Abstract
Assembly of spliceosomes involves a number of sequential steps in which small nuclear ribonucleoprotein particles (snRNPs) and some non-snRNP proteins recognize the splice site sequences and undergo various conformational rearrangements. A number of important intermolecular RNA-RNA duplexes are formed transiently during the process of splice site recognition. Various steps in the assembly pathway are dependent upon ATP hydrolysis, either for protein phosphorylation or for the activity of helicases, which may modulate the RNA structures. Major efforts have been made to identify proteins that interact with specific regions of the pre-mRNA during the stages of spliceosome assembly and catalysis by site-specific UV cross-linking. However, UV cross-linking is often inefficient for the detection of proteins that interact with base-paired RNA. Here we have used the complementary approach of methylene blue-mediated photo-cross-linking to detect specifically proteins that interact with the duplexes formed between pre-mRNA and small nuclear RNA (snRNA). We have detected a novel cross-link between a 65-kDa protein (p65) and the 5′ splice site. A range of data suggest that p65 cross-links to the transient duplex formed by U1 snRNA and the 5′ splice site. Moreover, although p65 cross-linking requires only a 5′ splice site within the pre-mRNA, it also requires ATP hydrolysis, suggesting that its detection reflects a very early ATP-dependent event during splicing.
Splicing of pre-mRNA occurs within the multicomponent spliceosome complex via two successive chemical steps (reviewed in references 33, 41, 47, and 63). Introns are excised and adjacent exons are spliced together, and this must be achieved with great accuracy, even though the introns may be many thousands of bases in length. Sequence elements at the splice sites themselves, as well as activating enhancer elements, provide the signals within the pre-mRNA that give rise to this specificity. The requirement for accuracy in determination of the precise splice junctions is reflected in a number of steps in which the primary consensus elements are recognized sequentially by separate protein or small nuclear RNA (snRNA) splicing factors. Work with both yeast and HeLa nuclear extracts has led to a model in which the spliceosome is assembled by sequential addition of small nuclear ribonucleoprotein particles (snRNPs) and other splicing factors and by substantial conformational rearrangements before the actual catalytic steps (reviewed in reference 58). A number of the steps leading to the formation of a catalytically competent spliceosome require ATP hydrolysis, either for protein phosphorylation by kinases or for the action of RNA helicases. Initial recognition of splice sites involves binding of U1 snRNP to the 5′ splice site, mediated by base pairing between the 5′ splice site consensus sequence and the 5′ end of U1 snRNA (62, 64, 84). At the 3′ end of the intron, the heterodimeric protein U2AF binds to the polypyrimidine tract via the RNA binding domains of the 65-kDa subunit (82). Both of these initial binding events can be assisted by proteins of the SR class (17, 29, 69). In yeast, the branch point sequence is initially recognized by the protein BBP (for branch point binding or bridging protein [known as SF1 or mBBP in mammalian systems]) (2, 3, 5). At this E, or commitment complex, stage, a bridging interaction can already occur across the intron (46), formed either by BBP (2) or by SR proteins (79). Subsequent formation of the A complex, or prespliceosome, involves ATP hydrolysis and the binding of 17S U2 snRNP to the branch point aided by the N-terminal domain of U2AF65 (74) and a DEAD-box helicase protein (21, 60, 75). Recognition of the branch point at this stage involves base pairing between the branch point sequence and a conserved region of U2 snRNA (53, 77, 83), with the branch point adenosine itself remaining bulged out of the intermolecular duplex (56). Formation of the B complex involves the binding of the U4/5/6 tri-snRNP to the A complex (30), a step that is also ATP dependent and that is activated by SR proteins (59). This is followed by conformational rearrangements. In particular, U4-U6 base pairing is disrupted, and U6 becomes base paired with both U2 snRNA (16, 42, 78) and with the intron part of the 5′ splice site sequence (26, 36, 68), replacing the U1-5′ splice site duplex and forming an RNA bridge, composed of U2 and U6 snRNAs, between the 5′ splice site and branch point (41, 51). In addition, U5 snRNA contacts bases just within the 5′ exon (49, 68). The spliceosome, poised for catalytic step 1, is now referred to as the C complex and is characterized by the presence of splicing intermediates.
With a view to understanding the detailed basis of this assembly process, major efforts have been made to characterize the protein contents of the E, A, B, and C complexes and to determine which spliceosome proteins contact various regions of the pre-mRNA (reviewed in references 34 and 76). In particular, UV cross-linking with pre-mRNAs containing a single labeled phosphate at various defined positions has helped to identify proteins that interact at or near the 5′ splice site, branch point, pyrimidine tract, and 3′ splice site AG, as well as at various exon positions, including exon enhancers (12, 13, 23, 24, 57, 69, 73, 80). A more elaborate survey of proteins in the vicinity of the branch point has involved the use of a photoactivable benzophenone group attached to a spacer arm, allowing the detection of proteins within a 15-Å range (40). The established pre-mRNA–protein contacts have been catalogued in reference 12. Although much useful information has been generated by these site-specific cross-linking experiments, the inherent properties of zero-length UV cross-linking may not allow the detection of all of the important interactions occurring at the various positions of the pre-mRNA. In particular, UV cross-linking is not as effective at detecting proteins that interact with fully based-paired RNA as with single-stranded RNA (ssRNA) (38). However, at important stages in the spliceosome cycle, the 5′ splice site is involved in such base-paired interactions with U1 and U6 snRNAs and the branch site with U2 snRNA. Although the specificity of these interactions is provided by RNA-RNA base pairing, it is likely that additional protein-RNA contacts stabilize and modulate the short intramolecular RNA-RNA duplexes. We have recently outlined the use of methylene blue (MB)-mediated cross-linking for the detection of proteins that interact with double-stranded RNA (dsRNA) (37, 38). MB cross-linking has a specificity that is complementary to UV cross-linking in that it preferentially detects proteins that interact with dsRNA, but not ssRNA, and it does not induce RNA-RNA cross-links. We therefore reasoned that MB cross-linking may allow the detection of proteins that interact with the transient duplexes that are formed between pre-mRNA and snRNA and that may have escaped detection in previous UV cross-linking investigations. By carrying out MB cross-linking during in vitro splicing reactions, we have detected a protein of ∼65 kDa that interacts with the 5′ splice site. The protein (p65) was not detected by UV cross-linking. MB cross-linking of p65 was transient; it occurred before complex A formation, persisted in complex A, and declined before step 1 of splicing. It required only a 5′ splice site within the pre-mRNA but was ATP dependent, suggesting that detection of p65 reflects a very early ATP-dependent step in splicing. Finally, p65 cross-linking occurred within U1 snRNP-containing complexes and was abolished by oligonucleotide-targeted destruction of the 5′ end of U1 snRNA, suggesting that p65 interacts at the 5′ splice site-U1 snRNA duplex.
MATERIALS AND METHODS
Chemicals and enzymes.
MB was purchased from BDL and used without further purification. It was dissolved as a stock solution in water at 2.5 mg ml−1. The concentration was determined by ɛ660 = 86,000 cm−1 M−1. Adenosine 5′-O-(3-thiotriphosphate) (ATP-γ-S) and adenylyl-imidodiphosphate (AMP-PNP) were purchased from Sigma. Enzymes were purchased from Promega, Boehringer Mannheim, and New England Biolabs and were used under the conditions suggested by the manufacturers. Antibodies against U1 70K protein and the snRNP 69-kDa protein were kind gifts from Reinhard Lührmann. Monoclonal antibody against U2AF65 was a generous gift from Maria Carmo-Fonseca, and Juan Valcárcel and Angela Kramer kindly provided polyclonal antibodies to SF1. The anti-tau monoclonal antibody (6) was from Boehringer.
Constructs.
The plasmids for transcribing the GC+DX, X23H, TM+1G, and HP1 (previously referred to as 23HP) RNAs were described previously (37, 61, 65–67). The Cons 5′ splice site transcript, containing a consensus 5′ splice site, and the transcript containing the 5′ splice site-complementary sequence were transcribed by SP6 RNA polymerase from plasmids generated by insertion of oligonucleotides with the following sequence in both orientations into the SphI site in pGEM4Z: CAG|GTAAGTAGGCCTATCGATCCATG. The consensus 5′ splice site is underlined, and the splice junction is shown by the vertical line. The GC hairpin was transcribed by SP6 RNA polymerase from a plasmid generated by insertion of oligonucleotides with the sequence GGG(CGG)6AATT(CCG)6CCC into the PvuII site of pSP70 vector. The insertions were confirmed by DNA sequencing. The sequences of the transcribed RNAs are given below.
In vitro transcription.
All 32P-labeled RNAs were synthesized from the corresponding linearized plasmids with SP6 RNA polymerase (Promega) by using an m7G(5′)ppp(5′)G dinucleotide primer (New England Biolabs). The RNAs were radiolabeled with the appropriate [α-32P]nucleotide triphosphate (400 Ci/mmol; Amersham) as indicated. High-specific-activity radiolabeled transcripts used for MB cross-linking were transcribed with 0.25 mM ATP, UTP, and CTP and 0.07 mM GTP and [α-32P]GTP. The transcripts used for in vitro splicing were radiolabeled to low specific activity by transcription with 0.25 mM ATP, GTP, and CTP and 0.15 mM UTP and [α-32P]UTP.
The consensus 5′ splice site RNA, the 5′ splice site cRNA, and the noncomplementary RNA (see Fig. 5 and 6C) had the respective sequences (5′ to 3′) GAATACGAAT TCGAGC TCGG TACCCGGGGATCCTC TAGAGTCGACCTGCAGGCATGCAGGTAAGTAGGCCTATCGATCCATGCAAGCT, GAATACGAAT TCGAGC TCGG TACCCGGGGATCCTCTAGAG TCGACC TGCAGGCATGGATCGATAGGCCTACTTACCTGCATGCAAGCT, and GAATACGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCT.
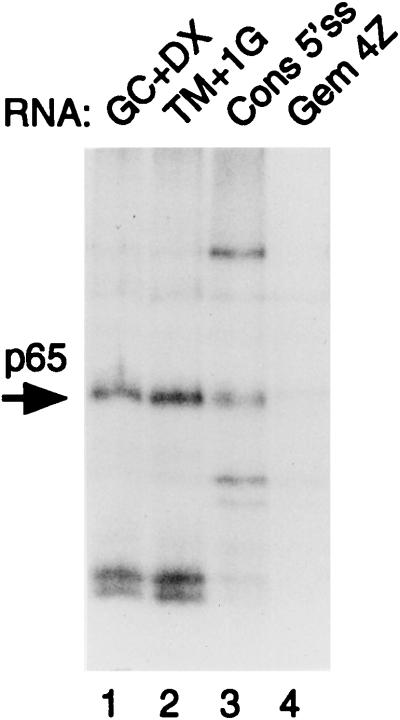
FIG. 5.
MB cross-linking of p65 to different 5′ splice sites. Plasmids were linearized so that transcripts contained 5′ splice sites but no branch point-3′ splice site elements. Transcripts were incubated with 40% HeLa nuclear extracts under splicing conditions for 15 min. Lane 1 contains GC+DX/XhoI. Lane 2 contains TM+1G/XhoI (same 5′ exon as GC+DX, but the 5′ splice site was changed from AAGGUACCC to AAGGUAGUG and the downstream intron sequences were from a synthetic polylinker). Lane 3 contains consensus 5′ splice site/HindIII. The RNA contains a consensus 5′ splice site (CAGGUAAGU) in the pGEM 4Z polylinker. Lane 4 contains the pGEM 4Z polylinker. p65 cross-linking was detected with the 5′ splice site-containing RNAs (lanes 1 to 3) but not the polylinker (lane 4).
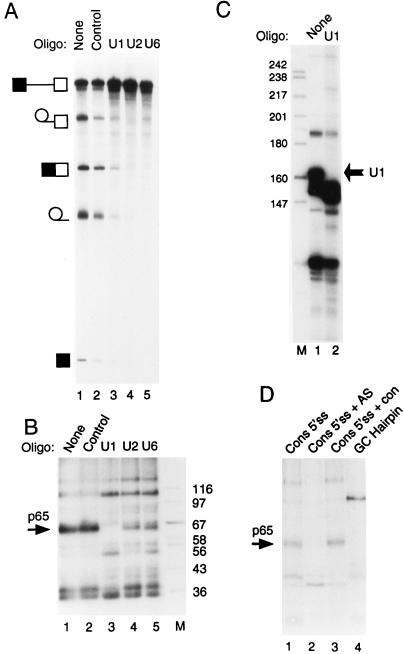
FIG. 6.
Cross-linking of p65 requires the 5′ end of U1 snRNA. (A) Splicing of GC+DX after oligonucleotide-directed RNase H destruction of snRNAs. Fifty percent HeLa nuclear extracts were incubated under normal splicing conditions in the presence of a DNA oligonucleotide and RNase H for 45 min. The transcript GC+DX was then added for a further 150 min of incubation. GC+DX was [32P]UTP labeled to low specific activity. Lanes: 1, no oligonucleotide-RNase H treatment; 2, control DNA oligonucleotide; 3, oligonucleotide against the 5′ end of U1 snRNA; 4, oligonucleotide against U2 snRNA; 5, oligonucleotide against U6 snRNA. Note that U2 and U6 targeting completely abolished splicing, while U1 destruction severely impaired splicing. (B) MB cross-linking of p65 after targeting of snRNAs. Fifty percent HeLa nuclear extracts were pretreated as in panel A. The transcript GC+DX [32P]GTP labeled to high specific activity was then added for a further 15 min of incubation. Lanes 1, no oligonucleotide-RNase H; 2, control oligonucleotide; 3, oligonucleotide against the 5′ end of U1 snRNA; 4, oligonucleotide against U2 snRNA; 5, oligonucleotide against U6 snRNA; M, protein size markers (kilodaltons). Note that p65 cross-linking is completely abolished after destruction of U1 snRNA (lane 3). (C) RNAs from untreated (lane 1) or a U1 oligonucleotide–RNase H-targeted extract (lane 2) were end labeled with 5′ [32P]pCp and then analyzed by electrophoresis and autoradiography. The intact U1 snRNA band in the untreated extract (indicated by the arrow) is undetectable (<1%) in the U1-targeted extract. (D) p65 does not cross-link to a nonspecific 5′ splice site duplex or to an RNA hairpin. RNAs were incubated with 40% HeLa nuclear extracts under normal splicing conditions for 15 min and then subjected to MB cross-linking. Lane 1, consensus 5′ splice site (Fig. 5). In lane 2, the consensus 5′ splice site was preannealed to a fivefold molar excess of unlabeled complementary RNA by heating at 65°C for 3 min and slow cooling to room temperature. In lane 3, the consensus 5′ splice site was incubated with a fivefold molar excess of a noncomplementary RNA at 65°C for 3 min and slowly cooled to room temperature. The nonradiolabeled RNA was transcribed from the same parental vector (pGEM 4Z) in the same direction, but lacking the 26-base 5′ splice site-containing insert. Lane 4, an RNA containing a 22-bp GC hairpin. Note that p65 cross-linking is inhibited by the 5′ splice site-RNA (compare lanes 1 and 2).
The complementary regions are underlined, and the 5′ splice site sequence is shown in boldface.
Single 32P-labeled transcripts.
For labelling the +1 position of the intron, the 5′-half RNAs were transcribed from template GC+DX linearized at StyI to produce a transcript terminating exactly at the 3′ end of TM exon 2. The 3′-half RNA was transcribed from PCR products. The upstream PCR oligonucleotide had an SP6 promoter. The 5′ and 3′ transcripts were trace-labeled with [α-32P]UTP (0.1 μCi in 40 μl of transcription mixture) to allow quantitation. GMP was used to initiate transcription of the 3′ RNAs, which were then dephosphorylated with alkaline phosphatase (37°C for 60 min) and subsequently phosphorylated with [γ-32P]ATP and T4 polynucleotide kinase (37°C for 30 min). Ten micrograms of yeast tRNA was added as a carrier. After ethanol precipitation, the transcripts were ligated with 5′-half transcripts by DNA oligonucleotide-mediated ligation (48). The ligation products were separated by 5% urea–polyacrylamide gel electrophoresis (PAGE), and the corresponding RNA band was recovered by soaking in 2.5 M NH4OAc overnight. The same procedures were used to generate intron +5 position-labeled RNA, except both the 5′ half and 3′ half of the transcripts were transcribed from PCR products. The oligonucleotides used for PCR amplification and for RNA ligation bridges are as follows. The oligonucleotides for the intron +1-labeled RNA 3′ half were 5′ GAATTTAGGTGACACTATAGTACCCAAAAAAAAAATTA 3′ and 5′ CTTGAGTTTCTTTTGCAGTGACACC 3′, and that for the bridge was 5′ CTCCTAATTTTTTTTTTGGGTACCTTGGCGGCGGTCTCG 3′. The oligonucleotide for the intron +5-labeled RNA 5′ half was made up of the SP6 promoter and 5′ ATACCTTGGCGGCGGTCT 3′, those for the 3′ half were 5′ GAATTTAGGTGACACTATAGTCCCAAAAAAAAAATTAGG 3′ and 5′ CTTGAGTTTCTTTTGCAGTGACACC 3′, and that for the bridge was 5′ CCTAATTTTTTTTTTTGGGACATACCTTGGCGGCGGTCTCG 3′.
In vitro splicing.
HeLa cell nuclear extracts were prepared with modifications of the method of reference 1. In vitro splicing was carried out under standard conditions (66). Briefly, 2 to 5 ng of RNA was incubated with 30 to 60% HeLa nuclear extracts in 10-μl splicing reaction mixtures at 30°C for the appropriate time. In all experiments, ATP was included to 0.5 mM, and creatine phosphate was included to 20 mM unless otherwise stated. For destruction of snRNAs, the DNA oligonucleotides (40 nM) and RNase H (0.87 U; Pharmacia) were incubated with nuclear extracts under splicing conditions for 45 min before RNAs were added. The DNA oligonucleotides used for targeting snRNAs were as follows. The control nonspecific oligonucleotide was 5′ GTGCACGTGCATGCACGTA 3′. The oligonucleotide for U1 snRNA was 5′ TGCCAGGTAAGTAT 3′, complementary to nucleotides 1 to 14. The oligonucleotide for U2 snRNA was 5′ ATAAGAACAGATACTACACTTGA 3′, complementary to nucleotides 27 to 49. The oligonucleotide for U6 snRNA was 5′ TCTCTGTATCGTTCC 3′, complementary to nucleotides 33 to 47.
The splicing mixtures were either proteinase K digested and analyzed by urea-PAGE or were used in MB cross-linking experiments (see below). To check the degree of digestion of U1 snRNA untreated or U1-targeted extracts, RNA was phenol extracted and ethanol precipitated. It was then 3′ end labeled with 5′ [32P]pCp and T4 RNA ligase in the presence of 10% dimethyl sulfoxide overnight at 4°C. Labeled RNAs were phenol extracted, ethanol precipitated, and analyzed by urea-polyacrylamide electrophoresis.
MB- and UV-induced cross-linking.
MB- and UV-induced cross-linking was carried out as previously reported (38). Briefly, 2 ng of MB in aqueous solution was added to 10-μl splicing reaction mixtures in a microtiter plate after incubation for the times indicated at 30°C. The microtiter plate was then placed on ice, and the samples were irradiated either with visible light for 20 min or with UV light in a UV cross-linker for 2× 8,600 μW (Stratagene). The light source for the MB cross-linking was a 40-W fluorescent tube mounted 4 to 5 cm above the samples. After irradiation, the samples were subjected to RNase digestion (RNase A at 1 μg μl−1, RNase T1 at 0.3 U μl−1, and RNase V1 at 0.035 U μl−1). To some reaction mixtures ascorbic acid was added. We have recently found that ascorbic acid is effective at quenching nonspecific protein-protein cross-linking and enhancing the specific dsRNA-protein cross-linking (37a). In these cases, ascorbic acid was added to a 1.5 to 5 mM final concentration immediately before illumination of the samples. Ascorbic acid was prebuffered with double the concentration of Tris-HCl (pH 8.0). Finally, the cross-linking results were analyzed by sodium dodecyl sulfate (SDS)-PAGE and autoradiography.
Glycerol gradients.
Control experiments were first carried out to ascertain that glycerol concentrations up to 30% and incubation on ice for up to 5 h did not interfere with p65 cross-linking. Two hundred-microliter splicing reaction mixtures containing 100 ng of [α-32P]GTP-labeled RNA, 2 mM MgCl2, 20 U of RNAsin, 0.5 mM ATP, 20 mM creatine phosphate, and 40% HeLa nuclear extracts were incubated at 30°C for the times indicated and then directly loaded onto 11-ml 10 to 30% glycerol gradients. The gradients were made with 2 mM MgCl2, 20 mM Tris-Cl (pH 7.5), and 80 mM KCl and precooled at 4°C. Centrifugation was carried out for 3 h at 40,000 rpm at 4°C in a Beckman SW40 rotor. Fifteen-drop fractions (∼0.5 ml) were collected at 4°C, and a 50-μl aliquot from each fraction was quantitated by Cerenkov counting. MB cross-linking was carried out with 400-μl aliquots from selected fractions with 2 ng of MB/μl and 5 mM sodium ascorbate (pH 7.5). Proteins were then precipitated by the sequential addition of 4 volumes of methanol, 1 volume of chloroform, and finally 3 volumes of water. Reaction mixtures were mixed thoroughly by vortexing before each reagent was added. The reaction mixture was then centrifuged for 5 min, and the upper layer was discarded, care being taken not to disturb the interface. Proteins were finally recovered by addition of 300 μl of methanol followed by 10 min of centrifugation. The pellet was resuspended, and the proteins were analyzed by SDS-PAGE (15% polyacrylamide).
Immunoprecipitations.
Immunoprecipitation of cross-linking reaction mixtures was carried out essentially as described before (37). Usually 30-μl reaction mixtures were incubated under specified conditions before dilution to 250 μl in NETS buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.05% Nonidet P-40). To perform immunoprecipitation under more stringent conditions, SDS was added to the reaction mixtures to 0.8% in order to disrupt U1 snRNPs before dilution with NETS buffer (final SDS concentration of less than 0.1%). We ascertained that the antibody-antigen interaction was stable in 0.1% SDS by demonstrating that U1 snRNA was coimmunoprecipitated by the U1 70K antibody.
RESULTS
MB and UV cross-linking of proteins to labeled pre-mRNA.
Since we planned to use MB cross-linking to probe for RNA-protein interactions during pre-mRNA splicing, it was important to ascertain that MB had no effects upon in vitro splicing reactions. Inclusion of MB at the concentrations used for cross-linking (0.2 or 2 ng/μl) or at 10-fold-higher concentrations had no detectable effects upon splicing reactions carried out in the absence of light (Fig. 1). We next compared MB and UV cross-linking by using GC+DX RNA, which is derived from the intron between rat α-tropomyosin (TM) exons 2 and 3 (66). This RNA contains no known internal stable secondary structure, increasing the likelihood of specifically detecting proteins that interact at the pre-mRNA–snRNA duplexes. Parallel time courses of splicing and MB and UV cross-linking were therefore carried out (Fig. 2). UV irradiation resulted in cross-linking of a large number of proteins (Fig. 2C). Particularly prominent bands at ∼54 and ∼35 kDa were observed at most time points. At later times (45 to 180 min), a high-molecular-mass band of ∼200 kDa was observed, possibly corresponding to the HeLa equivalent of the yeast PRP8 protein (12, 73). In contrast, and consistent with its preference for protein-dsRNA cross-linking, MB treatment resulted in detection of a much more limited array of proteins (Fig. 2B). Detection of these radiolabeled bands was dependent upon the presence of MB during illumination with visible light (data not shown). A protein of ∼130 kDa was observed immediately, but it disappeared within 5 min (Fig. 1B). A band of ∼35 kDa was detected at all time points and probably corresponds to the band strongly cross-linked by UV. This band probably corresponds to an SR protein. A very-high-molecular-mass band was also seen at the longest time point. In contrast, a band with mobility slightly higher than that of the 67-kDa marker (subsequently referred to as p65) appeared to interact with pre-mRNA in a manner that is more obviously consistent with a role in pre-mRNA splicing at the stages when the known intermolecular duplexes are formed. Cross-linking of p65 was transient, with a maximum signal at about 15 min. It subsequently decreased and was almost undetectable at 180 min. Comparison with the kinetics of splicing (maximal accumulation of intermediates at 45 min [Fig. 2A]) suggested that p65 cross-linking occurred before step 1 of splicing. Cross-linking to p65 also required ATP and Mg2+ (see Fig. 7). UV cross-linking did not detect p65 at any time point (Fig. 2C). p65 cross-linking was observed with RNA labeled with [α-32P]GTP (Fig. 2B), but not when MB cross-linking was carried out with [α-32P]CTP- or UTP-labeled RNA (data not shown). It was detected with lower intensity when the RNA was labeled with [α-32P]ATP. However, in a construct with a point mutation of the first G of the intron to A (G1→A), which allows step 1 to proceed, but not step 2 (61), the intensity of p65 cross-linking was greater with A label than with G label (data not shown), suggesting that cross-linking occurs at the 5′ splice site (see below).
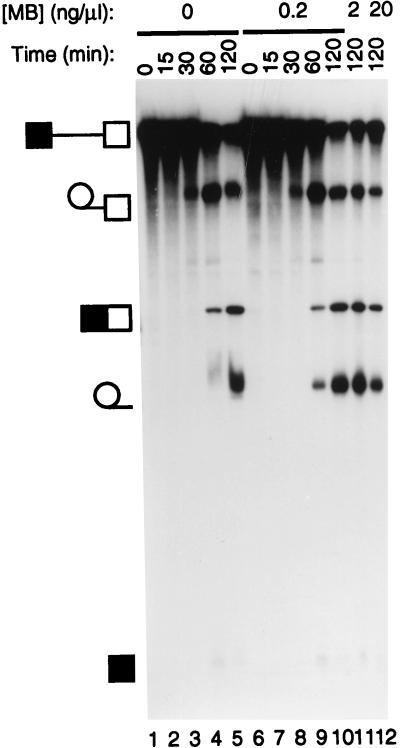
FIG. 1.
Splicing in the presence of MB. Splicing reactions were carried out with GC+DX RNA for the times indicated under normal conditions (lanes 1 to 5) or in the presence of MB at 0.2 ng/μl in lanes 6 to 10, 2 ng/μl in lane 11, and 20 ng/μl in lane 12. The identities of the various RNA bands are shown schematically to the left of the panel. MB did not interfere with splicing reactions even at a concentration 10-fold-higher than those used in any of the subsequent cross-linking experiments.
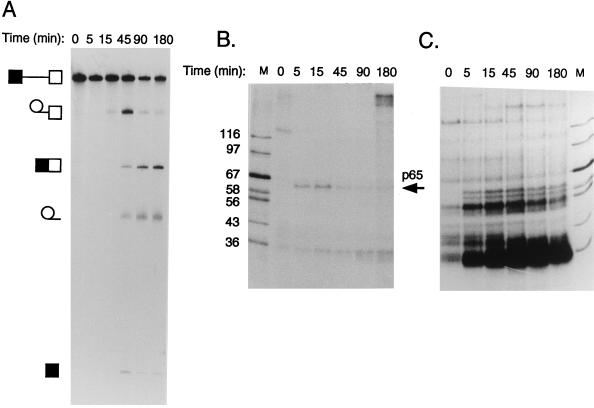
FIG. 2.
Kinetics of splicing and MB-mediated cross-linking. Results from a 3-h time course of splicing (A), MB cross-linking (B), and UV cross-linking (C) of GC+DX RNA are shown. Splicing was carried out with 60% HeLa nuclear extract for the times indicated. RNA was [32P]UTP labeled to low specific activity for splicing and was [32P]GTP labeled to high specific activity for cross-linking (see Materials and Methods). Lane M contains protein markers with sizes in kilodaltons indicated to the left of panel B. Note the ∼65-kDa band detected by MB but not UV cross-linking that appears transiently during the time course.
FIG. 7.
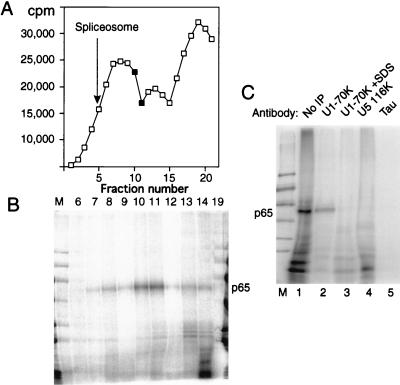
p65 cross-linking in U1 snRNP-containing complexes. Full-length labeled GC+DX RNA was incubated in nuclear extract for 15 min and then fractionated on a 10 to 30% glycerol gradient. (A) Profile of Cerenkov counts across the glycerol gradient. Fractions are numbered from 1 (heaviest) to 20 (lightest). Fully assembled spliceosomes, as indicated by the presence of splicing intermediates, were shown to peak in fraction 5 in control gradient fractionations. The complex that forms at time zero peaks at fraction 13. (B) MB cross-linking of gradient fractions from panel A. Cross-linked p65 was reproducibly maximal in fractions 10 and 11. Note that the lane containing fraction 19 has been contaminated by spillover of size markers. Nevertheless, this fraction can still be seen to be essentially free of cross-linked p65. (C) Labeled GC+DX XhoI RNA was incubated in nuclear extract and then MB cross-linked and electrophoresed by SDS-PAGE after no further treatment (lane 1) or after immunoprecipitation with monoclonal antibody H111 against U1 70K protein (lanes 2 and 3), antiserum against U5 116K protein (lane 4), or a control monoclonal antibody against tau protein (lane 5). In lane 3, the cross-linked sample was first pretreated with 0.8% SDS to disrupt U1 snRNPs before dilution into NETS buffer and immunoprecipitation with H111. p65 was immunoprecipitated by the U1 70K antibodies, but only if U1 snRNP remained intact (lane 2).
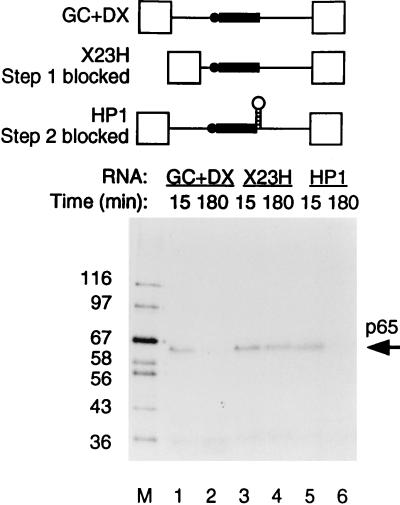
The timing of p65 cross-linking was investigated further by using two other forms of the TM intron. The transcript X23H contains the native intron between TM exons 2 and 3 in which the proximity of the branch and 5′ splice sites blocks spliceosome assembly before step 1 at the A complex stage (66). Transcript HP1 (previously referred to as 23HP [65, 67]) is derived from GC+DX by insertion of a stable 22-bp GC-rich hairpin between the polypyrimidine tract and 3′ splice site, which results in a step 2 block of splicing. Cross-linked p65 was observed with all of these RNAs after 15 min of incubation in nuclear extracts (Fig. 3, lanes 1, 3, and 5). The p65 band was not detected at 180 min either with GC+DX RNA or with HP1 (Fig. 3, lanes 2 and 6). However, the cross-link of p65 remained constant with X23H even up to 3 h (lane 4). These data therefore suggest that the p65 cross-link appears at or before the A complex stage of spliceosome assembly and that the subsequent decline in p65 cross-linking occurs after the A complex stage but before step 2 of splicing. Comparison of the kinetics of splicing and p65 cross-linking (Fig. 2A and B) suggests that the decline is actually before step 1. This is consistent with data indicating that p65 cross-linking occurs at the 5′ splice site-U1 snRNA duplex (see below), since this interaction is disrupted before C complex formation.
FIG. 3.
p65 cross-linking occurs before A complex formation and declines later. RNAs were incubated with 60% HeLa nuclear extract, and MB cross-linking was carried out at 15 and 180 min. X23H is the natural intron between TM exons 2 and 3; spliceosome assembly is blocked at the A complex stage because of the proximity of the branch point (shown schematically as the solid circle) to the 5′ splice site. HP1 is derived from GC+DX by insertion of a stable hairpin between the polypyrimidine tract (shown as the solid rectangle) and 3′ splice site, which blocks step 2 of splicing. Lane M, protein size markers (kilodaltons). p65 cross-linking with HP1 was identical to that with GC+DX (compare lanes 1 and 2 with 5 and 6), whereas with X23H, p65 cross-linking occurred but did not subsequently decline (lanes 3 and 4).
p65 cross-linking occurs at the 5′ splice site.
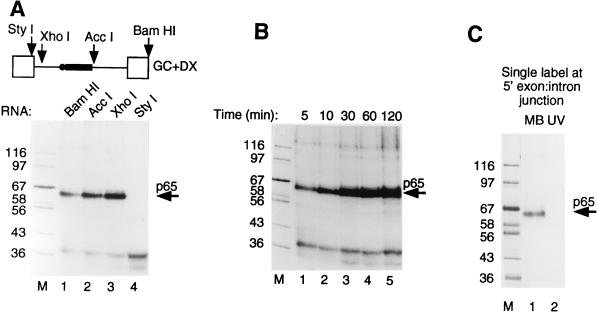
The preceding data showed that p65 could be cross-linked to pre-mRNA at an early stage in spliceosome assembly. Nevertheless, at the A complex stage, both the 5′ splice site and branch point have been recognized by base pairing with U1 and U2 snRNAs, respectively (41, 58), so p65 could potentially be interacting at either of these intermolecular duplexes. We next investigated the elements required for p65 cross-linking by making runoff GC+DX RNAs truncated at BamHI (full length, both steps of splicing), AccI (no 3′ splice site AG, step 1 occurs, no step 2), XhoI (5′ splice site and exon present), and StyI (5′ exon only). We investigated the interactions of p65 with these transcripts after 15 min in nuclear extract (Fig. 4A). MB cross-linking of p65 was observed with all transcripts containing a 5′ splice site (Fig. 4, BamHI, AccI, and XhoI in lanes 1 to 3, respectively). As described above, the time course of p65 cross-linking with BamHI and AccI RNAs was transient, decreasing at later time points (data not shown). In contrast, p65 cross-linking remained constant with XhoI RNA (Fig. 4B), consistent with our previous conclusion that the decline in p65 intensity occurs after the A complex stage. No p65 cross-linking was observed with StyI RNA, which terminates precisely at the exon-intron boundary (Fig. 4A, lane 4). The XhoI RNA, which cross-linked p65 efficiently, has only 34 nucleotides (nt) more than StyI RNA. Of these, 6 nt are the intron part of the 5′ splice site (GUACCC in this intron), while the remaining 28 are an artificial nonspecific spacer element. These data therefore indicate that a 5′ splice site is required for p65 cross-linking. In order to pinpoint precisely the site of p65 cross-linking, we made full-length GC+DX RNA with a single 32P label at the exon-intron junction (AAGp*GUACCC) by oligonucleotide-mediated RNA ligation (48). Splicing of the ligated RNA in HeLa nuclear extracts showed the correct sizes of the precursor, lariat intermediate, and lariat product (data not shown). MB cross-linking of this RNA after 15 min in nuclear extract produced a single clear p65 band (Fig. 4C, lane 1) which was not observed in parallel UV cross-linking (lane 2). This result demonstrates that p65 cross-linking occurs at the exon-intron junction. To test whether p65 cross-linking also occurs at other positions of the 5′ splice site, we made GC+DX RNA containing a single 32P label at the +5 position of the altered 5′ splice site sequence: AAGGUAUp*GU. The +5 single-labeled RNA spliced correctly, yet no cross-linking of p65 or any other proteins was detected (data not shown). We have not tested RNAs with single-labeled residues upstream of the intron-exon junction. However, taken together with the lack of cross-linking with RNAs truncated at the exon-intron boundary (Fig. 4A, StyI RNA) and the preferential cross-linking with G-labeled wild-type RNA but A-labeled G1→A mutant RNA, the data suggest that p65 MB cross-links with the 5′ splice site at the exon-intron junction.
FIG. 4.
Cross-linking of p65 occurs at the 5′ splice site. (A) MB cross-linking with different GC+DX RNAs truncated from the 3′ end by linearization at the restriction sites indicated in the top panel. In the upper panel, the RNA is shown schematically with exons as empty boxes, the intron as a line, the pyrimidine tract as the solid rectangle, and the branch point as the solid circle. Full-length BamHI RNA undergoes both steps of splicing, AccI lacks the 3′ splice site YAG and exon and goes through step 1 only, and XhoI contains only the 5′ exon and 5′ splice site and is blocked before A complex assembly, while StyI RNA contains only the 5′ exon. The RNAs were incubated with HeLa nuclear extracts under normal splicing conditions for 15 min before MB cross-linking. Note that cross-linking of p65 occurs with all RNAs except GC+DX/StyI, which lacks the 5′ splice site. (B) Time course of MB cross-linking with GC+DX/XhoI. Note that p65 cross-linking appears as normal but does not decline. (C) p65 cross-linking to RNA with a 5′ splice site-specific label. GC+DX RNAs containing a single 32P label at the 5′ exon-intron junction were incubated with 40% HeLa nuclear extracts under normal splicing conditions for 15 min before being subjected to cross-linking. Lane 1, MB cross-linking; lane 2, UV cross-linking. p65 MB cross-linked efficiently to RNA with the 5′ splice site label, but was not detected by UV cross-linking. Lane M, protein size markers (kilodaltons).
p65 cross-linking is not dependent on 5′ splice site context.
The preceding data show that p65 protein interacts with the 5′ splice site of the TM intron early in spliceosome assembly. The 5′ splice site in GC+DX is not a perfect consensus (AAG|GUAccc [nonconsensus bases shown in lowercase]). To rule out the possibility that p65 cross-linking is peculiar to this specific 5′ splice site, we made another TM-based transcript, TM+1G, which has the same exon sequence as GC+DX XhoI, but the 5′ splice site has been improved to the consensus (AAG|GUGAGU), and the sequences immediately downstream are comprised of a short polylinker (61). MB cross-linking with this RNA showed an identical p65 band (Fig. 5, lane 2). Therefore, cross-linking of p65 is not dependent on the specific 5′ splice site sequences of GC+DX. However, both GC+DX and TM+1G contain the same upstream exon sequences. To test whether p65 cross-linking is solely dependent upon the 5′ splice site, we inserted the consensus sequence (CAG|GUAAGU) into the SphI site of the pGEM+4Z polylinker. Cross-linking of p65 was observed with this RNA, but not with empty polylinker (Fig. 5, compare lanes 3 and 4). (Note that in lane 4, the very faint signal is from a band that is slightly larger than p65.) Thus p65 cross-linking requires only a 5′ splice site. We have also detected p65 cross-linking to the 5′ splice sites of adenovirus and human β-globin introns; however, in both of these cases, the results were complicated by another band migrating slightly slower than p65 (data not shown).
p65 cross-linking requires the 5′ end of U1 snRNA.
The preceding data demonstrated that p65 interacts at the 5′ splice site. Given the preference of MB cross-linking for mediating cross-linking of proteins to dsRNA, the likelihood is that p65 cross-links to a duplex formed between the 5′ splice site and an snRNA. The 5′ splice site is recognized early during spliceosome assembly via base pairing with the 5′ end of U1 snRNA (84). There is a later interaction involving U6 snRNA base pairing with the intron part of the 5′ splice site consensus (26, 36, 68). It has also been reported that U6 snRNA can interact with a short 5′ splice site RNA oligonucleotide (31, 32). To investigate the roles of snRNAs in the cross-linking of p65, we pretreated extracts with RNase H and short DNA oligonucleotides targeted to the 5′ end of U1 snRNA, the branch point-complementary region of U2 snRNA, and the conserved ACAGAG-containing region of U6 snRNA (see Materials and Methods). These are the regions of the snRNAs that base pair with the pre-mRNA, and oligonucleotide-targeted destruction has previously been shown to inhibit splicing (7, 8, 20, 35). Preincubation of nuclear extract with a control oligonucleotide and RNase H resulted in a modest decrease in splicing efficiency compared with that in nuclear extract that had been preincubated without oligonucleotide and RNase H treatment (Fig. 6A, compare lanes 1 and 2). Consistent with previous reports, destruction of U2 and U6 by oligonucleotides and RNase H abolished splicing (Fig. 6A, lanes 4 and 5) (7, 8, 20). In contrast, treatment with the U1 oligonucleotide severely diminished but did not completely abolish splicing (Fig. 6A, lane 3). Analysis of U1 snRNA in the control and the U1-targeted extract indicated that intact U1 snRNA was undetectable (>1%) after the oligonucleotide-RNase H cleavage (Fig. 6C). U1 snRNA-independent in vitro splicing has been reported previously and is promoted by SR proteins (14, 15, 70). Possibly the splicing enhancer within the 5′ exon of GC+DX pre-mRNA (43) facilitated the U1-independent splicing seen here. The snRNA-depleted extracts were then tested for p65 cross-linking after incubation with GC+DX RNA for 15 min. p65 cross-linking was abolished only in the U1 snRNA-targeted extract (Fig. 6B, lane 3). Targeting of U2 and U6 reduced the cross-linking intensity, but the effect was variable and never approached the complete abrogation of p65 cross-linking seen after U1 targeting (Fig. 6B, lanes 4 and 5). These data demonstrate that the 5′ end of U1 snRNA is required for p65 interaction. Considering that the 5′ end of U1 snRNA base pairs with the 5′ splice site at an early stage of spliceosome assembly and that MB cross-linking shows a high degree of preference for dsRNA-protein cross-linking, the most reasonable explanation for these data is that p65 interacts with the transient duplex formed between U1 snRNA and the 5′ splice site.
We carried out a control experiment to show that p65 cross-linking requires the specific 5′ splice site-U1 snRNA duplex. The consensus 5′ splice site RNA was incubated in nuclear extract either alone, after preannealing to a cRNA that forms a 32-bp duplex encompassing the consensus 5′ splice site, or after mock annealing with a similar RNA that lacks the complementary region (see Materials and Methods for RNA sequences). If p65 cross-linking merely requires the 5′ splice site to be in a base-paired duplex, then the cRNA-annealed sample should still support p65 cross-linking. On the other hand, if a duplex between the 5′ splice site and the specific complementary sequence in U1 snRNA is required, then the cRNA should inhibit p65 cross-linking as it competes with U1 for base pairing to the 5′ splice site sequence. While p65 cross-linking was seen with the 5′ splice site alone or the mock-annealed RNA (Fig. 6C, lanes 1 and 3), the cRNA completely suppressed it (lane 2). Thus p65 cross-linking requires the specific U1 snRNA-5′ splice site duplex. Finally, we incubated an RNA hairpin containing a 23-G-C-bp stem in the nuclear extract and subsequently subjected it to MB cross-linking. A 76-kDa protein was detected by cross-linking (Fig. 6C, lane 4). However, p65 did not cross-link to this hairpin, confirming that p65 is not a general nonspecific dsRNA binding protein.
p65 cross-linking occurs in U1 snRNP-containing complexes.
The preceding data showed that p65 cross-linking to pre-mRNA requires and occurs at a 5′ splice site and also requires the 5′ end of U1 snRNA. We suggest that the most likely interpretation is that p65 cross-links to the duplex formed between the 5′ splice site and U1 snRNA. MB cross-linking should be able to mediate cross-linking through either strand of such a duplex, and in theory, it should be possible to detect p65 cross-linking to the U1 snRNA partner of the intermolecular duplex. For various reasons, such an experiment is not technically feasible. Nevertheless, since the data to this point have pointed toward, but not directly demonstrated, the involvement of U1 snRNA, we sought to demonstrate that p65 cross-linking occurs in U1 snRNP-containing complexes.
Our first approach was to fractionate splicing complexes by sedimentation in 10 to 30% glycerol gradients followed by MB cross-linking. Figure 7A shows the profile of a glycerol gradient fractionation of a reaction mixture containing full-length GC+DX RNA after 15 min of incubation in nuclear extract. Figure 7B shows the results of MB cross-linking of gradient fractions. Cross-linked p65 was found to be maximal in fractions about halfway down the gradient (fractions 10 and 11). In comparison, control experiments showed that full spliceosomes were maximal in fraction 5 (indicated by an arrow in Fig. 7A) and that H complex, which forms immediately upon incubation, is centered around fraction 13. Very little p65 was observed in either the lighter fractions or in the heavier fractions that correspond to mature spliceosomes. A similar distribution of p65 was observed when the X23H RNA, which is blocked at the A complex stage, was separated on glycerol gradients (data not shown). Thus, p65 cross-linking occurs in complexes that are large enough to contain U1 snRNP and also other components. Interestingly, the mobility of the peak of p65 cross-linking, between the initial ∼22S peak and the 35S peak that contains the A complex, appears similar to that of the mammalian commitment complex (25a, 59). This experiment involved addition of MB only after the gradient separation of complexes. Therefore, the possibility that MB itself induces the association of p65 with the 5′ splice site can be ruled out.
To address whether p65-containing complexes also contain U1 snRNP, we carried out cross-linking followed by immunoprecipitation with monoclonal antibody H111, which is specific to U1 70K protein and which efficiently precipitates U1 snRNP (28). Cross-linked p65 was immunoprecipitated with the H111 antibody under conditions in which U1 snRNP remained intact (Fig. 7C, lane 2). However, if SDS was added to 0.8% to denature the U1 snRNP before dilution and immunoprecipitation, p65 was not recovered (lane 3). Thus p65 cross-linking occurs in a U1 snRNP-containing complex, but p65 is not the U1 70K protein itself. p65 was immunoprecipitated in this way after cross-linking to full-length GC+DX, to X23H RNA (data not shown), or to GC+DX XhoI RNA containing only a 5′ exon and 5′ splice site (Fig. 7C). It was not precipitated with a control monoclonal antibody to the microtubule-associated tau protein (lane 5) (6) or with antiserum to the U5 116-kDa protein (19, 37). These data therefore demonstrate that cross-linking of p65 to 5′ splice sites occurs in complexes that contain U1 snRNP.
p65 cross-linking requires ATP hydrolysis.
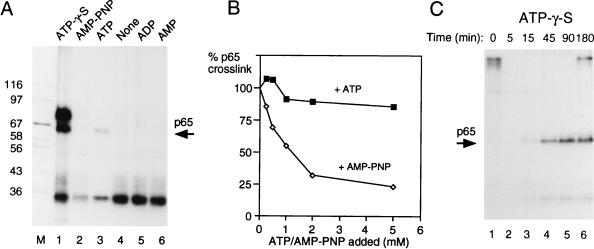
A number of steps in spliceosome assembly require ATP hydrolysis, either for phosphorylation of splicing factors or for driving conformational rearrangements via helicases (34, 76). The stable binding of U2 snRNP at the branch point to form complex A is the first identified ATP-dependent step in spliceosome assembly. However, an ATP-dependent transition between the E complex and a so-called E* complex has been observed in introns in which A complex assembly cannot occur because of the lack of a usable branch point (10). We investigated the dependency of p65 cross-linking upon ATP. MB cross-linking was carried out after incubation of the XhoI RNA (5′ splice site only) in the presence of ATP, ADP, AMP, or no adenine nucleotide. In contrast to all of the previous experiments, in which ATP was included to 0.5 mM and creatine phosphate was included to 20 mM, these experiments were carried out in the absence of creatine phosphate to avoid generation of ATP from the added nucleotides. In the absence of added nucleotides, p65 cross-linking was not observed (Fig. 8A, lane 4). Addition of 2 mM ATP (Fig. 8A, lane 3) resulted in p65 cross-linking. The intensity of p65 in Fig. 8 is less than that in previous figures, presumably due to the lack of creatine phosphate to regenerate ATP. Neither ADP, AMP, nor the nonhydrolyzable ATP analog AMP-PNP supported p65 cross-linking (lanes 5, 6, and 2, respectively), even after 3 h of incubation (data not shown). This suggests that ATP hydrolysis is required for p65 cross-linking. However, the inability of AMP-PNP to substitute for ATP could formally be due to a failure to bind rather than due to direct blockage of β-γ phosphodiester hydrolysis. To distinguish between these possibilities, p65 cross-linking was carried with 0.5 mM ATP and 20 mM creatine phosphate in the presence of increasing concentrations of AMP-PNP (added with an equimolar amount of MgCl2). As a control, in parallel reactions, increasing amounts of ATP plus MgCl2 were added. As shown in Fig. 8B, addition of AMP-PNP, but not ATP, led to inhibition of p65 cross-linking. The ability of AMP-PNP to act as an inhibitor of ATP-dependent p65 cross-linking suggests that it is able to compete with ATP for binding to the site that is responsible for p65 cross-linking. Therefore, the lack of p65 cross-linking in the presence of AMP-PNP is likely due to a requirement for ATP hydrolysis for p65 cross-linking.
FIG. 8.
Cross-linking of p65 requires ATP hydrolysis. (A) MB cross-linking of p65 with GC+DX/XhoI in the presence of different ATP analogs. The RNA was incubated with 40% nuclear extract in the presence of different ATP analogs for 15 min and then subjected to MB cross-linking. Creatine phosphate was omitted in all samples. All of the ATP analogs and ATP were added to a 2 mM final concentration. Lanes: 1, ATP-γ-S; 2, AMP-PNP; 3, ATP; 4, no nucleotides added; 5, ADP; 6, AMP; M, protein size markers (kilodaltons). (B) Inhibition of p65 cross-linking by AMP-PNP. Labeled X23H RNA (5 ng) was incubated in 40% nuclear extract with 0.5 mM ATP, 20 mM creatine phosphate, and 2 mM MgCl2 at 30°C for 30 min before MB cross-linking. Reaction mixtures were supplemented at time zero with additional ATP or AMP-PNP along with an equimolar amount of MgCl2 before incubation. p65 cross-linking was quantitated by phosphorimager and is plotted as the percentage of cross-linking in the absence of additional nucleotide (i.e., in the presence of the basal 0.5 mM ATP). (C) Three-hour time course of MB cross-linking of GC+DX/BamHI transcript in the presence of 2 mM ATP-γ-S with no creatine phosphate. Splicing was carried out with 60% HeLa nuclear extracts. Note that p65 cross-linking persists over the time course.
Next we tested the ATP analog ATP-γ-S. This analog can generally be hydrolyzed, although with slower kinetics than ATP. However, when used by protein kinases to phosphorylate proteins, phosphatases are subsequently unable to remove the thiophosphate groups from substrate proteins (72). With the XhoI RNA containing only a 5′ splice site, ATP-γ-S resulted in strong cross-linking of p65 (Fig. 8A, lane 1). In addition, a prominent cross-linked band at ∼70 kDa was observed. When a time course experiment was carried out with full-length BamHI GC+DX RNA in the presence of ATP-γ-S, two effects were observed (Fig. 8C). First, the kinetics of p65 appearance lagged behind that observed in the presence of ATP and creatine phosphate (compare Fig. 8C and 2B). Second, in the presence of ATP-γ-S, the intensity of p65 cross-linking did not diminish. In parallel experiments, we found that splicing was inhibited before step 1 in the presence of ATP-γ-S and that complex assembly, as assessed by glycerol gradients, was also delayed (data not shown). This was in contrast to previous experiments in which ATP-γ-S was reported to inhibit only step 2 of splicing (71), but is consistent both with the effects of protein phosphatase inhibitors tautomycin and microcystin LR, which inhibit before step 1 (44, 45), and with the effect of thiophosphorylation of U1 70K (72). Thus, these data show first that p65 appearance requires ATP hydrolysis. Second, they show that the decrease in p65 cross-linking is inhibited by ATP-γ-S. The second effect could indicate that the decline in p65 cross-linking involves either (i) a second ATP-dependent step that cannot use ATP-γ-S, (ii) dephosphorylation of proteins that were phosphorylated in an early step, or (iii) a step other than dephosphorylation that is sensitive to thiophosphate groups.
DISCUSSION
We have demonstrated here the application of MB cross-linking to identify a protein interacting at an snRNA–pre-mRNA duplex that was not detectable by UV cross-linking. Several lines of evidence suggest that p65 cross-links to the U1 snRNA-5′ splice site duplex. Within the pre-mRNA substrate, a 5′ splice site was necessary and sufficient for p65 cross-linking. With full-length RNAs, the kinetics of p65 appearance and the effects of mutations that block splicing at specific stages were consistent with an early and transient interaction. Site-specific labeling at the 5′ exon-intron junction demonstrated that cross-linking occurred at the 5′ splice site. Finally, p65 cross-linking required an intact 5′ end of U1 snRNA, was inhibited by an RNA containing a 5′ splice site-complementary sequence, and occurred in U1 snRNP-containing complexes. Given the preference of MB cross-linking for dsRNA, the simplest interpretation consistent with all of the data is that p65 cross-links to the U1 snRNA-5′ splice site duplex.
Identity of p65?
The identity of p65 is an open question. Candidate mammalian spliceosome proteins of approximately the appropriate size include U1 70K (55), U2AF65 (82), an Sm protein-associated 69-kDa protein (25), an ∼68-kDa proteolytic fragment of a novel pre-mRNA splicing factor (PSF) (24, 54), the 68-kDa SF1/mBBP (3), and the 17S U2 snRNP-associated SF3a66/SAP62 (4, 9). In addition, a recent large-scale effort to sequence spliceosome proteins via nanoelectropsray mass spectrometry has identified a number of novel proteins in the size range of p65 (42a). So far we have tested antibodies to U1 70K, U2AF65, p69, and SF1. None of these was able to immunoprecipitate p65, with the exception of the U1 70K antibody, and that was only able to precipitate p65 under conditions in which U1 snRNP was not disrupted. Although the immunoprecipitation data indicated that p65 is not U1 70K, the fact that it cross-links to the 5′ splice site before full E complex assembly, most likely to a U1 snRNA-5′ splice site duplex, and its coimmunoprecipitation with U1 70K suggest that p65 is a U1-associated protein, although it may not be as tightly associated as the core Sm proteins and the A, C, and 70K U1-specific proteins. Yeast U1 snRNP contains several additional specific proteins (18), including PRP39p (39) and PRP40p (27). PRP39p is a 75-kDa protein that facilitates binding of U1 snRNP to the 5′ splice site. It is possible that p65 is a member of this class of proteins and that in mammals they are more loosely associated with the snRNP. Other proteins that might be expected to have an early interaction with U1 are members of the SR family. These proteins are able to recruit and stabilize binding of U1 snRNP to the 5′ splice site (17, 29, 69). Binding of SF2 to both RNA and U1 70K protein is enhanced by its phosphorylation (81), which could be linked to the ATP dependence of p65 detection. However, the common SR proteins do not include a protein of 65 kDa (22). With the identity of p65 open to question, due caution is required in speculating about its function. Nevertheless, the specificity of its detection at the U1-5′ splice site duplex suggests that p65 may have a role in modulating this intermolecular RNA-RNA interaction, either by stabilizing it, or perhaps by flagging it for subsequent dissociation in a helicase-driven step.
ATP dependence of p65 cross-linking.
Cross-linking of p65 showed clear ATP dependence (Fig. 8). However, the cross-link appeared with RNA substrates containing only a 5′ splice site which are not capable of assembling any ATP-dependent complexes (46, 58). This is not necessarily inconsistent with the requirement of ATP for spliceosome assembly only at the E-to-A complex transition. First, it is possible that the ATP-dependent interaction of p65 at the 5′ splice site is not an essential interaction. Alternatively, it may be essential, but inhibiting it (by not adding ATP) may not block spliceosome assembly until a later step. An early ATP-dependent transition from the E complex to an E* complex has been described in introns in which A complex formation is blocked by the lack of a usable branch point sequence (10). U1 snRNP is destabilized in the E* complex; possibly, p65 cross-linking correlates with this destabilization step.
A number of steps during splicing consume ATP. These involve both protein phosphorylations, e.g., of U1 70K and SR proteins, and also conformational rearrangements, e.g., RNA helicase-driven RNA refolding reactions. It seems likely that the ATP dependence of p65 cross-linking is related to a protein phosphorylation step, since in yeasts and humans, the first action of a helicase occurs during recruitment of U2 snRNP to the branch point (21, 60, 75), yet p65 cross-linking occurs on substrates containing only a 5′ splice site. This need not imply that p65 itself is a phosphoprotein; rather a conformational rearrangement caused by phosphorylation of another protein may result in positioning of p65 adjacent to the U1-5′ splice site duplex. For instance, phosphorylation of SF2 enhances both RNA binding and interaction with U1 70K (81). However, the best candidate is phosphorylation of U1 70K. Protein phosphatase inhibitors differentially inhibit the two steps of splicing (44, 45), and ATP-γ-S has likewise been shown to inhibit splicing both before and after catalytic step 1 (72). The step 1 blockage was most likely due to thiophosphorylation of U1 70K, since U1-depleted extracts could be treated with ATP-γ-S and subsequent addition of U1 snRNP with prephosphorylated U1 70K allowed step 1 to occur at low levels, while thiophosphorylated U1 did not (72). Therefore, although the immunoprecipitation data suggest that p65 is not U1 70K, it appears plausible that the association of p65 with the 5′ splice site is controlled in part by the phosphorylation state of U1 70K.
Application of MB cross-linking to other interactions.
As outlined in the introduction, the U1 snRNA-5′ splice site duplex is only one of a number of intermolecular duplexes that form transiently during pre-mRNA splicing. It is likely that MB cross-linking could detect proteins that interact with the other duplexes. It is therefore pertinent to question why we did not detect other proteins at these duplexes. One obvious candidate is the U6-5′ splice site interaction. Most of our experiments used the GC+DX RNA, which has a 5′ splice site (AAGGUAccc) that would not be expected to base pair with U6—the nonconsensus CCC would not base pair with the U1 or U6 snRNAs. The RNA that we site-specifically labeled at intron position +5 had the sequence AAGGUAUGU, in which the final UGU would be expected to base pair well with U6 snRNA. Nevertheless, no cross-linking was observed (data not shown). It is possible that not all short RNA duplexes would be amenable to MB cross-linking. MB binds by intercalation, which causes some local distortion of the helix. In some cases, this could have the effect of disrupting the very RNA-protein interaction that one is trying to detect, particularly when the duplex is very short. It may be possible to detect specific cross-links by artificially extending and stabilizing the 5′ splice site-U6 duplex (15, 26). Similar reasons may have prevented detection of proteins at the branch point-U2 snRNA duplex. Our constructs had the branch point sequence ggCUAAC, which would form only 4 bp. However, it is less likely that we would have uncovered novel interactions at this site, since an extensive survey using a photoactivable group on a flexible linker arm has already been used to probe interactions within a 15-Å radius of the branch point (40). We have also attempted to detect proteins that would MB cross-link to the 3′ splice site during step 2 of splicing by using RNAs with a single-labeled phosphate at the 3′ exon-intron junction. The only suggested direct RNA-RNA interaction at the 3′ splice site AG dinucleotide is a non-Watson-Crick G-G base-pairing interaction that has been inferred on the basis of genetic suppression data (11, 52, 61). The adjacent exon bases contact both U5 and U2 snRNAs in yeast (49, 50); although RNA-RNA contacts occur in this region, they do not seem to involve simple Watson-Crick pairing and perhaps are not amenable to MB cross-linking. Perhaps unsurprisingly, we detected no MB cross-linking at this site (data not shown). We have recently used MB cross-linking to detect components of spliceosomes that had been arrested during step 2 of splicing by the insertion of a stable hairpin structure between the pyrimidine tract and the 3′ splice site. The majority of cross-linking in these reactions was to the intramolecular GC-rich hairpin within the pre-mRNA. We specifically detected the U5-specific 116-kDa protein cross-linking after step 1 of splicing. Again, this interaction was invisible to UV cross-linking (37). Given the positive data with the 5′ splice site-U1 snRNA duplex, it is likely that MB cross-linking could be used to identify proteins that interact with other transient duplexes during in vitro splicing or other in vitro processing reactions, such as rRNA processing or RNA editing, that involve transient intermolecular duplexes. In any case, the distinct specificity of MB cross-linking, with its preference for dsRNA, makes it a useful complementary approach that can readily be used with the same labeled RNA substrates alongside UV cross-linking.
ACKNOWLEDGMENTS
We thank Reinhard Lührmann, Bernhard Laggerbauer, and Tilmann Achsel for the U5 116K, U1 70K, and 69-kDa protein antibodies; Angela Kramer for SF1 antibody; and Maria Carmo-Fonseca and Juan Valcárcel for the U2AF65 antibody. We also thank Justine Southby and Gavin Roberts for detailed critical comments on the manuscript.
B.S. is a member of the CNRS and was the recipient of a Marie Curie Research Training Fellowship from the European Union and a NATO Fellowship during the course of this investigation. This work was supported by a grant from the Wellcome Trust (040375/Z/94/Z/PMG/RB) to C.W.J.S.
REFERENCES
- 1.Abmayr S M, Reed R, Maniatis T. Identification of a functional mammalian spliceosome containing unspliced pre-mRNA. Proc Natl Acad Sci USA. 1988;85:7216–7220. doi: 10.1073/pnas.85.19.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Arning S, Gruter P, Bilbe G, Kramer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 5.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 6.Binder L I, Frankfurter A, Rebhun L I. The distribution of tau in the mammalian central nervous system. J Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black D L, Chabot B, Steitz J A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 8.Black D L, Steitz J A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986;46:697–704. doi: 10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- 9.Brosi R, Groning K, Behrens S E, Lührmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 10.Champion-Arnaud P, Gozani O, Palandjian L, Reed R. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol Cell Biol. 1995;15:5750–5756. doi: 10.1128/mcb.15.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanfreau G, Legrain P, Dujon B, Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara M D, Palandjian L, Kramer R F, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;15:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crispino J D, Blencowe B J, Sharp P A. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 15.Crispino J D, Sharp P A. A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes Dev. 1995;9:2314–2323. doi: 10.1101/gad.9.18.2314. [DOI] [PubMed] [Google Scholar]
- 16.Datta B, Weiner A M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- 17.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabrizio P, Esser S, Kastner B, Lührmann R. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 19.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabrizio P, McPheeters D S, Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 21.Fleckner J, Zhang M, Valcarcel J, Green M R. U2AF65 recruits a novel human DEAD box protein required for the U2snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 22.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 23.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 24.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackl W, Lührmann R. Molecular cloning and subcellular localisation of the snRNP-associated protein 69KD, a structural homologue of the proto-oncoproteins TLS and EWS with RNA and DNA-binding properties. J Mol Biol. 1996;264:843–851. doi: 10.1006/jmbi.1996.0681. [DOI] [PubMed] [Google Scholar]
- 25a.Jamison S F, Crow A, Garcia-Blanco M A. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol. 1992;12:4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandels Lewis S, Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 27.Kao H-Y, Siliciano P G. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastner B, Kornstädt U, Bach M, Lührmann R. Structure of the small nuclear RNP particle U1: identification of the two structural protuberances with RNP-antigens A and 70K. J Cell Biol. 1992;116:839–849. doi: 10.1083/jcb.116.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 30.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 31.Konforti B B, Konarska M M. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- 32.Konforti B B, Koziolkiewicz M J, Konarska M M. Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell. 1993;75:863–873. doi: 10.1016/0092-8674(93)90531-t. [DOI] [PubMed] [Google Scholar]
- 33.Kramer A. The biochemistry of pre-mRNA splicing. In: Lamond A I, editor. Pre-mRNA processing. R. G. Austin, Tex: Landes Company; 1995. pp. 35–64. [Google Scholar]
- 34.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 35.Kramer A, Keller W, Appel B, Lührmann R. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- 36.Lesser C F, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z-R, Laggerbauer B, Lührmann R, Smith C W J. Crosslinking of the U5 snRNP-specific 116-kDa protein to RNA hairpins that block step 2 of splicing. RNA. 1997;3:1207–1219. [PMC free article] [PubMed] [Google Scholar]
- 37a.Liu, Z.-R., and C. W. J. Smith. Unpublished observations.
- 38.Liu Z-R, Wilkie A M, Clemens M J, Smith C W J. Detection of double-stranded RNA-protein interactions by methylene blue-mediated photo-crosslinking. RNA. 1996;2:611–621. [PMC free article] [PubMed] [Google Scholar]
- 39.Lockhart S R, Rymond B C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 41.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 42.Madhani H D, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 43.Mayer S A, Buvoli M, Nadal-Ginard B. Purine sequence elements in smooth muscle-specific α-tropomyosin exon 2 bind a novel 120 kD protein and activate 3′ splice site selection. J Cell Biochem Suppl. 1994;18C:116. [Google Scholar]
- 44.Mermoud J E, Cohen P, Lamond A I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mermoud J E, Cohen P T, Lamond A I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michaud S, Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 47.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA in the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- 48.Moore M J, Sharp P A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 48a.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 49.Newman A J, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 50.Newman A J, Teigelkamp S, Beggs J D. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsen T. RNA-RNA interactions in the spliceosome: unravelling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 52.Parker R, Siliciano P G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 53.Parker R, Siliciano P G, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 54.Patton J G, Porro E B, Galceran J, Tempst P, Nadal Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 55.Query C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 56.Query C C, Moore M J, Sharp P A. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- 57.Query C C, Strobel S A, Sharp P A. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 58.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 59.Roscigno R F, Garcia Blanco M A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 60.Ruby S W, Chang T H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 61.Scadden A D J, Smith C W J. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 64.Siliciano P G, Guthrie C. 5′ Splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 65.Smith C W J, Chu T T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith C W J, Nadal Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 67.Smith C W J, Porro E B, Patton J G, Nadal Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 68.Sontheimer E J, Steitz J A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 69.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarn W Y, Steitz J A. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 71.Tazi J, Daugeron M-C, Cathala G, Brunel C, Jeanteur P. Adenosine phosphorothioates (ATPalphaS and ATPgammaS) differentially affect the two steps of mammalian pre-mRNA splicing. J Biol Chem. 1992;267:4322–4326. [PubMed] [Google Scholar]
- 72.Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunel C, Lührmann R. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature. 1993;363:283–286. doi: 10.1038/363283a0. [DOI] [PubMed] [Google Scholar]
- 73.Teigelkamp S, Newman A J, Beggs J D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valcarcel J, Gaur R K, Singh R, Green M R. Interaction of U2AF-65 RS region with pre-mRNA of branchpoint and promotion base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 75.Wells S E, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Will C L, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 77.Wu J, Manley J L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 78.Wu J A, Manley J L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- 79.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 80.Wu S, Green M R. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 1997;16:4421–4432. doi: 10.1093/emboj/16.14.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao S-H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 82.Zamore P D, Patton J G, Green M R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 83.Zhuang Y, Weiner A M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 84.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]