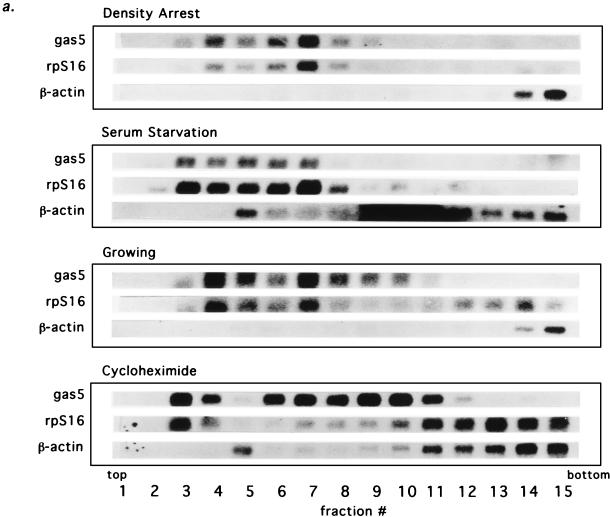
Messenger RNAs belonging to the 5′TOP gene family have been shown to shift from polysomes into submonosomal (or messenger ribonucleoprotein [mRNP]) particles during periods of growth arrest in a process which is reversed when cell growth resumes (reviewed in references 2 and 52). The classification of gas5 as a 5′TOP gene prompted us to ask whether its apparent upregulation in growth-arrested cells is correlated with a shift of its spliced RNA into stable submonosomal particles, where it does not undergo translation and degradation. Using density gradient centrifugation, we compared the distribution of gas5 RNA in extracts from growing NIH 3T3 cells with that of either serum-starved cells or cells whose growth was arrested at confluency. We also examined the gas5 transcript in cells that were growing but in which translation had been halted by cycloheximide treatment. Northern blots of RNA isolated from each gradient fraction were probed for spliced gas5 RNA and for rpS16 and β-actin mRNAs (Fig. 6a); 18S and 28S rRNAs were also analyzed (data not shown) to monitor the migration of 40S and 60S ribosomal subunits, as well as the monosome-polysome region. Quantitated data for the spliced gas5 RNA are shown in Fig. 6b.
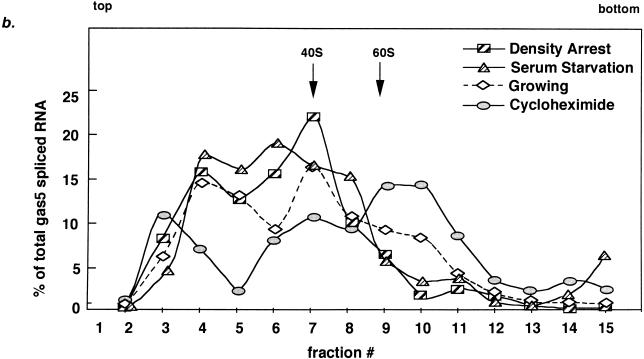
FIG. 6.
Spliced gas5 RNA shifts away from ribosomes during growth arrest. (a) Northern blots of RNA from 10 to 50% sucrose gradient fractions of extracts from NIH 3T3 cells density arrested, serum starved, growing, or treated with cycloheximide. The distributions of spliced gas5 RNA and rpS16 and β-actin mRNAs, along with 18S and 28S rRNAs (data not shown), were analyzed. The top, bottom, and fraction numbers for each gradient are indicated. The dark smear in fractions 9 to 12 of the β-actin profile during serum starvation is an artifact introduced during the blotting procedure; note that this profile otherwise mimics the β-actin profile during cycloheximide treatment. (b) Quantitation of Northern signals (by PhosphorImaging) of spliced gas5 RNA from density-arrested cells, serum-starved cells, growing cells, and cells treated with cycloheximide. The positions of ribosomal small subunits (40S) and large subunits (60S) were determined from the profiles of 18S and 28S rRNAs. For each fraction, the percentage of gas5 was determined with respect to the total amount of gas5 RNA in the gradient. The dotted line indicates that the normalized level of gas5 RNA in growing cells cannot be compared directly to that in density-arrested and cycloheximide-treated cells, since a portion of the spliced gas5 RNA (presumably that associated with ribosomes) has been degraded.
As expected for a 5′TOP transcript, over 90% of the spliced gas5 RNA is located in submonosomal fractions (3 through 7) when cell growth is halted by either density arrest or serum starvation (Fig. 6a and b). Even though the rate of transcription of the gas5 gene may be augmented during density arrest (Fig. 5b), the fraction of gas5 spliced RNA present in the submonosomal fractions is comparable to that observed during serum starvation. In growing cells, the spliced gas5 RNA is located mostly in submonosomal fractions (4 through 7) but is also present in fraction 10, which corresponds to 80S (monosomes) (Fig. 6a and b). In contrast, when elongation is inhibited by cycloheximide, 30% of the spliced gas5 RNA is trapped at approximately 80S, as expected for a message that contains only short ORFs. The rpS16 mRNA behaves comparably to gas5 RNA (Fig. 6a): in density-arrested or serum-starved cells, rpS16 mRNA is found predominantly in the submonosomal region and is absent from polysomes, whereas in growing cells it is distributed between the submonosomal and polysomal fractions (fractions 12 to 15). The non-5′TOP β-actin mRNA remains associated with heavy polysomes in growing and growth-arrested cells (Fig. 6a).
Previous analyses of 5′TOP mRNAs employed gradient conditions that optimally discriminate between polysomes and submonosomal particles (1, 25, 33, 44, 53, 65). For spliced gas5 RNA, which is likely to accommodate only a single ribosome due to its numerous stop codons, it was necessary to devise conditions that could distinguish between monosomes and submonosomal particles. By using 10 to 50% sucrose gradients and extended centrifugation times, we separated the submonosomal region into two distinct peaks. During density-arrested growth, increased levels of both spliced gas5 RNA and rpS16 mRNA are found predominantly in fractions that comigrate with 40S subunits (fraction 7), while levels in lighter mRNP particles (fraction 4) are less affected. Similar observations were made when the distribution of rp mRNAs from differentiated mouse myoblasts or rabbit reticulocyte lysate were analyzed by high-resolution sucrose gradients (27).
DISCUSSION
gas5 is a non-protein-coding multi-snoRNA host gene.
We have identified gas5 as the second member of the UHG class of snoRNA host genes. Ten box C/D snoRNAs (designated U74, U75, U76, U77 [human only], U44, U78, U79, U80, U47, and U81 [Fig. 1 and 2]) are encoded within the 11 introns of the gas5 gene; 8 of the 10 are novel species (U74 to U81). Each snoRNA exhibits extensive complementarity to 18S or 28S rRNA and is predicted to function in 2′-O-methylation of pre-rRNA (13, 39, 87). Alignment of the human and mouse gas5 intron-encoded snoRNAs reveals little variation within the regions exhibiting complementarity to rRNA and the box C, D′, and D sequences (Fig. 2), consistent with evidence suggesting that these are functionally relevant portions of the snoRNAs (13, 39). Considering the overall similarity between human and mouse gas5 genes, it was surprising to find that U77 is encoded within the fourth intron of human gas5 yet is not present in the mouse gene. In human cells, both U77 and U80 appear to be capable of targeting the same residue in 28S rRNA (A1552) for methylation; it is not known, however, whether they participate equally in this process. Since the two residues targeted by U80 (A1552 and G1597) are adjacent in the secondary structure of 28S rRNA (47), it seems that U80 could simultaneously guide methylation of these sites. If this is the case, it is unclear why a redundant mechanism to modify A1552 would be present in humans while absent in mice. (Northern analysis of RNA from NIH 3T3 cells [data not shown] indicates that a U77 homolog is not present in mouse.)
Similar to UHG, sequence comparison of human and mouse gas5 genes indicates that the only regions of conservation are their snoRNAs and 5′ end (Fig. 4a); in addition, the presence of only short ORFs and numerous stop codons suggests that gas5 does not generate a protein product (Fig. 4b). Since both the gas5 exons and non-snoRNA-containing intron segments range from 35 to 68% identity, these regions of the gene appear to have diverged at equivalent rates. (A similar range of identity is found for corresponding introns of mouse and human β-globin and rat and human β-actin genes.) To address whether the secondary structures of the gas5 and UHG spliced transcripts share any common structural features, these RNA sequences were analyzed by MulfFold (31); however, no conserved folds were found.
Recently, two additional non-protein-coding snoRNA host genes, called U17HG and U19HG, have been characterized (references 64 and 7, respectively). Similar to gas5 and UHG, the exon portions of U17HG are not conserved between human and mouse (64). The U17HG and U19HG spliced RNAs are polyadenylated and contain numerous stop codons; however, they do not appear to associate with polysomes (7, 64). Why do the spliced gas5 and UHG RNAs associate with ribosomes if they do not produce functional protein products? Previously, increased levels of UHG spliced RNA were observed after treatment of HeLa cells with translation inhibitors (84). The same response has been observed here for gas5 by using the translation inhibitors cycloheximide, pactamycin, and rapamycin (Fig. 5a). The presence of numerous stop codons in both spliced RNAs (Fig. 4) (84) suggests that nonsense-mediated decay—a process known to require active translation (49)—may be utilized by the cell to dispose of the exon portions of these transcripts. The fate of the nonconserved (and likely nonfunctional) peptide products which could be generated from the short ORFs of gas5 and UHG is not known.
gas5 is a member of the 5′TOP gene family.
The members of the 5′TOP gene family include all ribosomal proteins, as well as protein synthesis elongation factors and a number of genes without apparent ribosome-related functions (e.g., ATP synthase C subunit, nucleoside diphosphate kinase, and hnRNPA1) (see references 2, 11, and 52). Although the number of genes in this family is small, together the 5′TOP mRNAs can comprise >15% of the total mRNA in the cell. 5′TOP genes are classified according to their unusual pyrimidine-rich 5′-terminal sequence and also by whether their mRNAs accumulate in mRNP particles during arrested cell growth. Mapping the 5′ end of the gas5 transcript demonstrates that it commences at a cytidine residue followed by five pyrimidines (Fig. 3). Since gas5 contains sequence characteristics of the 5′TOP gene family, we investigated whether it is present in mRNP particles when cells are serum starved or arrested at confluency. Indeed, when the distribution of the spliced gas5 mRNA was analyzed by sucrose gradient centrifugation, it was found to accumulate in submonosomal fractions during growth arrest (Fig. 6).
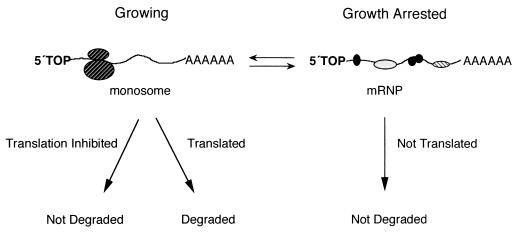
That gas5 is a member of the 5′TOP gene class explains the previously reported posttranscriptional accumulation of its spliced RNA in growth-arrested cells. In growing cells, active translation leads to rapid degradation of the spliced gas5 RNA, whereas inhibition of translation causes the level of gas5 transcript to rise. Likewise in growth-arrested cells, the spliced gas5 RNA accumulates, apparently because it is sequestered in mRNP particles and is not translated. Presented in Fig. 7 is a model which summarizes our findings. The observation of a growth-arrested submonosomal peak at 40S for both rpS16 and gas5 transcripts (Fig. 6) might indicate that these mRNPs can associate with the small ribosomal subunit but not engage the 60S to form active ribosomes. It is important to note that a principal difference between the regulation of spliced gas5 RNA and the protein-coding 5′TOP mRNAs is that translation of the latter does not result in rapid degradation.
FIG. 7.
Why is a non-protein-coding snoRNA host gene a growth-arrest-specific transcript? In growing cells, spliced gas5 RNA is translated and consequently degraded. When translation is inhibited, the levels of gas5 increase. Likewise, when cell growth is arrested, gas5 RNA shifts from monosomes into submonosomal particles, where it is sequestered from active ribosomes and therefore accumulates.
An additional characteristic of 5′TOP mRNAs is that their translation is specifically inhibited by the immunosuppressant rapamycin (32). Rapamycin has been shown to inhibit phosphorylation p70S6k, which in turn prevents phosphorylation of S6, possibly resulting in decreased affinity of 5′TOP mRNAs for the translation machinery (see reference 52). As expected, treatment of NIH 3T3 cells with rapamycin results in an increase in the level of gas5 spliced RNA, presumably because it not translated and degraded (Fig. 5a). Thus, it appears that degradation of the gas5 spliced RNA may be regulated through the p70S6k signal transduction cascade. According to our model, spliced gas5 RNA is expected to accumulate in nondividing cells. Indeed, an analysis of adult mouse tissues demonstrated that the gas5 transcript is present at high levels in brain, where the majority of cells are not dividing, and is present at low levels in liver, where cells are continuously dividing (16). An intriguing question is whether regulation of the spliced gas5 RNA through the p70S6k signalling pathway and its accumulation in certain tissues plays some unknown cellular role or whether the gas5 gene merely serves as a vehicle for the production of its intron-encoded snoRNAs.
In contrast to gas5, UHG does not accumulate in density-arrested or serum-starved cells and it is not sensitive to rapamycin (Fig. 5a and b). A number of studies have demonstrated that the 5′TOP sequences, along with adjacent downstream regions, are required to shift transcripts into submonosomal particles (3, 27, 42, 50). Since insertion of a single adenosine residue into a 5′ oligopyrimidine tract abrogates this shift (3, 42), the UHG transcript may not accumulate during growth arrest because its 5′-end sequence contains an adenosine residue at position +4. However, with exception of this adenosine, UHG contains an oligopyrimidine tract which is conserved between human and mouse. Interestingly, we observe modest increases in the levels of both U75 (encoded within gas5; Fig. 5b) and U22 (encoded within UHG; data not shown) when cells are density arrested, suggesting that these genes may be transcribed in a similar growth-dependent manner. However, we cannot exclude the unlikely possibility that snoRNAs are stabilized under these conditions.
snoRNA host genes possess characteristics of the 5′TOP gene family.
Strikingly, inspection of known snoRNA host genes reveals that they all exhibit characteristics of the 5′TOP gene class. Presented in Table 1 are transcription start sequences of host genes along with their corresponding snoRNAs. In contrast to the non-protein-coding host genes (gas5, UHG, U17HG, and U19HG), most host genes generate protein products in addition to their snoRNAs. Many snoRNAs are encoded within introns of ribosomal protein genes while others are found within introns of genes specifying translation factors or nucleolar proteins. The heat shock cognate 70 protein has been reported to localize to the nucleolus during heat shock (76), and the laminin binding protein/p40 has been reported to associate with ribosomes (70, 71). A ribosome-related function is not, however, obvious for the protein products of the ATP synthase β (56) and Q1Z7F5 genes (88). Despite their variety of functions, all snoRNA host genes possess at least some of the distinctive characteristics of 5′TOP genes: a 5′ oligopyrimidine tract, a cytidine transcription start site, and a short 5′UTR. The hsc70 transcript does not contain a strong oligopyrimidine tract—yet begins at a cytidine residue—and the eukaryotic initiation factors and ATP synthase β messages are reported to start with guanosine and adenosine residues, respectively (56, 60, 69), but these are located within oligopyrimidine sequences (Table 1).
TABLE 1.
snoRNA host genes contain 5′TOP sequences
| Host gene or protein | 5′TOP sequence (−5 to +10) | snoRNA | GenBank accession no. | Reference or source |
|---|---|---|---|---|
| gas5 | CTCGGCCTTTCGGAGab | U74–81, U44, U47 | X67267 | This work; 16 |
| UHG | TCGTTCTCATTTTTC | U22, U25–31 | U40580 | 84 |
| U17HGc | TCTCTCCTTTTTGGAb | U17 | D00591 | 64 |
| U19HG | CTGCGCCCTG | U19 | AJ224166 | 7 |
| rpS3 | TCCTTTCCTTd | U15 | D28344 | 35 |
| rpL1a | TTTCTCTTCCGTGGCa | U16, U18 | X06552 | 45 |
| rpS8 | GGTTTCTCTTTCCAG | U38–U40e | X67247 | M. Fried, direct submission |
| rpL5 | GCGGCCTTTTCCCCCa | U21 | D10737 | 36 |
| rpL7a | CCTTTCTCTCTCCTC | U24, U36ac | X61923 | 17 |
| rpL13a | TCCTCCTTTCCCAGGa | U32–U35 | X51528 | C. Sibille, direct submission |
| eIF4AI | ACTCCGCCCTAGATTa | U67 | M22873 | 69 |
| eIF4AII | CGCCTGTCTTTTCAGa | E3 | X14422 | 60 |
| EF-2 | GCGGTCTCTTCCGCCa | U37 | J03200 | 55 |
| EF-1β | CTTTTTCCTC | U51 | D28350 | 35 |
| Nucleolin | GCTGGCTTCGGGTGT | U20 | M60858 | 78 |
| hsc70 | GAAACCGGTGCTCAG | U14 | Y00371 | 21 |
| Laminin binding protein | CTCGACTTTCTTTGC | E2 | U43901 | 30 |
| ATP synthase β | CCTTCAGTCTCCACC | U59 | M27132 | 56 |
| Q1Z7F5 | AGCGCCTCTTTCCCT | U70 | M81806 | 88 |
Oligopyrimidine tracts are underlined. Transcription start sites (+1) are in boldface. Sequences are from human except those for Xenopus rpL1a, chicken rpL5, mouse rpL13a and eIF4A(I and II), hamster EF-2, and mouse gas5.
Transcription of gas5 and U17HG starts at adjacent C residues.
U17HG is located upstream of the RCC1 gene (64).
The thymidine transcription start site assigned for rpS3 cDNA may not reflect genomic sequence (35).
Two observations suggest that the 5′TOP sequences of snoRNA host genes play a role in addition to that in translational regulation. First, while all snoRNA host genes contain characteristics of the 5′TOP gene family (Table 1), they are not all translationally regulated in a 5′TOP growth-dependent manner. For example, eIF4AI and eIF4AII have been shown not to shift into mRNPs during growth arrest (28). Second, UHG possesses an absolutely conserved pyrimidine-rich 5′-end sequence but does not appear to be translationally regulated in the way gas5 is regulated. Since all snoRNA host transcripts release their snoRNAs in the nucleus, we suspect that a second role of 5′TOP sequences likely involves some nuclear process. One possibility is that 5′ oligopyrimidine tracts participate in the regulation of transcription of snoRNA-containing genes, balancing the synthesis of translation-associated components with the machinery that generates the ribosome. Alternatively, the pyrimidine-rich transcription start sites of snoRNA host genes could function by altering the composition of transcription initiation complexes to include factors that later assist in the splicing and release of snoRNAs. Examples of such communication between the transcription and mRNA processing machineries have been reported recently: splicing factors have been found to interact with the C-terminal domain of RNA polymerase II (reviewed in references 18 and 79) and the polyadenylation factor CPSF has been identified as a component of the TFIID transcription initiation complex (19). Our finding that all known snoRNA host genes contain 5′TOP sequences represents an additional step toward understanding the growth-dependent regulation of the protein synthesis machinery in vertebrate cells.
ACKNOWLEDGMENTS
We are grateful to Mei-Di Shu, Jahan Moslehi, and Zoe Bellows for their technical assistance and Timothy McConnell, Leo Otake, and Kazio Tycowski for their comments on the manuscript. We also thank Lennart Philipson, Tamas Kiss, and Witold Filipowicz (with Pawel Pelczar) for sharing unpublished data and Kazio Tycowski for his many helpful insights.
This work was supported by NIH grant GM-26154.
REFERENCES
- 1.Agrawal M G, Bowman L H. Transcriptional and translational regulation of ribosomal protein formation during mouse myoblast differentiation. J Biol Chem. 1987;262:4868–4875. [PubMed] [Google Scholar]
- 2.Amaldi F, Pierandrei-Amaldi P. TOP genes: a translationaly controlled class of genes including those coding for ribosomal proteins. In: Jeanteur P, editor. Progress in molecular and subcellular biology. Vol. 18. Berlin, Germany: Springer-Verlag; 1997. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 3.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Bally M, Hughes J, Cesareni G. SnR30: a new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:5291–5303. doi: 10.1093/nar/16.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bortolin M L, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli C, Henry Y, Gelunge J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4769–4775. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffarelli E, Fatica A, Prislei S, DeGregorio E, Frangepane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho-Vanegas O, Weighardt F, Ghigna C, Amaldi F, Riva S, Biamonti G. Growth-dependent and growth-independent translation of messengers for heterogeneous nuclear ribonucleoproteins. Nucleic Acids Res. 1997;25:3950–3954. doi: 10.1093/nar/25.19.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavaille J, Bachellerie J-P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 13.Cavaille J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 14.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of RNase MRP is required for normal processing of the ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciccarelli C, Philipson L, Sorrentino V. Regulation of expression of growth arrest-specific genes in mouse fibroblasts. Mol Cell Biol. 1990;10:1525–1529. doi: 10.1128/mcb.10.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coccia E M, Cicala C, Charlesworth C, Ciccarelli C, Rossi G B, Philipson L, Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo P, Yon J, Fried M. The organization and expression of the human L7a ribosomal protein gene. Biochim Biophys Acta. 1991;1129:93–95. doi: 10.1016/0167-4781(91)90218-b. [DOI] [PubMed] [Google Scholar]
- 18.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 19.Dantonel J-C, Murthy K G K, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 20.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dworniczak B, Mirault M E. Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate’ protein. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichler D C, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1995;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 23.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in pre-ribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 24.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 25.Geyer P K, Meyuhas O, Perry R P, Johnson L F. Regulation of ribosomal protein mRNA content and translation in growth-stimulated mouse fibroblasts. Mol Cell Biol. 1982;2:685–693. doi: 10.1128/mcb.2.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadjiolov A A. The nucleolus and ribosome biogenesis. Vienna, Austria: Springer-Verlag; 1985. [Google Scholar]
- 27.Hammond M L, Merrick W, Bowman L H. Sequences mediating the translation of mouse S16 ribosomal protein mRNA during myoblast differentiation and in vitro and possible control points for the in vitro translation. Genes Dev. 1991;5:1723–1736. doi: 10.1101/gad.5.9.1723. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, Hershey J W B. Translational initiation factor expression and ribosomal protein gene expression are repressed coordinately but by different mechanisms in murine lymphosarcoma cells treated with glucocorticoids. Mol Cell Biol. 1989;9:3679–3684. doi: 10.1128/mcb.9.9.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes J M X, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external spacer of yeast pre-ribosomal RNA and prevents formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackers P, Minoletti F, Belotti D, Clausse N, Sozzi G, Sobel M E, Castronovo V. Isolation from a multigene family of the active human gene of the metastasis-associated multifunctional protein 37LRP/p40 at chromosome 3p21.3. Oncogene. 1996;13:495–503. [PubMed] [Google Scholar]
- 31.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 32.Jeffries H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70S6k. EMBO J. 1997;12:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaspar R L R, White M W, Rhoads R E, Morris D R. Simultaneous cytoplasmic redistribution of ribosomal protein L32 mRNA and phosphorylation of eukaryotic initiation factor 4E after mitogenic stimulation of Swiss 3T3 cells. J Biol Chem. 1990;265:3619–3622. [PubMed] [Google Scholar]
- 34.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 35.Kato S, Sekine S, Oh S-W, Kim N-S, Umezawa Y, Abe N, Yokoyama-Kobayashi M, Aoki T. Construction of a human full-length cDNA bank. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 36.Kenmochi N, Maeda N, Tanaka T. The structure and complete sequence of the gene encoding chicken ribosomal protein L5. Gene. 1992;119:215–219. doi: 10.1016/0378-1119(92)90274-s. [DOI] [PubMed] [Google Scholar]
- 37.Kiss T, Bortolin M-L, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 39.Kiss-Laszlo Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of pre-ribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 41.Leader D J, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy S, Avni D, Hariharan N, Perry R P, Meyuhas O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang W Q, Fournier M J. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- 44.Loreni F, Amaldi F. Translational regulation of ribosomal protein synthesis in Xenopus cultured cells: mRNA relocation between polysomes and RNP during nutritional shifts. Eur J Biochem. 1992;205:1027–1032. doi: 10.1111/j.1432-1033.1992.tb16870.x. [DOI] [PubMed] [Google Scholar]
- 45.Loreni F, Ruberti I, Bozzoni I, Pierandrei-Amaldi P, Amaldi F. Nucleotide sequence of the L1 ribosomal protein gene of Xenopus laevis: remarkable homology among introns. EMBO J. 1985;4:3483–3488. doi: 10.1002/j.1460-2075.1985.tb04107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maden B E. Guides to 95 new angles. Nature. 1997;389:129–131. doi: 10.1038/38134. [DOI] [PubMed] [Google Scholar]
- 47.Maden B E H. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acids Res. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 48.Maden T. Click here for methylation. Nature. 1996;383:675–676. doi: 10.1038/383675a0. [DOI] [PubMed] [Google Scholar]
- 49.Maquat L. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 50.Mariottini P, Amaldi F. The 5′ untranslated region of mRNA for ribosomal protein S19 is involved in its translational regulation during Xenopus development. Mol Cell Biol. 1990;10:816–822. doi: 10.1128/mcb.10.2.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 52.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 53.Meyuhas O, Thompson E A, Perry R P. Glucocorticoids selectively inhibit translation of ribosomal protein mRNAs in P1798 lymphosarcoma cells. Mol Cell Biol. 1987;7:2691–2699. doi: 10.1128/mcb.7.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakanishi T, Kohno K, Ishiura M, Ohashi H, Uchida T. Complete nucleotide sequence and characterization of the 5′-flanking region of mammalian elongation factor 2 gene. J Biol Chem. 1988;263:6384–6391. [PubMed] [Google Scholar]
- 56.Neckelmann N, Warner C K, Chung A, Kudoh J, Minosha S, Fukuyama R, Maekawa M, Shimizu Y, Shimizu N, Liu J D, Wallace D C. The human ATP synthase β subunit gene: sequence analysis, chromosome assignment, and differential expression. Genomics. 1989;5:829–843. doi: 10.1016/0888-7543(89)90125-0. [DOI] [PubMed] [Google Scholar]
- 57.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 58.Nicoloso M, Caizergues-Ferrer M, Michot B, Azum M C, Bachellerie J-P. U20, a novel small nucleolar RNA, is encoded in an intron of the nucleolin gene in mammals. Mol Cell Biol. 1994;14:5766–5776. doi: 10.1128/mcb.14.9.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen P J, Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Brien C A, Wolin S L. A possible role for the 60-kDa Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- 62.Peculis B. RNA processing: pocket guides to ribosomal RNA. Curr Biol. 1997;7:480–482. doi: 10.1016/s0960-9822(06)00242-9. [DOI] [PubMed] [Google Scholar]
- 63.Peculis B A, Steitz J A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 64.Pelczar P, Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol Cell Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.Philipson, L. Personal communication.
- 64b.Philipson, L. Unpublished results.
- 65.Pierandrei-Amaldi P, Beccari E, Bozzoni I, Amaldi F. Ribosomal protein production in normal and anucleoate Xenopus embryos: regulation at the posttranscriptional and translational levels. Cell. 1985;42:317–323. doi: 10.1016/s0092-8674(85)80127-6. [DOI] [PubMed] [Google Scholar]
- 66.Prislei S, Michienzi A, Presutti C, Fragapane P, Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993;21:5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qu L H, Henry Y, Nicoloso M, Michot B, Azum M C, Renalier M H, Caizergues F M, Bachellerie J-P. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qu L H, Nicoloso M, Michot B, Azum M C, Caizergues F M, Renalier M H, Bachellerie J-P. U21, a novel small nucleolar RNA with a 13 nt complementarity to 28S rRNA, is encoded in an intron of ribosomal protein L5 gene in chicken and mammals. Nucleic Acids Res. 1994;22:4073–4081. doi: 10.1093/nar/22.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddy N S, Roth W W, Bragg P W, Wahba A J. Isolation and mapping of a gene for protein synthesis initiation factor 4A and its expression during differentiation of murine erythroleukemia cells. Gene. 1988;70:231–243. doi: 10.1016/0378-1119(88)90195-3. [DOI] [PubMed] [Google Scholar]
- 70.Rosenthal E T, Wordeman L. A protein similar to the 67 kDa laminin binding protein and p40 is probably a component of the translation machinery in Urechis caupo oocytes and embryos. J Cell Sci. 1995;108:245–256. doi: 10.1242/jcs.108.1.245. [DOI] [PubMed] [Google Scholar]
- 71.Satoh K, Narumi K, Sakai T, Abe T, Kikuchi T, Matsushima K, Sindoh S, Motomiya M. Cloning of 67-kDa laminin receptor cDNA and gene expression in normal and malignant cell lines of the human lung. Cancer Lett. 1992;62:199–203. doi: 10.1016/0304-3835(92)90096-e. [DOI] [PubMed] [Google Scholar]
- 72.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmitt M E, Clayton D A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider C, King R M, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 75.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 76.Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993;75:403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- 77.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA processing in eukaryotes. In: Zimmerman R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 469–490. [Google Scholar]
- 78.Srivastava M, McBride O W, Fleming P J, Pollard H B, Burns A L. Genomic organization and chromosomal localization of the human nucleolin gene. J Biol Chem. 1990;265:14922–14931. [PubMed] [Google Scholar]
- 79.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 80.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tollervey D, Guthrie C. Deletion of a yeast small nucleolar RNA gene impairs growth. EMBO J. 1985;4:3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;3:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 83.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tycowski K T, Shu M, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 85.Tycowski K T, Shu M-D, Steitz J A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 86.Tycowski K T, Shu M-D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 87.Tycowski K T, Smith C M, Shu M, Steitz J A. A small nucleolar RNA required for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van den Ouweland A M W, Kioschis P, Verdijk M, Tamanini F, Toniolo D, Poustka A, Van Oost B A. Identification and characterization of a new gene in the human Xq28 region. Hum Mol Genet. 1992;1:269–273. doi: 10.1093/hmg/1.4.269. [DOI] [PubMed] [Google Scholar]
- 89.Wyatt J R, Sontheimer E J, Steitz J A. Site-specific crosslinking of mammalian snRNPs to the 5′ splice site prior to the first step of pre-messenger RNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]