Abstract
We have recently reported that the geranylgeranyltransferase I inhibitor GGTI-298 arrests human tumor cells at the G1 phase of the cell cycle and increases the protein and RNA levels of the cyclin-dependent kinase inhibitor p21WAF1/CIP1. Here, we show that GGTI-298 acts at the transcriptional level to induce p21WAF1/CIP1 in a human pancreatic carcinoma cell line, Panc-1. This upregulation of p21WAF1/CIP1 promoter was selective, since GGTI-298 inhibited serum responsive element- and E2F-mediated transcription. A functional analysis of the p21WAF1/CIP1 promoter showed that a GC-rich region located between positions −83 and −74, which contains a transforming growth factor β-responsive element and one Sp1-binding site, is sufficient for the upregulation of p21WAF1/CIP1 promoter by GGTI-298. Electrophoretic mobility shift assays showed a small increase in the amount of DNA-bound Sp1-Sp3 complexes. Furthermore, the analysis of Sp1 transcriptional activity in GGTI-298-treated cells by using GAL4-Sp1 chimera or Sp1-chloramphenicol acetyltransferase reporter revealed a significant increase in Sp1-mediated transcription. Moreover, GGTI-298 treatment also resulted in increased Sp1 and Sp3 phosphorylation. These results suggest that GGTI-298-mediated upregulation of p21WAF1/CIP1 involves both an increase in the amount of DNA-bound Sp1-Sp3 and enhancement of Sp1 transcriptional activity. To identify the geranylgeranylated protein(s) involved in p21WAF1/CIP1 transcriptional activation, we analyzed the effects of the small GTPases Rac1 and RhoA on p21WAF1/CIP1 promoter activity. The dominant negative mutant of RhoA, but not Rac1, was able to activate p21WAF1/CIP1. In contrast, constitutively active RhoA repressed p21WAF1/CIP1. Accordingly, the ADP-ribosyl transferase C3, which specifically inhibits Rho proteins, enhanced the activity of p21WAF1/CIP1. Taken together, these results suggest that one mechanism by which GGTI-298 upregulates p21WAF1/CIP1 transcription is by preventing the small GTPase RhoA from repressing p21WAF1/CIP1 induction.
Small G proteins such as Ras, Rho, and Rac are intimately involved in signaling pathways that regulate mitogenesis (14, 25, 33). The role of Ras as a transducer of mitogenic signals from receptor tyrosine kinases to the nucleus is well established (14, 25, 33). Similarly, RhoA and Rac1 have been shown to be required for the G1-to-S-phase transition of the cell cycle during mitogenesis (29). It is therefore not surprising that these small G proteins are implicated in pathological conditions, such as cancer and certain cardiovascular diseases, where aberrant proliferation is involved. Indeed, oncogenic Ras mutations are found in 30% of all human tumors (2, 3). Furthermore, GTP-locked forms of Ras, RhoA, and Rac1 all cause uncontrolled proliferation and tumor growth (16, 32). Finally, elimination of oncogenic Ras by homologous recombination in human tumors with multiple genetic alternations inhibits their ability to grow in nude mice (37). Thus, elimination of oncogenic ras function alone is sufficient to reverse malignant transformation, and therefore pharmacological inhibition of small G-protein function would potentially be an excellent strategy for preventing or curing diseases in which aberrant proliferation is implicated. One approach that we have taken is to make pharmacological agents that inhibit prenylation of small G proteins, which is a lipid posttranslational modification required for their function (36).
Protein prenylation is catalyzed by three prenyl transferases that attach to carboxyl terminal cysteines either a farnesyl, by farnesyltransferase (FTase), or a geranylgeranyl, by geranylgeranyltransferase (GGTase) I and II (47). Whereas FTase and GGTase I recognize proteins that end with carboxyl-terminal CAAX (where C is cysteine, A is an aliphatic amino acid, and X is any amino acid) sequences, GGTase II catalyzes geranylgeranylation of proteins that end with CXC, XXCC, and CCXX sequences. FTase prefers CAAX sequences where X is methionine, serine, cysteine, or glutamine, whereas GGTase I prefers leucine or isoleucine at the X position. Among farnesylated proteins are H-Ras, K-Ras, N-Ras, and lamin B, and among geranylgeranylated proteins are Rac1, RhoA, and Rap1a (47). Although the X position of CAAX sequences determines whether a protein will be a substrate for FTase or GGTase I, there is some degree of cross-specificity between the two enzymes (47). For example, a member of the Rho family of small G proteins, RhoB, is known to be both farnesylated and geranylgeranylated under normal conditions (18). Furthermore, in human tumor cells that are treated with FTase inhibitors, K-Ras and N-Ras become geranylgeranylated (21, 34, 45).
We and others have made CAAX peptidomimetics that are potent inhibitors of FTase that are selective of FTase over GGTase I (9, 36). These agents are potent antagonists of oncogenic Ras processing and signaling and inhibit the growth of murine and human tumors in various animal models (9, 36). Furthermore, we have recently made CAAX peptidomimetics that are potent and selective for GGTase I over FTase and found these also to inhibit human tumor growth in nude mice (20, 26, 38, 42). Although the mechanisms by which FTase inhibitors and GGTase I inhibitors inhibit tumor growth are not known, there are several intriguing differences in their mechanisms of action. While FTase inhibitors induce apoptosis only when the cells are prevented from attaching to the substratum (19), GGTase I inhibitors induce apoptosis of attached cells (27). Furthermore, GGTase I inhibitors induce a G1 block in a large number of human tumor cell lines, whereas FTase inhibitors can either induce a G1 block or a G2/M enrichment or have no effect on cell cycle distribution (41). Finally, GGTase I, but not FTase, inhibitors block platelet-derived growth factor-dependent tyrosine phosphorylation of its receptors (26).
One possible mechanism by which cells arrest in G1 phase is mediated by cyclin-dependent kinase (CDK) inhibitors such as p21WAF1/CIP1. p21WAF1/CIP1 could mediate G1-phase arrest through inhibition of CDKs and possibly through inhibition of DNA replication (43, 46). The fact that inhibition of protein geranylgeranylation resulted in a G1-phase block in all cells we have evaluated prompted us to investigate the effects of GGTase I inhibitors on the cell cycle machinery. Recently, we have found that treatment of several human tumors with GGTI-298, a GGTase I inhibitor, induced an accumulation of p21WAF1/CIP1 (41). Here we show that the activation of p21WAF1/CIP1 by GGTI-298 occurs at the transcriptional level and that the promoter region involved contains a Sp1- and transforming growth factor β (TGF-β)-responsive element (TβRE). Furthermore, we have demonstrated that GGTI-298 increased the amount of Sp1 and Sp3 DNA binding and enhanced Sp1 transcriptional activity. Moreover, we show that the small GTPase RhoA, but not Rac1, represses p21WAF1/CIP1 transcription. Thus, our results suggest that one mechanism by which GGTI-298 upregulates p21WAF1/CIP1 transcription is by preventing the small GTPase RhoA from repressing p21WAF1/CIP1 induction.
MATERIALS AND METHODS
Plasmid constructs.
The p21WAF promoter deletion and mutant constructs were kindly provided by Xiao-Fan Wang (6). pSG4+Sp1N, pSG4+Sp1Q, pSG4+Sp1Δ, and pSG4+Sp1B-C express the GAL4-DNA binding domain (amino acids 1 to 147) fused to Sp1 transactivation domain (10). GAL4-VP16 expresses GAL4-DNA binding domain fused to the acidic activation domain (amino acids 411 to 454) of herpes simplex virus type 1 VP16 transcription factor. G5BCAT is a chloramphenicol acetyltransferase (CAT) reporter, which carries five GAL4-DNA binding sites upstream of E1B minimal promoter and the TATA box. pCMV-βgal and pSRE plasmids were provided by R. Jove, and 4XE2F-CAT was provided by W. D. Cress (Moffitt Cancer Research Center, University of South Florida). 6XSp1-CAT was previously described (1). The pcDNA3 expression vectors encoding for Rac1 wild type (Rac1-wt), Rac1-115I (activated), Rac1-17N (dominant negative), RhoA-wt, RhoA-63L (activated), and RhoA-19N (dominant negative) were constructed by inserting Rac1 and RhoA BamHI cDNA fragments from pzipNeo (16) into pcDNA3 (Invitrogen) at the BamHI site.
Tissue culture and transfection.
Panc-1 cells were grown in Dulbecco’s modified Eagle medium (DMEM) (GIBCO/BRL) supplemented with 10% fetal bovine serum (FBS). Panc-1 cells were transfected at 40% confluence with 6 μg of p21WAF1/CIP1, 4 μg of Rac1 or RhoA, and 0.5 μg of pCMV-βgal by the calcium phosphate precipitation method as described previously (1). DNA precipitates were removed 15 h after transfection, and the cells were replenished with fresh medium. Cells were harvested 30 h later and lysed in 200 μl of passive lysis buffer (Promega). Cell extracts were used for β-galactosidase, luciferase, and CAT assays. The thin-layer chromatography plates were scanned with a PhosphorImager, and the percentages of acetylated and nonacetylated forms of chloramphenicol were determined. All transfections were repeated a minimum of three times, and the standard deviations were calculated.
Electrophoretic mobility shift assay (EMSA).
Oligonucleotides corresponding to the wt p21WAF promoter sequences from −86 to −71 (GGTCCCGCCTCCTTG) and from −93 to −62 (GAGCGCGGGTCCCGCCTCCTTGAGGCGGGCCC) and their complementary sequences were synthesized and annealed. The sequence of the mutant competitor is GGTTATCTAGAACTG. Two picomoles of annealed wt oligonucleotides was end labeled with T4 kinase (Gibco BRL) and 50 μCi of [γ-32P]ATP. Nuclear extracts were prepared from both GGTI-298-treated and untreated Panc-1 cells. After two 24-h treatments with GGTI-298, cells in a 100-mm-diameter plate were washed three times with 4 ml of cold phosphate-buffered saline (PBS) and then harvested in 1 ml of TEN solution (40 mM Tris [pH 7.5], 1 mM EDTA [pH 8.0], 150 mM NaCl). Cells from two plates were combined into one conical tube and spun 10 min at 4,000 × g. The cell pellet was resuspended in 60 μl of hypotonic buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) and transferred to a microfuge tube. Three cycles of freezing-thawing were performed in dry ice/ethanol at 37°C. The nuclei (pellet) were recovered by centrifugation for 1 min at 14,000 × g. The nuclei were resuspended in 20 μl of buffer C (0.2 mM EDTA [pH 8.0], 20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 420 mM KCl, 25% glycerol, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, 1 mM DTT, 1 mM PMSF) and incubated 30 min at 4°C. Supernatants were clarified (nuclear extracts were obtained), and protein concentrations were determined.
Binding reactions were performed at room temperature (RT). The final volume of the binding reaction mixtures was 20 μl, in which 6 μg of nuclear extract, 1 μg of poly(dI-dC)/ml, unlabeled specific competitor, and 2 μl of 10× binding buffer (100 mM Tris [pH 7.5], 50 mM EDTA [pH 8.0], 10 mM MgCl2, 10 mM DTT, 50% glycerol, 250 mM NaCl) were combined and incubated 10 min at RT. Radiolabeled probe (40,000 counts per minute) was added, and incubation was resumed for 20 min at RT. For supershift assays with Sp1 and Sp3, 1 μl of Sp1- or Sp3-specific polyclonal antibody (Santa Cruz Biotechnology) was added to the binding reaction mixture, and incubation was resumed for 30 min at RT. Binding reaction products were resolved on 0.5× Tris-borate-EDTA buffer and 5.0% acrylamide gel at 100 V for 4 h at RT. The gels were subsequently dried and exposed for autoradiography.
ADP-ribosylation by Clostridium botulinum C3 exoenzyme.
C3 exoenzyme (Sigma) was introduced into cells by using Lipofectamine (Gibco BRL). Briefly, 10 μg of lyophilized C3 exoenzyme was resuspended in 2 ml of buffer (10 mM Tris-HCl [pH 7.5], 114 mM KCl, 15 mM NaCl, 5.5 mM MgCl2). C3 exoenzyme (2.5 μg, or 500 μl) was mixed with 500 μl of Opti-MEM (Gibco BRL) and 16 μl of PLUS reagent (Gibco BRL) for 15 min at RT. Meanwhile, in a separate tube, 12 μl of Lipofectamine was mixed with 1 ml of Opti-MEM. Next, the C3 exoenzyme and Lipofectamine mixtures were combined, and incubation was resumed for 15 min at RT. Next, cells were washed twice with 2 ml of Opti-MEM and then incubated with C3 exoenzyme mixture for 15 h at 37°C in 5% CO2. The medium was replaced by DMEM supplemented with 15% FBS and the incubation was resumed for 24 h. Cells were harvested and lysed in 200 μl of passive lysis buffer (Promega). Aliquots (20 μl each) of cell lysate were used for β-galactosidase and luciferase assays.
In vivo phosphorylation of Sp1 and Sp3.
Panc-1 cells (106) were treated with GGTI-298 (15 μM) for 30 h prior to incubation with ortho[32P]phosphate. Cells were washed twice with DMEM without phosphate (Gibco BRL) and then incubated in 2.5 ml of the same medium supplemented with 10% dialyzed FBS (Gibco BRL) for 1 h. After 2.5 mCi of phosphorus-32 (NEN Life Science Products) was added to each plate (1 mCi/ml), the incubation was resumed for 3 h. Afterward, cells were washed twice with ice-cold PBS and then lysed in 0.5 ml of immunoprecipitation (IP) buffer (30 mM HEPES [pH 7.5], 10 mM NaCl, 5 mM MgCl2, 25 mM NaF, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 2 mM sodium orthovanadate, 10 mg of aprotinin/ml, 10 mg of soybean trypsin inhibitor/ml, 25 mg of leupeptin/ml, 2 mM PMSF, 6.4 mg of phosphatase substrate/ml). Following centrifugation to remove cellular debris, 5-μl aliquots of cell lysate were used to determine protein concentration, and equal amounts of proteins were used for IP with Sp1 (1:200) and Sp3 (1:100) polyclonal antibodies (Santa Cruz Biotechnology). The IP was performed overnight at 4°C. Sp1 and Sp3 immunocomplexes were isolated using protein A-agarose beads (Santa Cruz Biotechnology). The beads were washed five times with IP buffer and finally were resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, followed by separation on SDS–8% polyacrylamide gel. Next, the gel was fixed in water-methanol-acetic acid (60%:30%:10%) for 1 h, dried, and exposed for autoradiography.
RESULTS
GGTI-298 upregulates p21WAF1/CIP1 promoter activity in human pancreatic tumor cells (Panc-1 cells).
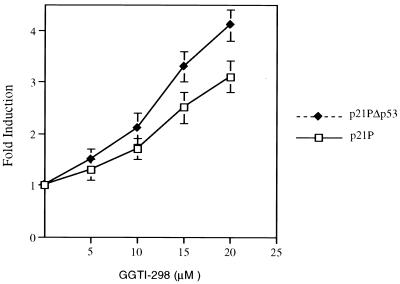
We have previously shown that GGTI-298 was able to arrest human tumor cells in the G1 phase of the cell cycle and induce the accumulation of p21WAF1/CIP1 (41). To evaluate whether p21WAF1/CIP1 was transcriptionally activated by GGTI-298 we analyzed the activity of its promoter in response to GGTI-298. We transiently transfected human pancreatic carcinoma cells, Panc-1 cells, with a luciferase reporter containing a full-length p21WAF1/CIP1 promoter and incubated cells with increasing doses of a GGTase I inhibitor (GGTI-298) for 36 h. The comparison of the relative luciferase activity of GGTI-298-treated cells with that of the untreated control cells showed an upregulation of the full-length promoter in a dose-dependent manner (Fig. 1). p21pΔp53, which contains the p21WAF1/CIP1 promoter lacking the p53 consensus site, was also upregulated by GGTI-298. The transcriptional activation of p21 was greater in the absence of the p53-binding site than in its presence (Fig. 1). These results demonstrate that the activation of p21WAF1/CIP1 promoter by GGTI-298 is mediated through a p53-independent pathway.
FIG. 1.
GGTI-298 upregulates p21WAF1/CIP1 promoter activity in human pancreatic tumor cells, Panc-1 cells, in a p53-independent manner. Panc-1 cells were transfected with 4 μg of p21P, which contains the full-length sequence of p21 promoter, or p21PΔp53, which is lacking the p53 consensus site and 0.5 μg of pCMV-βgal as described in Materials and Methods. At 15 h posttransfection, cells were incubated with increasing doses of GGTI-298 for 36 h. The fold induction was calculated by dividing the luciferase activity values of samples treated with GGTI-298 by the activity of untreated control samples. The samples were normalized for transfection efficiency against β-galactosidase activity. Bars represent standard deviations. The data are representative of three independent experiments.
GGTI-298 upregulates p21WAF1/CIP1 promoter through a region that contains a TGF-β-responsive element and Sp1-binding sites.
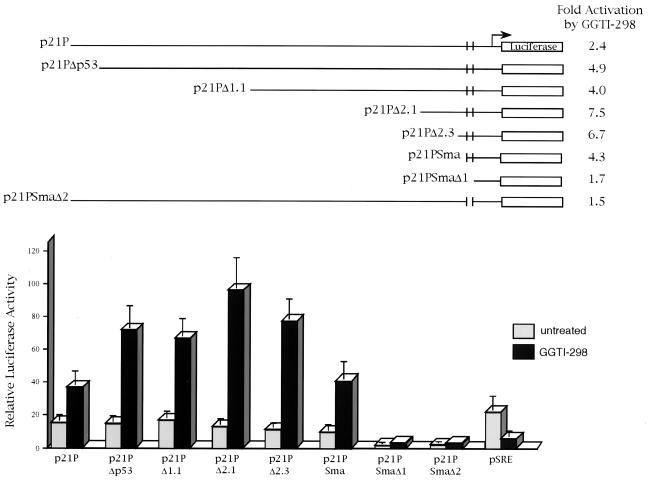
To pinpoint the region of the p21WAF1/CIP1 promoter that is upregulated by GGTI-298, we analyzed deletion mutants truncated in the 5-prime end of the promoter. As shown in Fig. 2, GGTI-298 activated by 2.4-fold the full-length promoter (p21P) and by 4.9-fold the promoter lacking the p53 consensus site (p21PΔp53). The constructs with deletions of 1.1 kb (p21PΔ1.1), 2.1 kb (p21PΔ2.1), and 2.3 kb (p21PΔ2.3) were activated 4-, 7.5-, and 6.7-fold, respectively. The construct p21PSma, which contains the sequences from −111 through the transcription initiation site, was activated 4.3-fold, suggesting that this region was sufficient for GGTI-298-mediated p21WAF1/CIP1 promoter upregulation. Deletion of the sequences between −111 and −62 (p21SmaΔ1) resulted in a decrease of the promoter basal activity and GGTI-298-mediated upregulation. Similarly, deletion of the sequences between −111 and −62 from the full-length promoter (p21PSmaΔ2) resulted in the loss of the promoter basal activity and the induction by GGTI-298. Thus, the sequences between −111 and −62, which contain a TGF-β-responsive element and two Sp1-binding sites, represent the minimal region for p21WAF1/CIP1 promoter basal activity and GGTI-298-mediated upregulation. In order to determine whether the upregulation by GGTI-298 was specific, we transiently transfected Panc-1 cells with a luciferase reporter that contains the serum responsive element (SRE) from the c-fos gene promoter. In contrast to the effects on p21WAF1/CIP1 promoter, GGTI-298 inhibited SRE-mediated transcription by threefold (Fig. 2).
FIG. 2.
Upregulation of p21WAF1/CIP1 promoter by GGTI-298 is mediated through a region that contains a TβRE and Sp1-binding sites. Panc-1 cells were transfected with the indicated p21P deletion constructs. At 15 h posttransfection, cells were incubated in either medium alone or medium containing GGTI-298 (15 μM) for 36 h as described in Materials and Methods. The fold induction was calculated by dividing the luciferase activity values of samples treated with GGTI-298 by the activity of untreated control samples. The samples were normalized for transfection efficiency against β-galactosidase activity. Panc-1 cells were also transfected with pSRE to determine specificity of GGTI-298. Each error bar represents the average deviation for three independent experiments. The construct map was adapted from Datto et al. (6).
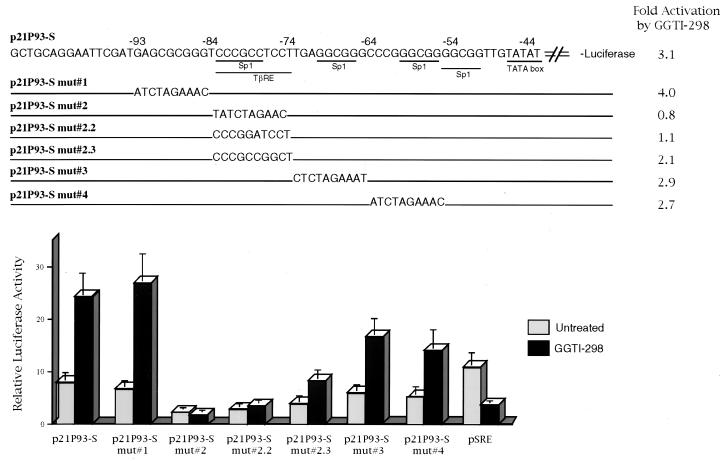
TGF-β/Sp1-responsive element between −83 and −74 is essential for p21WAF1/CIP1 promoter activity and upregulation by GGTI-298.
As described above, the analysis of p21WAF1/CIP1 promoter deletion mutants allowed us to identify the region between −111 and −62 as the minimal region for the upregulation by GGTI-298. This region of the promoter contains two Sp1-binding sites. The first Sp1 has previously been shown to be part of a TβRE. To further characterize the nucleotide sequence that is essential for GGTI-298-mediated upregulation, we analyzed a set of p21WAF1/CIP1 mutant constructs. p21P93-S, which contains the wt sequence from −93 to the transcription initiation site, was upregulated by 3.1-fold (Fig. 3). p21P 93-S 1, which is mutated in the sequences between −93 and −84, upstream of Sp1 and TβRE, was activated by fourfold. Similarly, constructs with mutations in Sp1-binding sites, sequences between −73 and −64 (p21P 93-S 3) and sequences between −63 and −54 (p21P 93-S 4), were activated 2.9- and 2.7-fold, respectively. In contrast, mutation of the Sp1 and TβRE sequences between −83 and −74 (p21P 93-S 2) resulted in a significant decrease of the promoter activity and GGTI-298-mediated upregulation (0.8-fold activation). Specifically, a two-nucleotide change, CC→GA, at positions −79 and −78 (p21P 93-S 2.2), which results in the alteration of Sp1 and TβRE, also abolished GGTI-298-mediated upregulation (1.1-fold activation). Furthermore, changing the nucleotides at position −77 and −76, TC→GG, which is the TβRE region that does not contain the Sp1-binding site, also reduced, from 3.1- to 2.1-fold, GGTI-298 activation (p21P 93-S 2.3) (Fig. 3). Thus, the region of Sp1 and TβRE between −83 and −74 is essential for the full response to GGTI-298.
FIG. 3.
Sp1- and TGF-β-responsive element at positions −83 to −78 is essential for GGTI-298-mediated upregulation of p21 promoter activity. Panc-1 cells were transfected with the indicated p21P mutant constructs. Starting at 15 h posttransfection, cells were incubated with GGTI-298 (15 μM) for 36 h. The fold induction was calculated by dividing the luciferase activity values of samples treated with GGTI-298 by the activity of untreated control samples. The samples were normalized for transfection efficiency against β-galactosidase activity. Each error bar represents the average deviation for three independent experiments. The construct map was adapted from Datto et al. (6). mut, mutation.
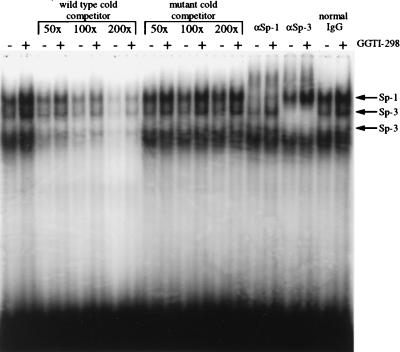
GGTI-298 increases Sp1- and Sp3-DNA binding to the sequence between −85 and −73 of the p21WAF1/CIP1 promoter.
The analysis of p21WAF1/CIP1 promoter mutants showed that Sp1 and TβRE sequences, from −83 to −74, were required for the upregulation mediated by GGTI-298. To determine the mechanism through which p21WAF1/CIP1 induction occurs, we performed EMSA using as a probe the sequence between −85 and −73 of p21WAF1/CIP1 promoter. The EMSA performed with nuclear extracts from both GGTI-298-treated Panc-1 cells and untreated Panc-1 cells and 32P-end-labeled Sp1 and TβRE (−85 to −73) probe revealed four specific bands (Fig. 4). The binding of these nuclear proteins could be competed by an excess of unlabeled wt −85 to −73 oligonucleotide. However, the mutant competitor, corresponding to the sequence from −85 to −73 of p21P 93-S 2, which was not upregulated by GGTI-298 (Fig. 3), was unable to compete for the binding of the retarded proteins (Fig. 4). Furthermore, the patterns of the retarded bands were similar whether nuclear extract from GGTI-298-treated samples or that from control samples was used. In contrast, the intensity of the retarded bands was increased in GGTI-298-treated samples (Fig. 4). The sequence from −85 to −73 that encompasses a TβRE was shown previously to bind to both Sp1 and Sp3. Supershift experiments in the presence of specific antibodies for Sp1 and Sp3 show shift of the top band with Sp1 antibody, whereas the second and third bands from the top were both shifted with Sp3 antibody. The pattern of the fourth band was unchanged by either Sp1 or Sp3 antibodies. None of the four bands shifted with normal rabbit immunoglobulin G. These results show the ability of GGTI-298 to enhance Sp1 and Sp3 DNA binding to the sequences from −85 to −73 of p21WAF1/CIP1 promoter (Fig. 4).
FIG. 4.
Sp1 and Sp3 interact with GGTI-298-responsive region. Nuclear extracts from GGTI-298-treated or untreated Panc-1 cells were incubated with a 32P-labeled probe corresponding to the sequence from −86 to −71 of the wt p21 promoter. Unlabeled wt or mutant competitors corresponding to the sequence from −86 to −71 of the wt p21 promoter and p21P93-S mut 2, respectively, were used. Polyclonal antibodies to either Sp1, Sp-3, or normal rabbit immunoglobulin G were included for supershift. Data are representative of two independent experiments.
GGTI-298 upregulates Sp1 transcriptional activity.
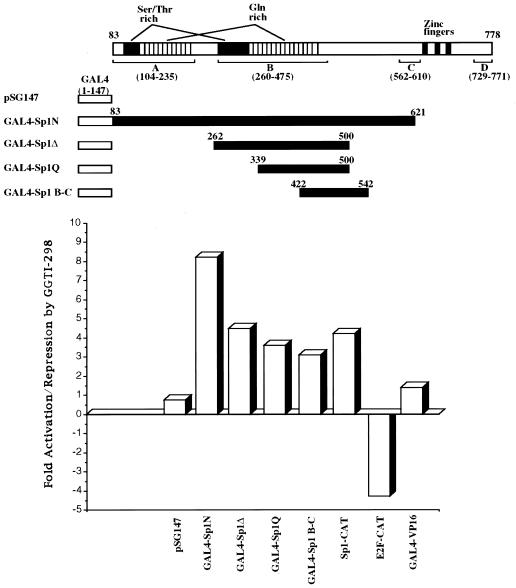
As we have shown above, GGTI-298 induces p21WAF1/CIP1 through a region that contains Sp1 and TβRE. To determine the effect of GGTI-298 on Sp1 transcriptional activity, we used chimeras which express GAL4-Sp1 fusions consisting of GAL4 DNA binding domain (amino acids 1 to 147) and Sp1 transactivation domain. The use of GAL4-Sp1 fusion proteins, containing different transactivation domains of Sp1, allows for analysis of the effect of GGTI-298 on Sp1-mediated transcription specifically, independent of Sp1 DNA binding activity. Panc-1 cells were transiently cotransfected with GAL4-Sp1 deletion constructs, G5BCAT reporters, which contain five GAL4-binding sites upstream of the E1B TATA box, and pCMV-βgal as an internal control for transfection efficiency. Cells were subsequently incubated with GGTI-298 (15 μM) for 36 h. After normalization for transfection efficiency, the samples were assayed for CAT activity. As shown in Fig. 5, the transcription mediated by GAL4-Sp1N (amino acids 83 to 621), GAL4-SpΔ (amino acids 262 to 500), GAL4-Sp1Q (amino acids 339 to 500), and GAL4-Sp1B-C (amino acids 422 to 542) was stimulated in response to GGTI-298. Thus, a region between amino acids 422 and 500 of Sp1 protein, shown to interact with TAFII110 (10), is sufficient to confer the stimulation by GGTI-298. However, these results do not rule out the possibility that regions outside the 422 to 500 region may contribute to the observed stimulation by GGTI-298. To determine the specificity of GGTI-298-mediated stimulation of Sp1 transcriptional activity, we cotransfected Panc-1 cells with GAL4-VP16, which expresses GAL4-DNA binding domain fused to the acidic activation domain (amino acids 411 to 454) of herpes simplex virus VP16 transcription factor. The effect on Sp1 was specific in that no effect of GGTI-298 on transcription mediated by GAL4-VP16 was observed (Fig. 5). This specificity was further demonstrated by showing that GGTI-298 downregulates E2F-mediated transcription in Panc-1 cells (Fig. 5). Furthermore, transcription mediated by Sp1-CAT reporter, which contains a repeat of six Sp1-binding sites, was also enhanced by GGTI-298. Taken together, these results show the ability of GGTI-298 to stimulate selectively Sp1-mediated transcription and suggest a model in which GGTI-298 upregulates p21WAF1/CIP1 by enhancing both Sp1 transcriptional activity and DNA binding.
FIG. 5.
GGTI-298 upregulates Sp1-transcriptional activity. Panc-1 cells were cotransfected with 1 μg of GAL4-Sp1 constructs, 4 μg of G5BCAT, 2 μg of Sp1-CAT or E2F-CAT, and 0.5 μg of pCMV-βgal. At 15 h posttransfection, cells were incubated with GGTI-298 (15 μM) for 36 h as described in Materials and Methods. Samples were normalized for transfection efficiency against β-galactosidase activity and then assayed for CAT activity. Thin-layer chromatography plates were scanned with a PhosphorImager, and the percentages of acetylated and nonacetylated forms of chloramphenicol were determined. The fold induction was calculated by dividing the CAT activity values of samples treated with GGTI-298 by the activity of untreated control samples. Data are representative of three independent experiments.
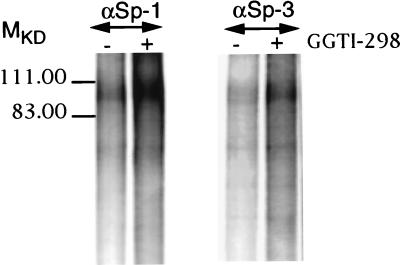
GGTI-298 mediates an increase in Sp1 and Sp3 phosphorylation.
As shown in Fig. 5, the transcriptional activity of GAL4-Sp1 fusion was specifically enhanced by GGTI-298. This result suggested that GGTI-298 might affect Sp1 posttranscriptional modification(s), such as phosphorylation. To determine whether GGTI-298 could affect the phosphorylation state of Sp1 and Sp3, GGTI-298-treated and -untreated cells were labeled with ortho[32P]phosphate as described in Materials and Methods. Cells were first treated with GGTI-298 (15 μM) for 30 h prior to labeling with ortho[32P]phosphate. Equal amounts of proteins were used for immunoprecipitation with Sp1 or Sp3 antibodies, followed by analysis of the immunocomplexes by SDS-PAGE. GGTI-298 treatment resulted in increased phosphorylation of both Sp1 and Sp3 (Fig. 6). The phosphorylation state of Sp1 in GGTI-298-treated cells was markedly higher than that of Sp3. Thus, phosphorylation of Sp1 and Sp3 could be one of the mechanisms leading to the enhancement of Sp1-transcriptional activity.
FIG. 6.
GGTI-298 mediates an increase in Sp1 and Sp3 phosphorylation. GGTI-298-treated and untreated Panc-1 cells were labeled with ortho[32P]phosphate as described in Materials and Methods. Equal amounts of proteins were used for IP with Sp1 (1:200) and Sp3 (1:100) polyclonal antibodies, followed by analysis of the immunocomplexes by SDS–8% PAGE. The gel was fixed, dried, and exposed for autoradiography. Data are representative of two independent experiments.
The small GTPase RhoA, but not Rac1, represses p21WAF1/CIP1 transcription.
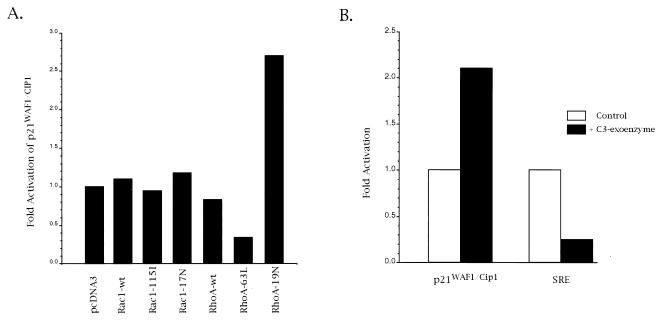
The upregulation of p21WAF1/CIP1 promoter by GGTI-298 suggested a role of geranylgeranylated proteins in p21WAF1/CIP1 regulation. Substrates for GGTase I, such as small GTPases RhoA and Rac1, play important roles in signal transduction and cell cycle regulation. To determine whether RhoA and Rac1 are involved in p21WAF1/CIP1 regulation, we cotransfected Panc-1 cells with p21WAF1/CIP1 promoter and Rac1 or RhoA expression vectors (Fig. 7). At 15 h posttransfection, cells were incubated in DMEM supplemented with 0.5% FBS for 24 h. Subsequently, cells were incubated with DMEM supplemented with 15% FBS, and the incubation was resumed for 24 h. Aliquots of cell lysate were assayed for β-galactosidase and luciferase assays. As shown in Fig. 7A, expression of the constitutively active RhoA (63L) resulted in a threefold repression of p21WAF1/CIP1 promoter activity. In contrast, the dominant negative mutant of RhoA (19N) had an opposite effect, in that its expression activated p21WAF1/CIP1 promoter by 2.5-fold. Neither the dominant negative mutant (Rac1-17N) nor the constitutively active Rac1 (Rac1-115I) had an effect on p21WAF1/CIP1 promoter. These results show the ability of RhoA to repress p21WAF1/CIP1 transcription.
FIG. 7.
The small GTPase RhoA is an upstream effector of p21WAF1/CIP1. (A) Panc-1 cells were cotransfected with 6 μg of p21WAF1/CIP1 promoter, 4 μg of Rac1 or RhoA, and 0.5 μg of pCMVβ-gal expression vectors. Rac1-115I and RhoA-63L vectors express constitutively active GTPases. Rac1-17N and RhoA-19N vectors express dominant negative mutants. Aliquots of cell lysate were subjected to β-galactosidase and luciferase assays. (B) Panc-1 cells were transfected with p21WAF1/CIP1 promoter, and at 15 h posttransfection cells were incubated in DMEM supplemented with 0.5% FBS for 24 h. Subsequently, cells were treated with C3 exoenzyme as described in Materials and Methods. Aliquots of cell lysate were analyzed for β-galactosidase and luciferase activities. Data are representative of at least three independent experiments.
To further demonstrate the involvement of RhoA in regulating p21WAF1/CIP1, we analyzed the activity of p21WAF1/CIP1 promoter in cells treated with C. botulinum C3 exoenzyme. C3 exoenzyme specifically ADP-ribosylates Rho proteins, which results in their inactivation. Panc-1 cells were transfected with p21WAF1/CIP1 promoter, and at 15 h posttransfection cells were incubated in DMEM supplemented with 0.5% FBS for 24 h. Subsequently, cells were treated with C3 exoenzyme as described in Materials and Methods. Aliquots of cell lysate were analyzed for β-galactosidase and luciferase activities. As shown in Fig. 7B, C3 exoenzyme mediated the activation of p21WAF1/CIP1 promoter. In contrast, SRE, which was shown to be positively regulated by Rho GTPases in response to serum, was downregulated by C3 exoenzyme. Taken together, these results demonstrate the involvement of Rho proteins in p21WAF1/CIP1 regulation.
DISCUSSION
Mutations in the ras oncogene and p53 tumor suppressor gene are the most frequently identified genetic alterations responsible for human cancers (for reviews see references 5, 2–4, and 22). Thus, recent drug discovery efforts have focused on developing pharmacological approaches to suppress ras oncogenic ability and/or to restore p53 function. One mechanism by which p53 keeps cells in check and prevents aberrant malignant growth involves induction of a G1 arrest that allows cells to repair DNA damage, initiate programmed cell death, or differentiate (7, 31). Often, the G1 arrest is mediated by p53-dependent transcriptional activation of the CDK inhibitor p21WAF1/CIP1 (8). It is believed that in about half of human cancers, this important p53-dependent induction of p21WAF1/CIP1 is not operational, due to the lack of functional p53. Thus, a desirable characteristic of novel anticancer agents is restoration of p21WAF1/CIP1 induction in the absence of functional p53 in human cancers.
Recently, we made a potent and selective GGTase I inhibitor, GGTI-298, that blocks human tumors in G1, induces apoptosis, and inhibits tumor growth in nude mouse xenografts (38, 41). Evaluation of the mechanism by which GGTase I inhibitors arrest cells in the G1 phase of the cell cycle revealed that GGTI-298 strongly induces p21WAF1/CIP1 accumulation in human tumors that lack both alleles of the p53 gene (41). In this study we used a human pancreatic carcinoma cell line, Panc-1, to demonstrate that GGTI-298 upregulates p21WAF1/CIP1 at the transcriptional level. Since p53 protein is one of the major transactivators of p21 promoter, we tested the effect of GGTI-298 on a p21 promoter that is lacking the p53-binding site. We found the p53 site to be dispensable for GGTI-298-mediated upregulation, suggesting a p53-independent mechanism. In contrast, a small region of the promoter (−84 to −74) comprising a TβRE that contains an Sp1-binding site was sufficient for upregulation by GGTI-298. A similar region (−93 to −44) that contains this TβRE was also shown to be the minimal region for TGF-β-, butyrate-, phorbol ester-, and okadaic acid-mediated upregulation of p21WAF1/CIP1 promoter (6, 28, 48). Sp1-binding sites similar to the one contained in the GGTI-298-responsive element are bound to a common set of ubiquitously expressed nuclear proteins which regulate the expression of a variety of genes, including those encoding p15INK4B, CYP11A, mdr1, α2(I) collagen, ornithine decarboxylase, pyruvate kinase M, and acetyl coenzyme A carboxylase (5, 17, 23, 39).
We found Sp1 and Sp3 DNA binding to the sequence from −84 to −74 of p21WAF1/CIP1 promoter to be enhanced by GGTI-298. Furthermore, GGTI-298 is capable of activating transcription from a CAT reporter plasmid that contains six Sp1-binding sites. Moreover, using chimera that express GAL4-DNA binding domain fused to Sp1 transactivation domain, we have shown that GGTI-298 is capable of activating specifically Sp1 transcriptional activity. In contrast, E2F- and SRE-mediated transcription were repressed. Taken together, these results show that two different mechanisms could lead to GGTI-298-mediated p21WAF1/CIP1 induction, one through the increase of Sp1 affinity for its binding site and the second through the stimulation of Sp1 transcriptional activity. Sp1 is a phosphoprotein that has been shown to be phosphorylated by DNA-dependent protein kinase (see reference 13 for a review). Interestingly, okadaic acid, a selective inhibitor of the serine-threonine phosphatase PP2A, was shown to induce p21WAF1/CIP1 (48), mediate hyperphosphorylation of Sp1 (35), and increase the transcriptional activity of Sp1 with a concomitant hyperphosphorylation of Sp1 (24). We have analyzed the phosphorylation states of Sp1 and Sp3 in response to GGTI-298 and found Sp1 and Sp3 to be highly phosphorylated in GGTI-298-treated cells compared to untreated cells. Interestingly, the increase in phosphorylation was observed only with the 106-kDa isoforms of Sp1 and Sp3, suggesting that specific isoforms may have different functions. Taken together, our results suggest that GGTI-298-mediated Sp1 phosphorylation may lead to the increase of both DNA-binding and transcriptional activity of Sp1.
The characterization of the signal transduction pathways that are involved in GGTI-298-mediated p21WAF1/CIP1 induction may lead to the identification of the proteins involved in p21WAF1/CIP1 regulation. First of all, GGTI-298 may affect cellular pathways that are used by TGF-β to trigger its growth-inhibiting effect. Indeed, TGF-β also upregulates p21 promoter activity through the same region as does GGTI-298 (−84 to −74) (6). It is interesting that the common α subunit of GGTase I and FTase has been shown by three independent groups to bind to and to be phosphorylated by TGF-β receptor (15, 40, 44). Furthermore, it has been suggested that GGTase I or FTase may be involved in mediating TGF-β signaling. Clues about the nature of the signal transduction pathways that are affected by GGTI-298 may also be obtained from the study of the GGTase I substrates that are involved in p21 regulation. Several small GTPases involved in signal transduction, such as the Rho family of proteins (i.e., RhoA, RhoB, CDC42Hs, and Rac1), are prenylated by GGTase I. Research at a number of laboratories over the past few years has revealed that the Rho GTPases play crucial roles in the G1/S transition of the cell cycle. For instance, injection of the constitutively active Cdc42, Rac1, and RhoA proteins in Swiss 3T3 fibroblasts was shown to stimulate cell cycle progression through the G1 phase and subsequent DNA synthesis, whereas injection of dominant negative forms of these GTPases blocked stimulation of DNA synthesis in response to serum (29). Rho proteins facilitate the progression from G1 to S phase in growth-stimulated cells by promoting the degradation of the CDK inhibitor p27Kip1 (12). Furthermore, the Rho family of GTPases has also been suggested to regulate cell proliferation by modulating transcription of specific genes, such as c-fos (11). We found the constitutively active small GTPase RhoA (63L) to downregulate p21WAF1/CIP1 promoter. In contrast, the dominant negative form of RhoA (19N) had an opposite effect in that it activated p21WAF1/CIP1 promoter. Neither the dominant negative mutant (Rac1-17N) nor the constitutively active Rac1 (Rac1-115I) had an effect on p21WAF1/CIP1 promoter, suggesting that Rac1 and RhoA may function through different pathways to control cell cycle progression. We further demonstrated the involvement of Rho proteins in regulating p21WAF1/CIP1 by showing the activation of p21WAF1/CIP1 by C. botulinum C3 exoenzyme, which specifically ADP-ribosylates and inactivates Rho proteins. These results show the ability of RhoA to regulate p21WAF1/CIP1, which could be one of the mechanisms by which RhoA controls cell growth. While this article was in revision, Olson and coworkers (30) reported that signals from Ras and Rho GTPases interact to regulate expression of p21WAF1/CIP1. The authors showed that induction of DNA synthesis by constitutively active Ras requires Rho signaling for the suppression of p21WAF1/CIP1 induction. These results are consistent with our findings and give further support to the idea that RhoA is the target for GGTI-298. Thus, the present study suggests that GGTI-298, which inhibits Rho proteins by preventing their geranylgeranylation, induces p21WAF1/CIP1 and a subsequent arrest in the G1 phase of the cell cycle. Furthermore, results from this study, coupled with our previous work (27, 41), also suggest that pharmacological agents capable of inhibiting protein geranylgeranylation restore cell growth arrest and apoptosis in cancer cells with a nonfunctional p53.
ACKNOWLEDGMENTS
We are grateful to X.-F. Wang (Duke University Medical Center) for supplying the p21WAF1/CIP1 constructs, C. Der (University of North Carolina at Chapel Hill) for RhoA and Rac1 pzip constructs, G. Gill (University of California, Berkeley) for GAL4 constructs, and P. D. Robbins (University of Pittsburgh) for Sp1 constructs.
This work was supported in part by Public Health Service Award CA-67771 from the National Cancer Institute. (S.M.S. and A.D.H.) and by ACS-IRG from the American Cancer Society (J.A.). The work was also supported in part by the Molecular Biology Facility at the H. Lee Moffitt Cancer Center and Research Institute.
REFERENCES
- 1.Adnane J, Shao Z, Robbins P D. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- 2.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 4.Collins K, Jacks T, Pavletich N P. The cell cycle and cancer. Proc Natl Acad Sci USA. 1997;94:2776–2778. doi: 10.1073/pnas.94.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornwell M M, Smith D E. SP1 activates the MDR1 promoter through one of two distinct G-rich regions that modulate promoter activity. J Biol Chem. 1993;268:19505–19511. [PubMed] [Google Scholar]
- 6.Datto M B, Yu Y, Wang X F. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 7.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 8.el-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 9.Gibbs J B, Oliff A. The potential of farnesyltransferase inhibitors as cancer chemotherapeutics. Annu Rev Pharmacol Toxicol. 1997;37:143–166. doi: 10.1146/annurev.pharmtox.37.1.143. [DOI] [PubMed] [Google Scholar]
- 10.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 12.Hirai A, Nakamura S, Noguchi Y, Yasuda T, Kitagawa M, Tatsuno I, Oeda T, Tahara K, Terano T, Narumiya S, Kohn L D, Saito Y. Geranylgeranylated rho small GTPase(s) are essential for the degradation of p27Kip1 and facilitate the progression from G1 to S phase in growth-stimulated rat FRTL-5 cells. J Biol Chem. 1997;272:13–16. [PubMed] [Google Scholar]
- 13.Jackson S, Gottlieb T, Hartley K. Phosphorylation of transcription factor Sp1 by the DNA-dependent protein kinase. Adv Second Messenger Phosphoprotein Res. 1993;28:279–286. [PubMed] [Google Scholar]
- 14.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 15.Kawabata M, Imamura T, Miyazono K, Engel M E, Moses H L. Interaction of the transforming growth factor-beta type I receptor with farnesyl-protein transferase-alpha. J Biol Chem. 1995;270:29628–29631. doi: 10.1074/jbc.270.50.29628. [DOI] [PubMed] [Google Scholar]
- 16.Khosravi-Far R, Solski P A, Clark G J, Kinch M S, Der C J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A P, Mar P K, Zhao B, Montgomery R L, Kang D C, Butler A P. Regulation of rat ornithine decarboxylase promoter activity by binding of transcription factor Sp1. J Biol Chem. 1995;270:4341–4348. doi: 10.1074/jbc.270.9.4341. [DOI] [PubMed] [Google Scholar]
- 18.Lebowitz P F, Casey J P, Prendergast G C, Thissen J A. Farnesyltransferase inhibitors alter the prenylation and growth-stimulating function of RhoB. J Biol Chem. 1997;272:15591–15594. doi: 10.1074/jbc.272.25.15591. [DOI] [PubMed] [Google Scholar]
- 19.Lebowitz P F, Sakamuro D, Prendergast G C. Farnesyl transferase inhibitors induce apoptosis of Ras-transformed cells denied substratum attachment. Cancer Res. 1997;57:708–713. [PubMed] [Google Scholar]
- 20.Lerner E C, Qian Y, Hamilton A D, Sebti S M. Disruption of oncogenic K-Ras4B processing and signaling by a potent geranylgeranyltransferase I inhibitor. J Biol Chem. 1995;270:26770–26773. doi: 10.1074/jbc.270.45.26770. [DOI] [PubMed] [Google Scholar]
- 21.Lerner E C, Zhang T, Knowles D B, Qian Y, Hamilton A D, Sebti S M. Inhibition of the prenylation of K-Ras, but not H- or N-Ras, is highly resistant to CAAX peptidomimetics and requires both a farnesyltransferase and a geranylgeranyltransferase I inhibitor in human tumor cell lines. Oncogene. 1997;15:1283–1288. doi: 10.1038/sj.onc.1201296. [DOI] [PubMed] [Google Scholar]
- 22.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Wu M, Welsh D. The role of the p53 tumour-suppressor gene in tumorigenesis. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J M, Nichols M A, Chandrasekharan S, Xiong Y, Wang X F. Transforming growth factor beta activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 24.Lin M C, Almus-Jacobs F, Chen H H, Parry G C, Mackman N, Shyy J Y, Chien S. Shear stress induction of the tissue factor gene. J Clin Investig. 1997;99:737–744. doi: 10.1172/JCI119219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick F. Ras-related proteins in signal transduction and growth control. Mol Reprod Dev. 1995;42:500–506. doi: 10.1002/mrd.1080420419. [DOI] [PubMed] [Google Scholar]
- 26.McGuire T F, Qian Y, Vogt A, Hamilton A D, Sebti S M. Platelet-derived growth factor receptor tyrosine phosphorylation requires protein geranylgeranylation but not farnesylation. J Biol Chem. 1996;271:27402–27407. doi: 10.1074/jbc.271.44.27402. [DOI] [PubMed] [Google Scholar]
- 27.Miquel K, Pradines A, Sun J, Qian Y, Hamilton A D, Sebti S M, Favre G. GGTI-298 induces G0-G1 block and apoptosis whereas FTI-277 causes G2-M enrichment in A549 cells. Cancer Res. 1997;57:1846–1850. [PubMed] [Google Scholar]
- 28.Nakano K, Mizuno T, Sowa Y, Orita T, Yoshino T, Okuyama Y, Fujita T, Ohtani-Fujita N, Matsukawa Y, Tokino T, Yamagishi H, Oka T, Nomura H, Sakai T. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J Biol Chem. 1997;272:22199–22206. doi: 10.1074/jbc.272.35.22199. [DOI] [PubMed] [Google Scholar]
- 29.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 30.Olson M F, Paterson H F, Marshall C J. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 31.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 32.Qiu R G, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quilliam L A, Khosravi-Far R, Huff S Y, Der C J. Guanine nucleotide exchange factors: activators of the Ras superfamily of proteins. Bioessays. 1995;17:395–404. doi: 10.1002/bies.950170507. [DOI] [PubMed] [Google Scholar]
- 34.Rowell C A, Kowalczyk J J, Lewis M D, Garcia A M. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- 35.Sanceau J, Kaisho T, Hirano T, Wietzerbin J. Triggering of the human interleukin-6 gene by interferon-gamma and tumor necrosis factor-alpha in monocytic cells involves cooperation between interferon regulatory factor-1, NF kappa B, and Sp1 transcription factors. J Biol Chem. 1995;270:27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- 36.Sebti S M, Hamilton A D. Inhibition of Ras prenylation: a novel approach to cancer chemotherapy. Pharmacol Ther. 1997;74:103–114. doi: 10.1016/s0163-7258(97)00014-4. [DOI] [PubMed] [Google Scholar]
- 37.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 38.Sun J, Qian Y, Hamilton A D, Sebti S M. Both farnesyltransferase and geranylgeranyltransferase I inhibitors are required for inhibition of oncogenic K-ras prenylation but each alone is sufficient to suppress human tumor growth in nude mouse xenografts. Oncogene. 1997;16:1467–1473. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- 39.Venepally P, Waterman M R. Two Sp1-binding sites mediate cAMP-induced transcription of the bovine CYP11A gene through the protein kinase A signaling pathway. J Biol Chem. 1995;270:25402–25410. doi: 10.1074/jbc.270.43.25402. [DOI] [PubMed] [Google Scholar]
- 40.Ventura F, Liu F, Doody J, Massague J. Interaction of transforming growth factor-beta receptor I with farnesyl-protein transferase-alpha in yeast and mammalian cells. J Biol Chem. 1996;271:13931–13934. doi: 10.1074/jbc.271.24.13931. [DOI] [PubMed] [Google Scholar]
- 41.Vogt A, Sun J, Qian Y, Hamilton A D, Sebti S M. The geranylgeranyltransferase-I inhibitor GGTI-298 arrests human tumor cells in G0/G1 and induces p21WAF1/CIP1/SDI1 in a p53-independent manner. J Biol Chem. 1997;272:27224–27229. doi: 10.1074/jbc.272.43.27224. [DOI] [PubMed] [Google Scholar]
- 42.Vogt A, Qian Y, McGuire T F, Hamilton A D, Sebti S M. Protein geranylgeranylation, not farnesylation, is required for the G1 to S phase transition in mouse fibroblasts. Oncogene. 1996;13:1991–1999. [PubMed] [Google Scholar]
- 43.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 44.Wang T, Danielson P D, Li B Y, Shah P C, Kim S D, Donahoe P K. The p21(Ras) farnesyltransferase alpha subunit in TGF-beta and activin signaling. Science. 1996;271:1120–1122. doi: 10.1126/science.271.5252.1120. [DOI] [PubMed] [Google Scholar]
- 45.Whyte D B, Kirschmeier P, Hokenberry T N, Nunez-Oliva I, James L, Catino J J, Bishop W R, Pai J K. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 46.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F L, Casey P J. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Grasso L, McClain C D, Gambel A M, Cha Y, Travali S, Deisseroth A B, Mercer W E. p53-independent induction of WAF1/CIP1 in human leukemia cells is correlated with growth arrest accompanying monocyte/macrophage differentiation. Cancer Res. 1995;55:668–674. [PubMed] [Google Scholar]