Abstract
The development of melanocytes, which are pigment-producing cells responsible for skin, hair, and eye color, is absolutely dependent on the action of the microphthalmia basic helix-loop-helix–leucine zipper (bHLH-LZ) transcription factor (Mi); mice lacking a functional Mi protein are entirely devoid of pigment cells. Mi has been shown to activate transcription of the tyrosinase, TRP-1, TRP-2, and QNR-71 genes through specific E-box elements, most notably the highly conserved M box. We investigated the mechanism which enables Mi to be recruited specifically to a restricted subset of E boxes in target promoters while being prevented from binding E-box elements in other promoters. We show both in vitro and in vivo that the presence of a T residue flanking a CATGTG E box is an essential determinant of the ability of Mi to bind DNA, and we successfully predict that the CATGTG E box from the P gene would not bind Mi. In contrast, no specific requirement for the sequences flanking a CACGTG E box was observed, and no binding to an atypical E box in the c-Kit promoter was detected. The relevance of these observations to the control of melanocyte-specific gene expression was highlighted by the fact that the E-box elements located in the tyrosinase, TRP-1, TRP-2, and QNR-71 promoters without exception possess a 5′ flanking T residue which is entirely conserved between species as diverse as man and turtle. The ability of Mi to discriminate between different E-box motifs provides a mechanism to restrict the repertoire of genes which are likely to be regulated by Mi and provides insight into the ability of bHLH-LZ transcription factors to achieve the specificity required for the precise coordination of transcription during development.
The development of an organism is dependent on a highly specific program of gene expression coordinated by signal transduction pathways acting to modulate the activity of transcription factors in response to environmental signals. A particularly important role is played by cell type-specific transcription factors, such as Pit-1 and MyoD, that act as “master regulators” of tissue-specific gene expression by binding specific elements in their target promoters. These master regulators frequently belong to transcription factor families which share highly related DNA binding specificities. Since all cell types will contain multiple members of these families, and since similar elements will be present in promoters which are not coordinately regulated, mechanisms to restrict the repertoire of factors able to bind any given sequence element must operate. Understanding how such specificity is generated is a major goal in developmental biology.
Melanocytes, which are pigment cells responsible for skin, hair, and eye color (34), afford an excellent system for studying the key events underlying cell type-specific gene expression. Mouse genetics has identified over 70 different genes which affect the melanocyte lineage, of which approximately 20 have been cloned. These include not only the genes involved directly in the genesis of pigment, such as tyrosinase or tyrosinase-related protein-1 (TRP-1), but also genes which are necessary for melanocyte survival or differentiation, including those encoding the c-Kit (28) and endothelin B (2, 22, 32) receptors or the transcription factors Pax3 (1, 6), Sox10 (31, 36), and microphthalmia (Mi) (21, 26, 39).
The basic helix-loop-helix–leucine zipper (bHLH-LZ) factor Mi (21, 26) plays a crucial role in the development of the melanocyte lineage; mice bearing mutations in the microphthalmia gene completely lack neural-crest-derived melanocytes (30, 37) and have small eyes due to aberrant formation of the retinal pigment epithelium (27). In addition, under some circumstances Mi can convert fibroblasts to cells expressing melanogenic markers (38). Based on transfection assays, Mi has been shown to activate the expression of the melanocyte-specific genes tyrosinase and TRP-1 (3, 14, 19, 42, 44) through an evolutionarily conserved 11-bp sequence termed the M box (25). The M box contains a core CATGTG E-box motif recognized by Mi and other members of the bHLH-LZ family of transcription factors.
Since neither tyrosinase nor TRP-1 is essential for the survival or differentiation of the melanocyte lineage, it is evident that Mi must target other, as yet uncharacterized genes. Given the significance of Mi as a master regulator of melanocyte development, the identification of these target genes is a major goal. However, as Mi is unlikely to regulate all promoters containing E-box elements, some mechanism must exist to enable Mi to be recruited specifically to a restricted subset of E boxes in target promoters while being prevented from binding E-box elements in other promoters. Understanding how this discrimination is achieved will provide significant insight into Mi function.
We investigated the mechanism which allows Mi to recognize a specific subset of CATGTG E-box elements. The results indicate that the presence of a 5′ flanking T residue is a critical determinant of Mi binding specificity both in vitro and in vivo. The ability of Mi to discriminate between different E-box motifs provides a mechanism to restrict the repertoire of genes which are likely to be regulated by Mi and provides an insight into the ability of bHLH-LZ transcription factors to achieve the specificity required for the precise coordination of transcription during development.
MATERIALS AND METHODS
DNA binding assays.
Standard DNA binding (band shift or electrophoretic mobility shift) assays were performed essentially as previously described (29) by using approximately 0.5 ng of labelled probe and increasing amounts (10, 50, and 250 ng) of unlabelled competitor oligonucleotide DNA. The full sequence of each oligonucleotide used is shown in Table 1.
TABLE 1.
Sequences of oligonucleotides used for DNA binding assays and as targets in the yeast CYC-lacZ reportersa
| Method description | Oligonucleotide sequence |
|---|---|
| DNA binding assays | |
| mTRP-1 M box | 5′-ctagaCAGTGGGGAGGGGAGTCATGTGCTGCCTAGTATt |
| mTRP-1 M box T→A | 5′-ctagaCAGTGGGGAGGGGAGaCATGTGCTGCCTAGTATt |
| mTRP-1 M box T→C | 5′-ctagaCAGTGGGGAGGGGAGcCATGTGCTGCCTAGTATt |
| mTRP-1 M box T→G | 5′-ctagaCAGTGGGGAGGGGAGgCATGTGCTGCCTAGTATt |
| CAC M box | 5′-ctagaCAGTGGGGAGGGGAGTCAcGTGCTGCCTAGTATt |
| hTyros InrE | 5′-gatctCTCAGAGCCAAGACATGTGATAATCACTGTAGTAGTa |
| hTyros InrE m1 | 5′-gatctCTCAGAGCCAAGACATGTGcTAATCACTGTAGTAGTa |
| mTyros InrE | 5′-ctagaCTTAGCCAAAACATGTGATAGTCACTCCAGGt |
| hTyros TDE | 5′-ctagaCTTGTGGAGATCATGTGATGACTTCCTGATTCCt |
| hTyros TDE m1 | 5′-ctagaCTTGTGGAGAaCATGTGATGACTTCCTGATTCCt |
| hTyros TDE m2 | 5′-ctagaCTTGTGGAGATCATGTGcTGACTTCCTGATTCCt |
| hTyros TDE m3 | 5′-ctagaCTTGTGGAGAaCATGTGcTGACTTCCTGATTCCt |
| c-Kit (E) | 5′-ctagaAGGGAGCACCTGCCAGGTGGCTGGCt |
| MB1 | 5′-ctagaCTCGAGGGGCCTGCCATGTGCACCCCGCCCGGGt |
| MB2 | 5′-ctagaCTCGAGGCACAAGACATGTGCTTAGCAAACGGGt |
| MB2 A→T | 5′-ctagaCTCGAGGCACAAGtCATGTGCTTAGCAAACGGGt |
| MB3 | 5′-ctagaCTCGAGACTACAGACACGTGCCACCACACCCAAt |
| P gene | 5′-ctagaAGCTGTGGGCGGGGCCATGTGGGCAGCTTTGTTt |
| Yeast expression | |
| M box | 5′-agatctctcgagCAGTGGGGAGGGAGTCATGTGCTGCCTAGTATCCAGTGGGGAGGGAGTCATGTGCTGCCTAGTATggatcc |
| c-Kit | 5′-agatctctcgagAGGGAGCACCTGCCAGGTGGCTGGCATAGGGAGCACCTGCCAGGTGGCTGGCggatcc |
| MB2 | 5′-agatctctcgagGCACAAGACATGTGCTTAGCAAACGGGATCGCACAAGACATGTGCTTAGCAAACGGGATggatcc |
| MB2 A→T | 5′-agatctctcgagGCACAAGtCATGTGCTTAGCAAACGGGATCGCACAAGtCATGTGCTTAGCAAACGGGATggatcc |
Only one strand is shown. The E-box elements are underlined, and additional bases used to create restriction enzyme sites at the termini are shown in lowercase letters. Lowercase is also used to indicate the positions of specific mutations referred to in the text. Arrows indicate residue substitutions.
Yeast manipulations.
Standard methods for yeast culture and transformation were followed (17). Saccharomyces cerevisiae W303-1a (a ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3) cultures were grown at 30°C in either YPD (1% yeast extract, 2% glucose, 1% peptone) or in minimal medium (0.67% yeast-nitrogen base, 1% glucose) supplemented with the appropriate amino acids (0.002%) as required. β-Galactosidase activity was assayed as described previously (20).
Yeast expression and reporter vectors.
Mi cDNAs encoding the (+) or (−) forms of the protein were inserted as BglII fragments into the unique BglII cloning site of the high-copy plasmid pKV701.VP16 and the low-copy plasmid pRS315KV, which are located between the GAL10 promoter and PGK terminator. In pKV701.VP16, the Mi protein is expressed with the VP16 transcription activation domain fused to the N terminus. The construction of the pRS315KV and pKV701.VP16 expression vectors has been described previously (20, 23), as have the Myc and Max expression vectors (7, 10). The lacZ reporter plasmids are based on pLSLS.CYC described by Crouch et al. (7). This plasmid contains a basal CYC promoter in which the two CATGTG E-box motifs have been mutated. Oligonucleotides containing specific sequence elements were inserted into a unique BglII cloning site located 5′ to the basal CYC promoter. The full sequences of the oligonucleotides used, each of which contains a duplicated element, are shown in Table 1.
Bacterial expression of MITF.
The glutathione S-transferase (GST)-Mi transcription factor (MITF) fusion protein expression vector, pGEX-MITF (43), was a gift from S. Shibahara. To produce purified GST fusion proteins, 20 to 40 ml of an overnight culture of BL21(DE3) pLysS cells containing a GST fusion protein vector was diluted 1:10 with Luria broth containing 100 μg of ampicillin/ml and cultured in a rotary shaker until the optical density at 600 nm reached 0.2 to 0.4. The expression of the GST-MITF fusion protein was induced for 4 h by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture to a final concentration of 200 μM. Cells were harvested by centrifugation at 5,000 to 7,000 rpm for 10 min at 4°C. The resultant pellet was resuspended in 2 to 5 ml of ice-cold NET buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl) and 1× proteinase inhibitor cocktail (Boehringer Mannheim), frozen on dry ice for 5 to 20 min, thawed on ice, and DNaseI treated on ice for 2 to 3 h. The supernatant was clarified by centrifugation at 4°C, mixed with 100 to 300 μl of glutathione Sepharose-4B beads (pre-equilibrated with NET buffer) (Pharmacia), and incubated at 4°C for 20 min on a roller. The beads were then washed three to four times with NET buffer and stored as a 50% slurry at 4°C in the presence of 0.1% thimerosal as a preservative. GST fusion protein was then eluted from the Sepharose-4B glutathione beads by first washing the beads once with 2 volumes of a mixture of 50 mM Tris-HCl (pH 8.0), 10 mM MgSO4, and 2 mM ATP for 30 min at room temperature. The beads were pelleted by centrifugation at room temperature and washed once with 2 volumes of a mixture of 100 mM Tris-HCl [pH 8.0], 120 mM NaCl, and 0.2% Triton X-100 for 30 min at room temperature and pelleted as described above. The bound protein was then serially eluted off the beads into 2 volumes of a mixture of 100 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.2% Triton X-100, and 25 mM glutathione. The protein concentration was then calculated by comparing against a bovine serum albumin standard on a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel. MITF protein was then purified from the GST-MITF bound to Sepharose-4B glutathione beads by means of thrombin cleavage (Sigma). Protein-bead complexes were washed once with 5 volumes of a mixture of 50 mM Tris-HCl [pH 7.4], 10 mM MgSO4, and 2 mM ATP, once with 5 volumes of a mixture of 100 mM Tris-HCl [pH 8.0], 120 mM NaCl, and 0.2% Triton X-100, and once with 5 volumes of thrombin buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM MgCl2, 2.5 mM CaCl2, 1 mM dithiothreitol). The fusion protein was cleaved by incubating overnight at 4°C in 0.5 to 2 volumes of thrombin buffer and 5 to 20 U of thrombin. The beads were then pelleted, and the supernatant was transferred to a new tube and stored at 4°C. Thrombin-cleaved human MITF protein had a usable shelf life of 3 to 4 days at 4°C for bandshift reactions. Both thrombin-cleaved- and GST-MITF fusion proteins bound DNA with the same specificity, though in general binding by the GST fusion protein was more readily observed.
Expression of USF-1.
The plasmid pT7Link.USF-1 (a gift from M.-D. Galibert) contains a USF-1 cDNA under the control of the T7 promoter and was transcribed and translated in vitro using rabbit reticulocyte lysate (Promega) in accordance with the manufacturer’s instructions. Reactions were incubated at 30°C for 1 h and used immediately for the DNA binding assays.
RESULTS
Mi discriminates between CATGTG E-box motifs.
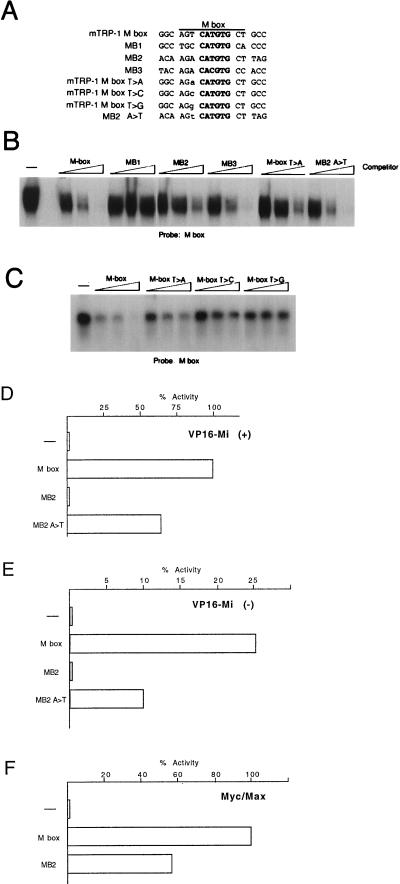
Mi plays an essential but poorly characterized role in melanocyte development. Although it can regulate tyrosinase and TRP-1 expression, it must also control the expression of genes involved in survival or proliferation of the melanocyte lineage. The identity of these genes is presently unknown, but since Mi is a member of the bHLH-LZ transcription factor family, it is likely that any Mi target gene will be regulated by E-box elements with the generic sequence CANNTG. Since Mi shares a related DNA binding domain with c-Myc and its dimerization partner Max, we initially considered the possibility that Mi and the Myc-Max heterodimer might regulate similar genes. It is known for example that Myc can regulate genes involved in both proliferation and apoptosis. One recently identified Myc target gene is that encoding the CDC25a phosphatase (12); up-regulation of cdc25a expression by Myc appears to correlate with the induction of apoptosis. Examination of the three E-box elements recognized by Myc (MB1, MB2, and MB3) located in the first and second introns of the cdc25a phosphatase gene, revealed that the MB2 element shares 10 of 11 bp with the known Mi target, the M box, which is conserved in all melanocyte-specific promoters analyzed to date. The striking similarity between MB2 and the M box strongly suggested that MB2, like the M box, would be a target for Mi. The sequence alignment between the 11-bp M box and the MB1, MB2, and MB3 elements from the cdc25a phosphatase gene is shown in Fig. 1A.
FIG. 1.
Mi discriminates between highly related E-box motifs. (A) E-box motifs used as probes and competitors. Only the E-box and flanking sequences are shown. The full sequence of each oligonucleotide is given in Table 1. (B) DNA-binding band shift (electrophoretic mobility shift) assay using bacterially expressed and purified Mi together with the M-box probe and the indicated competitor oligonucleotides at 10, 50, and 250 ng. Only the bound DNA is shown. (C) DNA-binding assay using in vitro transcribed and translated Mi and the M-box probe and indicated oligonucleotide competitors at 10, 50, and 250 ng. (D) Yeast one-hybrid assay using a CYC-lacZ reporter containing the indicated target elements, cotransformed with a vector expressing Mi(+) fused to the VP16 activation domain. (E) As for panel D but using a VP16-Mi(−) expression vector. (F) Yeast assay for activity of native Myc and Max proteins on the indicated CYC-lacZ-based reporters. Myc and Max were coexpressed in yeast with activation of transcription by Myc being absolutely dependent on Max expression, while Max alone does not activate transcription in this assay (not shown). Residue substitutions are indicated by “greater than” symbols. For example, T>A indicates a T-to-A substitution.
To determine whether Mi could bind the Myc binding sites from the cdc25a phosphatase gene, we performed a band shift assay using a radiolabelled M-box probe and competed for Mi binding by using either the M box or the MB1, MB2, or MB3 element. The results (Fig. 1B) obtained were surprising in two respects. First, although a CATGTG E-box element was present in both the M box and the MB1 element, no Mi binding to MB1 was observed. Second, the MB2 element bound Mi, but at least fivefold to 10-fold less efficiently than the M box, despite the fact that 10 of 11 bases are conserved between the two elements. In contrast, binding to MB3, which contains a CACGTG E box, was as efficient as binding to the M box. Thus, remarkably, Mi was able to discriminate between the M box and the MB1 and MB2 elements even though all three contained a CATGTG E-box motif. One clue as to the origin of the ability of Mi to distinguish between related E boxes lies in the fact that within the core binding sequence, the M box and MB2 differ only by a T-to-A transversion at position −4 relative to the center of the conserved CATGTG E box. It was possible therefore that this base played a key role in the ability of Mi to distinguish between these two sequences, particularly since the MB1 element which fails to bind Mi also differed from the M box at this position. However, since the MB3 CACGTG element bound Mi well but did not contain a T residue at the −4 position, it was also possible that the influence of specific flanking sequences was restricted to a CATGTG-type E-box element.
To test this possibility, we designed specific competitors in which the T residue at the −4 position of the M box was replaced with an A or in which the A residue in the equivalent position in MB2 was replaced with T. These mutated oligonucleotides (Fig. 1A) were also used as competitors in the band shift assay using the M-box probe. The results (Fig. 1B) demonstrate that the single A-to-T substitution in the MB2 element is sufficient to allow binding by Mi to a level equivalent to that of a wild-type (WT) M box. Similarly, Mi binding is reduced approximately fivefold to 10-fold using the M box T-to-A competitor. Thus, the presence of a T residue at the −4 position is a key determinant in the ability of Mi to distinguish between an M box and the MB2 site in vitro.
To investigate more systematically the requirement for the flanking residue, we also replaced the T present in the M box with either a C or a G and compared binding to that achieved using a WT M box or the M box T-to-A competitor. The results shown in Fig. 1C demonstrate that Mi was able to recognize efficiently only the WT M box and that replacement of the 5′ flanking T residue with either A, C, or G severely inhibited binding.
The results described above suggest that flanking sequences may determine whether Mi can bind a specific E-box element. However, these results were obtained in vitro, and it was possible that in vivo, the presence or absence of a 5′ T residue would play a less significant role. To determine whether the Mi could discriminate between different E-box elements in vivo, we made use of a yeast one-hybrid assay in which Mi was expressed as a VP16 activation domain-tagged protein, and its ability to activate reporters comprising specific E-box motifs upstream of a basal CYC-LacZ reporter was assessed. We have successfully used this assay previously to reconstitute Max-dependent transcription activation by Myc and to characterize the in vivo binding specificity of the Myc-Max complex (7, 10). Moreover, the yeast assay enables the binding specificity of Mi homodimers to be assessed in the absence any background generated by any other bHLH-LZ factors which may bind the M box or related sequences in mammalian cells.
Using an M-box reporter, expression of the VP16-tagged Mi resulted in efficient transcription activation (Fig. 1D). Using either an empty expression vector (not shown) or a reporter lacking an M box, no activation was observed, indicating that the activity of the M-box reporter was entirely dependent on both Mi and an appropriate Mi-binding site. In contrast, the MB2 reporter was essentially inactive both in the presence or absence of Mi, suggesting that the effect of the T residue at position −4 was even more critical in vivo than in vitro. Consistent with the results obtained in vitro, mutation of the 5′ flanking A residue present in the MB2 sequence to the T present in the M box enabled Mi to activate transcription from this sequence. Similar results were obtained using a non-VP16-tagged Mi (not shown), though activation of transcription was reduced and as such the assay was less sensitive.
The Mi protein exists in two forms, termed Mi(−) and Mi(+), which differ by an internal six amino acids resulting from the expression of a differentially spliced mRNA (20). The results described above were obtained by expression of the Mi(+) form of the protein. Given that it has been reported that the presence of these additional six amino acids may alter the DNA binding specificity of Mi (19), it was important to compare the ability of the Mi(+) or Mi(−) proteins to recognize the M-box and MB2 elements differentially. The result, shown in Fig. 1E, indicates that the Mi(−) protein activates an M box approximately three- to fourfold less well than the Mi(+) protein (compare activity to that shown in Fig. 1D), which is consistent with the lower level of activation by Mi(−) observed in mammalian cells (33). Nevertheless, as for the Mi(+) protein, Mi(−) was unable to activate transcription from the MB2 element but could bind an MB2 site in which the flanking A residue was changed to T (as in the M box). No activation was observed by expression of either protein by using an MB2 reporter. Thus the ability of Mi(+) and Mi(−) to discriminate between the M-box and MB2 elements was essentially indistinguishable.
To demonstrate that although the MB2 element was not targeted by Mi, it was nevertheless functional, we asked whether it could be activated by Myc-Max in yeast, as it had previously been reported to be a Myc-Max target element in mammalian cells. We have shown previously that activation by Myc in yeast is entirely dependent on Max, and that expression of either protein alone fails to activate transcription (7). To assay for the ability of Myc-Max to bind and activate transcription from the MB2 and M-box elements, vectors expressing both Myc and Max were introduced into yeast together with either the M-box or MB2 reporters, and the resulting levels of β-galactosidase activity were determined. The results are shown in Fig. 1F. In the absence of Myc-Max expression, no β-galactosidase activity from either reporter was detected (not shown). In contrast, expression of Myc-Max resulted in activation of both the M-box and MB2 reporters. No activation was observed using a reporter lacking either the M-box or MB2 element. This result is significant in two respects: first, it confirms that while the MB2 element cannot be activated by Mi, it is nevertheless fully competent to support transcription activation directed by the Myc-Max heterodimer; and second, the T residue flanking the CATGTG motif of the M box does not inhibit activation by the Myc-Max heterodimer. This was surprising since it had been shown previously that the ability of Myc-Max to bind a CACGTG E box is inhibited substantially by the presence of a 5′ T residue at the −4 position both in yeast and in mammalian cells (10, 35). Clearly, the inhibitory effect of the T residue on Myc-Max binding does not apply in the case of CATGTG E-box elements.
Conservation of specificity between melanocyte-specific E-box elements.
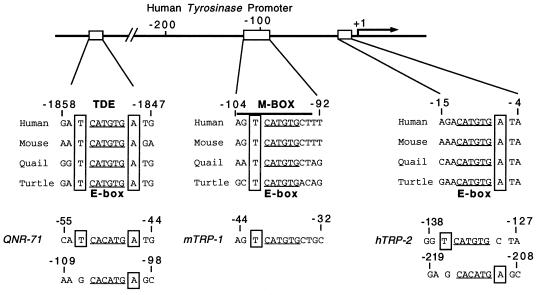
The data obtained so far strongly suggest that in vivo, the presence of a T residue immediately 5′ to the CATGTG motif is essential for DNA binding by either Mi(+) or Mi(−). Although the M box evidently represents a good target for Mi both in yeast and in mammalian cells, several other E-box elements present in melanocyte-specific promoters are known to be targets for Mi in mammalian transfection assays. If the presence of a 5′ flanking T residue were important, it might be expected that these E boxes would also contain a T at the −4 position and that the T residue would be conserved between species. We therefore aligned the E-box motifs from the M box, the initiator E box and the tyrosinase distal enhancer (TDE) E box identified in tyrosinase promoters from different species, as well as the TRP-1 and TRP-2 M-box elements and the E-box motifs from the quail QNR71 gene promoter (Fig. 2). Strikingly, the 5′ flanking T residue was present in the M box from the human, mouse, quail, and turtle tyrosinase genes as well as in the M-box motifs derived from the TRP-1 and TRP-2 genes. Similarly, a T residue at the −4 position was also present in the TDE from all four species, the TDE also being flanked on the 3′ side by a conserved A residue and being opposite a T on the bottom strand. At the tyrosinase initiator, no T residue was present on the 5′ −4 position, but the CATGTG E box did possess a conserved 3′ A residue opposite a T at the −4 position on the bottom strand. The striking conservation of the flanking T residues was also apparent in the QNR-71 gene promoter where both E-box elements, which are known targets for Mi (41), also conform to the consensus for binding by Mi. It is also satisfying that both E-box motifs in the QNR-71 promoter have also been shown to be Myc responsive (41), given that the yeast assays revealed that an M box can be regulated by the Myc-Max heterodimer.
FIG. 2.
Conservation of the E-box motifs within melanocyte-specific promoters and between species. The tyrosinase promoter contains three Mi-responsive CATGTG E-box motifs termed the TDE, the M box, and the initiator E box. All three elements from all four species possess a 5′ flanking T residue (boxed) at the −4 position on either the top strand (M box) the bottom strand (initiator E box) or both strands (TDE). The conservation of the 5′ flanking T residue is also seen in the Mi-responsive elements from the quail QNR71 promoter as well as in the TRP-1 M box, and the M box and upstream E box from the TRP-2 promoter. The numbers indicate the positions of the sequences shown relative to the transcription start sites in the cognate promoters.
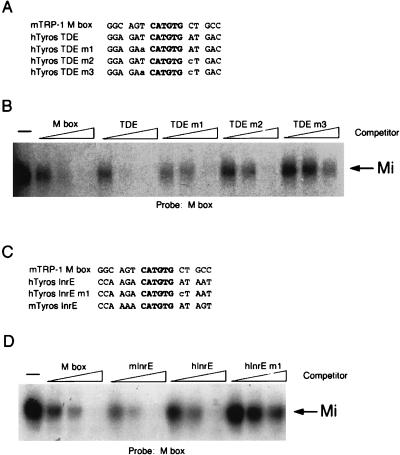
Taken together with the binding studies on the M-box and MB2 elements, the absolute conservation of a T residue flanking all E-box elements known to be targets for Mi strongly suggests that the presence of a T in this position enables Mi to discriminate between different E-box motifs in melanocytes. To test this further, we performed band shift assays using the M-box probe and WT or mutant competitors derived from the tyrosinase initiator or TDE (Fig. 3A and C). The results demonstrate that the efficiency of Mi binding to a CATGTG element correlates precisely with the presence of a 5′ T residue flanking the E box. Thus, a WT TDE, which contains a 5′ flanking T residue at the −4 position on both strands, competes for Mi binding approximately threefold better than an M box (Fig. 3B). Mutation of either T residue flanking the TDE to A or G, mutants m1 and m2, respectively, resulted in decreased DNA binding by Mi to a level seen using an M-box competitor. In contrast, a double mutant, m3, in which both flanking T residues are mutated, failed to bind Mi.
FIG. 3.
Mi binding to the TDE and initiator (Inr) require the presence of a 5′ flanking T residue. (A, C) E-box motifs used for probes and competitors in the DNA binding band shift assays (for complete sequences see Table 1). (B, D) Band shift assays using an M-box probe and indicated WT and mutant competitors at 10, 50, and 250 ng. Competitors were derived from the mouse TRP-1 M box, the human tyrosinase TDE (hTyros TDE), and the human and mouse initiator elements (hTyros InrE and mTyros InrE, respectively). Only the bound DNA is shown.
A similar pattern was observed using the tyrosinase initiator E-box sequence (Fig. 3D). Both the human and mouse initiator elements competed for Mi binding to the M box probe as well as the M box itself, whereas mutation of the T residue at the −4 position flanking the initiator E box on the bottom strand abolished its ability to bind Mi. In summary, the data obtained using the M box, TDE, and initiator all point to a requirement for a T residue located at the −4 position of a CATGTG E box if Mi is to bind DNA efficiently.
The MB3 element contains a CACGTG E box and binds Mi efficiently. To determine whether this was a peculiarity of the MB3 sequence or whether Mi could also bind CACGTG motifs found in other promoters, band shift assays using the M box probe together with E-box elements derived from the adenovirus major-late promoter, as well as the p53 and cyclin B1 promoters as competitors were performed. The results obtained (not shown) indicated that all the CACGTG E-box elements used were able to bind Mi, which is consistent with the previous observation that Mi could bind the CACGTG element in the adenovirus major-late promoter (19).
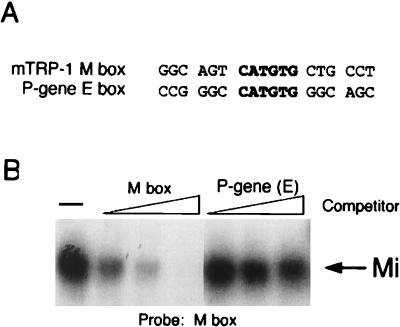
Mi does not bind the CATGTG E box in the P-gene promoter.
Since the tyrosinase, TRP-1, TRP-2, and QNR-71 promoters all contain Mi binding sites, it might be expected that this would be true for the promoters from other genes involved in melanogenesis. Mutations in the P gene, encoded by the pink-eyed dilution locus (15), result in oculocutaneous albinism, and the P gene itself is thought to encode a tyrosine transporter. The role of the P gene in melanogenesis, together with the fact that its promoter contains a CATGTG E box (24), has led to speculation that it too is regulated by the MITF. However, examination of the P-gene E box indicates that the CATGTG motif does not possess the critical flanking T residue (Fig. 4A) found to be essential for binding to the E-box elements in the tyrosinase, TRP-1, TRP-2, and QNR-71 promoters. On the basis of the experiments described above, we would predict that this element would not in fact bind Mi. To test this, we performed a band shift assay using an M-box probe and either the M-box or the P-gene element as competitors. The result (Fig. 4B) indicates that while the M box binds Mi efficiently, the P-gene CATGTG E box does not. Thus, the lessons learned from the analysis of Mi binding specificity on the tyrosinase and TRP-1 promoter elements enable accurate predictions to be made as to which E-box motifs are likely to be recognized by Mi.
FIG. 4.
Mi does not bind the E-box element in the P gene. (A) Oligonucleotides used as probes or competitors in the DNA binding band shift assay. Full sequences are given in Table 1. (B) Band shift assay using the M box probe and indicated competitors at 10, 50, and 250 ng. Only bound DNA is shown.
USF-1 binding specificity.
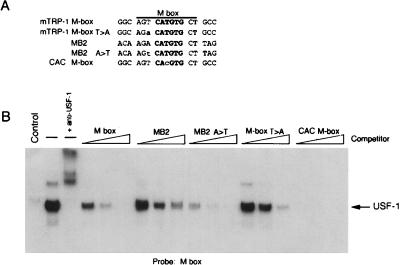
When the M box and tyrosinase initiator E box were first identified, DNA binding assays using melanoma cell nuclear extracts identified USF-1 as the predominant factor competent to bind these sequences in vitro (3, 25). Although subsequently modification of the assay conditions allowed the detection of Mi DNA-binding activity in similar nuclear extracts (5), it seems likely that both factors may be competent to bind the E-box motifs present in the melanocyte-specific promoters. Given the specificity of Mi binding to these elements, we also wished to determine whether DNA recognition by USF-1 was similar to or different from that of Mi. Understanding the precise requirements for USF-1 binding might provide clues as to whether Mi and USF-1 were likely to compete for binding in vivo. To this end, in vitro transcribed or translated USF-1 was used in a band shift assay together with an M-box probe and WT or mutant M box, or MB2 competitors. The results (Fig. 5) revealed that USF-1 possessed a remarkably similar binding specificity to Mi. In other words, USF-1 bound an MB2 element around fivefold less well than the M box, while mutating the 5′ flanking A residue to a T in MB2, or the 5′ flanking T residue in the M box to A, resulted in a fivefold increase or decrease in USF-1 binding, respectively. In addition, alteration of the CATGTG sequence at the core of the M box to CACGTG enabled USF-1 to bind this sequence at least fivefold to 10-fold better than the WT M box. These data suggest that Mi and USF might well compete for binding to melanocyte-specific promoters in vivo.
FIG. 5.
Efficient binding by USF-1 to a CATGTG E box requires a 5′ flanking T residue. (A) WT and mutant oligonucleotides used as probes and competitors. (B) In vitro transcribed-translated USF-1 was used in DNA binding band shift assay using an M-box probe and the indicated competitors at 10, 50, and 250 ng. Unprogrammed reticulocyte lysate was used as a control and the complex formed using the programmed lysate is supershifted by the addition of anti-USF-1 antibody. Only the bound DNA is shown.
We also attempted to use a VP16-USF chimera in a yeast one-hybrid assay to compare USF DNA binding specificity in vivo to that of Mi. Interestingly, using this assay, we were unable to observe any activation of E-box reporters, including the E box from the adenovirus major-late promoter, by the VP16-USF protein. Since the ability of USF to bind DNA in vivo appears to be regulated by phosphorylation (13), it is possible that the specific signal transduction pathway required for USF to bind DNA efficiently in vivo does not operate in yeast.
Mi homodimers do not bind an atypical E box in the c-Kit promoter.
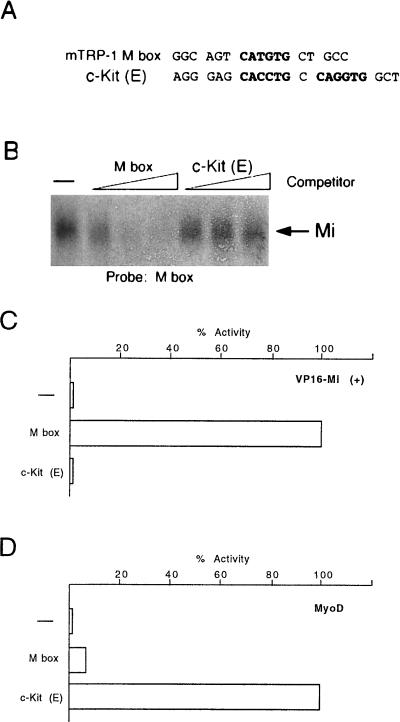
The results described above would indicate that Mi recognizes either a CACGTG E box or a highly specific subset of CATGTG E-box elements. However, a previous report (40) suggested that Mi may also bind an atypical E-box element, (CACGTGCCAGGTG) in the promoter for the c-Kit gene. Although it has recently been demonstrated that Mi is phosphorylated by mitogen-activated protein kinase in response to signalling via the c-Kit receptor tyrosine kinase (18), the ability of Mi to activate the gene encoding the receptor would provide a potential regulatory feedback loop. The ability of Mi to bind the c-Kit element would clearly increase the repertoire of genes which were likely candidates for regulation by Mi. In an attempt to understand better the ability of Mi to recognize this atypical E box and to identify which residues were required for Mi binding, we performed an initial band shift assay using the M-box probe and the c-Kit or M-box competitors. Surprisingly, under conditions where binding to the M box was readily apparent, we were unable to observe any binding by Mi to the c-Kit element (Fig. 6A and B). In the event that our inability to detect Mi binding to the c-Kit element was an artifact of our in vitro binding conditions, we also considered whether Mi could activate the atypical c-Kit E-box motif in the yeast one-hybrid assay. The c-Kit element was therefore cloned upstream from the CYC-LacZ reporter and its responsiveness to Mi compared to that of the M box. Consistent with the results of our in vitro binding assays, while Mi was able to active efficiently the M-box reporter, using either the Mi(+) (Fig. 6C) or Mi(−) (not shown) proteins, no activation from the c-Kit reporter was observed. The c-Kit E box was nevertheless functional since it was activated weakly though specifically by the bHLH factor MyoD (Fig. 6D), which failed to activate transcription via the M box. We conclude that under the conditions used here, Mi does not recognize the c-Kit sequence.
FIG. 6.
The atypical E-box elements in the c-Kit promoter do not bind Mi. (A) Oligonucleotides used as probe and competitors in the DNA binding assay. (B) Band shift assay using an M-box probe and bacterially expressed and purified Mi together with the indicated competitors at 10, 50, and 250 ng. (C) Yeast one-hybrid assay for binding of VP16 Mi to the M box and c-Kit E-box elements. (D) Yeast one-hybrid assay for binding of MyoD to the M box and c-Kit E-box elements. Yeast were transformed with the indicated CYC-lacZ reporters together with vectors expressing either an Mi-VP16 fusion protein or MyoD were assayed for β-galactosidase activity. The sequences of the oligonucleotides cloned into CYC-lacZ reporter plasmids are listed in Table 1.
DISCUSSION
Mi plays an essential but largely uncharacterized role in the development of the melanocyte lineage. Mi must therefore regulate genes required for the survival or maintenance of melanoblasts following their commitment to the melanocyte lineage. However, to date, the only genes which have been identified as targets for Mi are those such as tyrosinase, TRP-1, and TRP-2 which encode melanogenic enzymes and which, while playing a critical role in pigmentation, do not exert any influence on the survival or maintenance of the melanocyte lineage. The identity of the genes through which Mi mediates its developmental function is unknown, but presumably they are regulated by specific E-box elements. Given that any cell may contain multiple bHLH-LZ transcription factors each able to recognize an E-box motif, mechanisms must operate to preserve the specificity required to maintain the complex program of gene expression underlying development. This is as true for other cell types as it is for the melanocyte.
In attempting to understand the nature of these Mi-regulated genes, we initially considered that Mi might regulate at least a subset of those genes which are targets for the Myc-Max heterodimer. The cdc25A phosphatase gene appeared at first sight a particularly attractive candidate since Myc-Max was shown previously to regulate its expression via three E-box elements (12), one of which, MB2, exhibited a striking degree of identity to the known Mi target, the M box. However, in vitro binding assays established that in contrast to our expectations, Mi was unable to bind the MB1 element in vitro and bound MB2 poorly, despite the similarity to the M box, a result confirmed by using the yeast one-hybrid assay. Additional DNA binding assays demonstrated that the ability of Mi to discriminate between different CATGTG E-box elements was dictated by the nature of the base at position −4 relative to the center of the E box. For Mi to bind efficiently, the −4 position should be occupied by a T residue; that is, Mi will bind either TCATGTG, CATGTGA, or preferably TCATGTGA. Since a C in the −4 position does not allow binding by Mi, we presume that Mi makes a specific contact in the major groove with the methyl group present on a T residue. However, it is very difficult to say with any precision, since the basic regions of the bHLH and bHLH-LZ factors appear to recognize DNA in subtly different ways. The only positive clue to the requirement comes from the fact that in the USF crystal structure (9), Val8 in the basic region (the conserved Glu is at position 9) makes a van der Waals contact with a flanking T base. Since M contains an isoleucine in this position, it is possible that this residue makes a similar contact with the 5′ flanking T residue.
The results from our in vitro binding assays and the yeast one-hybrid assay suggesting that the ability of Mi to discriminate between CATGTG E boxes was determined by the presence or absence of a T residue at the −4 position were further substantiated by the alignment between all the E-box elements present in the promoters of the tyrosinase, TRP-1, TRP-2, and QNR-71 genes. In every case, the core CATGTG E-box motifs were flanked by a T residue at the −4 position. We went on to show that for both the initiator and the TDE, the presence of the T residue in this position was an essential determinant for M binding. The significance of this observation was underscored by the fact that for the tyrosinase promoter, the 5′ flanking T residue is entirely conserved between species as divergent as human and turtle, and that a second CATGTG E box in the tyrosinase enhancer, described by Yasumoto et al. (42), does not possess a T residue at the −4 position and is not responsive to Mi. Taken together, these observations strongly suggest that the ability of Mi to discriminate between different CATGTG E-box elements is dictated by the nature of the base at position −4 in vivo. One distinct advantage to the cell would be the fact that the ability of Mi to discriminate between different CATGTG E boxes would inevitably restrict the repertoire of E-box-containing genes regulated by Mi. Indeed, the absence of a flanking T residue adjacent to the CATGTG element in the P-gene promoter led us to predict accurately that this element would not be recognized by Mi.
Although our results clearly demonstrate that Mi can distinguish between different CATGTG E-box motifs, Mi appears to recognize CACGTG E boxes relatively indiscriminately. This raises an intriguing question, namely, if the tyrosinase, TRP-1, TRP-2, and QNR-71 promoters are regulated by Mi, why do their promoters not contain CACGTG E-box elements? Several explanations are plausible. It is possible, for example, that in vivo, E-box elements are recognized by different bHLH-LZ transcription factors at different times. The choice of which bHLH-LZ factor will be bound at any given time will be dictated by several factors including the relative abundance of each factor and whether their ability to bind DNA either alone or in cooperation with other transcription factors is regulated by specific signal transduction pathways. It is already known, for example, that the M-box and initiator E-box elements from the tyrosinase promoter are recognized both by Mi and by USF (3, 25, 42). DNA binding by USF appears to be regulated (13), as is the abundance of Mi whose expression is transiently increased by elevated cyclic AMP levels (4). Thus, in vivo, the various E-box elements in the tyrosinase and other promoters may under appropriate conditions exchange USF and Mi, and a high-affinity binding site for USF, such as a CACGTG-type E box, may hinder such an exchange.
Along the same lines, it is also possible that in vivo, CACGTG elements are simply not accessible to Mi owing to the presence of other bHLH-LZ factors able to recognize this sequence. Consequently the CATGTG elements found in the melanocyte-specific promoters may be recognized by a more restricted subset of bHLH-LZ factors with which Mi is better able to compete. In yeast for example, neither the bHLH factor Pho4 nor the bHLH-LZ factor Cpf1 are able to bind CATGTG E-box elements but will bind CACGTG motifs (16). Interestingly, the ability of Pho4 to bind a CACGTG E box is strongly inhibited by a T residue at the −4 position, while binding by Cpf1 is not (11). Similarly, binding by Myc-Max heterodimers, but not by Max-Max homodimers, is also inhibited by a T residue flanking a CACGTG motif in yeast and mammalian cells (10, 35), though as we have demonstrated here, this rule does not apply on a CATGTG E box. Nevertheless, the ability of bHLH-LZ factors to discriminate between different E-box motifs based on the nature of the 5′ flanking base appears to represent a common theme that is likely to underpin much of the specificity required for undertaking the program of gene expression mediated by bHLH-LZ factors during development.
In addition to these considerations, it is also possible that a CATGTG E box may contribute to binding other, non-bHLH-LZ factors. In this respect it has not escaped our attention that the 5′ half of an M box, 5′-AGTCAT may on the other strand represent a half site for members of the AP1 transcription factor families, with the T residue at the −4 position being at the core of this potential binding site. Whether the M box is in reality targeted by any of these factors acting alone or in conjunction with Mi is not currently known.
Of course, one criticism of the experiments which we have described here might be that the binding specificity of Mi both in vitro and in the yeast one-hybrid assays has been determined using Mi homodimers and that in vivo, DNA recognition by any Mi heterodimer might be different. Indeed, in vitro, Mi can bind DNA as a heterodimer with the bHLH-LZ transcription factors TFE3, TFEB, and TFEC (19). However, at the earliest times in the melanocyte lineage, when Mi function is clearly critical, there is no evidence that TFEB or TFE3 is expressed in these cells (27). Of course, we cannot rule out a role for Mi heterodimers later in melanocyte development. However, it is clear from the results of many studies on a variety of bHLH-LZ factors that DNA binding specificity is dictated by the basic region. The basic regions of TFE3, TFEB, and TFEC are identical to that of Mi and, although we have not examined this directly, we would not expect the binding specificity of any Mi heterodimer to be different from that of a Mi homodimer.
Finally, in contrast to previous work (40), we were unable to detect binding by Mi to the atypical E-box motif present in the c-Kit promoter either in vitro or in the yeast one-hybrid assay. Since our assay was performed under conditions where the M box was clearly recognized, the previous report of the interaction between Mi and this element may reflect the particular binding conditions used by those authors. That is not to say that these sequences do not act to regulate c-Kit expression in cells but simply that, in our view, they are unlikely to act as a target for Mi homodimers. Neither do we wish to imply that Mi cannot regulate the c-Kit promoter; regulation of c-Kit expression seems to be complex, with DNA rearrangements up to 100 kb away appearing to affect the activity of the c-Kit gene (8), and as such we cannot rule out any effect of Mi elsewhere in the sequences flanking the c-Kit gene.
In summary, we have provided for the first time a mechanism to explain how transcription activation by microphthalmia may be restricted to a specific subset of target genes. The insight gained from this work should prove invaluable in the search for genes other than tyrosinase, TRP-1, TRP-2, and QNR71 which are regulated by Mi. Moreover, although we have restricted our studies to Mi and the melanocyte lineage, it is likely that similar mechanisms operate to facilitate discrimination between different E-box elements by other members of the bHLH-LZ family of transcription factors in other cell types.
ACKNOWLEDGMENTS
We thank Ugur Yavuzer for performing the initial yeast one-hybrid assays, S. Shibahara for providing the GST-MITF expression vector, and Don Ayer for the yeast MyoD expression vector.
This work was supported by the Association for International Cancer Research and Marie Curie Cancer Care.
REFERENCES
- 1.Baldwin C T, Lipsky N R, Hoth C F, Cohen T, Mamuya W, Milunsky A. Mutations in PAX3 associated with Waardenburg syndrome type I. Hum Mutat. 1994;3:205–211. doi: 10.1002/humu.1380030306. [DOI] [PubMed] [Google Scholar]
- 2.Baynash A G, Hosoda K, Giaid A, Richardson J A, Emoto N, Hammer R E, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 3.Bentley N J, Eisen T, Goding C R. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotto C, Buscà R, Abbe P, Bille K, Aberdam E, Ortonne J-P, Ballotti R. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol Cell Biol. 1998;18:694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolotto C, Bille K, Ortonne J-P, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J Cell Sci. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butt J, Greenberg J, Winship I, Sellars S, Beighton P, Ramesar R. A splice junction mutation in PAX3 causes Waardenburg syndrome in a South African family. Hum Mol Genet. 1994;3:197–198. doi: 10.1093/hmg/3.1.197. [DOI] [PubMed] [Google Scholar]
- 7.Crouch D H, Fisher F, Clark W, Jayaraman P S, Goding C R, Gillespie D A. Gene-regulatory properties of Myc helix-loop-helix/leucine zipper mutants: Max-dependent DNA binding and transcriptional activation in yeast correlates with transforming capacity. Oncogene. 1993;8:1849–1855. [PubMed] [Google Scholar]
- 8.Duttlinger R, Manova K, Berrozpe G, Chu T Y, DeLeon V, Timokhina I, Chaganti R S, Zelenetz A D, Bachvarova R F, Besmer P. The Wsh and Ph mutations affect the c-kit expression profile: c-kit misexpression in embryogenesis impairs melanogenesis in Wsh and Ph mutant mice. Proc Natl Acad Sci USA. 1995;92:3754–3758. doi: 10.1073/pnas.92.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferré-D’Amaré A R, Pognonec P, Roeder R G, Burley S K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher F, Crouch D H, Jayaraman P S, Clark W, Gillespie D A, Goding C R. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12:5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher F, Goding C R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galaktionov K, Chen X, Beach D. CDC25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 13.Galibert M-D, Boucontet L, Goding C R, Meo T. Recognition of the E-C4 element from the C4 complement gene promoter by the upstream stimulatory factor-1 transcription factor. J Immunol. 1997;159:6167–6183. [PubMed] [Google Scholar]
- 14.Ganss R, Schutz G, Beermann F. The mouse tyrosinase gene. Promoter modulation by positive and negative regulatory elements. J Biol Chem. 1994;269:29808–29816. [PubMed] [Google Scholar]
- 15.Gardner J M, Nakatsu Y, Gondo Y, Lee S, Lyon M F, King R A, Brilliant M H. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science. 1992;257:1121–1124. doi: 10.1126/science.257.5073.1121. [DOI] [PubMed] [Google Scholar]
- 16.Goding, C. R., and J. Mellor. Unpublished observations.
- 17.Guthrie C, Fink G R. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press; 1991. [PubMed] [Google Scholar]
- 18.Hemesath T J, Price E R, Takemoto C, Badalian T, Fisher D E. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 19.Hemesath T J, Steingrimsson E, McGill G, Hansen M J, Vaught J, Hodgkinson C A, Arnheiter H, Copeland N G, Jenkins N A, Fisher D E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 20.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkinson C A, Moore K J, Nakayama A, Steingrimsson E, Copeland N G, Jenkins N A, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 22.Hosoda K, Hammer R E, Richardson J A, Baynash A G, Cheung J C, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 23.Jayaraman P S, Hirst K, Goding C R. The activation domain of a basic helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S T, Nicholls R D, Jong M T, Fukai K, Spritz R A. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics. 1995;26:354–363. doi: 10.1016/0888-7543(95)80220-g. [DOI] [PubMed] [Google Scholar]
- 25.Lowings P, Yavuzer U, Goding C R. Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol. 1992;12:3653–3662. doi: 10.1128/mcb.12.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore K J. Insight into the microphthalmia gene. Trends Genet. 1995;11:442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama A, Nguyen M T, Chen C C, Opdecamp K, Hodgkinson C A, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- 28.Nocka K, Majumder S, Chabot B, Ray P, Cervone M, Bernstein A, Besmer P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice. Genes Dev. 1989;3:816–826. doi: 10.1101/gad.3.6.816. [DOI] [PubMed] [Google Scholar]
- 29.O’Hare P, Goding C R. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 30.Opdecamp K, Nakayama A, Nguyen M T, Hodgkinson C A, Pavan W J, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- 31.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Préhu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Clayton-Smith J, Read A P, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburgs-Hirschprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 32.Puffenberger E G, Hosoda K, Washington S S, Nakao K, deWit D, Yanagisawa M, Chakravart A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 33.Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding C R. CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- 34.Silvers W K. The coat colors of mice. New York, N.Y: Springer-Verlag; 1979. [Google Scholar]
- 35.Solomon D L, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southard-Smith E M, Kos L, Pavan W. Sox10 mutation disrupts neural crest development in Dom Hirschprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 37.Steingrímsson E, Moore K J, Lamoreaux M L, Ferré-D’Amaré A R, Burley S K, Sanders Zimring D C, Skow L C, Hodgkinson C A, Arnheiter H, Copeland N G, Jenkins N A. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long J E, Meyers K A, Aaronson S A, Miki T. Ectopic expression of MITF, a gene for Waardenburgs syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14:50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 39.Tassabehji M, Newton V E, Read A P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 40.Tsujimura T, Morii E, Nozaki M, Hashimoto K, Moriyama Y, Takebayashi K, Kondo T, Kanakura Y, Kitamura Y. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood. 1996;88:1225–1233. [PubMed] [Google Scholar]
- 41.Turque N, Denhez F, Martin P, Planque N, Bailly M, Begue A, Stehelin D, Saule S. Characterization of a new melanocyte-specific gene (QNR-71) expressed in v-myc transformed quail neuroretina. EMBO J. 1996;15:3338–3350. [PMC free article] [PubMed] [Google Scholar]
- 42.Yasumoto K-I, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasumoto K-I, Yokayama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 44.Yavuzer U, Keenan E, Lowings P, Vachtenhein J, Currie G, Goding C R. The microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10:123–134. [PubMed] [Google Scholar]