ABSTRACT
Here, we report the draft genome sequence of Exiguobacterium sp. strain MMG028, isolated from Rose Creek, San Diego, CA, USA, assembled and analyzed by undergraduate students participating in a marine microbial genomics course. A genomic comparison suggests that MMG028 is a novel species, providing a resource for future microbiology and biotechnology investigations.
KEYWORDS: Exiguobacterium, bioremediation
ANNOUNCEMENT
To engage undergraduates in discovery-based research, novel marine bacteria were isolated and cultured. The bacterial genomes were sequenced, assembled, annotated, and analyzed by students in a marine microbial menomics (MMG) course at San Diego State University. Strain MMG028 was isolated from the water of Rose Creek Salt Marsh in San Diego, CA, USA (32.84022°N, 117.2220°W), on 30 August 2022 using a sterile cotton swab. A single colony was obtained on Marine Agar 2216 (BD Difco, Franklin Lakes, NJ, USA) and incubated at 28°C for 72 hours. Colonies were transferred to Marine Broth 2216 and incubated for 72 hours at 25°C before storage and DNA isolation.
Genomic DNA was extracted using a Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA). 16S rRNA gene amplification with primers 27F-1492R (1) and Sanger sequencing (Eton Biosciences, San Diego, CA, USA) identified the closest strain as Exiguobacterium sp. (94.98% ID, 0.0 E-value). DNA was submitted to the Microbial Genome Sequencing Center (Pittsburgh, PA USA) for library preparation (Illumina DNA Prep kit; San Diego, CA, USA) and whole-genome sequencing (NextSeq 550; Illumina), producing 2 × 150 bp paired-end reads (4.2M reads). Reads were trimmed using Trim Galore v.0.6.5 (2), assembled using Unicycler v0.4.8 (3), integrated into PATRIC v3.6.12 (4), and annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v5.1 (5), with default parameters. MMG028 has a 2.85 Mb genome, a total GC content of 47.75% with 16 contigs at 360.97× coverage, and an N50 value of 664,045 bp, with 3,015 predicted coding sequences. Default parameters were used except where otherwise noted.
A phylogenetic analysis revealed that strain MMG028 falls into the genus Exiguobacterium (Fig. 1), in the family Bacillaceae and class Bacilli (6–8). Comparing strain MMG028 with Exiguobacterium sp. strain AT1b yields an ANI value of 88.03 (4, 9, 10), a distance that is below the 95% threshold that delineates species (11), suggesting that MMG028 is a new species. We designate the current isolate as Exiguobacterium sp. strain MMG028.
Fig 1.
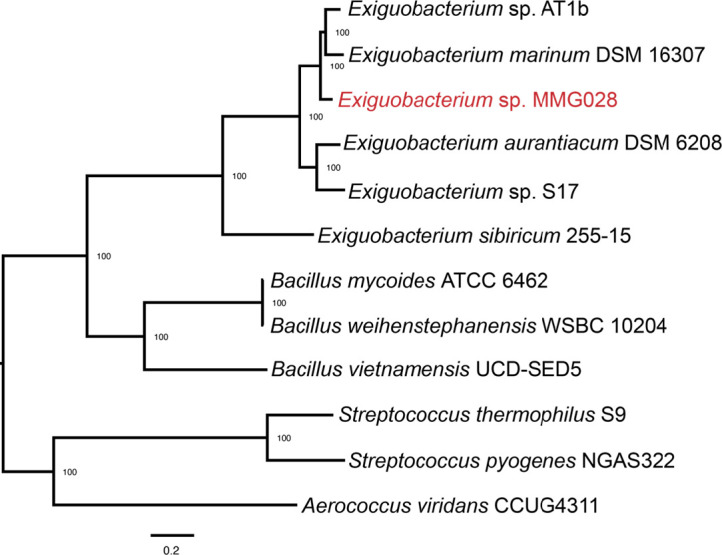
A maximum likelihood phylogeny constructed using the Codon tree method through PATRIC using 100 single-copy genes and proteins identified by PGFams (10, 12–18). The newly identified strain MMG028 is indicated in red. GenBank Accession numbers for the phylogenetic tree are as follows: Streptococcus thermophilus strain S9 (CP013939), Streptococcus pyogenes strain NGAS322 (CP010449), Aerococcus viridans strain CCUG4311 (CP014164), Exiguobacterium aurantiacum DSM 6208 (JNIQ00000000), Exiguobacterium marinum DSM 16307 (JHZT00000000), Bacillus mycoides ATCC 6462 (CP009692.1, CP009689.1, CP009691.1), Bacillus vietnamensis strain UCD-SED5 (LIXZ00000000), Exiguobacterium sibiricum 255-15 (CP001022, CP001024, CP001023), Exiguobacterium sp. AT1b (CP001615), Exiguobacterium sp. S17 (ASXD00000000), Bacillus weihenstephanensis strain WSBC 10204 (CP009746).
Species within and related to the Exiguobacterium genus have been found to adapt to diverse environments with the ability to withstand wide ranges of temperature, salinity, and pH (7). Features of other Exiguobacterium sp. strains include biosorption properties of heavy metals and other pollutants (19, 20). Strains isolated from marine environments have shown that the formation of a biofilm may be a key process for sequestering heavy metals (21). Analyses of Exiguobacterium sp. MMG028 with PATRIC v3.6.12 revealed several pathways involved in xenobiotic degradation including toluene, xylene, styrene, tetrachloroethene, and naphthalene (22, 23). As heavy metals and pollutants are accumulating in the environment due to human-induced industrial processes, the isolation and genome sequence of Exiguobacterium sp. MMG028 provides a new strain that may be a candidate for bioremediation.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (1942251, N.J.S.), the Gordon and Betty Moore Foundation (GBMF9344 to N.J.S.), and the Alfred P. Sloan Foundation, Sloan Research Fellowship (N.J.S.).
Contributor Information
Nicholas J. Shikuma, Email: nshikuma@sdsu.edu.
Frank J. Stewart, Montana State University, Bozeman, Montana, USA
DATA AVAILABILITY
The genome sequencing and assembly project for strain MMG028 has been deposited in DDBJ/EMBL/GenBank under BioProject number PRJNA716944, raw sequencing SRA accession number SRX19033163, and whole-genome sequencing genome accession number JAQJIN000000000.
REFERENCES
- 1. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krueger F. 2015. Trim Galore: a wrapper tool around cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files
- 3. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, et al. 2020. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. doi: 10.1093/nar/gkz943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crapart S, Fardeau M-L, Cayol J-L, Thomas P, Sery C, Ollivier B, Combet-Blanc Y. 2007. Exiguobacterium profundum sp. nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 57:287–292. doi: 10.1099/ijs.0.64639-0 [DOI] [PubMed] [Google Scholar]
- 7. Zhang D, Zhu Z, Li Y, Li X, Guan Z. 2021. Comparative genomics of exiguobacterium reveals what makes a cosmopolitan bacterium. mSystems 6:e0049921. doi: 10.1128/mSystems.00499-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kasana RC, Pandey CB. 2018. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol 38:141–156. doi: 10.1080/07388551.2017.1312273 [DOI] [PubMed] [Google Scholar]
- 9. Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. doi: 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL. 2013. Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, et al. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, Vonstein V, Wattam AR, Yoo H. 2016. Pattyfams: protein families for the microbial genomes in the PATRIC database. Front Microbiol 7:118. doi: 10.3389/fmicb.2016.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 18. Letunic I, Bork P. 2016. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manobala T, Shukla SK, Rao TS, Kumar MD. 2019. Uranium sequestration by biofilm-forming bacteria isolated from marine sediment collected from Southern coastal region of India. International Biodeterioration & Biodegradation 145:104809. doi: 10.1016/j.ibiod.2019.104809 [DOI] [Google Scholar]
- 20. Chauhan D, Agrawal G, Deshmukh S, Roy SS, Priyadarshini R. 2018. Biofilm formation by Exiguobacterium sp. DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC Adv 8:37590–37599. doi: 10.1039/c8ra06448b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SabaAndreasen R, Li Y, Rehman Y, Ahmed M, Meyer RL, Sabri AN. 2018. Prospective role of indigenous Exiguobacterium profundum PT2 in arsenic biotransformation and biosorption by planktonic cultures and biofilms. J Appl Microbiol 124:431–443. doi: 10.1111/jam.13636 [DOI] [PubMed] [Google Scholar]
- 22. Mishra S, Lin Z, Pang S, Zhang W, Bhatt P, Chen S. 2021. Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front Bioeng Biotechnol 9:632059. doi: 10.3389/fbioe.2021.632059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Qiao N, Sun F, Shao Z. 2008. Isolation, gene detection and solvent tolerance of benzene, toluene and xylene degrading bacteria from nearshore surface water and pacific ocean sediment. Extremophiles 12:335–342. doi: 10.1007/s00792-007-0136-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequencing and assembly project for strain MMG028 has been deposited in DDBJ/EMBL/GenBank under BioProject number PRJNA716944, raw sequencing SRA accession number SRX19033163, and whole-genome sequencing genome accession number JAQJIN000000000.


