Abstract
Induction of the prodynorphin gene has been implicated in medium and long-term adaptation during memory acquisition and pain. By 5′ deletion mapping and site-directed mutagenesis of the human prodynorphin promoter, we demonstrate that both basal transcription and protein kinase A (PKA)-induced transcription in NB69 and SK-N-MC human neuroblastoma cells are regulated by the GAGTCAAGG sequence centered at position +40 in the 5′ untranslated region of the gene (named the DRE, for downstream regulatory element). The DRE repressed basal transcription in an orientation-independent and cell-specific manner when placed downstream from the heterologous thymidine kinase promoter. Southwestern blotting and UV cross-linking experiments with nuclear extracts from human neuroblastoma cells or human brain revealed a protein complex of approximately 110 kDa that specifically bound to the DRE. Forskolin treatment reduced binding to the DRE, and the time course paralleled that for an increase in prodynorphin gene expression. Our results suggest that under basal conditions, expression of the prodynorphin gene is repressed by occupancy of the DRE site. Upon PKA stimulation, binding to the DRE is reduced and transcription increases. We propose a model for human prodynorphin activation through PKA-dependent derepression at the DRE site.
The level of expression of a given gene reflects the balance between positive and negative signals affecting the general transcription machinery via sequence-specific DNA-binding factors (17, 35, 40). Extracellular signals modify basal transcription by changing the expression and/or efficiency of transcriptional activators such as Fos, Jun, and cyclic AMP (cAMP)-responsive element (CRE) binding protein (CREB) (23, 31, 38) or transcriptional repressors such as inducible cAMP early repressor (ICER) and CRE modulator (CREM) (13, 35). Alternatively, extracellular signals can activate genes that are constitutively repressed through active derepression (19, 20, 39).
Learning and chronic pain adaptation involve profound changes in gene expression within the central nervous system, including prodynorphin gene expression (7, 33). Tonically, dynorphin peptides modulate the release of neurotransmitters from presynaptic terminals (42, 44) and so regulate brain function. Induction of the rat prodynorphin gene in different cells and tissues is associated with the activation of protein kinase A (PKA) and protein kinase C signal transduction pathways (7, 26, 41). In rat spinal cord neurons, early induction of c-Fos is followed by a substantial increase in prodynorphin mRNA levels (26, 33). Furthermore, antisense studies indicate that c-Fos participates in prodynorphin gene transactivation (22, 26). In cultured striatal neurons, however, induction of the rat prodynorphin gene has been associated exclusively with the phosphorylation of CREB upon dopamine D1 receptor activation (7).
Transcription of the rat prodynorphin gene is controlled by at least five regulatory elements distributed along 2 kb of the promoter region. Three elements have been described in the distal area of the promoter, at positions −1656, −1627, and −1543 relative to the transcription start site (11). These elements function as CREs (DynCRE1, -2, and -3), and binding to CREB results in repression of AP-1-mediated prodynorphin transactivation (8). The mechanism for this transrepression is not known, although competition by different nuclear complexes at the DynCRE3 element could be a possibility, since it has been shown that DynCRE3 also binds AP-1 nuclear complexes and through this interaction transactivates the prodynorphin gene (8, 29, 30). In the proximal promoter region, close to the transcription start site, a noncanonical AP-1 site (ncDynAP-1) and a CRE-like element (DynCRE4) are present (11, 33). The ncDynAP-1 site, located at position −257, is conserved in the human gene and binds Fos-Jun heterodimers in vitro. Through this interaction, the ncDynAP-1 site is responsible for the induction of prodynorphin in NCB20 cells after treatment with phorbol esters (33). The DynCRE4 element is located downstream from the transcription start site, centered at position +61, and participates in transcriptional induction via cAMP (11, 12, 28).
By deletion analysis and site-directed mutagenesis, we define the minimal inducible promoter responsible for basal and PKA-regulated transcription of the human prodynorphin gene in human neuroblastoma cells. We also demonstrate the functional importance of a downstream response element (DRE), which acts as a transcriptional silencer under basal conditions. Furthermore, we have identified a 110-kDa nuclear complex that binds specifically to the DRE in several human neuroblastoma cell lines under basal conditions. Upon PKA stimulation, binding to the DRE is reduced and the prodynorphin gene is transcribed. Our results suggest that prodynorphin transcription is actively repressed in human neuroblastoma cell lines and that PKA activation results in transcriptional derepression.
MATERIALS AND METHODS
Culture of cell lines.
The neuronal cell lines NB69, SK-N-MC, SK-NB(E), and SH-SY5Y were grown in Dulbecco’s modified Eagle’s medium (DMEM)–Ham F-12 medium supplemented with 10% fetal calf serum, 2 mM glutamine, and 50 μg of gentamicin per ml. DMEM containing the same supplements was used to grow HeLa and U373 cells.
Plasmid constructions.
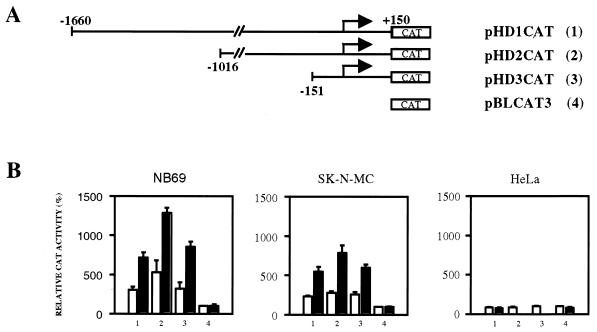
A 1.8-kb fragment (−1660 to +150) containing the regulatory region of the human prodynorphin gene (21) was prepared by PCR with human genomic DNA as a template and the specific primers 5′-CAAGCTTACAGATGAGCAATCAGAGGTTC-3′ and 5′-TTGGATCCCTGGGCAGCTTCTGGCCGAGCAGGTCGGTCGGAGGTG-3′. The PCR product was confirmed by sequencing on both strands and then cloned in the promoterless reporter vector pBLCAT3 (27) by using HindIII and BamHI primer restriction sites, generating reporter plasmid pHD1CAT. Deletion of a 644-bp BglII fragment from the 5′ end of the 1.8-kb fragment generated reporter plasmid pHD2CAT. Subsequent deletion up to position −150 by using a TaqI restriction site produced the reporter plasmid pHD3CAT containing 300 bp of the regulatory region (see Fig. 2A).
FIG. 2.
Deletion mapping of the human prodynorphin promoter. (A) Schematic representation of the 5′ deletions (no. 1 to 3) in the human promotor and the reporter vector pBLCAT3 (no. 4). (B) Transient transfections in the human neuroblastoma cells NB69 and SK-N-MC and in HeLa cells with the reporter plasmids 1 to 4 shown in panel A, corresponding to the numbers on the x axis. The results are expressed as CAT activity relative to that of empty reporter vector pBLCAT3 (no. 4) under basal conditions. Open bars represent values from untreated cultures, and solid bars represent values from forskolin-treated cultures. The transfection experiments were repeated seven times in duplicate.
Site-directed mutagenesis of the human prodynorphin promoter was performed directly on reporter plasmid pHD1CAT by using a non-PCR-based protocol (Transformer kit; Clontech). The oligonucleotide 5′-CCAAGCCGGAATCAAGGAGG-3′ (mDRE) introduced a single base mutation in the DRE, generating the reporter plasmid pHD1mDRECAT. The underlined letters depict the position of the mutated base.
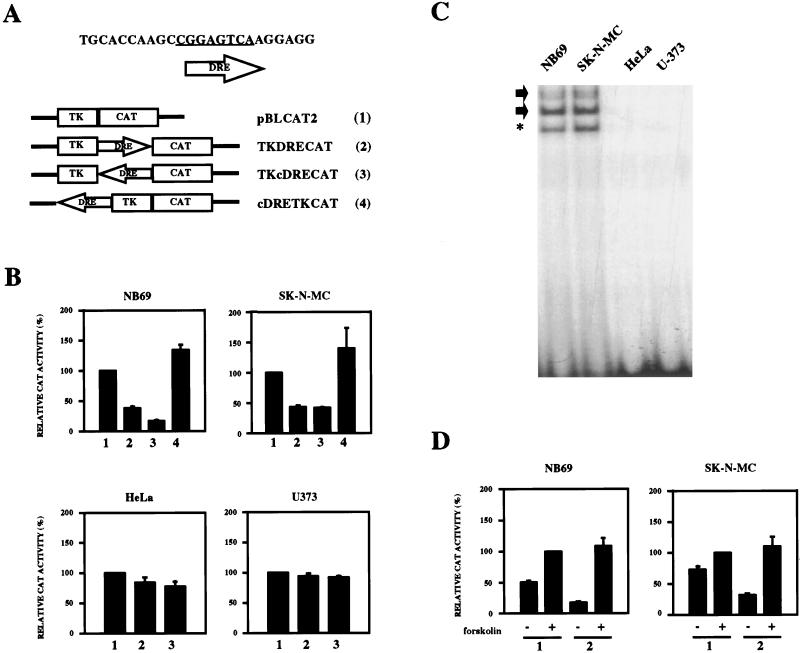
The oligonucleotide 5′-TCGACCAAGCCGGAGTCAAGGAGG-3′, containing the DRE sequence bearing XhoI or HindIII restriction sites, was cloned downstream or upstream of the thymidine kinase minimal promoter in the reporter vector pBLCAT2 (27). The resulting reporter plasmids pTKDRECAT (downstream), pTKcDRECAT (downstream complementary), and pcDRETKCAT (upstream complementary) (see Fig. 5A) were confirmed by sequencing.
FIG. 5.
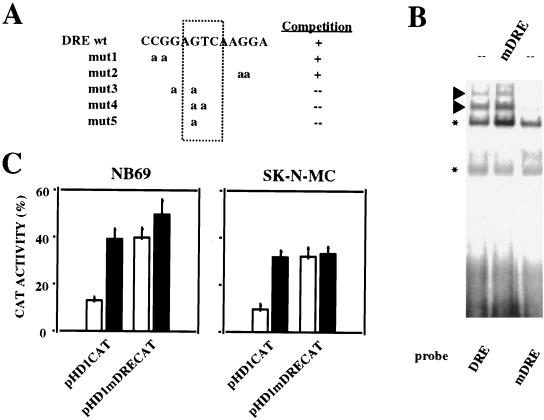
Repression of the heterologous promoter from the thymidine kinase gene by DRE. (A) Schematic representation of the DRE constructs used (no. 2 to 4) and the empty reporter vector pBLCAT2 (no. 1). (B) Effect of the DRE under basal conditions after transient transfections in NB69, SK-N-MC, HeLa, and U373 cell lines with the reporter plasmids 2 to 4 shown in panel A, corresponding to the numbers on the x axis. The results are expressed as CAT activity relative to that of the pBLCAT2 reporter (no. 1). (C) Electrophoretic mobility shift assay with the DRE as a probe and nuclear extracts from the indicated cell lines (top). Arrows indicate the DRE-specific retarded bands. Asterisks denote nonspecific retarded bands. (D) Effect of the DRE after transient transfections in forskolin-treated NB69 and SK-N-MC cells using the reporter plasmids 1 and 2 shown in panel A, corresponding to the numbers on the x axis. The results are expressed as CAT activity relative to that of the pBLCAT2 reporter after forskolin treatment. All transfection experiments were repeated four times in duplicate.
Primer extension analysis.
mRNA was extracted from NB69 and SK-N-MC cells (FastTrack; Invitrogen). Primer extension analysis was performed as described previously (4). The oligonucleotide 5′-GAAGCCGGAGTCAAGGAGGCCCCTG-3′ was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase and used as a primer for the extension. Labeled primer (1 pmol) was mixed with 250 ng of poly(A)+ RNA in annealing buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.4], 200 mM NaCl} and incubated for 12 h at 55°C. The annealed products were extended for 1 h at 42°C with Moloney murine leukemia virus reverse transcriptase (10 U) in the presence of 25 μg of actinomycin D per ml. The primer extension products were resolved in a denaturing 6% polyacrylamide gel together with a sequencing reaction mixture obtained with the same primer.
Transient transfection and CAT analysis.
Cells were seeded 8 h before transfection at a density of 2 × 106 cells per 100-mm-diameter dish. Ten micrograms of DNA, containing 3 μg of reporter plasmid, 1 μg of β-galactosidase expression vector (pCH110; Pharmacia), and 6 μg of carrier plasmid DNA, was coprecipitated with calcium phosphate and added to the NB69 cultures. For transfection of SK-N-MC cells, double the amount of plasmid DNA was used. Twelve hours later, the cells were washed, fresh medium was added, and the cells were left for an additional period of 24 h. Treatments (25 μM forskolin, 1 mM dibutyryl-cAMP, or 1 μM H89) were performed 6 h or 24 h before the NB69 or SK-N-MC cells, respectively, were harvested. Cells were lysed by three freeze-thaw cycles, and the protein concentration was determined (Bradford assay; Bio-Rad). The chloramphenicol acetyltransferase (CAT) activity was assayed in 100 μg of protein extract (18) and normalized with respect to β-galactosidase activity.
Electrophoretic mobility shift analysis.
Nuclear extracts were prepared as described previously (3). Cells (106) were incubated on ice in hypotonic buffer. The nuclei were pelleted and lysed with high-salt buffer. Nuclear proteins were quantified and extracts were immediately frozen in liquid nitrogen. Double-stranded oligonucleotides corresponding to the human DRE (5′-GAAGCCGGAGTCAAGGAGGCCCCTG-3′) and DREmut5 (mDRE) (5′-GAAGCCGGAATCAAGGAGGCCCCTG-3′ [mutation in boldface]) were labeled with [γ-32P]ATP and T4 polynucleotide kinase and used as probes. Nuclear proteins (5 to 10 μg) were incubated with radioactive oligonucleotide probe (100,000 cpm) for 20 min at room temperature in reaction buffer [10 mM HEPES (pH 7.9) 10% glycerol, 0.1 mM EDTA, 8 mM MgCl2, 1 mM dithiothreitol (DTT), 0.15 μg of poly(dI-dC) per ml]. Protein-DNA complexes were resolved in 5% nondenaturing polyacrylamide gels and visualized by autoradiography. For DNA competition experiments, a 3- to 30-fold excess of unlabeled double-stranded oligonucleotide was added 5 min before the probe. As cold competitors, the canonical CRE from the somatostatin gene (cCRE), 5′-CTTGGCTGACGTCAGAGA-3′, and the canonical AP-1 from the collagenase gene (cAP-1), 5′-AAGCTTGCATGACTCAGACAG-3′, were used. To assess nonspecific binding, cold double-stranded oligonucleotides containing the Sp1 site (5′-ATTCGATCGGGGCGGGGCG-3′ and the Oct1 site (5′-TGTCGAATGCAAATCACTAGAA-3′) were used in competition experiments. To define key nucleotides within the DRE sequence, the following mutated (boldface) double-stranded oligonucleotides were used in competition experiments: DREmut1, 5′-GAAGCAAGAGTCAAGGAGGCCCCTG-3′; DREmut2, 5′-GAAGCCGGAGTCAAAAAGGCCCCTG-3′; DREmut3, 5′-GAAGCCGAAATCAAGGAGGCCCCTG-3′; and DREmut4, 5′-GAAGCCGGAAACAAGGAGGCCCCTG-3′.
UV cross-linking.
Conditions for protein-DNA interaction were as described for electrophoretic mobility shift experiments. After incubation of nuclear extracts with the DRE probe, the reaction mixture was UV cross-linked for 30 min by using a high-intensity UV lamp at a distance of 3.5 cm at room temperature (4). An equal volume of Laemmli buffer (×2; 100 mM Tris [pH 6.8], 200 mM DTT, 4% sodium dodecyl sulfate [SDS], 0.2% bromphenol blue, and 20% glycerol) was added, and samples were boiled. The UV cross-linked products were resolved in SDS–10% polyacrylamide gels and visualized by autoradiography. Competition was performed by adding a 30-fold excess of cold DRE oligonucleotide 5 min before the probe. In control experiments with proteinase K (3 μg), the enzyme was added during the incubation.
Southwestern analysis.
Southwestern analysis was carried out essentially as described previously (4). Nuclear proteins (50 μg) were resolved in SDS–10% polyacrylamide gels and transferred to nitrocellulose membranes. The blots were renatured in phosphate-buffered saline for 5 min at room temperature and blocked with 3% nonfat dry milk in TNED buffer (40 mM Tris [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT) for 1 h. The binding assay was performed for 2 h at room temperature in TNED buffer containing salmon sperm DNA (10 μg/ml) and 106 cpm of double-stranded DRE oligonucleotide per ml labeled with [α-32P]dCTP by nick translation. Unbound probe was removed by washing three times with 20 ml of the same buffer for 10 min each at room temperature. The blots were exposed for autoradiography. In competition experiments, a 50-fold excess of unlabeled oligonucleotides containing cAP-1, cCRE, DRE, or mutated DRE was used.
RNA analysis.
Extraction of total RNA, reverse transcription (RT)-PCR, and Southern blotting were performed essentially as described previously (33). The specific primers used to amplify human prodynorphin mRNA were 5′-TGGCAGGGGCTGGTCCTGGCTGCCTGC-3′ and 5′-TTATGCATCAAAAAGCTCTCCAGAGTA-3′. To control for the amount of RNA in each sample, β-actin was amplified.
RESULTS
The human prodynorphin gene uses two major transcription start sites.
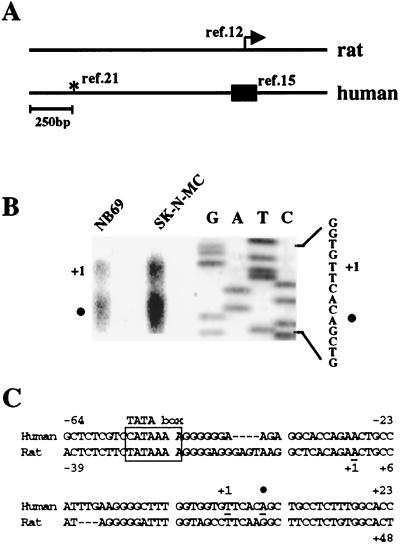
Sequence analysis of the human (15) and rat (12) prodynorphin transcripts indicated that the transcription start sites in both genes are located within the same region (Fig. 1A, box and arrow, respectively), which is different from the site initially assigned for the human gene (21) (Fig. 1A, asterisk). To map the human prodynorphin gene, we first established the transcription start site by primer extension analysis with mRNA from NB69 and SK-N-MC cells, human neuroblastoma cell lines that express prodynorphin (8). In both cell lines, we found that the human prodynorphin gene uses two major transcription start sites (Fig. 1B), separated by 5 nucleotides, which have a location equivalent to the transcription start sites in the rat prodynorphin gene. The sequence ATAAA, located 50 bp upstream from the initiation of the longer transcript, is a putative TATA box in the human prodynorphin gene (Fig. 1C).
FIG. 1.
The transcription start site in the human prodynorphin gene. (A) Alignment of the rat and human prodynorphin genes. The arrow indicates the transcription start site in the rat gene (12). The box encompasses the region in the human gene recently proposed to contain the start of transcription for the human gene (15), and the asterisk denotes a position proposed earlier (21). (B) Primer extension analysis of human prodynorphin transcripts using mRNA from NB69 and SK-N-MC cells. The first transcription start site is represented by +1, and the solid circle indicates a second transcription start site 5 bp downstream. A sequencing reaction with the primer used for the extension analysis is shown to the right. (C) Nucleotide sequence of the region in the human prodynorphin gene containing the transcription start site (+1 and solid circle) shown in relation to a putative TATA box (boxed) located at −50 bp. For comparison, the corresponding region of the rat gene is shown.
Basal and inducible transcription of the human prodynorphin gene requires the 5′ untranslated region.
To analyze regulatory mechanisms that control the expression of the human prodynorphin gene, we cloned a 1.8-kb genomic fragment encompassing the promoter region +150 to −1660 and prepared a set of reporter constructs containing the entire fragment (pHD1CAT) and 5′ deletions linked to a CAT reporter gene (Fig. 2A). Transient transfections were performed with NB69 and SK-N-MC cells. In both cell lines, under basal conditions, a threefold increase in acetylation was obtained with the pHD1CAT construct compared to the empty vector pBLCAT3 (Fig. 2B). The 5′ deletion constructs, pHD2CAT and pHD3CAT, still retained acetylation values, similar to pHD1CAT and significantly higher than pBLCAT3 (Fig. 2B). Upon PKA stimulation, which induces prodynorphin expression (7, 26, 41), increased transactivation occurred with the pHD1CAT to -3CAT constructs. These results point to the 300-bp fragment (−150 to +150) included in pHD3CAT as the minimal inducible promoter for the human prodynorphin gene. Interestingly, transfection with the pHD1CAT to -3CAT constructs in HeLa cells, a cell line devoid of prodynorphin expression, resulted in acetylation values not different from those obtained with pBLCAT3 under either basal or forskolin-stimulated conditions (Fig. 2B).
The minimal inducible promoter contains the DRE.
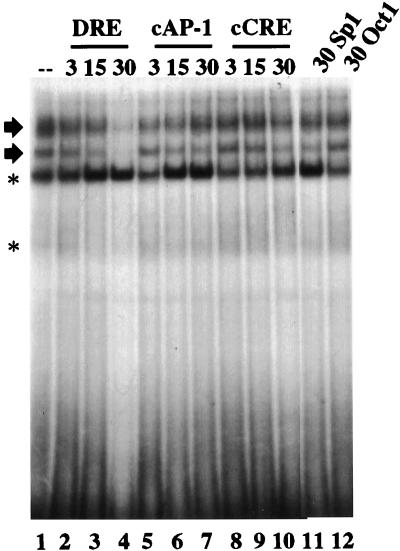
Sequence analysis of the minimal inducible promoter showed that the last 4 bases of the rat DynCRE4 (GTCA), a sequence involved in the transcriptional activation of the rat prodynorphin gene (11, 12, 28), were conserved in the human prodynorphin gene at position +40. With NB69 nuclear extracts, an oligonucleotide encompassing nucleotides from +30 to +50 of the human gene generated several retarded bands (Fig. 3, lane 1). The two upper bands were competed by the element itself (Fig. 3, lanes 2 to 4) and were not affected by a 30-fold excess of oligonucleotide containing the Sp1 site or the Oct1 site (Fig. 3, lanes 11 and 12), indicating the specificity of the protein-DNA interaction. Moreover, competition with increasing concentrations of cold cCRE or cAP-1 did not modify the appearance of the specific retarded bands (Fig. 3, lanes 5 to 10). Based on these results, we named the putative regulatory sequence centered at +40, DRE.
FIG. 3.
Electrophoretic mobility shift analysis of the DRE from the human prodynorphin gene. Results from competition assays of the DRE retarded bands with the indicated fold excess (3-, 15-, and 30-fold) of cold competitors (top) are shown. The two specific DRE retarded bands are indicated by arrows. Nonspecific retarded bands are indicated by asterisks.
To further investigate the functional relevance of the human DRE, we performed site-directed mutagenesis of the DRE within the reporter plasmid pHD1CAT. To select a base for the mutation of the DRE, we prepared a series of variants of the DRE oligonucleotide containing substitutions of selected nucleotides with adenosines. We analyzed their ability to compete the DRE retarded bands in electrophoretic mobility shift assays. The DRE mutants 1 and 2 were still able to compete the DRE retarded bands as efficiently as cold wild-type DRE, while mutants 3 to 5 had lost this capability (Fig. 4A). These results indicate that the central single base substitution G→A in the mutated oligonucleotide DREmut5 is sufficient to alter the functionality of the DRE site. Electrophoretic mobility shift assays performed with the DREmut5 oligonucleotide (hereafter mDRE) showed a considerable reduction in the intensity of the two uppermost retarded bands compared with wild-type DRE, and cold mDRE did not compete the DRE-specific retarded bands (Fig. 4B). After transient transfections in NB69 or SK-N-MC cells, increased CAT activity was observed with the mutated reporter plasmid pHD1mDRECAT, similar to that seen with the wild-type pHD1CAT after forskolin treatment, and no further induction was obtained by PKA stimulation with the mutated DRE construct (Fig. 4C). We interpret these results as a repression of basal transcription at the DRE site. Upon PKA activation, or mutation of the DRE, the repression is relieved and transcription is activated. Since forskolin is not able to transactivate the construct containing the mutated DRE, we hypothesize that PKA-mediated prodynorphin induction is initiated through derepression at the DRE site. Based on alignment of the human (21), pig (21), rat (12), and mouse (34) prodynorphin promoters, together with data from the competition using DRE mutant oligonucleotides, we propose the sequence PuNGTCAPuPuG as the consensus DRE.
FIG. 4.
Site-directed mutagenesis of the DRE on the human prodynorphin promoter. (A) Sequence of the different DRE mutant oligonucleotides aligned with the wild-type DRE. Their ability to compete the specific DRE retarded bands is shown to the right. (B) Electrophoretic mobility shift assay showing the retardation pattern with the wild-type DRE as a probe (lane 1) and the effect on the specific retarded bands (arrowheads) of the selected mutation DREmut5 (renamed mDRE [lane 3]). Competition with 30-fold excess of cold mutated DRE (mDRE) is shown in lane 2. Asterisks denote nonspecific retarded bands. (B) Transient transfections in NB69 and in SK-N-MC cells were performed with plasmids containing either the wild-type promoter (pHD1CAT) or the promoter mutated at the DRE site (pHD1mDRECAT). CAT activity obtained from each reporter plasmid under basal conditions (open bars) and after forskolin treatment (solid bars) is shown. The transfection experiments were repeated four times in duplicate.
The DRE acts as a transcriptional silencer and mediates PKA-dependent transcriptional derepression.
To determine whether the DRE can repress transcription from a heterologous promoter, we subcloned the DRE sequence in both orientations downstream from the minimal promoter of the thymidine kinase (tk) gene in the reporter vector pBLCAT2, generating reporter plasmids pTKDRECAT and pTKcDRECAT (Fig. 5A). Transient transfections in NB69 and SK-N-MC cells revealed that insertion of the DRE site downstream from the tk minimal promoter, independent of the orientation, significantly reduced the basal transcription obtained with the tk minimal promoter alone (Fig. 5B). The same experiment performed with HeLa cells or U373 human glioma cells failed to show any difference in transcription between the pTKDRECAT or pTKcDRECAT and pBLCAT2 reporter plasmids (Fig. 5B). Consistent with these results, nuclear extracts from HeLa and U373 cells failed to generate the specific DRE retarded bands in electrophoretic mobility shift assays that are readily observed with nuclear extracts from NB69 or SK-N-MC cells (Fig. 5C). Thus, transcriptional repression by DRE is cell specific and involves specific nuclear proteins present in NB69 and SK-N-MC cells that interact with the DRE sequence. However, the DRE did not influence basal transactivation when placed upstream from the tk minimal promoter in reporter plasmid pcDRETKCAT (Fig. 5A and B). These results indicate that the DRE represses transcription in a manner independent of the orientation and promoter context, but only when placed downstream from the TATA box. To test the hypothesis that PKA-dependent transcription of the human prodynorphin gene is mediated through derepression at the DRE, NB69 and SK-N-MC cells were transfected with reporter plasmid pTKDRECAT and treated with forskolin. Consistent with the notion of transcriptional derepression, in both cell lines under stimulated conditions, CAT activity from the reporter pTKDRECAT was similar to that obtained with the vector pBLCAT2 (Fig. 5D).
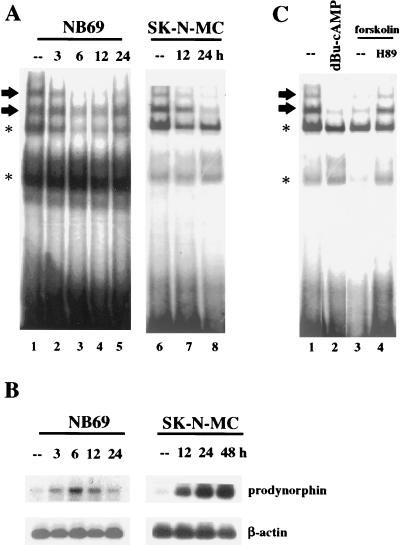
To further investigate the derepression activity, we tested whether the appearance of the specific DRE retarded bands was modified as a result of PKA stimulation. Nuclear extracts were prepared from the two neuroblastoma cell lines under basal conditions and at different times following forskolin treatment. Our results clearly showed that the intensity of the specific DRE retarded bands was reduced upon forskolin treatment (Fig. 6A). Interestingly, in each cell line, the time course for the decreased intensity of the DRE bands closely correlated with the time course of prodynorphin mRNA induction (Fig. 6B). Furthermore, the decrease in intensity of the specific DRE retarded band after forskolin was reversed by H89, a selective inhibitor of PKA (Fig. 6C, compare lanes 3 and 4), and was mimicked by treatment with the stable analog of cAMP, dibutyryl-cAMP (Fig. 6C, compare lanes 1 to 3). Together, these findings support a mechanism by which, under basal conditions, a nuclear repressor occupies the DRE site. Upon PKA induction, the repressor is released from the DRE and transcription can proceed.
FIG. 6.
Blocking of binding to the DRE after PKA activation correlates with the increase in prodynorphin expression in NB69 and SK-N-MC cells. (A) Electrophoretic mobility shift assays showing reduction in the DRE-specific retarded bands (arrows) at the indicated times (top) after forskolin treatment. (B) Southern blot after RT-PCR showing increase in prodynorphin expression in NB69 and SK-N-MC cells at the indicated times (top) after forskolin treatment. Amplification of β-actin was performed in parallel as a control for the mRNA input. (C) Electrophoretic mobility shift assay showing reduction in the DRE-specific retarded bands (arrows) by dibutyryl-cAMP (lane 2) or forskolin (lane 3) treatments at the 6-h time point with the NB69 cell line. The effect of forskolin treatment blocked by H89 is shown in lane 4. Asterisks in panels A and C denote nonspecific retarded bands.
A 110-kDa nuclear protein complex binds specifically to the DRE.
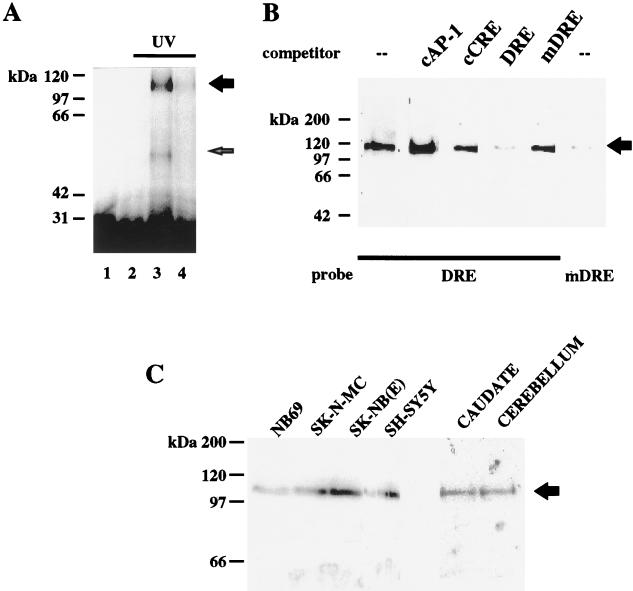
Our results indicate that the DRE behaves as a stimulus-dependent transcriptional silencer and controls both basal and forskolin-induced transcription. To characterize the nuclear protein or proteins that bind to the DRE, we performed UV cross-linking of nuclear extracts from NB69 cells incubated with the DRE probe. Two specific bands with apparent molecular masses of 110 and 55 kDa, respectively, were obtained after UV irradiation (Fig. 7A, lane 3). The intensity of these bands was reduced when the protein-DNA interaction was performed in the presence of proteinase K or when a 30-fold excess of cold DRE was added (Fig. 7A, lanes 2 and 4, respectively). Omission of the UV irradiation resulted in the absence of specific bands (Fig. 7A, lane 1).
FIG. 7.
Characterization of the nuclear activity that binds to the DRE site. (A) NB69 nuclear extracts were UV cross-linked after incubation with the DRE probe and electrophoresed. Three micrograms of proteinase K (lane 2) or a 30-fold excess of cold DRE (lane 4) abolished or reduced, respectively, the specific autoradiographic signal (lane 3). Without UV irradiation, no specific band was observed (lane 1). (B) Southwestern analysis with NB69 nuclear extracts. A protein complex of 110 kDa was obtained with the DRE probe. The autoradiographic band was competed with an excess of cold DRE but not by cold mDRE (top). A probe containing the mutated DRE seriously reduced the autoradiographic signal. (C) Southwestern analysis with the DRE as a probe and nuclear extracts from the neuroblastoma cell lines indicated (top) and human brain samples. The arrow indicates the specific band obtained for the 110-kDa protein complex. Protein molecular mass markers are shown to the left.
In order to validate this finding, we performed Southwestern analysis with nuclear extracts from NB69 cells and the DRE oligonucleotide as a probe. Binding to a protein complex of 110 kDa was observed reproducibly (Fig. 7B, lane 1). The intensity of the 110-kDa band was specifically reduced upon competition with an excess of cold DRE oligonucleotide, but was not modified by an excess of cold oligonucleotide containing the canonical sequences for CRE or the AP-1 site (Fig. 7B, lanes 2 to 4). Furthermore, no reduction in intensity was observed upon competition with the mutated DRE oligonucleotide (Fig. 7B, lane 5), although when the mutated DRE oligonucleotide was used as a probe, a faint 110-kDa band was still observed (Fig. 7B, lane 6). This is in agreement with results obtained by electrophoretic mobility shift assays (Fig. 4A) and suggests that the mutated DRE may retain some residual binding activity in vitro. The 110-kDa DRE binding protein complex was also detected in nuclear extracts from SK-N-MC, SK-NB(E), and SH-SY5Y human neuroblastoma cell lines (Fig. 7C). Conversely, the 110-kDa band was absent in nuclear extracts from HeLa and U373 cells (data not shown). Interestingly, Southwestern analysis of nuclear extracts prepared from human caudate and cerebellum also showed the 110-kDa band corresponding to the DRE binding protein (Fig. 7C). These results support the hypothesis that the 110-kDa protein complex that binds specifically to the DRE site corresponds to the repressor activity, which, under basal conditions, is responsible for transcriptional silencing of genes containing the DRE element.
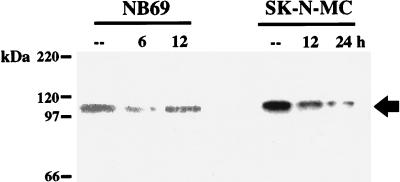
To test whether the 110-kDa DRE binding protein complex indeed corresponds to the factor which reduces its binding to the DRE upon PKA stimulation, we performed Southwestern experiments using nuclear extracts from forskolin-treated NB69 or SK-N-MC cells. In agreement with the time course for unbinding from the DRE (Fig. 6A) and the time course for prodynorphin induction (Fig. 6B) after forskolin, a substantial reduction in intensity of the 110-kDa band was observed 6 or 24 h after PKA stimulation of NB69 or SK-N-MC cells, respectively (Fig. 8). In NB69 cells, binding to the 110-kDa band returned to control values 12 h following forskolin (Fig. 8), corresponding to the time when prodynorphin mRNA levels have returned to basal values (Fig. 6B). Conversely, in SK-N-MC cells, in which prodynorphin mRNA levels remained elevated up to 48 h after forskolin (Fig. 6B), binding to the 110-kDa band stayed reduced 24 h after treatment, the longest time point analyzed (Fig. 8).
FIG. 8.
PKA activation blocks binding of the DRE probe to the 110-kDa protein complex. The results represent Southwestern analysis with nuclear extracts from NB69 and SK-N-MC cells at different times after forskolin treatment. The arrow indicates the specific band obtained for the 110-kDa protein complex.
DISCUSSION
Using deletion mapping and site-directed mutagenesis, we have characterized the DRE site, an intragenic regulatory element that regulates basal and PKA-dependent induction of the human prodynorphin gene. Our results suggest a novel mechanism that involves PKA-dependent transcriptional derepression by differential binding of the DRE binding protein complex to the DRE.
In an early report about the human prodynorphin gene, the transcriptional start site was assigned based on its proximity to a putative TATA box (21). Later, Douglass et al. (12) determined the transcription start site in the rat gene and, upon comparison with the human gene, concluded that the postulated transcription start site in the human gene was incorrect. Instead, they found a region in the human promoter with close sequence similarity to the rat prodynorphin transcription start site (12). More recently, RNase protection analysis also identified this region as the start of transcription in the human gene (15). Our data for the human transcription start site agree with these more recent predictions (12, 15). Moreover, we found that the human gene contains two transcription start sites, as has been previously described for the rat gene (12). The existence of several sites for transcriptional initiation may reflect the presence of a strong secondary structure in this region, but its functional significance, if any, is presently not understood.
The human DRE is related to rat DynCRE4, an asymmetric CRE-like sequence (CGTCA) first described in the rat prodynorphin promoter that participates in transcriptional activation by cAMP (11, 12, 28). Like DynCRE4, the human DRE is intragenic and is located in close proximity to the transcription start site. However, the human DRE contains the sequence GGAGTCA, which more closely resembles an AP-1 sequence (TGAGTCA) in which the initial T is replaced by a G (underlined). Nevertheless, competition experiments with AP-1 or CRE cold oligonucleotides in either electrophoretic mobility shift or Southwestern assays did not modify the binding to the DRE probe, suggesting that the DRE behaves neither as a canonical CRE nor as an AP-1 site. Instead, the DRE probe forms two cell-specific slowly migrating retarded bands with nuclear extracts from prodynorphin-expressing human neuroblastoma cells. The central G residue in the DRE is essential for the binding to the element in vitro and for the regulatory action of the DRE in transient transfection experiments, further indicating the specificity of the DRE-protein interaction. We have defined a preliminary DRE consensus (PuNGTCAPuPuG) based on sequence alignment of prodynorphin genes and nucleotide substitutions. However, a final consensus has to await the identification of DRE sequences present in other genes. Importantly, reduction in the binding to the DRE upon PKA stimulation correlates well with an increased transactivation of the prodynorphin promoter and an increased expression of prodynorphin, indicating that the DRE functions as a transcriptional silencer. Supporting this notion, the DRE can repress transcription in an orientation-independent manner when placed downstream from the transcription start site of a heterologous promoter. However, the DRE shows a position-dependent repressor activity, being totally ineffective when placed upstream of the TATA box. A similar effect has been described recently for the SNOG site, a negative regulatory element that confers neuron-specific expression to the GAP-43 gene and is also present in promoters of other neuron-specific genes, such as those coding for SNAP-25 and nNOS (43). Since the position downstream from the transcription start site appears to be essential for the function of DRE, occupancy of this site by specific repressor proteins may interfere with the formation of the transcription initiation complex or with its conversion to a functional elongation complex and thus inhibit transcription. Such mechanisms have been demonstrated to operate in the repression of the H5 histone gene (16). Furthermore, a position-dependent transcriptional function has been shown recently for the neuron-restrictive silencer (NRS) element (25, 32) in the regulation of the nicotinic acetylcholine receptor gene (5). In neuronal tissue, the NRS activates transcription if placed in close proximity to or downstream from the TATA box, but silences the expression of the gene in neurons when located further upstream (5). Interestingly, in nonneuronal tissue, the NRS consistently behaves as a transcriptional silencer (5).
Two independent experimental techniques, UV cross-linking and Southwestern analysis, have allowed the initial characterization of the DRE binding activity, a 110-kDa nuclear complex that specifically binds to the DRE. In addition, a weaker 55-kDa band observed after UV cross-linking was competed with unlabeled DRE. It is tempting to speculate that the 110-kDa complex represents a dimer of the 55-kDa protein and that these two forms generate the two DRE-specific retarded bands. The DRE binding activity has been detected reproducibly in the prodynorphin-expressing cell lines tested, as well as in caudate samples from human brain. The latter finding further strengthens the functional significance of the DRE binding activity, since it is associated with the high level of expression of the prodynorphin gene in the caudate in vivo (2). However, the 110-kDa nuclear activity was observed also in human cerebellum. Since prodynorphin is expressed at very low levels in rat cerebellum (2), this suggests that the DRE binding protein complex may be involved in the regulation of other genes having DRE or DRE-like elements in their promoters.
The increased activity of the prodynorphin promoter containing a mutation in the DRE implicates DRE binding activity in the repression of prodynorphin transcription under basal conditions. Unlike other repressor factors that restrict transcription of their target genes to cells in which these genes are not expressed (6, 24, 36, 37, 43), the DRE binding activity was detected in cell lines and a brain region where prodynorphin is expressed. This indicates that the DRE binding activity does not confer cell-specific expression, but rather determines gene inducibility in a cell-specific manner. Derepression of the peripherin gene occurs in a cell-specific manner in NGF-differentiated PC12 cells through protein-protein interactions that displace the repressor NF1-L from the negative regulatory element NRE located in the proximal promoter of the gene. However, NF1-L is also detectable in undifferentiated PC12 cells and in nonneuronal cells, where it silences transcription of the peripherin gene (1, 39).
PKA activation in human neuroblastoma cells transactivates the prodynorphin gene, and the time course of this induction correlates with a reduction in binding to the DRE. The mechanism of PKA-mediated derepression at the DRE is presently unknown. However, one could speculate that phosphorylation by PKA reduces the affinity of the DRE binding protein for the DRE, accounting for the derepression. Alternatively, PKA-dependent phosphorylation of other nucleoproteins able to interact with the DRE binding protein may play a role in prodynorphin derepression. Recently, mechanisms of derepression have been documented in which changes in the protein components of a DNA-protein complex lead to unbinding from the regulatory element (1, 39). Release of repressor proteins, allowing transactivators to bind to a regulatory element, has been described in the elastin and Pit1, or GHF1, genes. The IGF-I-mediated increase in elastin transcription occurs via a mechanism of derepression that involves activation of a retinoblastoma control element by nuclear complexes which contain the Rb protein following the abrogation of Sp3 repressor binding (9). In the case of the Pit1/GHF1 gene, the release of AP-1 complexes allows the Pit1/GHF1 protein to act at an enhancer site close to the AP-1 site, thus causing upregulation of the gene (10). Furthermore, a mechanism of derepression mediated by distal regulatory elements has been proposed in the rat prodynorphin gene (8). In this case, transactivation of the rat promoter by AP-1 nuclear complexes requires derepression by phosphorylation of the CREB protein.
Taken together, our results suggest that PKA-dependent transcriptional derepression acting at the DRE is responsible for the induction of the prodynorphin gene in human neuroblastoma cells. Recently, using expression cloning with the DRE as a probe, we have isolated a cDNA encoding a protein that binds to DRE as a 110-kDa complex. A manuscript describing these results has been submitted elsewhere. Further studies involving the cDNA coding for the DRE binding protein should lead to better understanding of the mechanism of the PKA-induced changes in prodynorphin gene expression.
ACKNOWLEDGMENTS
We thank N. S. Foulkes and W. A. Link for critical reading of the manuscript, J. Chowen for correction of English style and grammar, and I. DomPablo and D. Campos for technical assistance.
This work has been supported by grants from DGICYT (PB95-0099), CAM (I+D0049/94), Europharma S.A., and Janssen-Cilag S.A. to J.R.N.
REFERENCES
- 1.Adams A D, Choate D M, Thompson M A. NF1-L is the DNA-binding component of the protein complex at the peripherin negative regulatory element. J Biol Chem. 1995;270:6975–6983. doi: 10.1074/jbc.270.12.6975. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez G, Fairén A, Douglass J, Naranjo J R. Expression of the prodynorphin gene in the developing and adult cerebral cortex of the rat: an in situ hybridization study. J Comp Neurol. 1990;300:287–300. doi: 10.1002/cne.903000302. [DOI] [PubMed] [Google Scholar]
- 3.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1995. [Google Scholar]
- 5.Bessis A, Champtiaux N, Chatelin L, Changeux J-P. The neuron-restrictive silencer element: a dual enhancer/silencer crucial for patterned expression of a nicotine receptor gene in the brain. Proc Natl Acad Sci USA. 1997;94:5906–5911. doi: 10.1073/pnas.94.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 7.Cole R L, Konradi C, Douglass J, Hyman S E. Neuronal adaptation to amphetamine and dopamine: molecular mechanisms of prodynorphin gene regulation in rat striatum. Neuron. 1995;14:813–823. doi: 10.1016/0896-6273(95)90225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins-Hicok J, Lin L, Spiro C, Laybourn P J, Tschumper R, Rapacz B, McMurray C T. Induction of the rat prodynorphin gene through Gs-coupled receptors may involve phosphorylation-dependent derepression and activation. Mol Cell Biol. 1994;14:2837–2848. doi: 10.1128/mcb.14.5.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conn K J, Rich C B, Jensen D E, Fontanilla M R, Bashir M M, Rosenbloom J, Foster J A. Insulin-like growth factor-1 regulates transcription of elastin gene through a putative retinoblastoma control element. J Biol Chem. 1996;271:28853–28860. doi: 10.1074/jbc.271.46.28853. [DOI] [PubMed] [Google Scholar]
- 10.Delhase M, Castrillo J L, de la Hoya M, Rajas F, Hooghe-Peters E L. AP-1 and Oct-1 transcription factors down-regulate the expression of the human PIT1/GHF1 gene. J Biol Chem. 1996;271:32349–32358. doi: 10.1074/jbc.271.50.32349. [DOI] [PubMed] [Google Scholar]
- 11.Douglass J, McKinzie A A, Pollock K M. Identification of multiple DNA elements regulating basal and protein kinase A-induced transcriptional expression of the rat prodynorphin gene. Mol Endocrinol. 1994;8:333–344. doi: 10.1210/mend.8.3.8015551. [DOI] [PubMed] [Google Scholar]
- 12.Douglass J, McMurray C T, Garrett J E, Adelman J P, Calvetta L. Characterization of the rat prodynorphin gene. Mol Endocrinol. 1989;3:2070–2078. doi: 10.1210/mend-3-12-2070. [DOI] [PubMed] [Google Scholar]
- 13.Foulkes N S, Sassone-Corsi P. Transcription factors coupled to the cAMP-signalling pathway. Biochim Biophys Acta. 1996;1288:101–121. doi: 10.1016/s0304-419x(96)00025-x. [DOI] [PubMed] [Google Scholar]
- 14.Geijer T, Bergh J, Terenius L. Expression of prodynorphin in human small cell lung carcinoma cell lines. Regul Pept. 1991;34:181–188. doi: 10.1016/0167-0115(91)90177-i. [DOI] [PubMed] [Google Scholar]
- 15.Geijer T, Telkov M, Terenius L. Characterization of human prodynorphin gene transcripts. Biochem Biophys Res Commun. 1995;215:881–888. doi: 10.1006/bbrc.1995.2546. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Cuadrado A, Rousseau S, Renaud J, Ruiz-Carrillo A. Repression of the H5 histone gene by a factor from erythrocytes that binds to the region of transcription initiation. EMBO J. 1992;11:1857–1866. doi: 10.1002/j.1460-2075.1992.tb05237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich J A, Cutter G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 18.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 20.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 21.Horikawa S, Takai T, Toyosato M, Takahashi H, Noda M, Kakidani H, Kobo T, Hirose T, Inayama S, Hayashida H, Miyata T, Numa S. Isolation and structural organization of the human preproenkephalin B gene. Nature. 1983;306:611–614. doi: 10.1038/306611a0. [DOI] [PubMed] [Google Scholar]
- 22.Hunter J C, Woodburn V L, Durieux C, Pettersson E K E, Poat J A, Hughes J. c-fos antisense oligodeoxynucleotide increases formalin-induced nociception and regulates preprodynorphin expression. Neuroscience. 1995;65:485–492. doi: 10.1016/0306-4522(94)00500-5. [DOI] [PubMed] [Google Scholar]
- 23.Karin M, Smeal T. Control of transcription factor by signal transduction pathways: the beginning to the end. Trends Biochem Sci. 1992;17:418–422. doi: 10.1016/0968-0004(92)90012-x. [DOI] [PubMed] [Google Scholar]
- 24.Kemp L M, Dent C L, Latchman D S. Octamer motif mediates transcriptional repression of HSV immediate early genes and octamer-containing cellular promoters in neuronal cells. Neuron. 1990;4:215–222. doi: 10.1016/0896-6273(90)90096-x. [DOI] [PubMed] [Google Scholar]
- 25.Kraner S D, Chong J A, Tsay H-J, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 26.Lucas J J, Mellström B, Colado M I, Naranjo J R. Molecular mechanism of pain: serotonin1A receptor agonists trigger transactivation by c-Fos of the prodynorphin gene in spinal cord neurons. Neuron. 1993;10:599–611. doi: 10.1016/0896-6273(93)90163-l. [DOI] [PubMed] [Google Scholar]
- 27.Luckow B, Schütz G. CAT construction with multiple unique restriction sites for the functional analysis of eukaryotic promoter and regulatory elements. Nucleic Acids Res. 1987;13:5490–5491. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMurray C T, Devi L, Calavetta L, Douglass J. Regulated expression of the prodynorphin gene in the R2C Leydig tumor cell line. Endocrinology. 1989;124:49–59. doi: 10.1210/endo-124-1-49. [DOI] [PubMed] [Google Scholar]
- 29.Messersmith D J, Kim D J, Gu J, Dubner R, Iadarola M J. c-Jun activation of DynCRE 3 site in the prodynorphin promoter. Mol Brain Res. 1996;40:15–22. doi: 10.1016/0169-328x(96)00029-0. [DOI] [PubMed] [Google Scholar]
- 30.Messersmith D J, Gu J, Dubner R, Douglass J, Iadarola M J. Basal and inducible transcriptional activity of an upstream AP-1/CRE element (DYNCRE3) in the prodynorphin promoter. Mol Cell Neurosci. 1994;5:238–245. doi: 10.1006/mcne.1994.1028. [DOI] [PubMed] [Google Scholar]
- 31.Morgan J I, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 32.Mori N, Schoenherr C, Vandenberh D J, Anderson D J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 33.Naranjo J R, Mellström B, Achaval M, Sassone-Corsi P. Molecular pathways of pain: Fos/Jun-mediated activation of the prodynorphin gene through a noncanonical AP-1 site. Neuron. 1991;6:607–617. doi: 10.1016/0896-6273(91)90063-6. [DOI] [PubMed] [Google Scholar]
- 34.Naranjo, J. R. Unpublished data.
- 35.Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 36.Schoenherr C J, Anderson D J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 37.Schoenherr C J, Anderson D J. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 38.Segal R A, Greenberg M E. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 39.Thompson M A, Lee E, Lawe D, Gizang-Ginsberg E, Ziff E B. Nerve growth factor-induced derepression of peripherin gene expression is associated with alterations in proteins binding to a negative regulatory element. Mol Cell Biol. 1992;12:2501–2513. doi: 10.1128/mcb.12.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 41.Ventura C, Pintus G, Vaona I, Bennardini F, Pinna G, Tadolini B. Phorbol ester regulation of opioid peptide gene expression in myocardial cells. J Biol Chem. 1995;270:30115–30120. doi: 10.1074/jbc.270.50.30115. [DOI] [PubMed] [Google Scholar]
- 42.Wagner J J, Terman G W, Chavkin C. Endogenous dynorphins inhibit excitatory neurotransmission and block LTP induction in the hippocampus. Nature. 1993;363:451–454. doi: 10.1038/363451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber J R M, Skene J H P. Identification of a novel repressive element that contributes to neuron-specific gene expression. J Neurosci. 1997;17:7583–7593. doi: 10.1523/JNEUROSCI.17-20-07583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisskopf M G, Zalutsky R A, Nicoll R A. The opioid peptide dynorphin mediates heterosynaptic depression of hippocampal mossy fiber synapses and modulates long-term potentiation. Nature. 1993;362:423–427. doi: 10.1038/362423a0. [DOI] [PubMed] [Google Scholar]