INTRODUCTION
The worldwide spread of antibiotic-resistant bacteria, collectively called the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp) pathogens, has become a major public health concern because of the shortage of effective antimicrobial agents available for treatment.1 Increasing resistance to β-lactams (penicillins, cephems, monobactams, and carbapenems), fluoroquinolones, and aminoglycosides has become a serious clinical threat because of their heavy use in the treatment of gram-negative infections. Gram-negative bacteria have developed resistance to b-lactams, aminoglycosides, and fluoroquinolones through the production of various β-lactamases, aminoglycoside-modifying enzymes (AMEs)/16S ribosomal RNA (rRNA) methyltransferases (MTases), and substituting key amino acid residues in the QRDRs (quinolone resistance–determining regions) of DNA gyrase (GyrA) and topoisomerase IV (ParC), respectively.2
In the 1980s, the use of aminoglycosides became increasingly avoided because of ototoxicity and nephrotoxicity, and was subsequently replaced with β-lactams and fluoroquinolones, which had less toxicity and broader antibacterial spectra. However, with the rapid increase of β-lactam–resistant and fluoroquinolone-resistant bacteria, the clinical usefulness of aminoglycosides has now been revisited, together with an improvement in their safety through optimized dosing regimens, as an effective choice in the combined drug therapy against a range of resistant gram-negative bacterial infections.3
Streptomycin, produced by Streptomyces griseus, was the first clinically introduced aminoglycoside, reported by Jones and colleagues4 in 1944, followed by neomycin, which was discovered from Streptomyces fradiae by Waksman and Lechevalier in 1949.5 In 1957, Umezawa and colleagues6 reported kanamycin from Streptomyces kanamyceticus, which proved to be effective in treating tuberculosis. Subsequently, gentamicin (1963) and tobramycin (1967) were identified from soil Actinomycetes.7 Kasugamycin, composed of an inositol, an amino sugar, and an amidine carboxylic acid, was also discovered by Umezawa and colleagues,8 in the 1960s, and this aminoglycoside was used in large amounts in agriculture to treat and prevent rice blast, but not in humans because of its toxicity. Semisynthetic aminoglycosides, such as amikacin (1972, kanamycin A–based), arbekacin (1973, kanamycin B/dibekacin-based), and isepamicin (1977, gentamicin B–based), which have potent activity against both gram-negative and gram-positive bacteria, were further developed.7
Clinically available aminoglycosides are structurally classified into 2 major classes: those with a 2-deoxystreptamine (2-DOS) core moiety and those without (eg, streptomycin) (Fig. 1). In addition, aminoglycosides with a 2-DOS core moiety are divided into subgroups, 4,5-disubstituted 2-DOS (neomycin, ribostamycin, paromomycin) and 4,6-disubstituted 2-DOS (kanamycin, gentamicin, tobramycin, amikacin, arbekacin, isepamicin), based on the substituent linkage position (see Fig. 1).
Fig. 1.

Core elements of aminoglycosides and aminoglycoside structures.
ACTION OF AMINOGLYCOSIDES AND AMINOGLYCOSIDE RESISTANCE MECHANISMS
Aminoglycosides with a 2-DOS core primarily bind to helix 44 of the 16S rRNA comprising bacterial 30S ribosomal subunits. Aminoglycoside binding causes various disruptions in protein synthesis: disturbing transfer RNA (tRNA) translocation, lowering translational fidelity, interfering with the ribosome subunit mobility, disturbing ribosome recycling, and interfering with the formation of intersubunit bridges.9–14 Aminoglycosides also bind to, and may disturb protein synthesis at, helix 69 of the 23S rRNA in 50S ribosomal subunits.12,14,15
Bacteria resist aminoglycosides through a variety of intrinsic and acquired resistance mechanisms.16 Base mutations within the A site of 16S rRNA, amino acid substitutions in ribosomal proteins, and activated efflux pumps are classic intrinsic aminoglycoside resistance mechanisms in pathogenic bacteria. Production of AMEs, either intrinsic or acquired, is the most common aminoglycoside resistance mechanism. AMEs are divided into 3 groups: acetyltransferase (AAC), phosphotransferase (APH), and adenylyltransferase (AAD or ANT). These AMEs modify NH3 or OH groups at several positions in aminoglycosides, using cofactors, acetyl–coenzyme A or ATP, thereby deactivating them. The most clinically significant resistance mechanism is acquired 16S rRNA methyltransferase (MTase), because these confer high-level and broad-spectrum aminoglycoside resistance by adding a CH3 group to specific residues within the A site of 16S rRNA using S-adenosylmethionine (SAM) as the cofactor (Fig. 2). The binding affinity of certain aminoglycosides to the CH3-added 16S rRNA is predicted to be significantly reduced compared with that of the original 16S rRNA, resulting in high-level aminoglycoside resistance.
Fig. 2.

Mechanisms of methylation of G1405 and A1408 residues in 16S rRNA by aminoglycoside resistance 16S rRNA MTases.
Globally Distributed N7-G1405 16S Ribosomal RNA Methyltransferases ArmA, RmtB, and RmtC
The 16S rRNA MTase gene armA was first identified together with blaCTX-M3 on plasmid pCTX-M3 of Citrobacter freundii isolated in 1996, followed by documentation in 2007,17 and was subsequently found on plasmid pIP1204 of K pneumoniae in 2000.18 The rmtB and rmtC genes were found on the plasmids of Serratia marcescens and Proteus mirabilis, respectively, isolated in Japan in the first half of the 2000s.19,20 Since then, these 3 16S rRNA MTases have been identified globally, primarily found in Enterobacterales, including Escherichia coli, Klebsiella spp, Enterobacter spp, S marcescens, Citrobacter spp, Proteus spp, and Salmonella spp, isolated from various sources, including humans, livestock, companion animals, and wastewater.21,22 Regarding glucose-nonfermenting gram-negative bacteria, armA has mainly been identified in A baumannii, whereas rmtB/rmtC have rarely been found in that species.23–26 So far, a few P aeruginosa clinical isolates have been reported to carry these 3 16S rRNA MTase genes.25,27,28 The spread of these MTases has thus far been limited to gram-negative bacteria and has not reached clinically important pathogenic gram-positive bacteria, including Staphylococcus spp and Enterococcus spp, although the engineered introduction of these MTase genes could confer a high level of aminoglycoside resistance to S aureus, as well as in gram-negative bacteria.29 One of the clinically concerning issues of these MTase producers is that they tend to also show resistance to β-lactams, fluoroquinolones, polymyxins, and fosfomycin in addition to aminoglycosides through various antimicrobial resistance genetic determinants; for example, β-lactamase genes (blaSHV, blaCTX-M, blaKPC, blaNDM, blaCMY, blaDHA, and blaOXA), fluoroquinolone-resistance genes (qnr, aac(6′)-Ib-cr, qep, and nucleic mutations in QRDRs of gyrA/parC), colistin-resistance gene (mcr), and the fosfomycin-resistance gene (fosA).21,22,30,31
ArmA, RmtB, and RmtC 16S rRNA MTases can confer high-level resistance to 4,6-disubstituted 2-DOS (see Fig. 1). The levels of aminoglycoside resistance conferred by these MTases are high (eg, both amikacin and gentamicin have minimum inhibitory concentrations [MICs] ≥256 μg/mL) compared with those conferred by AMEs.18–20 These increased MIC values are good indicators of the production of aminoglycoside-resistance 16S rRNA MTases relating to Enterobacterales, Acinetobacter spp, and P aeruginosa, and can be applied to the initial screening of 16S rRNA MTase producers (discussed later).
ArmA, RmtB, and RmtC 16S rRNA MTases share only modest amino acid identities with each other (up to 30%), but show high three-dimensional structural similarities (Fig. 3).32,33 These MTases add a CH3 group to the N7 position of G1405 in 16S rRNA, which causes steric hindrance to the substituent at 3″ position of ring III of 4,6-disubstituted 2-DOS, using SAM as a cofactor.19,29,33 Moreover, although 16S rRNA MTases recognize the mature 30S ribosomal subunit consisting of 16S rRNA and ribosomal proteins, they do not methylate naked 16S rRNA alone, or mature 70S ribosomes.29,33
Fig. 3.
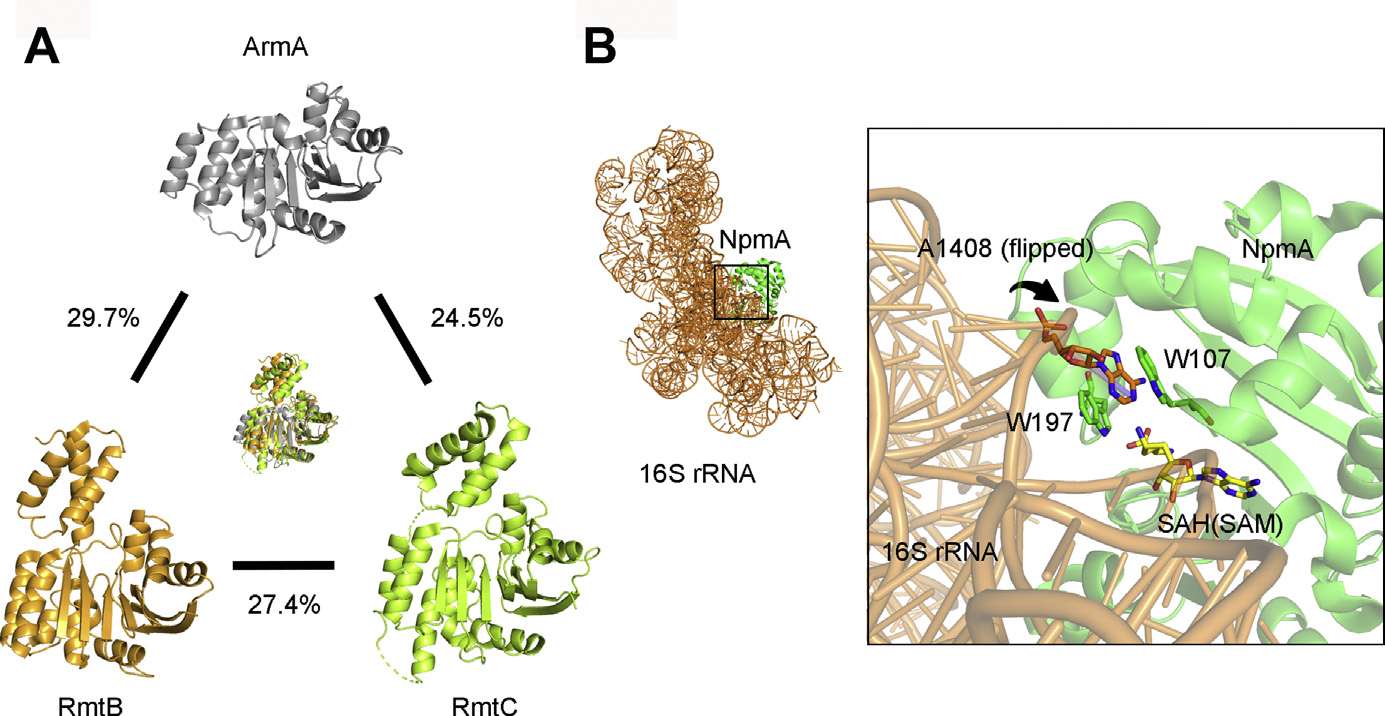
(A) Three-dimensional structures of ArmA (gray), RmtB (orange), and RmtC (light green). These figures were rendered with Protein Data Bank (PDB) data (PDB identifier [ID], 3FZG, 3FRH, and 6PQB). The percentages indicate amino acid identities. (B) Binding mode between 16S rRNA (orange) and NpmA (green). The S-adenosyl-l-homocysteine (SAH) molecule is shown in yellow sticks and the A1408 residue in orange sticks. The 2 tryptophan residues (W107 and W197) of NpmA are shown in green sticks. This figure was rendered with PDB data (PDB ID, 4OX9).
N7-G1405 MTases confer resistance to 4,6-disubstituted 2-DOS but not to 4,5-disubstituted 2-DOS or other aminoglycosides (eg, streptomycin and spectinomycin) (Fig. 4). The difference in specificity toward aminoglycosides can be explained based on the binding modes between aminoglycosides and 16S rRNA (see Fig. 4). The N7 position of the G1405 residue is closest to and oriented toward the substituent at 3″ position in the ring III of 4,6-disubstituted 2-DOS. The introduction of a CH3 residue at the N7 position may lead to a steric clash and/or electrostatic repulsion against the side chain of ring III, leading to reduced binding affinities of aminoglycosides and resulting in increased 4,6-disubstituted 2-DOS resistance. In contrast, ring III and ring IV of 4,5-disubstituted 2-DOS are normally far away from the N7 position of G1405 (see Fig. 4), and the introduction of CH3 by MTase does not disturb their binding, resulting in almost no change in the MIC values of 4,5-disubstituted 2-DOS (see Fig. 4). Other aminoglycosides, such as streptomycin, that bind to 16S rRNA without interacting with the G1405 position could still bind to 16S rRNA with m7G1405 and show normal activity.
Fig. 4.

Molecular models of binding mode between neomycin B/kanamycin A and wild G1405/m7G1405 in 16S rRNA. These figures were rendered based on crystal structures (PDB ID, 2ESI and 2ET4). Basic residues and aminoglycoside molecules are depicted in silver and green sticks, respectively, and the orange dashed lines indicate hydrogen bonds. The red translucent circle indicates the predicted position of the steric clash between the residue and aminoglycoside. MIC values were cited from references.20 (From Wachino J, Yamane K, Shibayama K, et al. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother 2006;50(1):178–84 https://doi.org/10.1128/AAC.50.1.178-184.2006; with permission.)
Sporadic N7-G1405 16S Ribosomal RNA Methyltransferases
Contrary to ArmA, RmtB, and RmtC, the spread of RmtA, RmtD, RmtE, RmtF, RmtG, and RmtH largely remain regional. RmtA has been reported in P aeruginosa clinical isolates from East Asian countries, Japan, and South Korea.34–36 Recently, an RmtA-producing K pneumoniae was also isolated in Switzerland, the first identification of RmtA in a species other than P aeruginosa.37
RmtD (RmtD1 to RmtD3) has mainly been found in P aeruginosa and Enterobacterales isolated in South America, Argentina, Chile, and Brazil. Notably, Tada and colleagues38 and Urbanowicz and colleagues39 reported RmtD3-producing clinical isolates of P aeruginosa from Myanmar and Poland, respectively, indicating that RmtD-group MTase may be starting to spread outside South America.
The number of reports for RmtE (RmtE1 to RmtE3) producers are also limited, 3 from the United States (all E coli),40–43 1 from China (E coli),44 and the last 1, recently, from Myanmar (P aeruginosa).45 rmtE-group genes have also been deposited in the GenBank from A baumannii and Enterobacter cloacae complex under accession numbers MH572011 and LC511997, respectively.
RmtF is the most prevalent MTase, after ArmA/RmtB/RmtC. The first identification of RmtF was in K pneumoniae isolates in La Réunion Island in 2011,46 followed by Enterobacterales from India,47–49 United Kingdom,47,50 Nepal,51 South Africa,52 United States,53 Australia,54 Egypt,55 Switzerland,56 and Ireland,50 and P aeruginosa from Nepal.57 One of the clinical risks associated with RmtF producers is that they frequently coproduce NDM-group metallo-β-lactamase, which confers carbapenem resistance.
RmtG producers (all Klebsiella spp), which often coproduce Klebsiella pneumoniae carbapenemase (KPC), have been reported from Chile,58 United States,59,60 Brazil,61,62 India,48 and Switzerland.63 The sources of RmtH producers are limited, 1 in K pneumoniae from a patient who had been injured in Iraq,64 and the other also in K pneumoniae from a newborn admitted to a hospital in Lebanon.65 The enzymatic functions of RmtA, RmtD, RmtE, RmtF, RmtG, and RmtH are likely the same as those of ArmA, RmtB, and RmtC, in that they methylate the N7 position of G1405.66
N1-A1408 16S Ribosomal RNA Methyltransferase
NpmA was first identified in a clinical isolate of E coli (sequence type 131) in Japan.67 This NpmA-producing E coli was identified through selection for high-level resistance to apramycin, a veterinary aminoglycoside. NpmA causes a flip of A1408 from h44 in 30S ribosomal subunits (see Fig. 3B)68 and modifies the N1-A1408 position in 16S rRNA (and N1-G1408 in 16S rRNA69). The N1-A1408 position is proximal to ring I of 4,5-disubstituted, 4,6-disubstituted 2-DOS and apramycin (Fig. 5). Methylation at the N1 position of A1408 can confer broader aminoglycoside resistance than that of the N7-G1405 MTases because the spatial position of ring I remains the same regardless of the structures of aminoglycosides, at least for 4,5-disubstituted, 4,6-disubstituted 2-DOS and apramycin (see Fig. 5). Kanazawa and colleagues70 recently reported that the introduction of a CH3 group at the N1 position of A1408 prevents the formation of a pseudopair between the ring I of aminoglycosides and the A1408, especially the positively charged N1 atom that electrically prevents the binding of aminoglycosides carrying the NH31 in ring I (eg, amikacin, gentamicin). Nevertheless, they modeled the mode of binding between aminoglycosides with a 6′-OH group in ring I (eg, paromomycin) and m1A1408 and showed that this class of aminoglycoside might still be active against NpmA producers. The extent of MIC increase of paromomycin was, in fact, limited to only 4-fold by NpmA production (see Fig. 5). Therefore, 4,5-disubstituted 2-DOS with the ring I 6′-OH group may be a good starting point for designing the next generation of aminoglycosides that would be active against 16S rRNA MTase producers.70
Fig. 5.

Molecular models of binding mode between neomycin B/paromomycin/gentamicin C1a and wild A1408/m1A1408 in 16S rRNA. The figures were rendered based on crystal structures (PDB ID, 2ET4, 5ZEM, 5ZEJ, and 2ET3). Basic residues and aminoglycoside molecules are depicted in orange and green sticks, respectively, and orange dashed lines indicate hydrogen bonds. The red translucent circle indicates the predicted position of the steric clash between the residue and aminoglycoside. MIC values were cited from references.67 (From Wachino J, Shibayama K, Kurokawa H, et al. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 2007;51(12):4401–9; with permission.)
The E coli npmA gene was flanked by 2 copies of IS26 elements and located on 115-kb transferable IncF plasmids. Since the first report of npmA in 2007, reports of npmA are still rare compared with those of N7-G1405 MTase genes, such as armA, rmtB, and rmtC.71,72 Notably, NpmA2, which has 1 amino acid difference compared with NpmA, was recently identified from Clostridioides difficile (discussed later).
Fitness Costs by 16S Ribosomal RNA Methyltransferase Production in Bacteria
ArmA/RmtB/RmtC are widespread, whereas reports of NpmA have been limited. To explore the difference, some researchers focused on the relationship between the fitness costs of aminoglycoside resistance 16S rRNA MTases and their distribution. Aminoglycoside-resistance 16S rRNA MTases modify the G1405 or A1408 positions, which are close to endogenously methylated residues, C1402 by RsmI and C1407 by RsmF, in E coli. Endogenous methylation at G1405 and A1408 may affect the normal process of housekeeping methylation at C1402 and C1407 positions and reduce optimal ribosomal function. G1405 methylation by ArmA production impeded the methylation of the C1402 position, but not C1407, and resulted in growth impairment.73 In contrast, RmtC impedes methylation at C1407 but is not associated with fitness cost.74 Although NpmA interfered with the endogenous C1407 methylation, it does not affect cell fitness.73 Ishizaki and colleagues75 recently investigated the fitness cost incurred by NpmA production as well and showed low growth rate and cell survival for engineered E coli producing NpmA. Overall, aminoglycoside-resistance MTases might affect cell fitness cost, but it remains difficult to attribute the difference in the prevalence of N7-G1405 MTase (ArmA, RmtB, RmtC)/N1-A1408 MTase (NpmA) to the fitness costs their production incurs. NpmA confers a lower level of resistance to amikacin and gentamicin compared with N7-G1405 MTases, making it difficult to detect NpmA producers when using frank amikacin and gentamicin resistance as the screening criteria. Screening with apramycin resistance may facilitate identification of more N1-A1408 MTase producers.
Origin of Acquired 16S Ribosomal RNA Methyltransferase Gene
As described earlier, 9 types of acquired N7-G1405 MTases (ArmA, RmtA-RmtH) and N1-A1408 MTase, have thus far been identified in pathogenic gram-negative bacteria. Aminoglycoside-producing Actinomycetales innately possess aminoglycoside-resistance 16S rRNA MTase genes as a self-defense mechanism22; however, their G 1 C contents are high, indicating that they are unlikely the direct origin of 16S rRNA MTases of pathogenic bacteria, which have much lower G 1 C contents. Since the first report of N7-G1405 MTases early in the first decade of the 2000s, likely ancestor proteins have not been identified for any of the N7-G1405 MTases. However, Marsh and colleagues76 recently reported that the potential origin of acquired N1-A1408 16S rRNA MTase gene npmA might be the chromosomally encoded 16S rRNA MTase gene carried by some C difficile strains. Near-identical nucleotide sequences (99%–100%) were observed between the acquired npmA in E coli and the chromosomal gene of C difficile. NpmA-producing C difficile showed a higher level of resistance to aminoglycosides compared with non–NpmA-producing C difficile, suggesting that NpmA is associated with aminoglycoside resistance in C difficile. However, aminoglycosides are not used in the treatment of C difficile infections, and susceptibility breakpoints for C difficile have not been defined either. The genetic environment of E coli npmA showed little similarity to those found in C difficile, whereas some C difficile isolates shared 99% genetic identity with each other within the 3-kb regions surrounding npmA. It is noteworthy that not every C difficile strain has npmA on its chromosome. The npmA gene found in some C difficile isolates might also have been derived from other bacteria.
Screening Methods for 16S Ribosomal RNA Methyltransferase Producers
N7-G1405 MTase producers show high-level resistance to 4,6-disubstitited 2-DOS, such as arbekacin, amikacin, and gentamicin. The MIC values of these 3 aminoglycosides for N7-G1405 MTase producers are mostly greater than 256 μg/mL, and no growth-inhibitory zone is observed around the disks containing these aminoglycosides by the disk diffusion test. Routine microdilution susceptibility testing performed in clinical microbiology laboratories does not generally include high concentrations of aminoglycosides. Thus, a practical approach for screening of potential N7-G1405 MTase producers would be to identify isolates resistant to both amikacin and gentamicin and subject them to manual susceptibility testing, which includes high concentrations of aminoglycosides. This screening strategy is applicable for Enterobacterales, Acinetobacter spp, and P aeruginosa. In contrast, it must be borne in mind that some nonfermenting gram-negative bacteria, Pseudomonas spp, Burkholderia spp, and Stenotrophomonas maltophilia and Achromobacter xylosoxidans innately show high levels of aminoglycoside resistance and should not be misidentified as aminoglycoside-resistance 16S rRNA MTase producers.
Because only N1-A1408 MTase producers have so far been detected, it is difficult to discuss screening methods for N1-A1408 MTase producers. In addition, in contrast with N7-G1405 MTase producers, resistance levels toward amikacin and gentamicin conferred by N1-A1408 MTases are similar to those conferred by AMEs. The most remarkable phenotype of N1-A1408 MTase producer is high-level apramycin resistance. The first E coli strain producing NpmA was identified through growth on agar plates containing 500 μg/mL apramycin, whereas almost all other tested clinical isolates could not grow on it (Wachino and colleagues, unpublished data, 2005). The only exception was AAC(3)-IV producers, which could also grow on agar plates containing higher concentration of apramycin.
New Aminoglycoside: Plazomicin
Plazomicin (PLZ), initially known as ACHN-490, is a new, semisynthetic, next-generation aminoglycoside (Fig. 6). PLZ is categorized as one of the essential medicines in the World Health Organization (WHO) model list in 2019.77 This aminoglycoside was developed by Achaogen Co Ltd in 2009 by adding hydroxylaminobutyric acid to sisomicin at the 1 position and the 2-hydroxyethyl group at the 6′ position. PLZ was approved in 2018 by the Food and Drug Administration (FDA) for the treatment of complicated urinary tract infections and acute pyelonephritis. PLZ was designated to avoid modification by a variety of clinically relevant AMEs, thus its effectiveness is expected to be greater than conventional aminoglycosides for aminoglycoside-resistant pathogens producing AMEs (see Fig. 6).
Fig. 6.
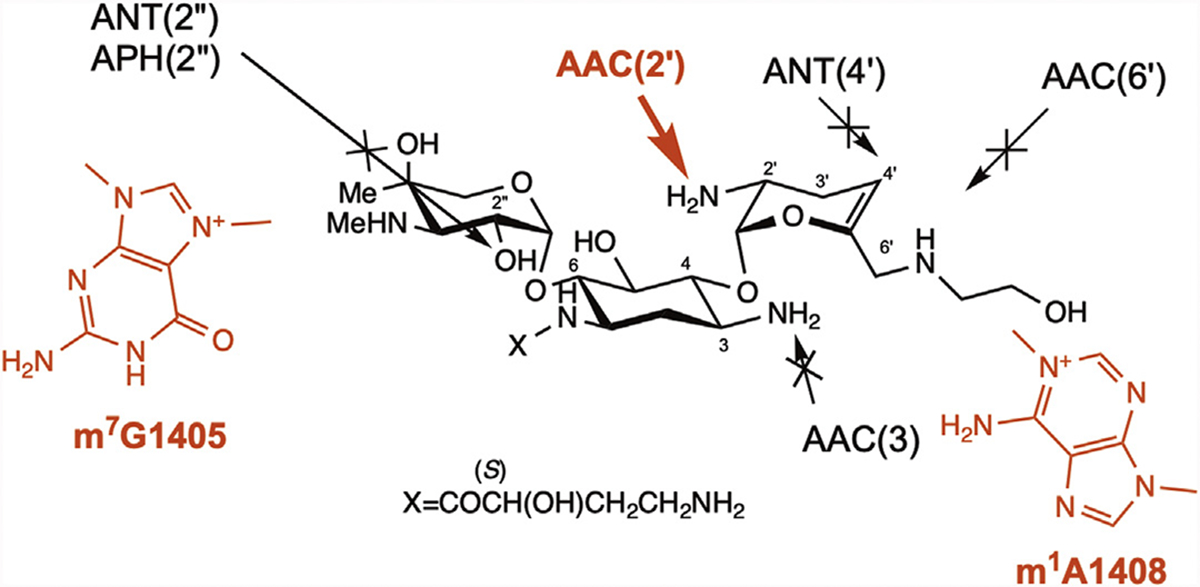
Structure of plazomicin and modification targets of AMEs.
PLZ has shown high potency in in vitro susceptibility testing against gram-negative bacteria.78 The susceptibility percentage of E coli, K pneumoniae, Klebsiella aerogenes, Klebsiella oxytoca, E cloacae complex, and S marcescens to PLZ ranged from 97.6% to 100%, and the MIC90 (MIC required to inhibit the growth of 90% of organisms) values of PLZ for these Enterobacteriaceae were 0.5 to 1 μg/mL, which is less than the clinical breakpoint of 2 μg/mL approved by FDA.78 Overall, PLZ was as potent as or superior to other aminoglycosides, including amikacin, gentamicin, and tobramycin. Compared with these Enterobacteriaceae, PLZ was less active against P mirabilis and Morganella morganii, with 44.3% and 66.7% susceptibility, respectively, and MIC90 values of 4 μg/mL for both, similar or inferior to amikacin, gentamicin, and tobramycin.78 Compared with Enterobacterales, glucose-nonfermenting gram-negative pathogens, including P aeruginosa and A baumannii, were less susceptible to PLZ, with MIC90 values of 16 and 8 μg/mL, respectively.78 The activity of PLZ was also significantly lower against S maltophilia, with an MIC90 value of greater than 64 μg/mL,78 although this organism also showed natural resistance to other aminoglycosides. It is also noteworthy that PLZ is highly active against ESBL-producing E coli and K pneumoniae, carbapenemase-producing Enterobacteriaceae, and colistin-resistant Enterobacteriaceae, with susceptibility rates of greater than 90%.78
Mechanisms of Plazomicin Resistance
PLZ showed potent activity against clinically relevant AME-producing bacteria, as expected, but a small portion of the tested Enterobacterales strains were highly resistant to PLZ.79 All these resistant bacteria were reported to be 16S rRNA MTase producers. PLZ has the same ring III structure as sisomicin, whose MIC values are very high for N7-G1405 MTase producers. Thus, it is reasonable that N7-G1405 MTase producers show cross-resistance to PLZ (MIC>256 μg/mL) (see Fig. 6). N1-A1408 MTase producers also confer PLZ resistance (MIC>256 μg/mL),80 probably through the 2-hydroxyethyl group at the 60 position of ring I interfering with m1A1408 (see Fig. 6).
Cox and colleagues80 recently reported the detailed behavior of various AMEs toward PLZ. As expected, production of most AMEs tested, including AAC(3)-Ia, AAC(3)-II, AAC(3)-IV, AAC(6′)-Ib, AAC(6′)-Ib-cr, AAC(6′)-Ie-APH(2″)-Ia, AAC(6′)-Ii, ANT(2″)-Ia, ANT(4′)-Ia, APH(2″)-IIa(-Ib), APH(2″)-IVa(-Id), APH(3′)-IIIa, APH(3″)-Ia, APH(4)-Ia, APH(6)-Ia, and APH(9)-Ia in engineered E coli BW25113 strain (originally PLZ MIC 2 μg/mL) conferred no or only slight resistance (2-fold to 4-fold increase in MIC) to PLZ. In contrast, only AAC(2′)-Ia showed a 16-fold increase in PLZ MIC. Cox and colleagues80 modeled the complex structure of AAC(2′)-Ia and PLZ and confirmed the binding mode between them. AAC(2′)-Ia production is likely one of the causes for PLZ resistance, as observed in organisms such as Providencia stuartii that possess chromosomally encoded aac(2′)-Ia genes.
SUMMARY
Gram-negative bacteria with high-level aminoglycoside resistance caused by the production of 16S rRNA MTases have spread globally and across a variety of environments since their first identification in the early 2000s, and new variants of the 16S rRNA MTase have since emerged. This development is further complicated by the fact that 16S rRNA MTase producers often carry other clinically relevant resistance genes, including carbapenemase genes (eg, blaNDM and blaKPC) and colistin-resistance genes (mcr). The threat of 16S rRNA MTase in the emergence and spread of extensive drug resistance and pandrug resistance among pathogenic gram-negative bacteria therefore should not be underestimated.
KEY POINTS.
Aminoglycoside-resistant gram-negative pathogens producing 16S ribosomal RNA (rRNA) methyltransferase (MTase) have emerged and spread globally.
16S rRNA MTase-producing gram-negative pathogens tend to show a multidrug-resistance profile against β-lactams and fluoroquinolones.
16S rRNA MTase producers resist the newly approved aminoglycoside, plazomicin.
Treatment options are limited for infections caused by multidrug-resistant 16S rRNA MTase producers.
ACKNOWLEDGMENTS
This research was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research C). The work by Y. Doi was supported by grants from the National Institutes of Health (R01AI104895, R21AI135522).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest associated with this article.
REFERENCES
- 1.Mulani MS, Kamble EE, Kumkar SN, et al. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 2019;10:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair JM, Webber MA, Baylay AJ, et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 2015;13(1):42–51. [DOI] [PubMed] [Google Scholar]
- 3.Serio AW, Keepers T, Andrews L, et al. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal Plus 2018;8(1). 10.1128/ecosalplus.ESP-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones D, Metzger HJ, Schatz A, et al. Control of gram-negative bacteria in experimental animals by streptomycin. Science 1944;100(2588):103–5. [DOI] [PubMed] [Google Scholar]
- 5.Waksman SA, Lechevalier HA. Neomycin, a new antibiotic active against streptomycin-resistant bacteria, including tuberculosis organisms. Science 1949;109(2830):305–7. [DOI] [PubMed] [Google Scholar]
- 6.Umezawa H, Ueda M, Maeda K, et al. Production and isolation of a new antibiotic: kanamycin. J Antibiot (Tokyo) 1957;10(5):181–8. [PubMed] [Google Scholar]
- 7.Becker B, Cooper MA. Aminoglycoside antibiotics in the 21st century. ACS Chem Biol 2013;8(1):105–15. [DOI] [PubMed] [Google Scholar]
- 8.Umezawa H, Hamada M, Suhara Y, et al. Kasugamycin, a new antibiotic. Antimicrob Agents Chemother (Bethesda) 1965;5:753–7. [PubMed] [Google Scholar]
- 9.Hirokawa G, Kiel MC, Muto A, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J 2002;21(9):2272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 2001;70:415–35. [DOI] [PubMed] [Google Scholar]
- 11.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem 2005;74:129–77. [DOI] [PubMed] [Google Scholar]
- 12.Wasserman MR, Pulk A, Zhou Z, et al. Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions. Nat Commun 2015;6:7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garneau-Tsodikova S, Labby KJ. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 2016;7(1):11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halfon Y, Jimenez-Fernandez A, La Rosa R, et al. Structure of Pseudomonas aeruginosa ribosomes from an aminoglycoside-resistant clinical isolate. Proc Natl Acad Sci U S A 2019. 10.1073/pnas.1909831116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Pulk A, Wasserman MR, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol 2012;19(9):957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 1997;5(6):234–40. [DOI] [PubMed] [Google Scholar]
- 17.Golebiewski M, Kern-Zdanowicz I, Zienkiewicz M, et al. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother 2007;51(11):3789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother 2003;47(8):2565–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi Y, Yokoyama K, Yamane K, et al. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother 2004;48(2):491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachino J, Yamane K, Shibayama K, et al. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother 2006;50(1):178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doi Y, Wachino JI, Arakawa Y. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin North Am 2016;30(2):523–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wachino J, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 2012;15(3):133–48. [DOI] [PubMed] [Google Scholar]
- 23.Wachino JI, Jin W, Kimura K, et al. Intercellular transfer of chromosomal antimicrobial resistance genes between Acinetobacter baumannii strains mediated by prophages. Antimicrob Agents Chemother 2019;63(8). 10.1128/AAC.00334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamane K, Wachino J, Doi Y, et al. Global spread of multiple aminoglycoside resistance genes. Emerg Infect Dis 2005;11(6):951–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tada T, Miyoshi-Akiyama T, Kato Y, et al. Emergence of 16S rRNA methylase-producing Acinetobacter baumannii and Pseudomonas aeruginosa isolates in hospitals in Vietnam. BMC Infect Dis 2013;13:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bado I, Papa-Ezdra R, Delgado-Blas JF, et al. Molecular characterization of carbapenem-resistant Acinetobacter baumannii in the intensive care unit of Uruguay’s University Hospital identifies the first rmtC gene in the species. Microb Drug Resist 2018;24(7):1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurung M, Moon DC, Tamang MD, et al. Emergence of 16S rRNA methylase gene armA and cocarriage of blaIMP-1 in Pseudomonas aeruginosa isolates from South Korea. Diagn Microbiol Infect Dis 2010;68(4):468–70. [DOI] [PubMed] [Google Scholar]
- 28.Mohanam L, Menon T. Emergence of rmtC and rmtF 16S rRNA methyltransferase in clinical isolates of Pseudomonas aeruginosa. Indian J Med Microbiol 2017;35(2):282–5. [DOI] [PubMed] [Google Scholar]
- 29.Wachino J, Shibayama K, Kimura K, et al. RmtC introduces G1405 methylation in 16S rRNA and confers high-level aminoglycoside resistance on Gram-positive microorganisms. FEMS Microbiol Lett 2010;311(1):56–60. [DOI] [PubMed] [Google Scholar]
- 30.Lupo A, Saras E, Madec JY, et al. Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J Antimicrob Chemother 2018;73(4):867–72. [DOI] [PubMed] [Google Scholar]
- 31.Bartoloni A, Sennati S, Di Maggio T, et al. Antimicrobial susceptibility and emerging resistance determinants (blaCTX-M, rmtB, fosA3) in clinical isolates from urinary tract infections in the Bolivian Chaco. Int J Infect Dis 2016;43:1–6. [DOI] [PubMed] [Google Scholar]
- 32.Nosrati M, Dey D, Mehrani A, et al. Functionally critical residues in the aminoglycoside resistance-associated methyltransferase RmtC play distinct roles in 30S substrate recognition. J Biol Chem 2019. 10.1074/jbc.RA119.011181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt E, Galimand M, Panvert M, et al. Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases. J Mol Biol 2009;388(3):570–82. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama K, Doi Y, Yamane K, et al. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 2003;362(9399):1888–93. [DOI] [PubMed] [Google Scholar]
- 35.Yamane K, Doi Y, Yokoyama K, et al. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2004;48(6):2069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin JS, Kwon KT, Moon DC, et al. Emergence of 16S rRNA methylase rmtA in colistin-only-sensitive Pseudomonas aeruginosa in South Korea. Int J Antimicrob Agents 2009;33(5):490–1. [DOI] [PubMed] [Google Scholar]
- 37.Poirel L, Schrenzel J, Cherkaoui A, et al. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J Antimicrob Chemother 2011;66(8):1730–3. [DOI] [PubMed] [Google Scholar]
- 38.Tada T, Shimada K, Mya S, et al. A new variant of 16S rRNA Methylase, RmtD3, ina clinical isolate of Pseudomonas aeruginosa in Myanmar. Antimicrob Agents Chemother 2018;62(1). 10.1128/AAC.01806-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urbanowicz P, Izdebski R, Baraniak A, et al. Pseudomonas aeruginosa with NDM-1, DIM-1 and PME-1 β-lactamases, and RmtD3 16S rRNA methylase, encoded by new genomic islands. J Antimicrob Chemother 2019;74(10):3117–9. [DOI] [PubMed] [Google Scholar]
- 40.Lee CS, Hu F, Rivera JI, et al. Escherichia coli sequence type 354 coproducing CMY-2 cephalosporinase and RmtE 16S rRNA methyltransferase. Antimicrob Agents Chemother 2014;58(7):4246–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CS, Li JJ, Doi Y. Complete sequence of conjugative IncA/C plasmid encoding CMY-2 β-lactamase and RmtE 16S rRNA methyltransferase. Antimicrob Agents Chemother 2015;59(7):4360–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis MA, Baker KN, Orfe LH, et al. Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob Agents Chemother 2010;54(6):2666–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Pacey MP, Doi Y. Chromosomal 16S ribosomal RNA Methyltransferase RmtE1 in Escherichia coli sequence type 448. Emerg Infect Dis 2017;23(5):876–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia J, Sun J, Li L, et al. First report of the IncI1/ST898 conjugative plasmid carrying rmtE2 16S rRNA Methyltransferase gene in Escherichia coli. Antimicrob Agents Chemother 2015;59(12):7921–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tada T, Hishinuma T, Watanabe S, et al. Molecular characterization of multidrug-resistant Pseudomonas aeruginosa isolates in Hospitals in Myanmar. Antimicrob Agents Chemother 2019;63(5). 10.1128/AAC.02397-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galimand M, Courvalin P, Lambert T. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother 2012;56(7):3960–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidalgo L, Hopkins KL, Gutierrez B, et al. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J Antimicrob Chemother 2013;68(7):1543–50. [DOI] [PubMed] [Google Scholar]
- 48.Filgona J, Banerjee T, Anupurba S. Incidence of the novel rmtF and rmtG methyltransferases in carbapenem-resistant Enterobacteriaceae from a hospital in India. J Infect Dev Ctries 2015;9(9):1036–9. [DOI] [PubMed] [Google Scholar]
- 49.Rahman M, Prasad KN, Pathak A, et al. RmtC and RmtF 16S rRNA Methyltransferase in NDM-1-Producing Pseudomonas aeruginosa. Emerg Infect Dis 2015;21(11):2059–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor E, Sriskandan S, Woodford N, et al. High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int J Antimicrob Agents 2018;52(2):278–82. [DOI] [PubMed] [Google Scholar]
- 51.Tada T, Miyoshi-Akiyama T, Dahal RK, et al. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int J Antimicrob Agents 2013;42(4):372–4. [DOI] [PubMed] [Google Scholar]
- 52.Rubin JE, Peirano G, Peer AK, et al. NDM-1-producing Enterobacteriaceae from South Africa: moving towards endemicity? Diagn Microbiol Infect Dis 2014;79(3):378–80. [DOI] [PubMed] [Google Scholar]
- 53.Lee CS, Vasoo S, Hu F, et al. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol 2014;52(11):4109–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidjabat HE, Townell N, Nimmo GR, et al. Dominance of IMP-4-producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother 2015;59(7):4059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamal D, Fernandez-Martinez M, Salem D, et al. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis 2016;43:17–20. [DOI] [PubMed] [Google Scholar]
- 56.Mancini S, Poirel L, Tritten ML, et al. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J Antimicrob Chemother 2018;73(3):821–3. [DOI] [PubMed] [Google Scholar]
- 57.Tada T, Shimada K, Satou K, et al. Pseudomonas aeruginosa Clinical Isolates in Nepal Coproducing Metallo-β-Lactamases and 16S rRNA Methyltransferases. Antimicrob Agents Chemother 2017;61(9). 10.1128/AAC.00694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poirel L, Labarca J, Bello H, et al. Emergence of the 16S rRNA methylase RmtG in an extended-spectrum-β-lactamase-producing and colistin-resistant Klebsiella pneumoniae isolate in Chile. Antimicrob Agents Chemother 2014;58(1):618–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bueno MF, Francisco GR, O’Hara JA, et al. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 2013;57(5):2397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu F, Munoz-Price LS, DePascale D, et al. Klebsiella pneumoniae sequence type 11 isolate producing RmtG 16S rRNA methyltransferase from a patient in Miami, Florida. Antimicrob Agents Chemother 2014;58(8):4980–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramos PI, Picao RC, Almeida LG, et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 2014;15:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Passarelli-Araujo H, Palmeiro JK, Moharana KC, et al. Molecular epidemiology of 16S rRNA methyltransferase in Brazil: RmtG in Klebsiella aerogenes ST93 (CC4). An Acad Bras Cienc 2019;91(suppl 1):e20180762. [DOI] [PubMed] [Google Scholar]
- 63.Mancini S, Poirel L, Corthesy M, et al. Klebsiella pneumoniae co-producing KPC and RmtG, finally targeting Switzerland. Diagn Microbiol Infect Dis 2018;90(2):151–2. [DOI] [PubMed] [Google Scholar]
- 64.O’Hara JA, McGann P, Snesrud EC, et al. Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother 2013;57(5):2413–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beyrouthy R, Robin F, Hamze M, et al. IncFIIk plasmid harbouring an amplification of 16S rRNA methyltransferase-encoding gene rmtH associated with mobile element ISCR2. J Antimicrob Chemother 2017;72(2):402–6. [DOI] [PubMed] [Google Scholar]
- 66.Correa LL, Witek MA, Zelinskaya N, et al. Heterologous expression and functional characterization of the exogenously acquired aminoglycoside resistance methyltransferases RmtD, RmtD2, and RmtG. Antimicrob Agents Chemother 2016;60(1):699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wachino J, Shibayama K, Kurokawa H, et al. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 2007;51(12):4401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunkle JA, Vinal K, Desai PM, et al. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proc Natl Acad Sci U S A 2014;111(17):6275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zelinskaya N, Witek MA, Conn GL. The pathogen-derived aminoglycoside resistance 16S rRNA methyltransferase NpmA possesses dual m1A1408/m1G1408 specificity. Antimicrob Agents Chemother 2015;59(12):7862–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanazawa H, Baba F, Koganei M, et al. A structural basis for the antibiotic resistance conferred by an N1-methylation of A1408 in 16S rRNA. Nucleic Acids Res 2017;45(21):12529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeganeh Sefidan F, Mohammadzadeh-Asl Y, Ghotaslou R. High-level resistanceto aminoglycosides due to 16S rRNA methylation in Enterobacteriaceae isolates. Microb Drug Resist 2019. 10.1089/mdr.2018.0171. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Z, Lan F, Liu M, et al. Evaluation of automated systems for aminoglycosides and fluoroquinolones susceptibility testing for Carbapenem-resistant Enterobacteriaceae. Antimicrob Resist Infect Control 2017;6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lioy VS, Goussard S, Guerineau V, et al. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA 2014;20(3):382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gutierrez B, Escudero JA, San Millan A, et al. Fitness cost and interference of Arm/Rmt aminoglycoside resistance with the RsmF housekeeping methyltransferases. Antimicrob Agents Chemother 2012;56(5):2335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishizaki Y, Shibuya Y, Hayashi C, et al. Instability of the 16S rRNA methyltransferase-encoding npmA gene: why have bacterial cells possessing npmA not spread despite their high and broad resistance to aminoglycosides? J Antibiot (Tokyo) 2018;71(9):798–807. [DOI] [PubMed] [Google Scholar]
- 76.Marsh JW, Pacey MP, Ezeonwuka C, et al. Clostridioides difficile: a potential source of NpmA in the clinical environment. J Antimicrob Chemother 2019;74(2):521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.World Health Organization Model List of Essential Medicines, 21st List, 2019. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 78.Saravolatz LD, Stein GE. Plazomicin: a new aminoglycoside. Clin Infect Dis 2019. 10.1093/cid/ciz640. [DOI] [PubMed] [Google Scholar]
- 79.Castanheira M, Deshpande LM, Woosley LN, et al. Activity of plazomicin compared with other aminoglycosides against isolates from European and adjacent countries, including Enterobacteriaceae molecularly characterized for aminoglycoside-modifying enzymes and other resistance mechanisms. J Antimicrob Chemother 2018;73(12):3346–54. [DOI] [PubMed] [Google Scholar]
- 80.Cox G, Ejim L, Stogios PJ, et al. Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect Dis 2018;4(6):980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


