Abstract
Previously, we have shown that TAL1 and the LIM-only protein gene (LMO) are regularly coactivated in T-cell acute lymphoblastic leukemia (T-ALL). This observation is likely to relate to the findings that TAL1 and LMO are highly synergistic in T-cell tumorigenesis in double-transgenic mice. To understand the molecular mechanisms of functional synergy between TAL1 and LMO in tumorigenesis and transcriptional regulation, we tried to identify downstream target genes regulated by TAL1 and LMO by a subtractive PCR method. One of the isolated genes, that for retinaldehyde dehydrogenase 2 (RALDH2), was regularly expressed in most of the T-ALL cell lines that coexpressed TAL1 and LMO. Exogenously transfected TAL1 and LMO, but not either alone, induced RALDH2 expression in a T-ALL cell line, HPB-ALL, not expressing endogeneous TAL1 or LMO. The RALDH2 transcripts in T-ALL were, however, mostly initiated within the second intron. Promoter analysis revealed that a GATA site in a cryptic promoter in the second intron was essential and sufficient for the TAL1- and LMO-dependent transcriptional activation, and GATA3 binds to this site. In addition, forced expression of GATA3 potentiated the induction of RALDH2 by TAL1 and LMO, and these three factors formed a complex in vivo. Furthermore, a TAL1 mutant not binding to DNA also activated the transcription of RALDH2 in the presence of LMO and GATA3. Collectively, we have identified the RALDH2 gene as a first example of direct transcriptional target genes regulated by TAL1 and LMO in T-ALL. In this case, TAL1 and LMO act as cofactors for GATA3 to activate the transcription of RALDH2.
In various types of leukemia, specific recurrent chromosomal translocations are frequently observed and potential oncogenic transcription factors have been identified from the chromosomal breakpoints (46). These transcription factors are mostly involved in normal hematopoietic cell differentiation and growth. In T-cell acute lymphoblastic leukemia (T-ALL), frequent chromosomal abnormalities are found in the TAL1 (also called SCL or TCL5) locus (5, 9, 10, 14). Ectopic expression of TAL1, which is not normally expressed in T cells (44, 57), is observed in ∼60% of T-ALL patients (4). TAL1 encodes at least two alternative isoforms, full-length TAL1α and N-terminally truncated TAL1β, having a basic helix-loop-helix (bHLH) motif found in a number of transcription factors involved in the regulation of cell differentiation (6). TAL1 dimerizes with ubiquitous bHLH E-proteins (E47, E12, and HEB) (20, 22), and the heterodimers bind to the E-box motif (CANNTG), most preferably to AACAGATGGT (21). However, no downstream target genes have been identified. It is thus uncertain whether TAL1–E-protein heterodimers regulate transcription through binding to this preferred E-box.
LMO1 (RBTN1 or TTG1) and LMO2 (RBTN2 or TTG2) are also genes originally identified from recurrent chromosomal breakpoints in T-ALL (7, 34, 49). Their expression in T-ALL is ectopic, like that of TAL1 (15, 18, 35, 49). They encode highly related LIM-only class proteins (LMO), which consist of only two tandemly repeated LIM domains. The LIM domain is a cysteine-rich zinc finger-like motif present in certain homeodomain transcription factors and some kinases and cytoskeletal proteins, and it appears to mediate protein-protein interactions (45, 51). LMO are nuclear proteins (35, 61) and are considered to be involved in transcriptional regulation, even though they do not have DNA binding activity.
Targeted-disruption experiments revealed that both TAL1 and LMO2 are essential for embryogenesis, as mutant mice die at embryonic day 9.5 due to the absence of yolk sac erythropoiesis (48, 52, 61). Since TAL1 and LMO2 physically associate in the erythroid cell lineage (56, 59), these transcription factors are likely to regulate erythroid cell differentiation and growth by forming a complex.
Previously we have shown that TAL1 is regularly coexpressed with LMO1 or LMO2 in T-ALL (41). Furthermore, transgenic mice ectopically expressing TAL1 in T cells did not efficiently develop tumors (13, 27, 47), whereas double-transgenic mice expressing TAL1 and LMO rapidly developed leukemia (2, 30). These results suggest that not only in the regulation of erythroid cell differentiation but also in the development of T-ALL, TAL1 and LMO act synergistically, most probably by forming a complex (30). However, no downstream target genes regulated by TAL1 and LMO have been identified. Thus, the molecular mechanism by which these factors regulate transcription in the erythroid lineage and T-ALL remains mostly unknown.
The fact that chromosomal abnormalities involving TAL1 and LMO are highly restricted to T-ALL suggests that some other T-cell-specific cofactor(s) is involved in the oncogenic function of TAL1 and LMO (41). In the erythroid lineage, LMO2 physically interacts with GATA1 as well as TAL1 and bridges these factors to form a larger complex in vivo (42). GATA1 was first identified as a protein binding to the conserved regulatory elements among many erythroid cell-specific genes (55). Targeted disruption of GATA1 blocks differentiation of erythroid precursors (16, 43), implying that TAL1, LMO2, and GATA1 cooperatively regulate transcription in the erythroid lineage. Although GATA1 is not expressed in the T-cell lineage, another member of the GATA-binding protein family, GATA3, is expressed (19, 28, 33). We have shown that GATA3 physically interacts with LMO in vitro and is a potent cofactor for TAL1 and LMO in transactivation of an artificial reporter gene in a T-ALL cell line (41).
To understand the mechanism of T-cell tumorigenesis and transcriptional regulation by TAL1 and LMO, we have been focusing on downstream target genes of these factors. Previously, we have shown that the gene for a T-ALL-specific tumor marker, TALLA1 (54), is likely to be one such gene. TALLA1 is regularly coexpressed with TAL1 and LMO in T-ALL cell lines (41). Coexpression of exogeneous TAL1 and LMO1, but not either alone, strongly induced TALLA1 in a T-ALL cell line, HPB-ALL, not expressing endogeneous TAL1 or LMO (41). However, the mechanism of induction of TALLA1 by TAL1 and LMO remains unknown. In this study, we show that the gene for retinaldehyde dehydrogenase 2 (RALDH2) (63) is also induced in HPB-ALL cells by coexpression of exogeneous TAL1 and LMO and that RALDH2 is regularly expressed in most T-ALL-derived cell lines ectopically coexpressing TAL1 and LMO. Promoter analysis revealed that a transcriptional complex containing TAL1, LMO, and GATA3 binds to a GATA site in a cryptic promoter in the second intron of the RALDH2 gene and regulates transcription through this site. Thus, the RALDH2 gene is a direct target gene regulated by TAL1 and LMO in T-ALL, and in this case TAL1 and LMO act as cofactors for GATA3.
MATERIALS AND METHODS
Cell culture.
Hematopoietic cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. For details of each T-ALL cell line, see reference 54. HPB-ALL clones stably expressing TAL1α and/or LMO1 were described previously (41). 293E and COS cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum.
PCR-based subtraction.
Suppression-subtractive hybridization between parental HPB-ALL cells and HPB-ALL cells stably expressing TAL1α and LMO1 was carried out by using a PCR-Select cDNA subtraction kit (Clontech) according to the protocol recommended by the manufacturer. Obtained PCR products were digested with RsaI and ligated with AD1 adapter. AD1-ligated tester cDNA (300 pg) and driver cDNA (300 ng) were hybridized and amplified by using AD1S primer. PCR products were digested with RsaI and ligated with AD2 adapter. AD2-ligated tester cDNA (10 pg) and driver cDNA (300 ng) were hybridized and amplified by using AD2S primer. Amplified fragments were cloned into pCRII (Invitrogen). All PCRs were carried out with Pfu DNA polymerase (Stratagene). Adapter sequences were as follows: AD1S, 5′-CAGCTCCACAACCTACATCATTCCGT-3′; AD1A, 5′-ACGGAATGATGT-3′; AD2S, 5′-GTCCATCTTCTCTCTGAGACTCTGGT-3′; and AD2A, 5′-ACCAGAGTCTCA-3′. AD1S-AD1A and AD2S-AD2A were annealed, yielding AD1 and AD2, respectively.
Isolation of RALDH2 cDNA and genomic clones.
A Jurkat cDNA library (Clontech) was screened by colony hybridization, using as a probe 32P-radiolabeled cDNA fragments obtained by subtractive PCR. Four clones were analyzed by DNA sequencing, and then 5′ rapid amplification of cDNA ends (RACE) was performed with Molt4 and K562 poly(A)+ RNAs. Amplified products were cloned into pCRII (Invitrogen), and several clones were sequenced. Full-length cDNAs were amplified from Molt4 and K562 poly(A)+ RNAs by reverse transcription-PCR (RT-PCR) with primers designed from the sequence of 5′RACE products and the Jurkat cDNA, and several clones were sequenced.
Genomic clones were obtained by screening a human genomic library by using the 32P-labeled 5′ portion of the Molt4 RALDH2 cDNA fragment as a probe. The inserts of positive clones were subcloned into pBluescriptII (Stratagene) and sequenced from both directions.
Northern blotting and RT-PCR.
Total RNA was prepared from various cell lines by using Trizol reagent (Life Technologies, Inc.). Poly(A)+ RNA was purified with an mRNA purification kit (Pharmacia). Two micrograms of each poly(A)+ RNA was fractionated by electrophoresis on a 1% agarose gel containing formaldehyde and blotted onto a nylon membrane. Filters were hybridized with 32P-labeled probes by using QuikHyb hybridization solution (Stratagene) and washed at 60°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate.
RT-PCR was performed essentially as described previously (41). Amplification was carried out by denaturation at 94°C for 1 min (3 min in the first cycle), annealing at 65°C for 1 min, and extension at 72°C for 1 min (3 min in the last cycle). The number of cycles was 28 for F1-R1, 37 for F2-R1, 30 for F3-R1, and 35 for detection of E2A and RALDH2 in transiently transfected HPB-ALL cells. Primer sequences were as follows: F1, 5′-AACAACGAGTGGCAGAACTCAGAGAG-3′; F2, 5′-TTCAGTTGTGCCTCTTCCTCTCTAAC-3′; F3, ACAAAGCAGTGCAGGCAGCC-3′; R1, ATCGAAAGGTTTTGATGACGCCCTGC-3′; RALDH2 in transiently transfected HPB-ALL cells, 5′-AGGAGATACTGGATGTGTCTGCTAGC-3′ and 5′-CTCACAGTGTCTTCTGCAATGCAAGC-3′; and E2A, 5′-CAGCAGGGTTTCCAGGCCTGAGGTGC-3′ and 5′-GCTGCTGTGCGACTCAGTGAAGTGGG-3′.
Transfection and luciferase assay.
Expression vectors for TAL1, LMO1, LMO2, and GATA3 were as described previously (41). The GATA3 KRR mutant (pc-KRR) was kindly provided by Astar Winoto (53). The EcoRI fragment of pc-KRR was inserted into pMIKneo, yielding pMIKneoGATA3KRR. The GATA3CFM mutant was constructed by PCR with mutant primers changing Cys(317) and Cys(320) to Gly. The TAL1βBM mutant was also constructed by PCR changing Arg-Glu-Arg(20–22) to Ala-Ala-Ala. To make a series of RALDH2-T reporter plasmids, each genomic fragment was amplified by PCR with 5′-GAGGAGCTCCCTTCTCCACACTGAACCAAGAGAG-3′ and the following primers: −1.7-luc, 5′-GAGGGTACCGAGTGTAGGTTTGGAGTGATGTAGG-3′; −308-luc, 5′-GAGGGTACCGAGTGTTCCCTGTCTATAATCCAGCC-3′; −267-luc, 5′-GAGGGTACCTGTGAAGTTCAAGAAGCAGACAAGG-3′; −231-luc, 5′-GAGGGTACCTAGATAAAAGATTTCCTATGAAATAA-3′; −228-luc, 5′-GAGGGTACCATAAAAGATTTCCTATGAAATAACTG-3′; −207-luc, 5′-GAGGGTACCAACTGCCTTCAAACAGCAGAGCAGCA-3′; −181-luc, 5′-GAGGGTACCAACATATGCTCTCAGTACACCACTAC-3′; and −142-luc, 5′-GAGGGTACCACTTTTTTCATGACAGTGGATGGTTC-3′. Amplified fragments were inserted into the KpnI-SacI site of the luciferase reporter plasmid pGV-B (TOYO Ink, Tokyo, Japan). To make tk-luc, the herpes simplex virus (HSV) thymidine kinase (tk) minimal promoter (36) was amplified by PCR with 5′-GAGAGATCTCAGTCGGGGCGGCGCGGTCC-3′ and 5′-GAGAAGCTTCGGTCGCTCGGTGTTCGAGG-3′, digested with BglII and HindIII, and cloned into the same site of pGV-B. To make GATA×3-tk-luc, two synthetic oligonucleotides, 5′-CGTAGATAAAAGTAGATAAAAGTAGATAAAAGAGCT-3′ and 5′-CTTTTATCTACTTTTATCTACTTTTATCTACGGTAC-3′, were annealed and cloned into the KpnI-SacI site of tk-luc. Recipient cells (107) were transfected by electroporation with various combinations of plasmids together with 0.1 μg of pRL-CMV (Promega). After 24 h, cell lysates were prepared and assayed by using the Dual-luciferase reporter assay system (Promega). Luciferase activity was normalized with Renilla luciferase activity.
Immunoprecipitation.
Two synthetic oligonucleotides, 5′-AATTGCCACCAT GGACTACAAGGACGACGACGACAAGGAATTCCCGGGTCGACA-3′ and 5′-CTAGTGTCGACCCGGGAATTCCTTGTCGTCGTCGTCCTTGTAGTCCATGGTGGC-3′, were annealed and cloned into the EcoRI-SpeI site of pMIKneo, yielding pMIKneoFLAG. Two synthetic oligonucleotides, 5′-AATT GCCACCATGGACTACCCATACGACGTCCCAGACTACGCTGAATTCC CGGGTCGACA-3′ and 5′-CTAGTGTCGACCCGGGAATTCAGCGTAGTCTGGGACGTCGTATGGGTAGTCCATGGTGGC-3′, were annealed and cloned into the EcoRI-SpeI site of pMIKneo, yielding pMIKneoHA. cDNAs of TAL1β, TAL1βBM, LMO1, LMO2, GATA3, GATA3CFM, and E47S were amplified by PCR with the following primer sets: TAL1β, 5′-GAGGAATTCATGGAGATTACTGATGGTCC-3′ and 5′-GAGGTCGACGGATCCTCACCGAGGGCCGGCTCCATC-3′; TAL1βBM, 5′-GAGGAATTCATGGAGATTACTGATGGTCC-3′ and 5′-GAGGAATTCTGATCCTGGTGGCCCAGACCCATCAC-3′; LMO1, 5′-GAGGAATTCATGATGGTGCTGGACAAGGA-3′ and 5′-GAGGTCGACGGATCCCGTTACTGAACTTGGGATTC-3′; LMO2, 5′-GAGGAATTCATGTCCTCGGCCATCGAAAG-3′ and 5′-GAGGTCGACGGATCCCCTATATCATCCCATTGATC-3′; GATA3 and GATA3CFM, 5′-GAGGAATTCATGGAGGTGACGGCGGACCAGCCGCG-3′ and 5′-GAGGAATTCGTGAGCATCGAGCAGGGCTCTAACCC-3′, and E47S, 5′-GAGGAATTCAGTACGGACGAGGTGCTGTC-3′ and 5′-GAGGAATTCCTCGTCCCACGGAGGCATAC-3′. TAL1β, LMO1, and LMO2 fragments were digested with EcoRI and SalI and cloned into the same site of pMIKneoFLAG and pMIKneoHA. TAL1βBM, GATA3, GATA3CFM, and E47S fragments were digested with EcoRI and cloned into the same site of pMIKneoFLAG and pMIKneoHA. 293E cells (2 × 106) were transfected with various combinations of 5 μg of each plasmid by using Lipofectamine Plus reagent (GIBCO BRL). After 48 h, cells were lysed in 600 μl of low-stringency Nonidet P-40 (NP-40) buffer (10 mM HEPES [pH 7.6], 250 mM NaCl, 5 mM EDTA, 0.1% NP-40) (56) and immunoprecipitated with anti-FLAG antibody (Kodak). After washing five times with cold low-stringency NP-40 buffer, immunoprecipitates were analyzed by immunoblotting with anti-FLAG or antihemagglutinin (anti-HA) (Boehringer Mannheim).
Electrophoretic mobility shift assay (EMSA).
Cell lysates of transfected 293E or COS cells were prepared by using low-stringency NP-40 buffer as described above. Nuclear extracts of HPB-ALL cells were prepared as described previously (12). Double-stranded oligonucleotide probes were end labeled by using [γ-32P]ATP and T4 polynucleotide kinase. One microliter of cell lysates or 5 μg of nuclear extracts was incubated with or without competitors (100 ng), anti-FLAG monoclonal antibody (MAb) (2.8 μg), or anti-GATA3 MAb (0.2 μg) (Santa Cruz Biotechnology, Inc.) for 5 min at room temperature and then with radiolabeled probes (2 × 104 cpm/ng; 0.5 ng/reaction mixture) for another 20 min at room temperature in 20 μl of a solution consisting of 20 mM HEPES (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and 0.5 μg (293E and COS cell lysates) or 2 μg (HPB-ALL nuclear extract) of poly(dI-dC). The samples were electrophoresed on a 5% polyacrylamide gel with TAE buffer (7 mM Tris-HCl [pH 7.5], 3 mM sodium acetate, and 1 mM EDTA) at 120 V for 1.5 h. Gels were dried and subjected to autoradiography. Oligonucleotide probes were as follows: TAL1CS, 5′-ACCTGAACAGATGGTCGGCT-3′ and 5′-AGCCGACCATCTGTTCAGGT-3′; TCR-GATA, 5′-GTTAGAGATAGCATCGCCCC-3′ and 5′-GGGGCGATGCTATCTCTAAC-3′; RALDH2-GATA, 5′-GGCCCCTTTTGTAGATAAAAGATTTCCGGG-3′ and 5′-CCCGGAAATCTTTTATCTACAAAAGGGGGCC-3′; and RALDH2-GATAM, 5′-GGCCCCTTTTGTTCTAGAAAGATTTCCGGG-3′ and 5′-CCCGGAAATCTTTCTAGAACAAAAGGGGCC-3′.
Nucleotide sequence accession numbers.
The nucleotide sequences of human RALDH2 cDNA are listed in the DDBJ/EMBL/GenBank database under accession no. AB015226, AB015227, and AB015228. The sequence in Fig. 3A is listed in the DDBJ/EMBL/GenBank database under accession no. AB015229.
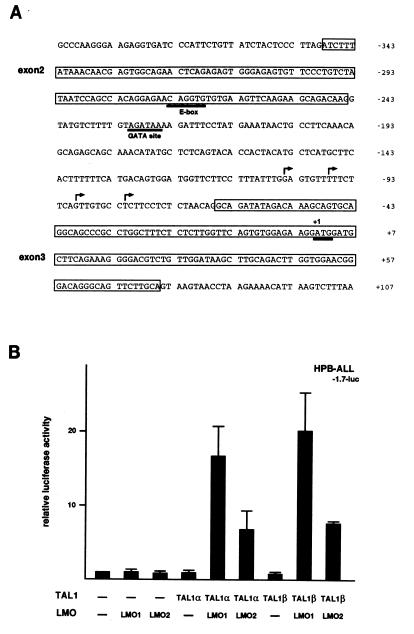
FIG. 3.
Induction of the RALDH2-T promoter by TAL1 and LMO. (A) Nucleotide sequence around the promoter region of RALDH2-T. Exon sequences are boxed. A consensus GATA site, an optimal E-box, and the putative initiation codon of RALDH2-T are underlined. Major transcriptional initiation sites are indicated by arrows. (B) HPB-ALL cells were cotransfected with 20 μg of a luciferase reporter plasmid containing the 1.7-kb genomic fragment of the RALDH2-T promoter (−1.7-luc), 0.1 μg of pRL-CMV for normalization of transfection efficiency, and 5 μg each of expression plasmids without inserts (−) or with the indicated inserts. Relative luciferase activity compared to that of the reporter plasmid alone was determined. Means and standard deviations for three independent experiments are shown.
RESULTS
RALDH2 is induced by TAL1 and LMO in T-ALL.
To identify target genes regulated by ectopically coexpressed TAL1 and LMO in T-ALL, we used a T-ALL-derived cell line, HPB-ALL, not expressing TAL1 or LMO and its subline stably expressing transfected TAL1α and LMO1 (41). Using suppression-subtractive hybridization followed by two cycles of subtractive PCR (see Materials and Methods), we obtained several fragments of genes whose expression was strongly upregulated in the subline coexpressing TAL1α and LMO1 (data not shown). Four of these fragments were derived from the same gene. Since the encoded amino acid sequence is 98% identical to mouse RALDH2 (63), this gene is the human counterpart of the RALDH2 gene (see below).
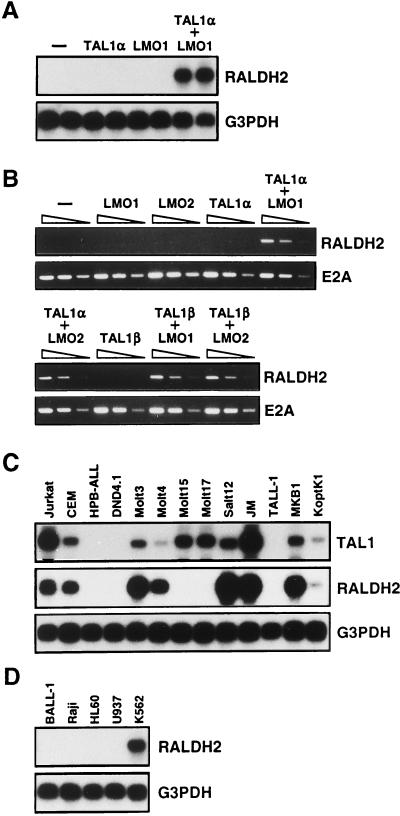
To examine the roles of TAL1α and LMO1 in the induction of RALDH2, we analyzed the expression of RALDH2 in a panel of HPB-ALL clones stably expressing transfected TAL1α or LMO1 or both (41). As shown in Fig. 1A, the vector control clones and those expressing either TAL1α or LMO1 alone did not express RALDH2 at all, whereas those coexpressing both TAL1α and LMO1 strongly expressed it. To rule out possible clonal variations, we also examined HPB-ALL cells transiently transfected with TAL1 and LMO expression vectors (Fig. 1B). Consistent with the results obtained with stable clones, only HPB-ALL cells cotransfected with TAL1α and LMO1 expressed RALDH2. Furthermore, even the N-terminally truncated isoform TAL1β induced RALDH2 expression in the presence of LMO1 or LMO2. These results strongly suggest that RALDH2 is a transcriptional downstream target gene of TAL1 and LMO and that both TAL1 and LMO are required for induction of RALDH2.
FIG. 1.
Induction of RALDH2 by TAL1 and LMO in T-ALL cell lines. (A) Northern blot analysis for RALDH2 in HPB-ALL clones. HPB-ALL cells were transfected with the indicated combinations of expression vectors, and stable transformants were isolated. Two clones were analyzed for each combination. Poly(A)+ RNA (2 μg each) was electrophoresed, blotted onto a filter, and hybridized with the 32P-labeled 3′ untranslated region of RALDH2 cDNA obtained by subtractive PCR. The same filter was reprobed for glyceraldehyde-3-phosphate dehydrogenase (G3PDH) as an internal control. (B) RT-PCR analysis for RALDH2 in HPB-ALL cells transiently expressing the indicated combinations of TAL1 and LMO. HPB-ALL cells were cotransfected with 15 μg each of the indicated expression vectors and 1 μg of pRC/CMV-luc by electroporation. Total RNA was prepared 20 h after transfection and subjected to RT-PCR analysis for RALDH2 and E2A (control). The amounts of total RNA used were 10, 2, and 0.4 ng from left to right. Amplification products were electrophoresed on 2% agarose and stained with ethidium bromide. Transfection efficiency was tested by luciferase assay and varied within threefold. (C and D) Northern blot analysis for RALDH2 in T-ALL-derived cell lines (C) and other hematopoietic cell lines (D). Poly(A)+ RNA (2 μg each) was examined as described for panel A. BALL-1 and Raji, B-cell lines, HL60, promyelocytic cell line; U937, monocytoid cell line; K562, erythroleukemia cell line.
To examine whether expression of RALDH2 is regularly associated with ectopic expression of TAL1 and LMO in T-ALL, we performed Northern blot analysis of 13 T-ALL cell lines (54). As shown in Fig. 1C, HPB-ALL, DND4.1, and TALL-1 cells, which express neither TAL1 nor LMO (41), did not express RALDH2 either. In contrast, except for Molt15 and Molt17, all of the T-ALL cell lines that co-express TAL1 and either LMO1 or LMO2 (41) also expressed RALDH2. Although Molt15 and Molt17 cells, which were the only T-ALL cell lines with the CD4− CD8− double-negative phenotype (54), did not express RALDH2 despite coexpression of TAL1 and LMO2, these results strongly support the idea that RALDH2 is indeed induced by coactivation of TAL1 and LMO in the majority of T-ALL. Among other hematopoietic cell lines tested, K562, which is an erythroleukemia-derived cell line endogenously expressing TAL1 and LMO2, also expressed RALDH2 (Fig. 1D).
Transcription of RALDH2 in T-ALL starts from a T-ALL-specific promoter.
Full-length cDNA clones of RALDH2 were obtained from Molt4 and K562 cells. The coding sequence of cDNA obtained from K562 cells showed a striking homology to the mouse RALDH2 gene, and the deduced amino acid sequence was 98% identical to that of mouse RALDH2. In contrast, the 5′ portion of cDNA obtained from Molt4 cells was 237 bp shorter than that of K562 cells, and the 5′-terminal 28-bp sequence was not present in the K562 cDNA. Analysis of the genomic sequence of RALDH2 revealed that the 28-bp sequence was derived from the second intron. All of the clones obtained from Molt4 cells by the 5′RACE method contained the same short 5′ portion, suggesting that transcription was initiated from a cryptic promoter within the second intron of the RALDH2 gene in Molt4 cells (Fig. 2A). This was further confirmed by primer extension analysis and RNase protection assay (data not shown). These analyses also revealed that there were several transcriptional initiation sites within ∼30 bp around the boundary between the second intron and third exon of the RALDH2 gene.
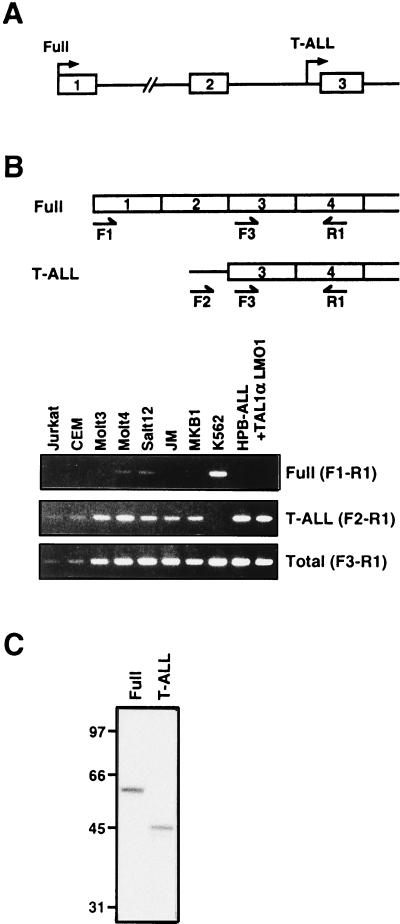
FIG. 2.
Structures of the RALDH2 gene and mRNA. (A) Schematic illustration of the RALDH2 gene. Boxes and lines correspond to exons and introns, respectively. The transcription start sites of the full-length RALDH2 and the T-ALL-type RALDH2 (RALDH2-T) are marked by arrows. (B) 5′ structure of RALDH2 mRNA in T-ALL cell lines. Two types of RALDH2 cDNA and the positions of primers used for RT-PCR to distinguish each type are shown schematically at the top. Total RNA samples prepared from the indicated cell lines were subjected to RT-PCR. Amplification products were electrophoresed on 2% agarose and stained with ethidium bromide. Two clones were analyzed for HPB-ALL cells stably transfected with TAL1α and LMO1. (C) In vitro translation from two types of RALDH2 mRNA. Both types of RALDH2 cDNA were transcribed in vitro and translated in rabbit reticulocyte lysate in the presence of [35S]methionine. Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography.
To test which types of message were transcribed in other T-ALL cell lines, RALDH2 transcription was examined by RT-PCR with primer sets specific for the full-length type (F1-R1), the 5′-truncated type (F2-R1), or both (F3-R1) (Fig. 2B). As expected, K562 cells expressed only the full-length type. In contrast, although some full-length type messages were also detected in Molt4 and SALT12 cells, the major transcripts in T-ALL-derived cell lines were of the truncated type. Furthermore, HPB-ALL clones stably expressing transfected TAL1α and LMO1 also expressed only the truncated-type mRNA (Fig. 2B). These results suggest that ectopic expression of TAL1 and LMO in T-ALL induces transcription mostly if not exclusively from the intronic promoter of the RALDH2 gene. Furthermore, this alternatively initiated type of mRNA was not detected in normal human tissues (data not shown), suggesting that this type of mRNA is highly specific for T-ALL. The first AUG codon of T-ALL-specific mRNA is in the same frame of the full-length RALDH2 mRNA, suggesting that the former encodes an N-terminally truncated protein. To examine this, we performed in vitro translation experiments. As shown in Fig. 2C, 56 and 46-kDa proteins were translated from the full-length mRNA and the T-ALL-specific mRNA, respectively. This suggests that the N-terminally truncated 46-kDa protein is produced in T-ALL cells. We thus designated this N-terminally truncated form RALDH2-T (T-ALL type).
A GATA site in the RALDH2-T promoter is required for induction by TAL1 and LMO.
To understand the mechanism of regulation of the RALDH2-T promoter by TAL1 and LMO (Fig. 3A), we constructed a luciferase reporter gene containing the 1.7-kb genomic fragment upstream of the first ATG codon of RALDH2-T (−1.7-luc [see Fig. 4A]) and transfected it with or without TAL1 and LMO into HPB-ALL cells. As shown in Fig. 3B, either TAL1 or LMO alone did not induce the transcription from the reporter. However, TAL1 and LMO1 or LMO2 transactivated the reporter 18- and 8-fold, respectively. TAL1α and TAL1β had similar activities in this assay. These results were in accordance with the induction of the endogeneous RALDH2-T in HPB-ALL cells by TAL1 and LMO, suggesting that the 1.7-kb genomic fragment contains a regulatory element(s) responsible for the induction of RALDH2-T by TAL1 and LMO in T-ALL. As shown in Fig. 3A, a TATA box-like sequence is not seen in the promoter region, but the major initiation site matches the consensus initiator (Inr) sequence (24). There are an E-box (CAGGTG) known to be recognized by TAL1-E2A heterodimers (60) at position −273 (ATG is +1) and a consensus GATA site (AGATAA) (29, 37) at position −239.
FIG. 4.
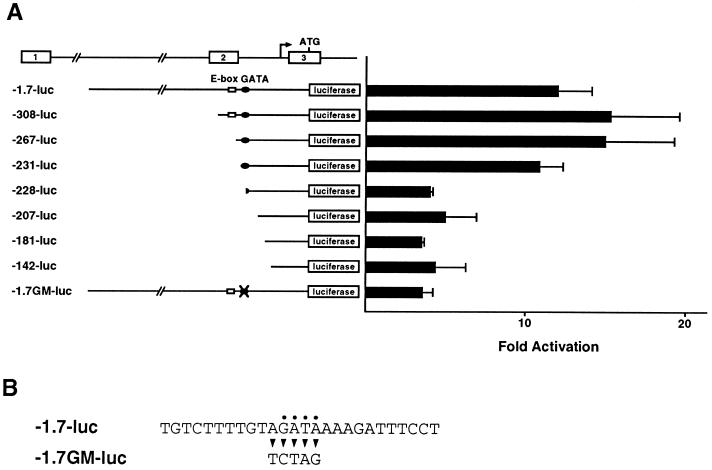
Requirement of a GATA site but not an E-box in the RALDH2-T promoter for induction by TAL1 and LMO. (A) HPB-ALL cells were cotransfected with one of the indicated RALDH2-T reporter plasmids and expression vectors for TAL1α and LMO1. The results show fold activation in luciferase activity from the reporter alone. Means and standard deviations for three independent experiments are shown. (B) Mutations introduced in −1.7GM-luc.
To identify responsive elements in the RALDH2-T promoter, we constructed a series of 5′ deletion mutants and examined activation of these reporters by TAL1α and LMO1 in HPB-ALL cells (Fig. 4A). Unexpectedly, deletion of the E-box did not affect the response (compare −308-luc with −267-luc). Mutation of the E-box also had little effect on the transcription from the reporter (data not shown). In contrast, the deletion removing 3 bases in the GATA site abrogated the induction of the reporter by TAL1α and LMO1 (compare −231-luc with −228-luc). Further deletions to position −142 (∼50 bp upstream of transcription start sites) and the deletion downstream of the start site had little further effect (Fig. 4A and data not shown). To specifically address the role of the GATA site at position −239, this element was changed to a restriction site (−1.7GM-luc [Fig. 4B]). The mutation of the GATA site effectively abrogated the response. These results clearly indicate that the GATA site at position −239 in the RALDH2-T promoter is essential for induction by TAL1 and LMO.
GATA3 is involved in induction of RALDH2-T by TAL1 and LMO.
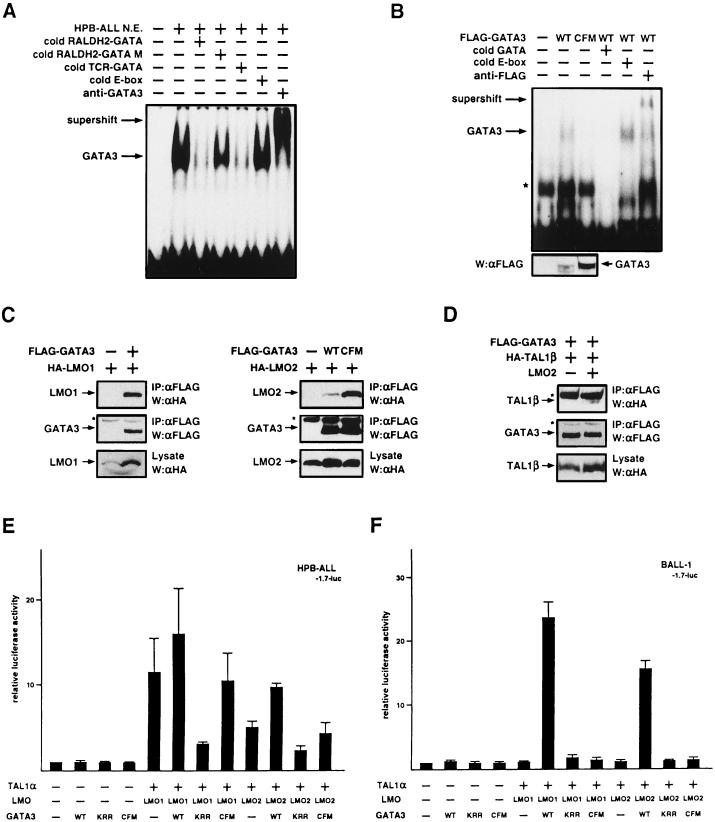
Next, we examined the binding of nuclear proteins to the GATA site in the RALDH2-T promoter. Nuclear extracts were prepared from HPB-ALL cells and used for EMSA. As shown in Fig. 5A, binding to the GATA site was observed. This binding was competed out by a 200-fold excess of the same cold oligonucleotide but not by the same amount of the mutant oligonucleotide corresponding to the sequence of the GATA mutant reporter (−1.7GM-luc [Fig. 4B]), suggesting that the binding was specific for the GATA site. The binding was also competed out by the T-cell receptor alpha (TCRα) GATA site, suggesting that the binding protein is a GATA-binding protein family member. Among six members identified in the GATA-binding protein family, only GATA3 is known to be expressed in the T-cell lineage (19, 28, 33). Thus, we used an anti-GATA3 MAb for supershift experiments. The anti-GATA3 MAb indeed supershifted the binding complex (Fig. 5A), indicating that the binding protein is GATA3. The same shift band was observed even with nuclear extracts from an HPB-ALL clone stably expressing TAL1α and LMO1, suggesting that TAL1α and LMO1 did not directly bind to this site and that the binding of GATA3 to this site was not appreciably enhanced by TAL1α and LMO1 (data not shown).
FIG. 5.
Involvement of GATA3 in induction of RALDH2-T by TAL1 and LMO. (A) GATA3 binds to the GATA site in the RALDH2-T promoter. EMSA was carried out by using nuclear extracts prepared from HPB-ALL cells and a labeled oligonucleotide spanning the GATA site in the RALDH2-T promoter. Cold competitors in 200-fold excess or anti-GATA3 MAb was added as indicated. (B) The GATA3-CFM mutant does not bind to DNA. COS cells were transfected with wild-type (WT) or mutant GATA3 expression vectors. EMSA was carried out by using cell lysates from transfected COS cells and a labeled oligonucleotide spanning the GATA site in the TCRα enhancer. Cold competitors in 200-fold excess or anti-FLAG MAb was added as indicated. Expression of GATA3 was confirmed by Western blotting (W) with anti-FLAG MAb (αFLAG). The asterisk indicates nonspecific binding. (C) Interaction between GATA3 and LMO1 or LMO2 in vivo. Cell lysates from 293E cells transiently transfected with expression vectors for the indicated tagged proteins were precipitated with anti-FLAG MAb. Immunoprecipitates (IP) and cell lysates were immunoblotted with anti-FLAG or anti-HA MAb as indicated. The asterisk indicates immunoglobulin H. (D) Complex formation between TAL1, LMO2, and GATA3. 293E cells were transiently transfected with expression vectors for FLAG-GATA3 and HA-TAL1 with or without that for LMO2, and cell lysates were immunoprecipitated with anti-FLAG MAb. Immunoprecipitates and cell lysates were immunoblotted with anti-FLAG or anti-HA MAb as indicated. The asterisk indicates immunoglobulin H or L. (E and F) Effect of GATA3 on induction of the RALDH2-T promoter by TAL1 and LMO. HPB-ALL (E) and BALL-1 (F) cells were cotransfected with 15 μg of a luciferase reporter containing the 1.7-kb genomic fragment of the RALDH2-T promoter (−1.7-luc), 0.1 μg of pRL-CMV for normalization of transfection efficiency, and 5 μg each of expression plasmids without inserts (−) or with the indicated inserts. Relative luciferase activity compared to that of the reporter alone was determined. Means and standard deviations for three independent experiments are shown.
Next we examined whether overexpression of GATA3 could affect the activation of the RALDH2-T promoter by TAL1 and LMO. As shown in Fig. 5E, overexpression of GATA3 alone in HPB-ALL cells did not enhance −1.7-luc reporter expression. However, its induction by TAL1α and LMO was further upregulated by overexpression of GATA3, especially with the combination of TAL1α and LMO2. Furthermore, a dominant negative GATA3 (KRR) (53) effectively suppressed induction of the reporter by TAL1α and LMO. These results strongly support the idea that GATA3 is involved in the induction of the RALDH2-T promoter by TAL1 and LMO. This was further confirmed by the same transfection assays with a B-ALL-derived cell line, BALL-1 (54) (Fig. 5F). We confirmed by RT-PCR that BALL-1 cells did not express TAL1, LMO1, LMO2, or GATA3 (data not shown). In this cell line, the −1.7-luc reporter gene was not activated even by cotransfection of TAL1α and LMO. GATA3 alone had no effect either. However, cotransfection of GATA3, TAL1α, and LMO activated the reporter by ∼20-fold. This further supports the idea that GATA3 is involved in the transactivation of the RALDH2-T promoter by TAL1 and LMO.
These results led us to examine the physical interactions between TAL1, LMO, and GATA3. TAL1 had been reported to interact with LMO in vivo (56, 59). This was also confirmed by our experiments (see Fig. 6B). FLAG-tagged GATA3 and HA-tagged LMO1 or LMO2 were transiently expressed in 293E cells. As shown in Fig. 5C, HA-LMO1 as well as HA-LMO2 was coimmunoprecipitated with FLAG-GATA3 from the cell lysates by an anti-FLAG MAb, suggesting that both LMO1 and LMO2 were capable of binding to GATA3 in vivo. On the other hand, direct interaction between TAL1β and GATA3 was not observed (Fig. 5D). However, in the presence of LMO2, HA-TAL1β was coimmunoprecipitated with FLAG-GATA3, suggesting that LMO2 is capable of bridging TAL1 and GATA3 to form a complex in vivo (Fig. 5D). Collectively, these observations strongly suggest that a complex consisting of TAL1, LMO, and GATA3 is formed in T-ALL cells and that the complex induces transcription from the RALDH2-T promoter.
FIG. 6.
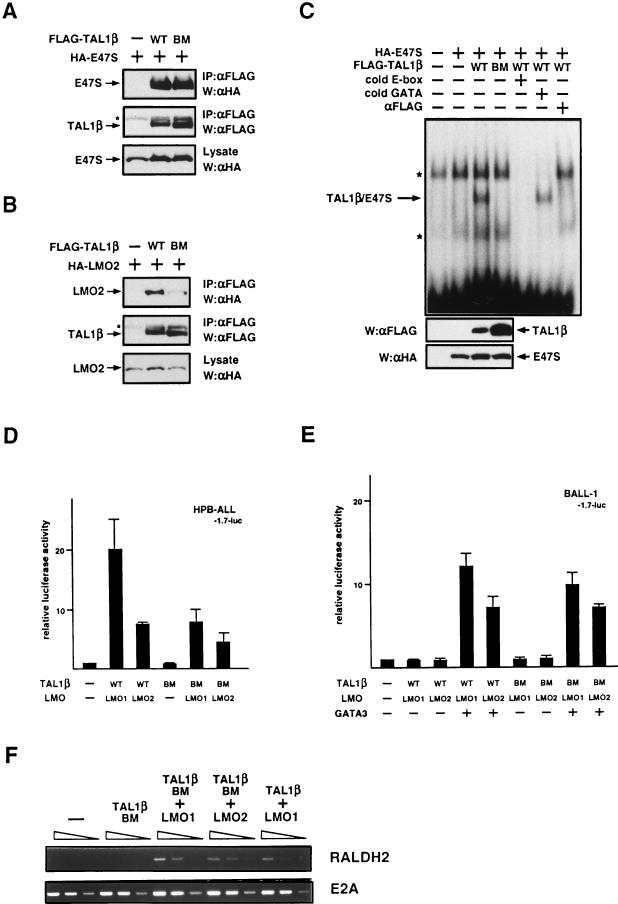
No requirement of DNA binding activity of TAL1 for induction of RALDH2-T. (A and B) TAL1β-BM can interact with E47 and LMO2. Cell lysates from 293E cells transiently transfected with expression vectors for each tagged proteins were immunoprecipitated (IP) with anti-FLAG MAb (αFLAG). Immunoprecipitates and cell lysates were immunoblotted (W) with anti-FLAG or anti-HA MAb as indicated. The asterisk indicates immunoglobulin L. WT, wild type. (C) TAL1β-BM does not bind to DNA. 293E cells were transfected with expression vectors for wild-type or mutant TAL1β and E47S. EMSA was carried out by using cell lysates from transfected 293E cells and the labeled consensus E-box oligonucleotide. Cold competitors in 200-fold excess or anti-FLAG MAb was added as indicated. Expression of TAL1β and E47S was confirmed by Western blotting with anti-FLAG MAb. Asterisks indicate nonspecific binding. (D and E) TAL1β-BM induces transcription from the RALDH2-T reporter in collaboration with LMO and GATA3. HPB-ALL (D) and BALL-1 (E) cells were cotransfected with 20 μg (D) or 15 μg (E) of a luciferase reporter containing the 1.7-kb genomic fragment of the RALDH2-T promoter (−1.7-luc), 0.1 μg of pRL-CMV for normalization of transfection efficiency, and 5 μg each of expression plasmids without inserts (−) or with the indicated inserts. Relative luciferase activity compared to that of the reporter alone was determined. Means and standard deviations for three independent experiments are shown. (F) TAL1β-BM induces endogeneous RALDH2-T expression with LMO in HPB-ALL cells. HPB-ALL cells were cotransfected with 15 μg each of indicated expression vectors and 1 μg of pRC/CMV-luc by electroporation. Total RNA was prepared 20 h after transfection and subjected to RT-PCR analysis for RALDH2 and E2A (control). The amounts of total RNA used were 10, 2, and 0.4 ng from left to right. Amplification products were electrophoresed on 2% agarose and stained with ethidium bromide. Transfection efficiency was tested by luciferase assay and varied within threefold.
DNA binding activity of GATA3 is required for transactivation of the RALDH2-T promoter.
Thus far, we have shown that a GATA site in the RALDH2-T promoter is essential for the induction by TAL1 and LMO and that GATA3 acts in collaboration with TAL1 and LMO. Therefore, we next tested whether the DNA binding activity of GATA3 was necessary for induction of the RALDH2-T promoter. For this purpose, we constructed a GATA3 mutant lacking the DNA binding activity. In the case of GATA1, the C-terminal Zn finger is required for DNA binding activity, and a mutant with cysteines in this region changed to glycines could not bind to DNA but still interacted with other transcription factors, such as SP1 (38). Like this GATA1 mutant, the C-terminal Zn finger of GATA3 was mutated to generate GATA3-CFM. Whole-cell lysates of COS cells transiently expressing FLAG-GATA3 or FLAG-GATA3-CFM were used for EMSA to test the DNA binding activity. As shown in Fig. 5B, FLAG-GATA3 bound to the GATA site of the TCRα enhancer region (19) and was supershifted by the anti-FLAG MAb. On the other hand, FLAG-GATA3-CFM could not bind to the same oligonucleotide even though proteins were detected by Western blotting. It was also demonstrated that GATA3-CFM interacted with LMO2 in vivo by coprecipitation assays (Fig. 5C). These results confirmed that GATA3-CFM could form a complex with TAL1 and LMO but could not bind to DNA. After these experiments, we tested whether GATA3-CFM was capable of activating the RALDH2-T promoter in collaboration with TAL1 and LMO. When GATA3-CFM was transfected with TAL1α and LMO, it did not enhance the transcription from the reporter −1.7-luc in HPB-ALL cells or induce the transcription in BALL-1 cells (Fig. 5E and F). Thus, the DNA binding activity of GATA3 is necessary for the transactivation of the RALDH2-T promoter by the complex containing GATA3, TAL1, and LMO.
The DNA binding activity of TAL1 is dispensable for induction of RALDH2-T.
Next we tested whether the DNA binding activity of TAL1 is required for activation of the RALDH2-T promoter. The basic region of bHLH-type transcription factors is known to be the DNA binding domain (8, 11). Therefore, we mutated three amino acids, in the basic region, RRR, which are highly conserved in various bHLH proteins (40), to alanines (AAA) and designated this mutant TAL1β-BM (for basic region mutant). To test whether TAL1β-BM could still interact with E47, a partner of the heterodimer, or with LMO, coprecipitation experiments were performed. As shown in Fig. 6A, HA-tagged E47S was coprecipitated with FLAG-TAL1β-BM as well as with wild-type FLAG-TAL1β. HA-tagged LMO2 was also coprecipitated with FLAG-TAL1β-BM, although the interaction appeared to be weaker than that of LMO2 and wild-type TAL1β (Fig. 6B). These results confirmed that TAL1β-BM was still capable of binding to E47 and LMO2. Next we examined the DNA binding activity of TAL1β-BM. As shown in Fig. 6C, binding to the consensus E-box (21) by the lysate of 293E cells expressing both FLAG-TAL1β and HA-E47S was observed. This binding was inhibited by addition of anti-FLAG MAb, confirming that the complex contained FLAG-TAL1β. The binding was competed out by addition of the cold E-box oligonucleotide but not by the cold GATA site oligonucleotide, confirming that the TAL1-E47 heterodimer was capable of binding to the consensus E-box but not to the GATA site in the RALDH2-T promoter. In contrast, no specific binding by the lysate of 293E cells expressing FLAG-TAL1β-BM and HA-E47S was observed, even though the protein expression was confirmed by Western blotting. These results indicated that TAL1β-BM was capable of forming a heterodimer with E47S but that the resulting dimer did not efficiently bind to DNA.
After these experiments, we examined whether TAL1β-BM was still capable of inducing the RALDH2-T promoter in collaboration with LMO and GATA3. As shown in Fig. 6D, TAL1β-BM induced the transcription from the RALDH2-T reporter −1.7-luc in HPB-ALL cells in the presence of LMO1 or LMO2, although less efficiently than the wild-type. Similarly, TAL1β-BM in the presence of LMO and GATA3 activated the transcription by ∼10-fold in BALL-1 cells, as TAL1β did (Fig. 6E). Furthermore, transiently expressed TAL1β-BM and LMO induced the expression of endogeneous RALDH2 in HPB-ALL cells (Fig. 6F). These results clearly demonstrated that the DNA binding activity of TAL1, in contrast to that of GATA3, is not necessary for the activation of the RALDH2-T promoter. Thus, TAL1 acts as a cofactor for GATA3 in the induction of the RALDH2-T promoter.
TAL1, LMO, and GATA3 activate transcription through the GATA site.
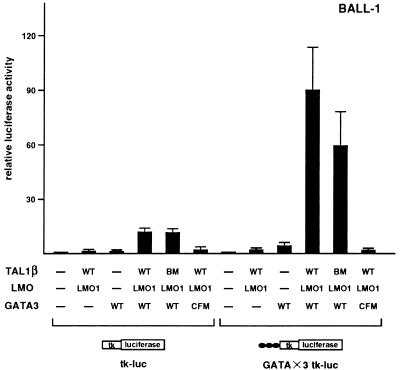
To further demonstrate that the GATA site in the RALDH2-T promoter was sufficient for the induction of transcription by the combination of TAL1, LMO, and GATA3, it was tandemly repeated three times and linked to a heterologous HSV tk minimal promoter (36), making GATA×3-tk-luc. This reporter plasmid was cotransfected into BALL-1 cells with various combinations of expression plasmids for TAL1β, LMO1, GATA3, and their mutants. As shown in Fig. 7, although parental tk-luc weakly responded to the combination of TAL1β, LMO1, and GATA3, GATA×3-tk-luc was further activated by ∼7-fold, indicating that the combination of TAL1β, LMO1, and GATA3 was indeed capable of activating the transcription through the GATA site alone. As the case of −1.7-luc, the transcription from GATA×3-tk-luc was activated even by TAL1β-BM in collaboration with LMO1 and GATA3 but not by GATA3-CFM even in the presence of TAL1β and LMO1. These results further demonstrate that TAL1 and LMO induce the transcription of the RALDH2-T promoter through the GATA site at position −239 by acting as cofactors for GATA3.
FIG. 7.
Transactivation of an artificial reporter consisting of three tandem repeats of the GATA site by TAL1, LMO, and GATA3. BALL-1 cells were cotransfected with 15 μg of an artificial luciferase reporter (GATA×3-tk-luc or tk-luc), 0.1 μg of pRL-CMV for normalization of transfection efficiency, and 5 μg each of expression plasmids without inserts (−) or with the indicated inserts. tk-luc contains only the HSV tk minimal promoter. GATA×3-tk-luc contains three copies of the GATA site from the RALDH2-T promoter linked to the minimal HSV tk promoter. Relative luciferase activity compared to that of the reporter alone was determined. Means and standard deviations for three independent experiments are shown. WT, wild type.
DISCUSSION
We have identified the RALDH2 gene as a transcriptional target gene for TAL1 and LMO in T-ALL. RALDH2 was strongly induced in HPB-ALL cells upon stable as well as transient coexpression of TAL1 and LMO. Most T-ALL-derived cell lines that ectopically coexpress TAL1 and LMO also express RALDH2. A T-ALL-specific alternative promoter present in the second intron of the RALDH2 gene, termed the RALDH2-T promoter, was transactivated by TAL1 and LMO through binding of a TAL1-LMO-GATA3 complex to a GATA site in the RALDH2-T promoter. These results strongly suggest that the RALDH2 gene is a first direct downstream gene for the abberantly activated transcription factors TAL1 and LMO in T-ALL.
Functional synergy between TAL1 and LMO.
Targeted disruption of either TAL1 or LMO2 resulted in embryonic lethality by the absence of yolk sac erythropoiesis (48, 52, 61). In erythroid cells, TAL1 and LMO2 are coexpressed and are thought to regulate erythropoiesis by forming a oligomeric complex with GATA1, Ldb1, and E2A (60). TAL1 and LMO are also synergistic in T-cell tumorigenesis in double-transgenic mice (2, 30). This suggests that a complex consisting of TAL1 and LMO also plays an important role in the development of T-cell leukemia. In the present study, we have shown that both TAL1 and LMO are required for abberant expression of RALDH2-T in T-ALL (Fig. 1 and 3). Although the role of RALDH2-T in the development of T-ALL is unknown (see below), complex formation between TAL1 and LMO is likely to be important in the regulation of expression of target genes involved in tumorigenesis.
The roles of LMO1 and LMO2 in normal development are thought to be different, since these genes are expressed in different tissues (15, 18, 35, 49). In fact, LMO2 but not LMO1 is coexpressed with TAL1 in the hematopoietic lineage (56). The structures of LMO1 and LMO2 are, however, very similar, and both are capable of interacting with TAL1 and GATA proteins in vivo (Fig. 5C) (42, 56, 59). Both are capable of inducing RALDH2 in collaboration with TAL1 (Fig. 1 and 3). Previously, we have shown that TAL1 is regularly coexpressed with either LMO1 or LMO2 in most T-ALL cell lines (41). These results suggest that LMO1 and LMO2 are both capable of playing a similar role in T-ALL oncogenesis.
Since target genes regulated by TAL1 and LMO2 in erythroid cells have not been identified, their transcriptional activity has not been clarified. However, by using erythroid precursor-derived cell lines, TAL1 and LMO2 were both shown to maintain erythroid cells in an immature state and promote self-renewal (17, 58). Thus, the complex of TAL1 and LMO may promote expression of genes involved in cell growth in the erythroid cell lineage. This may relate closely to their roles in T-cell tumorigenesis.
Mechanism of transcriptional regulation of the RALDH2-T promoter by TAL1 and LMO.
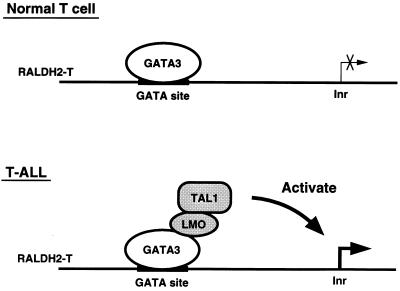
When we identified RALDH2-T as a target gene induced by TAL1, we expected that an E-box in the promoter region of RALDH2-T was an important element (Fig. 3A). However, the results obtained by transient-transfection experiments using a series of reporter genes with deletions and point mutations clearly demonstrated that a GATA site at position −239 but not an E-box at position −273 was necessary for the induction by TAL1 and LMO (Fig. 4). Our data further showed that GATA3 was capable of binding to the GATA site (Fig. 5A) and was involved in the transactivation by TAL1 and LMO (Fig. 5E and F). The combination of TAL1, LMO, and GATA3 activated the transcription even from an artificial reporter gene linked to three tandem repeats of the GATA site (Fig. 7), suggesting that the GATA site alone is sufficient for transactivation by TAL1 and LMO in the presence of GATA3. The observations that the TAL1-E47 heterodimer did not bind to the GATA site (Fig. 6C) and that TAL1β-BM lacking the DNA binding activity was still capable of inducing RALDH2-T like the wild-type TAL1β in the presence of LMO and GATA3 (Fig. 6D to F) further indicate that the direct binding of TAL1 to the promoter is not required. Together with the fact that the LMO family proteins are thought to have no DNA binding activity, this indicates that both TAL1 and LMO appear to act as cofactors for GATA3. As illustrated in Fig. 8, GATA3 is likely to bind to the GATA site in the RALDH2-T promoter region in normal T cells. Importantly, however, GATA3 alone does not activate transcription from the RALDH2-T promoter. Only when TAL1 and LMO are ectopically coexpressed in T-ALL, a large transcriptional complex containing TAL1, LMO, and GATA3 that is capable of activating the RALDH2-T promoter is formed. Thus, GATA3 may act as a DNA-binding platform for LMO and TAL1 (Fig. 8).
FIG. 8.
Model of transcriptional activation of the RALDH2-T promoter by TAL1, LMO, and GATA3. In normal T cells, GATA3 binds to the GATA site in the RALDH2-T promoter but does not activate transcription. When TAL1 and LMO are ectopically expressed in T-ALL, a large complex containing TAL1, LMO, and GATA3 is formed on the GATA site in the RALDH2-T promoter to activate transcription from a downstream initiator (Inr) (see Discussion).
Why are both TAL1 and LMO required for the induction of RALDH2-T expression in T cells? LMO can interact with GATA3 in the absence of TAL1 in vivo (Fig. 5C), but expression of LMO alone in HPB-ALL cells did not induce transcription from the RALDH2-T promoter (Fig. 3). Thus, although it has been reported that LMO1 and LMO2 contain transactivation domains in the N termini (32, 50), LMO itself cannot act as a coactivator for GATA3. The role of LMO in the induction of RALDH2-T expression may be to bridge TAL1 and GATA3 to form a complex. Since TAL1 does not interact with GATA3 directly (Fig. 5D), the TAL1-E47 heterodimer may act as a transcriptional coactivator for GATA3 only in the presence of LMO.
Various transcription factors are known to act synergistically with GATA family proteins. For example, SP1 and EKLF regulate the globin gene locus control region with GATA1 (38). In the regulation of endothelin-1 gene expression in endothelial cells, GATA2 and AP1 act cooperatively (26). In these cases, GATA family proteins and other factors are known to cooperate by binding to respective binding sites in the regulatory regions. However, when one of these sites is mutated, two factors are still capable of activating transcription through the remaining site. In other words, one factor which does not bind to DNA is still capable of collaborating with the other DNA-binding factors by acting as a cofactor. Similar observations have been reported for bHLH-type transcription factors. Myogenic factor MyoD and MADS box factor MEF2 cooperatively activate transcription through each binding site reciprocally (39). Notably, there is a potential E-box besides the GATA site in the RALDH2-T promoter (Fig. 3). Deletion or mutation of this E-box, however, did not affect the induction by TAL1 and LMO (Fig. 4 and data not shown). Moreover, the existence of the E-box did not complement the mutation in the GATA site for transcriptional activation by TAL1 and LMO (Fig. 4). Even in the presence of TAL1 and LMO, GATA3-CFM lacking DNA binding activity could not transactivate the RALDH2-T promoter (Fig. 5E and F), whereas a similar mutant, GATA1 disr.Cf, was still capable of acting as a cofactor for SP1 in the globin gene enhancer (38). Furthermore, the TAL1-LMO-GATA3 complex could not activate transcription through the three tandem repeats of the most preferred E-box element (AACAGATGGT) linked to tk-luc in BALL-1 cells (data not shown). These results suggest that, in contrast to the other cases described above, the TAL1-LMO-GATA3 complex activates transcription from the RALDH2-T promoter only through the GATA site.
Why does GATA3 activate RALDH2-T transcription only by forming a complex with TAL1 and LMO? In the experiments using GATA×3-tk-luc in BALL-1 cells, only sevenfold activation was observed even though three tandem repeats of the element were used (Fig. 7). This may suggest that other cis-acting elements in the RALDH2-T promoter are still involved in full induction by TAL1, LMO, and GATA3. Preliminary data indeed revealed that the selective deletion of the 50-bp fragment downstream from the GATA site also decreased the induction of RALDH2-T as did the GATA site mutation (data not shown). This suggests that there is a downstream cis-acting element(s) that acts synergistically with the GATA site in the induction of RALDH2-T by the TAL1-LMO-GATA3 complex. TAL1 and LMO may mediate synergism between GATA3 and an unknown factor(s) that binds to the downstream element(s). The TAL1-LMO-GATA3 complex in T-ALL may contain Ldb1 (also called NL1 or CLIM2) (1, 3, 25), like the TAL1-LMO2-GATA1 complex in erythroid cells (60). It was shown that Ldb1 was required for synergistic activation of the αGSU promoter by P-Lim and P-Otx (3). Thus, Ldb1 may play a role in cooperation between the TAL1-LMO-GATA3 complex and another factor(s) binding to the downstream regions of the RALDH2-T promoter in T-ALL. As shown in Fig. 1C, Molt15 and Molt17 cells, which express TAL1, LMO2, and GATA3 (41), did not express RALDH2-T. Since these cell lines strongly express TALLA1 (54), which is also apparently induced by TAL1 and LMO in T-ALL (41), TAL1 and LMO2 are likely to be functional in these cell lines. This also suggests that some factor(s) other than GATA3 and not expressed in Molt15 and Molt17 cells, which are the only CD4− CD8− double-negative lines among the T-ALL cell lines examined (54), is involved in the transactivation of the RALDH2-T promoter. Further studies are needed to elucidate the exact mechanism of RALDH2-T induction in T-ALL.
Role of RALDH2-T in T-ALL.
RALDH2 was identified as an enzyme converting retinal to retinoic acid (63). In fact, RALDH2 is expressed in the trunk region of the developing embryo, where retinoic acid is produced (63). It has been shown that retinoic acid inhibits activation-induced apoptosis of T cells (23, 62). One possibility is that ectopically expressed RALDH2-T inhibits apoptosis of T cells by generating retinoic acid. To detect retinoic acid in T-ALL cell lines, we transfected a luciferase reporter gene containing a retinoic acid response element (31). Even in the presence of retinal as a substrate, activation of the reporter was not observed (data not shown). One possibility is that the N-terminally truncated RALDH2-T protein may not have the enzymatic activity. Alternatively, RALDH2-T may convert another substrate or act as a dominant negative factor for other enzymes. The role of RALDH2-T in the development of T-ALL thus remains to be determined. Besides its potential role in T-ALL, RALDH2-T may also provide a useful marker for diagnosis and monitoring of T-ALL, since this type of RALDH2 transcript could not be detected in normal tissues even by RT-PCR (data not shown). Furthermore, the RALDH2-T promoter or its GATA site may be exploited for driving gene expression specifically in T-ALL for therapeutic purposes.
ACKNOWLEDGMENTS
We thank K. Maruyama for pMIKneo and pMIKhyg, Astar Winoto for pc-KRR, Tetsuya Yoshida for helpful discussions, and Yorio Hinuma and Masakazu Hatanaka for constant support and encouragement.
REFERENCES
- 1.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 2.Aplan P D, Jones C A, Chervinsky D S, Zhao X, Ellsworth M, Wu C, McGuire E A, Gross K W. An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with LMO1 to generate T-cell malignancies in transgenic mice. EMBO J. 1997;16:2408–2419. doi: 10.1093/emboj/16.9.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 4.Bash R O, Hall S, Timmons C F, Crist W M, Amylon M, Smith R G, Baer R. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A Pediatric Oncology Group study. Blood. 1995;86:666–676. [PubMed] [Google Scholar]
- 5.Begley C G, Aplan P D, Denning S M, Haynes B F, Waldmann T A, Kirsch I R. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard O, Lecointe N, Jonveaux P, Souyri M, Mauchauffe M, Berger R, Larsen C J, Mathieu-Mahul D. Two site-specific deletions and t(1;14) translocation restricted to human T-cell acute leukemias disrupt the 5′ part of the tal-1 gene. Oncogene. 1991;6:1477–1488. [PubMed] [Google Scholar]
- 7.Boehm T, Foroni L, Kaneko Y, Perutz M F, Rabbitts T H. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan T J, Chakraborty T, Olson E N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll A J, Crist W M, Link M P, Amylon M D, Pullen D J, Ragab A H, Buchanan G R, Wimmer R S, Vietti T J. The t(1;14)(p34;q11) is nonrandom and restricted to T-cell acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1990;76:1220–1224. [PubMed] [Google Scholar]
- 10.Chen Q, Cheng J T, Tasi L H, Schneider N, Buchanan G, Carroll A, Crist W, Ozanne B, Siciliano M J, Baer R. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 1990;9:415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R L, Cheng P F, Lassar A B, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elwood N J, Begley C G. Reconstitution of mice with bone marrow cells expressing the SCL gene is insufficient to cause leukemia. Cell Growth Differ. 1995;6:19–25. [PubMed] [Google Scholar]
- 14.Finger L R, Kagan J, Christopher G, Kurtzberg J, Hershfield M S, Nowell P C, Croce C M. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc Natl Acad Sci USA. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foroni L, Boehm T, White L, Forster A, Sherrington P, Liao X B, Brannan C I, Jenkins N A, Copeland N G, Rabbitts T H. The rhombotin gene family encode related LIM-domain proteins whose differing expression suggests multiple roles in mouse development. J Mol Biol. 1992;226:747–761. doi: 10.1016/0022-2836(92)90630-3. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green A R, DeLuca E, Begley C G. Antisense SCL suppresses self-renewal and enhances spontaneous erythroid differentiation of the human leukaemic cell line K562. EMBO J. 1991;10:4153–4158. doi: 10.1002/j.1460-2075.1991.tb04993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg J M, Boehm T, Sofroniew M V, Keynes R J, Barton S C, Norris M L, Surani M A, Spillantini M G, Rabbits T H. Segmental and developmental regulation of a presumptive T-cell oncogene in the central nervous system. Nature (London) 1990;344:158–160. doi: 10.1038/344158a0. [DOI] [PubMed] [Google Scholar]
- 19.Ho I C, Vorhees P, Marin N, Oakley B K, Tsai S F, Orkin S H, Leiden J M. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu H L, Cheng J T, Chen Q, Baer R. Enhancer-binding activity of the Tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol Cell Biol. 1991;11:3037–3042. doi: 10.1128/mcb.11.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu H L, Huang L, Tsan J T, Funk W, Wright W E, Hu J S, Kingston R E, Baer R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol Cell Biol. 1994;14:1256–1265. doi: 10.1128/mcb.14.2.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu H L, Wadman I, Baer R. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc Natl Acad Sci USA. 1994;91:3181–3185. doi: 10.1073/pnas.91.8.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata M, Mukai M, Nakai Y, Iseki R. Retinoic acids inhibit activation-induced apoptosis in T cell hybridomas and thymocytes. J Immunol. 1992;149:3302–3308. [PubMed] [Google Scholar]
- 24.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurata L W, Kenny D A, Gill G N. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawana M, Lee M E, Quertermous E E, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol Cell Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelliher M A, Seldin D C, Leder P. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIα. EMBO J. 1996;15:5160–5166. [PMC free article] [PubMed] [Google Scholar]
- 28.Ko L J, Yamamoto M, Leonard M W, George K M, Ting P, Engel J D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991;11:2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson R C, Lavenir I, Larson T A, Baer R, Warren A J, Wadman I, Nottage K, Rabbits T H. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 31.Lefebvre P, Gaub M P, Tahayato A, Rochette-Egly C, Formstecher P. Protein phosphatases 1 and 2A regulate the transcriptional and DNA binding activities of retinoic acid receptors. J Biol Chem. 1995;270:10806–10816. doi: 10.1074/jbc.270.18.10806. [DOI] [PubMed] [Google Scholar]
- 32.Mao S, Neale G A, Goorha R M. T-cell proto-oncogene rhombotin-2 is a complex transcription regulator containing multiple activation and repression domains. J Biol Chem. 1997;272:5594–5599. doi: 10.1074/jbc.272.9.5594. [DOI] [PubMed] [Google Scholar]
- 33.Marine J, Winoto A. The human enhancer-binding protein Gata3 binds to several T-cell receptor regulatory elements. Proc Natl Acad Sci USA. 1991;88:7284–7288. doi: 10.1073/pnas.88.16.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire E A, Hockett R D, Pollock K M, Bartholdi M F, O’Brien S J, Korsmeyer S J. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuire E A, Davis A R, Korsmeyer S J. T-cell translocation gene 1 (Ttg-1) encodes a nuclear protein normally expressed in neural lineage cells. Blood. 1991;77:599–606. [PubMed] [Google Scholar]
- 36.McKnight S L, Gavis E R, Kingsbury R, Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981;25:385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- 37.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 40.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 41.Ono Y, Fukuhara N, Yoshie O. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or −2 and induces TALLA1, a highly specific tumor marker of T-ALL. J Biol Chem. 1997;272:4576–4581. doi: 10.1074/jbc.272.7.4576. [DOI] [PubMed] [Google Scholar]
- 42.Osada H, Grutz G, Axelson H, Forster A, Rabbits T H. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D’Agati V, Orkin S H, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 44.Pulford K, Lecointe N, Leroy-Viard K, Jones M, Mathieu-Mahul D, Mason D Y. Expression of TAL-1 proteins in human tissues. Blood. 1995;85:675–684. [PubMed] [Google Scholar]
- 45.Rabbitts T H, Boehm T. LIM domains. Nature (London) 1990;346:418. doi: 10.1038/346418a0. [DOI] [PubMed] [Google Scholar]
- 46.Rabbitts T H. Chromosomal translocations in human cancer. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 47.Robb L, Rasko J E, Bath M L, Strasser A, Begley C G. scl, a gene frequently activated in human T cell leukaemia, does not induce lymphomas in transgenic mice. Oncogene. 1995;10:205–209. [PubMed] [Google Scholar]
- 48.Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey R P, Metcalf D, Begley C G. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royer-Pokora B, Loos U, Ludwig W D. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11) Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 50.Sanchez-Garcia I, Axelson H, Rabbitts T H. Functional diversity of LIM proteins: amino-terminal activation domains in the oncogenic proteins RBTN1 and RBTN2. Oncogene. 1995;10:1301–1306. [PubMed] [Google Scholar]
- 51.Schmeichel K L, Beckerle M C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 52.Shivdasani R A, Mayer E L, Orkin S H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature (London) 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 53.Smith V M, Lee P P, Szychowski S, Winoto A. GATA-3 dominant negative mutant. Functional redundancy of the T cell receptor alpha and beta enhancers. J Biol Chem. 1995;270:1515–1520. doi: 10.1074/jbc.270.4.1515. [DOI] [PubMed] [Google Scholar]
- 54.Takagi S, Fujikawa K, Imai T, Fukuhara N, Fukudome K, Minegishi M, Tsuchiya S, Konno T, Hinuma Y, Yoshie O. Identification of a highly specific surface marker of T-cell acute lymphoblastic leukemia and neuroblastoma as a new member of the transmembrane 4 superfamily. Int J Cancer. 1995;61:706–715. doi: 10.1002/ijc.2910610519. [DOI] [PubMed] [Google Scholar]
- 55.Tsai S F, Martin D I, Zon L I, D’Andrea A D, Wong G G, Orkin S H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature (London) 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 56.Valge-Archer V E, Osada H, Warren A J, Forster A, Li J, Baer R, Rabbitts T H. The LIM protein RBTN2 and the basic helix-loop-helix protein TAL1 are present in a complex in erythroid cells. Proc Natl Acad Sci USA. 1994;91:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visvader J, Begley C G, Adams J M. Differential expression of the LYL, SCL and E2A helix-loop-helix genes within the hemopoietic system. Oncogene. 1991;6:187–194. [PubMed] [Google Scholar]
- 58.Visvader J E, Mao X, Fujiwara Y, Hahm K, Orkin S H. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadman I, Li J, Bash R O, Forster A, Osada H, Rabbitts T H, Baer R. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbitts T H. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, Vacchio M S, Ashwell J D. 9-cis-retinoic acid inhibits activation-driven T-cell apoptosis: implications for retinoid X receptor involvement in thymocyte development. Proc Natl Acad Sci USA. 1993;90:6170–6174. doi: 10.1073/pnas.90.13.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao D, McCaffery P, Ivins K J, Neve R L, Hogan P, Chin W W, Drager U C. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]