ABSTRACT
We report the draft genomes of four Kluyveromyces marxianus isolates obtained from the elaboration process of henequen (Agave fourcroydes) mezcal, a Mexican alcoholic beverage. The average nucleotide identity analysis revealed that isolates derived from agave plants are distinct from those from other environments, including agave fermentations.
KEYWORDS: Kluyveromyces marxianus, agave, biodiversity
ANNOUNCEMENT
Kluyveromyces marxianus is a thermotolerant yeast with a fast growth rate and the ability to metabolize a wide range of carbohydrates making it a promising cell factory for industrial biotechnology (1–3). K. marxianus has been frequently isolated from dairy products (4) and other habitats such as fermented beverages (5), plants and fruits (6, 7), and sugarcane mills (8, 9), among others. Isolates from agave and associated fermentations may constitute a new clade within the K. marxianus species (4). Among the 21 K. marxianus genomes available in NCBI only two correspond to strains from agave: UFS-Y2791 from an agave plant (Schabort, D. T., Letebele, P. K., Steyn, L., Kilian, S. G. and duPreez, J. C., unplished data) and SLP1 from spontaneous mezcal fermentation (5). Here, we present the draft genomes of four K. marxianus isolates isolated from the elaboration process of henequen mezcal as previously described (10). Henequen (Agave fourcroydes) is an agave species native to the Yucatan Peninsula.
DNA was prepared from overnight cultures in yeast extract-peptone-dextrose broth at 30°C and 150 rpm (10) using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research) following the manufacturer’s instructions. DNA quality and purity were assessed by 0.7% (wt/vol) agarose gel electrophoresis in 1× TBE buffer, and UV absorbance measurements were performed on a Nanodrop 2000 spectrophotometer (Thermo Scientific). DNA was quantified using a Qubit 3.0 fluorometer (Life Technologies). Paired-end genomic DNA libraries were constructed using the TruSeq Nano kit (Illumina) according to the manufacturer’s instructions. Libraries’ quality and quantity were verified using a 2100 BioAnalyzer (Agilent Technologies). Sequencing was performed on the Illumina HiSeq 2500 platform through the standard rapid-sequencing protocol to generate 150-bp paired-end reads.
Reads’ quality was assessed with FastQC v0.11.9 (11). The adapters and low-quality bases were discarded using Trimmomatic v.0.39 with default parameters (12). De novo genome assemblies were generated using Velvet v.1.2.10 (kmer 37) (13) and Spades v.3.12.0 (kmers 21, 33, 55, 77, and 99) (14), and the obtained assemblies were merged with Metassembler v.1.5 using the Spades contigs as primary assembly (15). Assemblies’ quality was assessed using QUAST v.4.1 (16). Gene prediction was performed with Funannotate (v.1.8.14) using Kluyveromyces lactis as the training species (17). Assemblies’ completeness was evaluated with BUSCO v.5.4.7 using the saccharomycetes_odb10 database (18). Average nucleotide identity (ANI) analysis was calculated with pyani v0.2 using the ANIb method (19). The heatmap was built in R with ggplot2 and pheatmap.
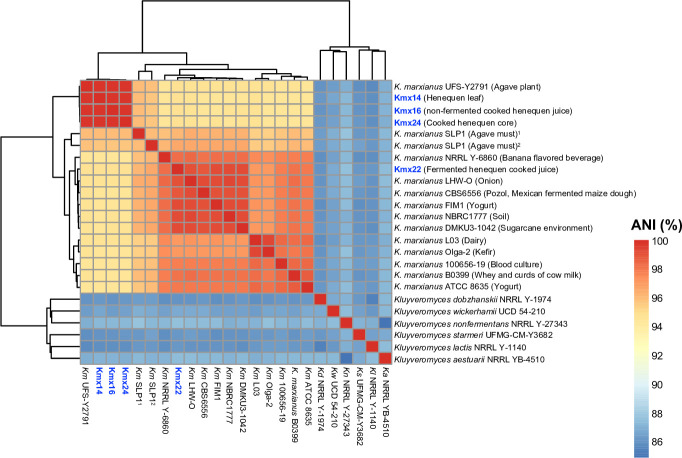
Table 1 details the sequencing data, assemblies’ statistics, BUSCO scores, and ANI values. Isolates UFS-Y2791, Kmx14, Kmx16, and Kmx24 from agave plant, henequen leaf, non-fermented henequen cooked juice, and cooked henequen core formed a separate group with ANI values greater than 99% between each other (Fig. 1). Interestingly, isolates Kmx22 and SLP1 from fermented henequen cooked juice and mezcal fermentations, respectively, did not belong to this group and exhibited more relatedness to K. marxianus isolates from dairy and other environments. These data confirm that there is further yeast diversity to be accessed in agave environments (4) in a similar way to what has been described for cactus yeasts (20).
TABLE 1.
General features of the sequenced genomes
| Isolate | Origin | No. of reads | No. of contigs | Total length (Mb) | Coverage (×) | GCcontent (%) | N50 (kb) | L50 (kb) | BUSCO scores (%) | ANI score (%)c | ANI coverage (%)c | No. of predicted genes | No. of proteins | GenBank assembly accession no. | SRA accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca | Db | |||||||||||||||
| Kmx14 | Henequen leaf | 4,395,815 | 215 | 10.6 | 116 | 39.99 | 87,645 | 37 | 98.9 | 0.3 | 94.5 | 94.9 | 4,923 | 4,744 | https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_029873725.1/ GCA_029873725.1 | SRR23105050 |
| Kmx16 | Non-fermented henequen cooked juice | 4,686,443 | 219 | 10.5 | 124 | 40.04 | 87,096 | 36 | 98.7 | 0.1 | 94.6 | 94.2 | 4,916 | 4,747 | GCA_029873675.1 | SRR23105048 |
| Kmx22 | Fermented henequen cooked juice | 5,405,732 | 396 | 10.6 | 142 | 40.11 | 45,399 | 74 | 99.4 | 0.1 | 99.2 | 96.9 | 4,935 | 4,773 | GCA_029873665.1 | SRR23105047 |
| Kmx24 | Cooked henequen core | 3,024,326 | 218 | 10.6 | 79 | 39.99 | 78,647 | 40 | 99.1 | 0.1 | 94.5 | 95 | 4,922 | 4,743 | GCA_029873655.1 | SRR23105046 |
C, completeness.
D, duplication level.
ANI against the reference genome K. marxianus DMKU3-1042.
Fig 1.
Heatmap of ANI values. The isolates sequenced here are indicated in blue color. The Kluyveromyces genomes were downloaded from NCBI: K. marxianus UFS-Y2791 (GenBank accession number: GCA_001692465.1), K. marxianus SLP1 alternate-pseudohaplotype (GCA_021014395.1), K. marxianus SLP1 principal pseudohaplotype of diploid (GCA_021014425.1), K. marxianus NRRL Y-6860 (GCA_002356615.1), K. marxianus LHW-O (GCA_003046155.1), K. marxianus CBS6556 (GCA_016625955.1), K. marxianus FIM1 (GCA_001854445.2), K. marxianus NBRC 1777 (GCA_001417835.1), K. marxianus DMKU3-1042 (GCA_001417885.1), K. marxianus L03 (GCA_008000265.1), K. marxianus Olga-2 (GCA_016584165.1), K. marxianus 100656–19 (GCA_902364165.1), K. marxianus B0399 (GCA_001660455.1), K. marxianus ATCC 8635 (GCA_017309885.1), K. dobzhanskii NRRL Y-1974 (GCA_003705805.2), K. wickerhamii UCD 54–210 (GCA_000179415.1), K. nonfermentans NRRL Y-27343 (GCA_003670155.1), K. starmeri UFMG-CM-Y3682 (GCA_008973615.1), K. lactis NRRL Y-1140 (GCA_000002515.1), and K. aestuarii NRRL YB-4510 (GCA_003707555.1).
ACKNOWLEDGMENTS
This research was supported by the Universidad Autónoma Metropolitana-Unidad Cuajimalpa in Mexico City (Research project 87 S210-21 “Caracterización y potencial de aplicación de levaduras y bacterias autóctonas de México, DCNI-05-210-21) and the CONAHCyT research grant CB-2010-01 156451 awarded to S.L.B.
Contributor Information
Sylvie Le Borgne, Email: sylvielb@cua.uam.mx.
Jason E. Stajich, University of California Riverside, Riverside, California, USA
DATA AVAILABILITY
The genome assembly generated in this study and the reads are deposited under BioProject ID PRJNA904382 at the NCBI.
REFERENCES
- 1. Fonseca GG, Heinzle E, Wittmann C, Gombert AK. 2008. The yeast Kluyveromyces marxianus and its biotechnological potential. Appl Microbiol Biotechnol 79:339–354. doi: 10.1007/s00253-008-1458-6 [DOI] [PubMed] [Google Scholar]
- 2. Lane MM, Morrissey JP. 2010. Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol Rev 24:17–26. doi: 10.1016/j.fbr.2010.01.001 [DOI] [Google Scholar]
- 3. Bilal M, Ji L, Xu Y, Xu S, Lin Y, Iqbal HMN, Cheng H. 2022. Bioprospecting Kluyveromyces marxianus as a robust host for industrial biotechnology. Front Bioeng Biotechnol 10:851768. doi: 10.3389/fbioe.2022.851768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortiz-Merino RA, Varela JA, Coughlan AY, Hoshida H, da Silveira WB, Wilde C, Kuijpers NGA, Geertman J-M, Wolfe KH, Morrissey JP. 2018. Ploidy variation in Kluyveromyces marxianus separates dairy and non-dairy isolates. Front Genet 9:94. doi: 10.3389/fgene.2018.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arellano-Plaza M, Gschaedler-Mathis A, Noriega-Cisneros R, Clemente-Guerrero M, Manzo-Ávalos S, González-Hernández JC, Saavedra-Molina A. 2013. Respiratory capacity of the Kluyveromyces marxianus yeast isolated from the mezcal process during oxidative stress. World J Microbiol Biotechnol 29:1279–1287. doi: 10.1007/s11274-013-1291-7 [DOI] [PubMed] [Google Scholar]
- 6. Lachance MA. 1995. Yeast communities in a natural tequila fermentation. Antonie Van Leeuwenhoek 68:151–160. doi: 10.1007/BF00873100 [DOI] [PubMed] [Google Scholar]
- 7. Lachance MA. 1971. Kluyveromyces Van der Walt, p 471–481. In Kurtzman CP, Fell JW, Boekhout T (ed), The yeasts, 5th ed. Elsevier, Amsterdam. [Google Scholar]
- 8. Lertwattanasakul N, Kosaka T, Hosoyama A, Suzuki Y, Rodrussamee N, Matsutani M, Murata M, Fujimoto N, Tsuchikane K, Limtong S, Fujita N, Yamada M. 2015. Genetic basis of the highly efficient yeast Kluyveromyces marxianus: complete genome sequence and transcriptome analyses. Biotechnol Biofuels 8:47. doi: 10.1186/s13068-015-0227-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki T, Hoshino T, Matsushika A. 2014. Draft genome sequence of Kluyveromyces marxianus strain DMB1, isolated from sugarcane bagasse hydrolysate. Genome Announc 2:e00733-14. doi: 10.1128/genomeA.00733-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lappe-Oliveras P, Avitia M, Sánchez-Robledo SD, Castillo-Plata AK, Pedraza L, Baquerizo G, Le Borgne S. 2023. Genotypic and phenotypic diversity of Kluyveromyces marxianus isolates obtained from the elaboration process of two traditional Mexican alcoholic beverages derived from agave: pulque and henequen (Agave fourcroydes) mezcal. J Fungi (Basel) 9:795. doi: 10.3390/jof9080795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrews S. 2010. FastQC - a quality control tool for high throughput sequence data. Babraham Bioinformatics. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 12. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zerbino DR. 2010. Using the velvet de novo assembler for short‐read sequencing technologies. Curr Protoc Bioinformatics 11:5.1-11.5.12. doi: 10.1002/0471250953.bi1105s31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 15. Wences AH, Schatz MC. 2015. Metassembler: merging and optimizing de novo genome assemblies. Genome Biol 16:207. doi: 10.1186/s13059-015-0764-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmer JM, Stajich J. 2020. Funannotate V1.8.1: eukaryotic genome annotation. Zenodo. Available from: https://zenodo.org/records/4054262. Retrieved 20 Nov 2023. [Google Scholar]
- 18. Manni M, Berkeley MR, Seppey M, Zdobnov EM. 2021. BUSCO: assessing genomic data quality and beyond. Curr Protoc 1:e323. doi: 10.1002/cpz1.323 [DOI] [PubMed] [Google Scholar]
- 19. Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- 20. Starmer WT, Schmedicke RA, Lachance MA. 2003. The origin of the cactus-yeast community. FEMS Yeast Res 3:441–448. doi: 10.1016/S1567-1356(03)00056-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome assembly generated in this study and the reads are deposited under BioProject ID PRJNA904382 at the NCBI.