Abstract
Lemnoideae, commonly referred to as the duckweed, are aquatic plants found worldwide. Wolffia species are known for their extreme reduction in size and complexity, lacking both roots and leaves, and they hold the distinction of being the smallest plants among angiosperms. Interestingly, it belongs to the Araceae family, despite its apparent morphological differences from land plants in the same family. Traditional morphological methods have limitations in classifying these plants, making molecular-level information essential. The chloroplast genome of Wolffia arrhiza is revealed that a total length of 169,602 bp and a total GC content of 35.78%. It follows the typical quadripartite structure, which includes a large single copy (LSC, 92,172 bp) region, a small single copy (SSC, 13,686 bp) region, and a pair of inverted repeat (IR, 31,872 bp each) regions. There are 131 genes characterized, comprising 86 Protein-Coding Genes, 37 Transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Moreover, 48 simple sequence repeats and 32 long repeat sequences were detected. Comparative analysis between W. arrhiza and six other Lemnoideae species identified 12 hotspots of high nucleotide diversity. In addition, a phylogenetic analysis was performed using 14 species belonging to the Araceae family and one external species as an outgroup. This analysis unveiled W. arrhiza and Wolffia globosa as closely related sister species. Therefore, this research has revealed the complete chloroplast genome data of W. arrhiza, offering a more detailed understanding of its evolutionary position and phylogenetic categorization within the Lemnoideae subfamily.
Subject terms: Evolution, Genetics
Introduction
Lemnoideae, commonly known as duckweed, is a monocotyledonous aquatic plant belonging to the Araceae family1,2. It exhibits a growth pattern of floating freely or being submerged3,4. It is extensively distributed worldwide, with a particular prevalence in tropical and subtropical regions1,5–7. It thrives in freshwater ponds, rivers, and various other aquatic environments. These were classified into a total of five genera and 38 species: Spirodela SCHLEID (containing 2 species), Landoltia LES & D. J. CRAWFORD (comprising 1 species), Lemna L. (with 14 species), Wolffiella HEGELM (including 10 species), and Wolffia SCHLEID (encompassing 11 species). These findings were initially documented by Landolt in 19861,8,9. Of these, the Wolffia genus is notable for being the smallest angiosperm plant in the world, measuring merely 1 mm in diameter, and it does not possess stems or roots. Instead, it has spherical fronds10. It features a level upper surface that hovers above the water's top layer, with parallelly arranged stomata (Fig. 1). Additionally, this plant generates infrequently small flowers with only one stamen and pistil, which emerge from a hole on the upper side of the frond. However, the main method of propagation is vegetative reproduction11,12. This is the way in which a new leaf bud emerges from the reproductive pouch of the parent leaf, undergoing gradual growth and eventual separation13. Consequently, Wolffia has the capacity to rapidly double its population in as little as 2–3 days14,15 in the optimal conditions such as temperatures within the range of 20–30 °C and a pH level spanning from 5.0 to 7.016,17. This exceptional ability establishes Wolffia as one of the fastest-growing plants globally.
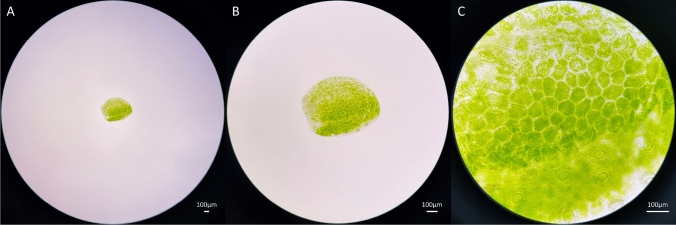
Figure 1.
Microscopic photos of Wolffia arrhiza taken at (A) 40×, (B) 100×, and (C) 400× magnification. The majority is composed of chloroplast, and stomata can be seen in the lower part of the specimen. The scale bar in the lower right of each figure is 100μm for each magnification.
Because of its rapid growth, high reproductive rate, and straightforward cultivation and harvesting, it was regarded as an ideal candidate for an experimental model plant in numerous fields. Wolffia arrhiza, specifically, has the capacity to absorb and cleanse excessive nitrogen and phosphorus that may result in eutrophication in aquatic environments18. Furthermore, its nutritional potential is noteworthy, as 40% of its dry weight is constituted by protein, and it contains notable levels of amino acids, calcium, magnesium, and vitamins that hold significance in the human diet19. These characteristics have captured interest, leading to research into the application of W. arrhiza in fields such as wastewater management20, components of human food21 or animal feed22.
The morphological classification of these duckweed species is exceptionally challenging due to their significantly diminished size and simplified intricacy. Accordingly, molecular taxonomy has been vital for species classification, and to establish the phylogenetic relationships within the Lemnoideae subfamily, the chloroplast barcode has been adopted and utilized23–25. In contrast to the nuclear genome, the chloroplast genome presents advantages in species classification owing to its smaller genome size, haploid inheritance, conserved structure, and slower mutation rate26,27. This enabled to the classification of Lemnoideae within the Araceae family, alongside land plants that have considerable morphological dissimilarities28. Moreover, it has been indicated that Landoltia, previously grouped within the Spirodela genus, is now a novel and separate genus, distinct from both Spirodela and Lemna29. Thus, in order to establish a robust basis for gaining insight into the genetic variation and its placement in the phylogenetic tree of Lemnoideae subfamily, it is imperative to gather more chloroplast information from a diverse of duckweed species9,30.
In this study, the full chloroplast genome of W. arrhiza was assembled from whole genome sequencing data from MGI DNBSEQ-G50 second-generation sequencing platform. The overview for conducting research is as follows: (i) characterization of the complete chloroplast genome of W. arrhiza; (ii) a comparative analysis of chloroplast genome data using six species available from NCBI; (iii) Analyzing evolution progression and phylogenetic relationship. The goal is to improve the clarify of relationships within Lemnoideae and establish a fundamental basis for a coherent classification system.
Results
Chloroplast genome characteristics of W. arrhiza
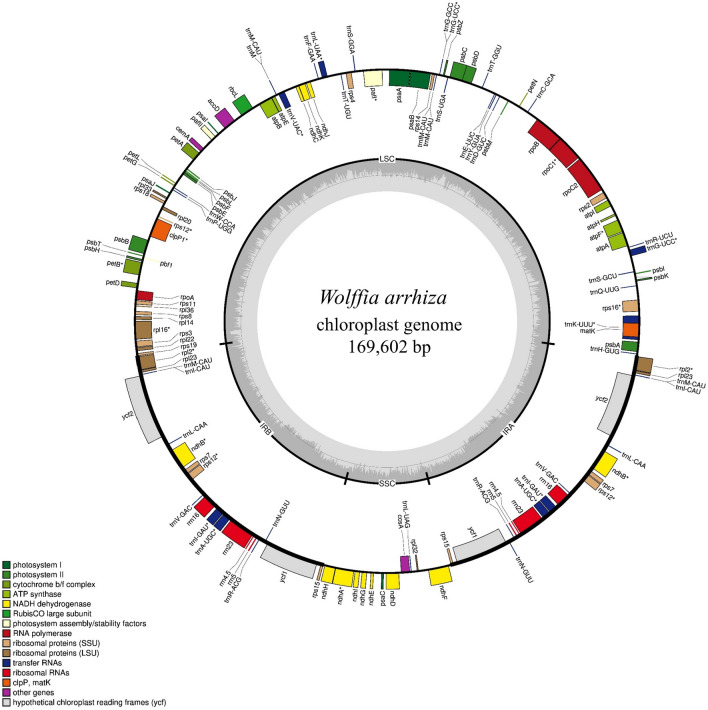
The chloroplast genome of W. arrhiza is 169,602 bp in the quadripartite structure with one large single copy (LSC) region of 92,172 bp, one small single copy (SSC) region of 13,686 bp, and a pair of inverted repeat (IR) regions of 31,872 bp each (Fig. 2). The total guanine and cytosine (GC) content of the chloroplast genome is 35.78%. The relative occupation ratio of the LSC, SSC, and IR regions in the chloroplast genome were 33.63%, 30.79%, and 39.97%, respectively. It contains a total of 131 predicted genes, which are divided into three groups: 86 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The entire set of genes exhibited a GC content of 38.52%. Within this, PCGs demonstrated a GC content of 37.08%, tRNA genes had a GC content of 52.50%, and rRNA genes exhibited a GC content of 54.69%. The LSC region contained a total of 83 genes, comprising 61 PCGs and 22 tRNA genes. In the SSC region, there were 11 genes, including 10 PCGs and one tRNA gene. The IR regions consisted of 36 genes, with seven PCGs, seven tRNA genes, and four rRNA genes duplicated (Table 1). Additionally, the rps12 gene is a trans-spliced gene that exons found in both the LSC and IRs, while the rps19 gene extended across two regions between the LSC and IRb.
Figure 2.
The gene map of the chloroplast genome of Wolffia arrhiza. The map identifies three distinct regions: the large single copy region (LSC), the small single copy region (SSC), and the inverted repeat A/B regions (IRA/B). Additionally, the innermost dark gray track represents the GC contents. Genes on the inner side of the map are transcribed counterclockwise, while those on the outer side are transcribed clockwise.
Table 1.
Chloroplast genome structure and feature of Wolffia arrhiza.
| Genome feature | Length (bp)/Numbers | GC content (%) | |
|---|---|---|---|
| Structure length | Total | 169,602 | 35.78 |
| LSC region | 92,172 | 33.63 | |
| SSC region | 13,686 | 30.79 | |
| IR (a/b) region | 31,872 | 39.97 | |
| Counts of genes in different categories | Genes | 131 | 38.52 |
| PCGs | 86 | 37.08 | |
| tRNA | 37 | 52.50 | |
| rRNA | 8 | 54.69 | |
| Detailed counts of genes in different regions | LSC region | 61 PCGs, 22 tRNA | – |
| SSC region | 10 PCGs, 1 tRNA | – | |
| IR (a/b) region | 7 PCGs, 7 tRNA, 4 rRNA | – | |
According to the chloroplast genome annotation of W. arrhiza, 112 unique genes were categorized into four functional groups. There were 59 transcription and translation-related genes, 46 photosynthesis-related genes, five biosynthesis-related genes, and two genes whose functions were unidentified (Table 2).
Table 2.
Genetic classification of the chloroplast genome of Wolffia arrhiza.
| Category (number) | Group (number) | Gene name |
|---|---|---|
| Transcription and translation (59) | Ribosomal RNAs (4) | rrn16*, rrn23*, rrn4.5*, rrn5* |
| Transfer RNAs (30) | trnA-UGC*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU*, trnI-GAU*, trnK-UUU, trnL-CAA*, trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU*, trnP-UGG, trnQ-UUG, trnR-ACG*, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC*, trnV-UAC, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Small subunit of ribosome (SSU) (12) | rps2, rps3, rps4, rps7*, rps8, rps11, rps12*, rps14, rps15*, rps16, rps18, rps19 | |
| Large subunit of ribosome (LSU) (9) | rpl2*, rpl14, rpl16, rpl20, rpl22, rpl23*, rpl32, rpl33, rpl36 | |
| DNA-dependent RNA polymerase (4) | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis (46) | Photosystem I (7) | psaA, psaB, psaC, psaI, psaJ, pafI, pafII |
| Photosystem II (15) | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbT, psbZ, pbf1 | |
| Subunit of cytochrome (6) | petA, petB, petD, petG, petL, petN | |
| ATP synthase (6) | atpA, atpB, atpE, atpF, atpH, atpI | |
| RubisCO (1) | rbcL | |
| NADH dehydrogenase (11) | ndhA, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Biosynthesis (5) | Maturase (1) | matK |
| ATP-dependent Protease (1) | clpP1 | |
| Acetyl-CoA-carboxylase (1) | accD | |
| Envelop membrane protein (1) | cemA | |
| C-Type cytochrome synthesis (1) | ccsA | |
| Unknown (2) | Hypothetical chloroplast reading frames(ycf) (2) | ycf1*, ycf2* |
Duplicated genes are denoted by an asterisk (*).
Concurrently, a total of 17 unique intron genes were detected, and they were distributed across the LSC (11), IR (5), and SSC (1, ndhA) regions. It was comprised 11 PCGs and 6 tRNA genes. Among these, 15 genes (atpF, ndhA, ndhB, petB, rpl16, rpl2, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) had a single intron each, while the remaining 2 genes (clpP1, pafI) contained two introns each (Table 3).
Table 3.
Introns and exons length information of the Wolffia arrhiza.
| Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|
| atpF | LSC | 144 | 839 | 402 | ||
| clpP1* | LSC | 71 | 811 | 295 | 656 | 243 |
| ndhA | SSC | 551 | 916 | 532 | ||
| ndhB* | IR | 777 | 703 | 756 | ||
| pafI* | LSC | 126 | 750 | 226 | 789 | 155 |
| petB | LSC | 6 | 752 | 642 | ||
| rpl16 | LSC | 9 | 1575 | 399 | ||
| rpl2* | IR | 391 | 660 | 431 | ||
| rpoC1 | LSC | 453 | 744 | 1620 | ||
| rps12* | LSC, IR | 114 | 540 | 232 | – | 26 |
| rps16 | LSC | 40 | 1025 | 200 | ||
| trnA-UGC* | IR | 37 | 801 | 36 | ||
| trnG-UCC | LSC | 24 | 619 | 48 | ||
| trnI-GAU* | IR | 42 | 804 | 35 | ||
| trnK-UUU | LSC | 36 | 2545 | 43 | ||
| trnL-UAA | LSC | 37 | 507 | 50 | ||
| trnV-UAC | LSC | 38 | 615 | 37 |
Duplicated genes are denoted by an asterisk (*). The duplicated 3' ends of the rps12 gene, which is trans-spliced, are found in the IR regions with the 5' end of the gene located in the LSC region.
Repeat sequences analysis
The web application Misa successfully identified a total of 48 Simple Sequence Repeats, SSRs, with lengths ranging from 10 to 16 bp. There was a total of 42 mononucleotide and 6 dinucleotide repeat types observed, all composed of A or T bases. There were 26 mononucleotides consisting solely of A and 16 mononucleotides consisting solely of T. Likewise, four dinucleotides comprised of AT repeats and two dinucleotides comprised of TA repeats. Among the identified SSRs, the LSC region contains the highest number, accounting for the majority (72.92%) with a total of 35 SSRs. The SSC region hosts seven SSRs (14.58%), while the IR region holds six SSRs (12.5%). At the same time, the Intergenic spacer (IGS) region presents the largest number of SSRs, totaling 40 (83.3%) of the total SSRs. Five SSRs (10.42%) were found in introns, and the remaining three SSRs (6.25%) were in PCG regions. Notably, each of the introns within the petB, rps16, trnK-UUU, pafI, and clpP1 genes contained one SSR. Moreover, SSRs within PCG were observed in one instance within the rpoB gene and one each in the ycf1 genes of IRa and IRb (Table 4).
Table 4.
The types of SSRs in Wolffia arrhiza and their corresponding regions and locations.
| Repeat type | Repeat unit | Number of repeats | Repeat length | Number of SSRs | Region | Location |
|---|---|---|---|---|---|---|
| Mono-nucleotide | A | 10 | 10 | 18 | LSC(15) | IGS(12) |
| Intron(3, petB, rps16, trnK-UUU) | ||||||
| SSC(3) | IGS(3) | |||||
| 11 | 11 | 5 | LSC(2) | IGS(2) | ||
| SSC(1) | IGS(1) | |||||
| IRb(2) | IGS(1) | |||||
| PCG(1, ycf1) | ||||||
| 13 | 13 | 3 | LSC(2) | IGS(2) | ||
| IRb(1) | IGS(1) | |||||
| T | 10 | 10 | 9 | LSC(7) | IGS(5) | |
| Intron(1, pafI) | ||||||
| PCG(1, rpoB) | ||||||
| SSC(2) | IGS(2) | |||||
| 11 | 11 | 5 | LSC(3) | IGS(2) | ||
| Intron(1, clpP1) | ||||||
| IRa(2) | IGS(1) | |||||
| PCG(1, ycf1) | ||||||
| 12 | 12 | 1 | LSC(1) | IGS(1) | ||
| 13 | 13 | 1 | IRa(1) | IGS(1) | ||
| Di-nucleotide | AT | 6 | 12 | 3 | LSC(3) | IGS(3) |
| 7 | 14 | 1 | LSC(1) | IGS(1) | ||
| TA | 6 | 12 | 1 | SSC(1) | IGS(1) | |
| 8 | 16 | 1 | LSC(1) | IGS(1) | ||
| Total | 48 |
LSC(35, 72.92%) SSC(7, 14.58%) IR(6, 12.5%) |
IGS(40, 83.3%) Intron(5, 10.42%) PCG(3, 6.25%) |
A total of 32 long repeat sequences were detected using the REPuter web application. Among these repeats, there were 16 forward repeats (F), 1 reverse repeat (R), and 15 palindromic repeats (P). The lengths exhibited a distribution ranging from 30 to 69 bp, and among them, a unique palindromic repeat measuring 31,872 bp in length was identified. Out of these, 13 were exclusively located within the LSC region (40.63%), while 6 were uniquely situated in the IR region (18.75%). Additionally, 7 were suspended across both IRa and IRb (21.87%), with the remaining 6 spanning across the LSC and IR (18.75%), covering two structural regions. Furthermore, there were a total of 12 repeats solely present within a single PCG (37.5%), and all these repeats were located in the ycf2 gene. There was also one repeat that distributed across both the intron and the PCG (3.12%), and it was the longest repeat in terms of length. This was present across a total of 20 genes. One repeat was identified in both the IGS and the PCG (3.12%), and PCG corresponded to the pbf1 gene. There were three repeats spanning two PCGs (9.38%), and all these PCGs were identified as tRNA genes. In addition, six repeats were observed, spanning across introns and the IGS (18.75%), with four of them positioned in introns within the pafI gene, and the other two in the petB gene. The remaining nine repeats, containing the only reverse repeat that was detected, were exclusively located in the IGS (28.13%) (Table 5).
Table 5.
The types of long repeat in Wolffia arrhiza and their corresponding regions and locations.
| Repeats match type | Repeats length range | Repeats length | Number of repeats set | Region of repeats set | Location of repeats set |
|---|---|---|---|---|---|
| F | 30–39 | 30 | 4 | LSC(3) | IGS(2) |
| PCG(1, trnS(GCU)-trnS(UGA)) | |||||
| LSC-IRb(1) | Intron-IGS(1, pafI) | ||||
| 31 | 3 | LSC(1) | IGS(1) | ||
| IRa(1) | PCG(1, ycf2) | ||||
| IRb(1) | PCG(1, ycf2) | ||||
| 32 | 1 | LSC(1) | IGS(1) | ||
| 34 | 1 | LSC(1) | IGS(1) | ||
| 36 | 1 | LSC-IRa(1) | Intron-IGS(1, petB) | ||
| 37 | 2 | IRa(1) | PCG(1, ycf2) | ||
| IRb(1) | PCG(1, ycf2) | ||||
| 39 | 1 | LSC-IRb(1) | Intron-IGS(1, pafI) | ||
| 40–49 | 41 | 2 | IRa(1) | PCG(1, ycf2) | |
| IRb(1) | PCG(1, ycf2) | ||||
| 60–69 | 63 | 1 | LSC(1) | IGS(1) | |
| P | 30–39 | 30 | 2 | LSC(1) | IGS(1) |
| LSC-IRa(1) | Intron-IGS(1, pafI) | ||||
| 31 | 4 | LSC(2) | IGS(1) | ||
| IGS-PCG(1, pbf1) | |||||
| IRa-IRb(2) | PCG(2, ycf2) | ||||
| 32 | 2 | LSC(2) |
PCG(2, trnS(GCU)-trnS(GGA) / trnS(UGA)-trnS(GGA)) |
||
| 36 | 1 | LSC-IRb(1) | Intron-IGS(1, petB) | ||
| 37 | 2 | IRa-IRb(2) | PCG(2, ycf2) | ||
| 39 | 1 | LSC-IRa(1) | Intron- IGS(1, pafI) | ||
| 40–49 | 41 | 2 | IRa-IRb(2) | PCG(2, ycf2) | |
| 70~ | 31,872 | 1 | IRa-IRb(1) |
Intron-PCG(1, rps7-ycf2-ndhB-ycf1-trnA(UGC)-rpl2-rpl23-trnI(CAU)-rrn5-rrn23-rrn16-trnN(GUU)-trnV(GAC)-trnR(ACG)-rps12-rrn4.5-rps15-trnI(GAU)-trnL(CAA)- trnM(CAU) |
|
| R | 30–39 | 32 | 1 | LSC(1) | IGS(1) |
In the case of differing regions or locations between the repeats, they are connected using '-' and indicated as a repeat set. The genes follow the same pattern. F for forward repeat, R for reverse repeat, and P for palindromic repeat.
Codon usage
A total of 86 PCGs and their CDS were extracted from the chloroplast genome of W. arrhiza. These sequences have a combined length of 84,507 bp and consist of 28,169 codons. Leucine (Leu) was the most commonly encoded amino acid, comprising 10.50% of the total with 2959 codons. Conversely, Cysteine (Cys) was the least frequently encoded amino acid, making up only 1.10% of the total with 310 codons. The RSCU values for each codon fell within the range of 0.3 (CGG, Arg) to 2.01 (AGA, Arg). Out of a total of 30 codons with a high frequency of usage (RSCU > 1), except for UUG (Leu), 29 of these preferred synonymous codons ended with A or U(T) nucleotides. For the 32 codons with RSCU < 1, the 29 codons ended with C or G nucleotide, excluding CUA(Leu), AUA(Ile), and UGA(TER). Additionally, the terminator most preferred was UAA, showing an RSCU value of 1.60. In contrast, the codons AUG (Met) and UGG (Trp) demonstrated an RSCU value of 1, suggesting there is no bias as they each encode only one amino acid (Table S1).
Comparison of chloroplast genomes within Lemnoideae
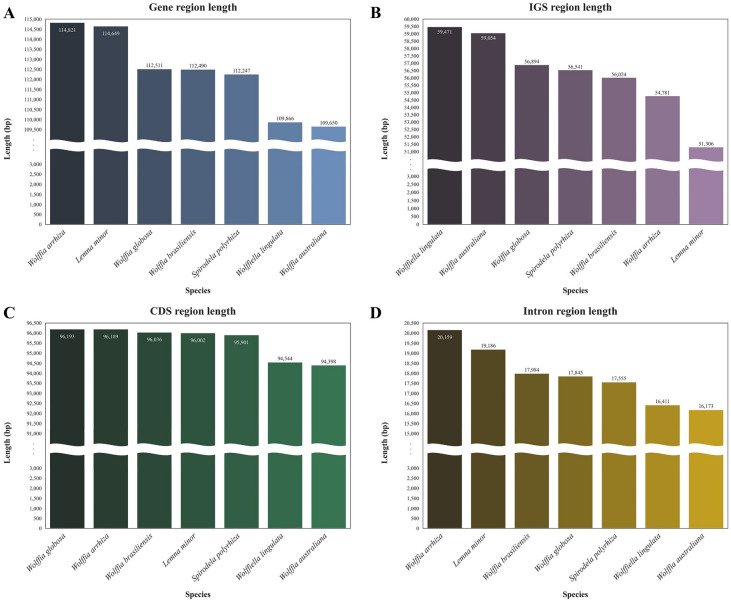
The lengths of genes and IGS regions were compared among the chloroplast genomes of seven species of duckweed within the Lemnoideae subfamily (Fig. 3). The gene regions exhibited a range in length from 109,650 bp to 114,821 bp. Among the seven Lemnoideae species, W. arrhiza possessed the longest gene region with 114,821 bp (Fig. 3A). On the other hand, IGS regions had lengths that ranged from 51,306 bp to 59,471 bp, with W. arrhiza possessing the second shortest IGS region at 54,781 bp, following Lemna minor (Fig. 3B). The gene regions were further categorized into coding sequences (CDS) and intron regions. It was observed that CDS regions had a length range from 94,398 bp to 96,193 bp. Notably, W. arrhiza possessed the second-longest CDS region, measuring 96,189 bp, which was 4 bp shorter than W. globosa (Fig. 3C). Conversely, the lengths of intron regions ranged from 16,173 bp to 20,159 bp, with W. arrhiza having the longest intron region at 20,159 bp (Fig. 3D).
Figure 3.
Length for each region of the Lemnoideae, including Wolffia arrhiza. (A) Gene region length (B) IGS region length (C) CDS region length (D) Intron region length. The X-axis represents the species names, while the Y-axis depicts the lengths of the regions. The arrangement of each graph is based on the ascending order of region lengths.
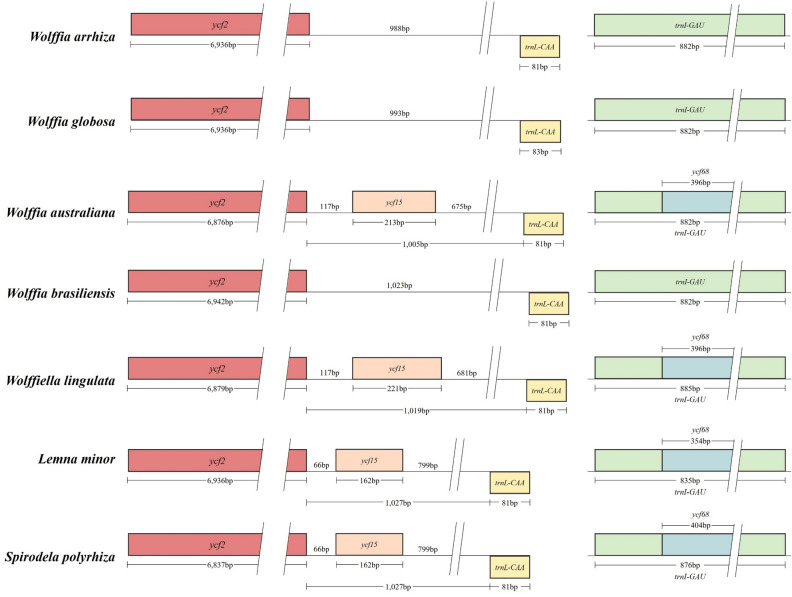
To gain further insight into these changes, an analysis was conducted on events such as insertions, deletions, duplications, and intron changes in genes across Lemnoideae species (Table S2). Compared to other Lemnoideae species, W. arrhiza was represented by the genes pafI, pafII, clpP1, and pbfI, which are synonymous with the ycf3, ycf4, clpP, and psbN genes in other species. When comparing other genes, the most significant alterations were the deletion events of pseudogenes ycf68 and ycf15 in the IR region (Fig. 4). Upon a more detailed examination, it was observed that the gene ycf68, which had perfect overlap with trnI-GAU in other species, was deleted in W. arrhiza, leaving only trnI-GAU. However, another deleted gene, ycf15, which has been alone in other species, underwent deletion in W. arrhiza and sequences remained at an IGS region. The length between ycf2 and trnL-CAA flanking ycf15 in W. australiana, Wolffiella lingulata, Lemna minor, and Spirodela polyrhiza were 1005 bp, 1019 bp, 1027 bp, and 1027 bp, respectively. In W. arrhiza, W. globosa, and W. brasiliensis, where ycf15 was deleted, the IGS length between ycf2 and trnL-CAA was 988 bp, 993 bp, and 1023 bp, respectively.
Figure 4.
Comparative analysis of alterations resulting from the ycf68, and ycf15 deletion in Wolffia arrhiza. This represents one of the IR regions, and the other IR region exhibits a same aspect with reverse complementarity. The squares represent genes, where those transcribed on the forward strand are positioned at the top of the line, and those transcribed on the reverse strand are located at the bottom of the line. The numbers adjacent to the squares represent the lengths of individual genes, while the numbers above the lines are the lengths of IGS between each gene. ycf2 is depicted by the color red, ycf15 by orange, trnL-CAA by yellow, trnI-GAU by green, and ycf68 by blue.
To identify variations in introns, the gap between the longest and shortest length values for each species within the same gene was calculated (Table S2). As a result, it was determined that the lengths of the petB and rpl16 genes in W. arrhiza are 1400 bp and 1983 bp, respectively. These lengths exceed twice the sizes observed in other species, where they typically range from 642 to 701 bp for petB and 411 bp (the exception of Lemna minor, which has a length of 1714 bp) for rpl16. When analyzing the exons and introns of these genes in each species, the exons exhibit consistent lengths across all species (ranging from 642 to 654 bp for petB and 408–411 bp for rpl16). However, in W. arrhiza, the introns are notably longer, with the petB gene containing a 752 bp intron and the rpl16 gene containing a 1575 bp intron (Table S3).
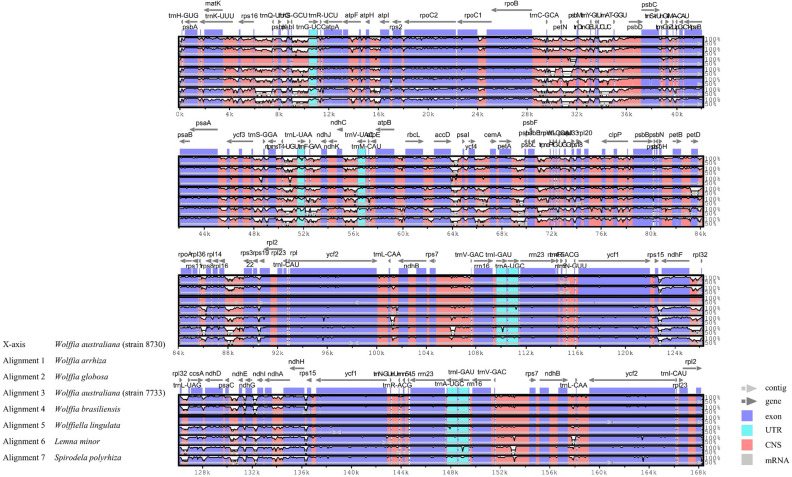
Sequence homology among the chloroplast genomes of seven species within the Lemnoideae subfamily was assessed and visualized via the shuffle-LAGAN mode in mVista. The annotation data relied upon the reference strain, W. australiana (strain 8730). As a result, it was established that the chloroplast genome sequence of duckweed maintains a high degree of sequence conservation, with very few regions exhibiting sequence identity below 90% (Fig. 5). In detail, the IR region showed a higher level of preservation when contrasted with the LSC and SSC regions. In addition, the mutation rate was greater in the IGS region in contrast to the PCG region. The majority of PCGs were generally well-preserved, but significant variations were observed in some PCGs, including matK, rpoC2, ndhF, cssA, ndhD, and ndhH. In contrast to the PCG regions, the non-coding regions showed a relatively higher mutation rate in numerous locations. Within non-coding regions, intergenic regions displayed the highest variability rate. Upon visual examination of the figure, the most notable segments appeared to be trnC(GCA)-petN, petN-psbM, and trnE(UUC)-trnT(GGU).
Figure 5.
Analyzed the chloroplast genome sequences of seven Lemnoideae species, including Wolffia arrhiza, using mVista, with Wolffia australiana as the reference. The X-axis represents the coordinates of the chloroplast genome sequence position, while the Y-axis indicates the range of sequence identity from 50 to 100%. The direction and position of the genes are depicted by the gray arrows on the graph. The graph's shaded colors have the following meanings: the dark blue regions correspond to protein coding sequences (CDS), the pink regions represent Conserved Non-Coding Sequences (CNS), and the light-blue regions indicate UTRs.
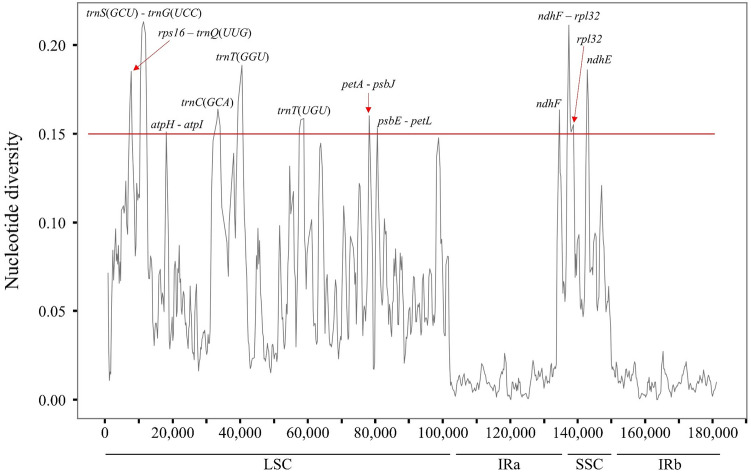
To clearly identify the variable regions within the mVista results, a sliding window analysis was executed using DnaSP v.6.10 software, followed by the calculation of nucleotide diversity values (π, Pi). There were 769 nucleotide diversity point observed, with values ranging from 0.00000 to 0.21294, and an average value of 0.04589 (Fig. 6). The nucleotide diversity value was highest (0.21294) in the LSC region, while the IR region had the lowest value (0.00048), excluding zero. In this regard, the IR region exhibited significantly lower variability compared to the LSC and SSC regions. Among them, 12 locations demonstrated high Pi values greater than 0.15. Eight of them were found in the LSC region, while four were located in the SSC region. Within the LSC region, 5 locations were detected in intergenic regions including rps16-trnQ(UUG), trnS(GCU)-trnG(UCC), atpH-atpI, petA-psbJ, psbE-petL, while 3 locations were found in the coding regions of trnC(GCA), trnT(GGU), and trnT(UGU). In the SSC region, one of the four positions was located in the intergenic region of ndhF-rpl32, whereas the other three were found in the coding regions of ndhF, rpl32, and ndhE. The coding region and non-coding region with the highest nucleotide diversity values were trnT(GGU) (0.18841) and trnS(GCU)-trnG(UCC) (0.21294), respectively, located in the LSC.
Figure 6.
Nucleotide diversity of chloroplast genome sequences in Lemnoideae, including Wolffia arrhiza. The X-axis represents the alignment sequence's position, while the Y-axis indicates the values for nucleotide diversity. The use of a hyphen to connect two genes signifies a non-coding region, whereas the representation of a single gene indicates a coding region.
To delve deeper into the nucleotide diversity results, SNPs and InDels were analyzed in seven Lemnoideae species, utilizing Wolffia australiana as a reference. This revealed 17,269 SNPs and 2030 InDels. The majority of SNPs appeared in the IGS regions (51.91%), followed by exon regions (35.20%) and intron regions (12.89%) (Table S6). These findings aligned with earlier results (Fig. 6), particularly noting that the IGS in trnS(GCU)—trnG(UCC) represented 5.30% of the total IGS region, and the IGS in ndhF—rpl32 comprised 4.03% of the total IGS region. Additionally, InDels were predominantly distributed in IGS regions (76.35%), with lesser occurrences in intron (17.10%) and exon regions (6.55%) (Table S7). Most were short InDels of 10 base pairs or fewer, accounting for 80.59% of the total, and one long InDel of 1000 bp was detected. Similarly with SNPs aspect, the IGS regions in trnS(GCU)—trnG(UCC), and ndhF—rpl32, constituted 4.54% and 1.86% of the total IGS region, respectively.
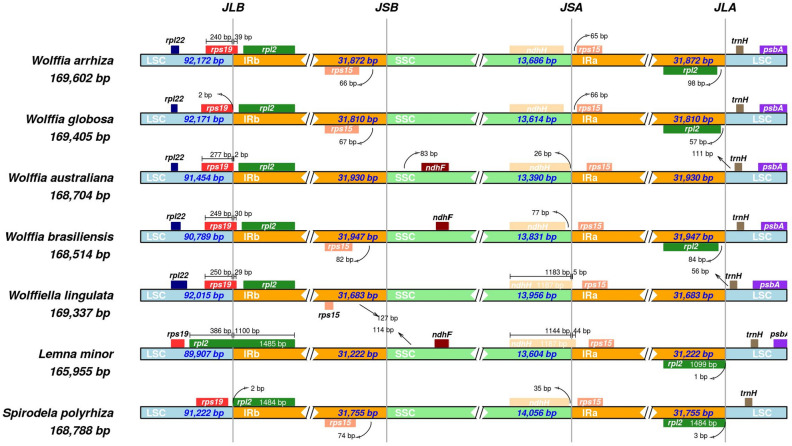
The gene distribution at the boundaries of the LSC/SSC and IR regions in the chloroplast genomes of the seven species was compared using IRscope. Overall, the distribution of genes at each boundary region appeared to be similar, with rpl22, rps19, rpl2, rps15, ndhF, ndhH, trnH, and psbA. However, it was observed that the rpl2 gene is found solely in the IRb region and is absent in the IRa region of W. australiana and Wolffiella lingulata (Fig. 7, Table S5). Although not shown in the figure due to their location at the boundaries and greater distance, other genes did not undergo any loss. Nevertheless, variations were noted in the association between genes and the boundary lines. The JLB (LSC/IRB) boundary displayed three different configurations: positioned within the rps19 gene, within the rpl2 gene, or within IGS between the rps19 and rpl2 genes. For W. arrhiza, the JLB boundary can be found within the rps19 gene. The rps19 gene spans 240 bp in the LSC region, and the remaining 39 bp extend into the IRB region. Similar cases are apparent in the W. australiana, W. brasiliensis, and Wolffiella lingulata. These species have respectively occupied 277 bp, 249 bp, and 250 bp within the LSC region, along with extensions of 2 bp, 30 bp, and 29 bp into the IRB. In the case of Lemna minor, it was observed that the boundary of the JLB was located within the rpl2 gene. The rpl2 gene spanned 1100 bp within the IRB region, with the remaining 386 bp extending towards the LSC region. W. globosa and Spirodela polyrhiza were both found to have the JLB boundary positioned within the IGS between the rps19 and rpl2 genes. Additionally, the rps19 and rpl2 genes were identified within the LSC and IRB regions, respectively. In the instance of the JSA (SSC/IRA) boundary, it presented in two different cases, with one found within the ndhH gene and the other within the IGS region lying between the ndhH and rps15 genes. For W. arrhiza, the ndhH and rps15 genes were contained within their respective SSC and IRA regions, rather than extending beyond them. This same pattern was also observed in W. globosa, W. australiana, W. brasiliensis, and Spirodela polyrhiza. Nevertheless, for Wolffiella lingulata and Lemna minor, the boundary of the JSA was positioned within the ndhH gene, with each extension 1183 bp and 1144 bp into the SSC region, while the remaining 5 bp and 44 bp entered the IRA region.
Figure 7.
Comparing the boundaries of chloroplast genome regions in seven species, including Wolffia arrhiza, from the Lemnoideae subfamily: LSC, IRs, and SSC. The junctions between each pair of genomic regions are indicated as JLB (LSC/IRB), JSB (SSC/IRB), JSA (SSC/IRA), and JLA (LSC/IRA). Genes transcribed on the forward strand are depicted above the line, whereas genes transcribed on the reverse strand are exhibited below the line. Furthermore, the numbers above the genes signify the gap between the gene's start or end and the region's boundary.
Phylogenetic analysis
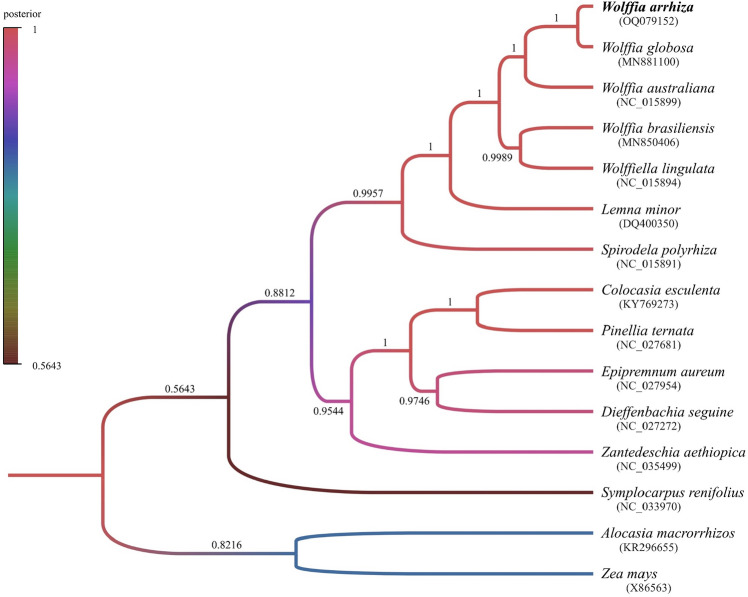
To explore the phylogenetic relationship of W. arrhiza, a phylogenetic tree was created that included a total of 15 species. This species set comprised 7 species from the Lemnoideae subfamily, which includes W. arrhiza, and 7 species within the Araceae family to which Lemnoideae subfamily belongs, along with one outgroup species, Zea mays. Excluding genes in the IR, there are 44 PCGs shared between them (Table S4). In addition, based on the results from Prottest, the CpREV + G + I model was determined to be the best fit model for explaining protein evolution across the 15 species. The Bayesian analysis was performed using BEAST v1.10.4 software, employing 50,000,000 Markov Chain Monte Carlo (MCMC) chains with the previously identified shared PCG and the most suitable model. Consequently, the phylogenetic tree was constructed, with Bayesian posterior probability values that ranged from 0.5643 to 1 (Fig. 8). The tree can be divided into three parts: W. arrhiza—Spirodela polyrhiza, Colocasia esculenta—Symplocarpus renifolius, and the outgroup. W. arrhiza is classified as part of the W. arrhiza—Spirodela polyrhiza section, belonging to the Lemnoideae subfamily. It exhibits the nearest evolutionary relationship with W. globosa.
Figure 8.
Bayesian phylogenetic tree of Araceae species based on the chloroplast genome data. The colors of the branches in the tree represent the Bayesian posterior probability, as indicated by the color bar. The numerical values displayed on the branches represent precise posterior values. At the bottom of each species name is the genbank accession number.
Discussion
The complete chloroplast genome of W. arrhiza was assembled using the short reads from MGI DNBSEQ-G50 next-generation sequencing platform, revealing a common circular quadripartite genomic structure. Despite W. arrhiza is an aquatic plant, it exhibited a similar pattern to land plants, having a length of 169,602 bp falling within the range of 130–170 kbp and a GC content of around 35.78%, which is in proximity to the average of 36.3% in land plants31. Additionally, the analysis of SSRs indicated that in W. arrhiza, SSRs are composed exclusively of A or T nucleotides, predominantly distributed in the LSC region out of the four regions (LSC, SSC, IRs) and in the IGS among the three locations (IGS, PCG, intron) (Table 4). A strong A/T bias and high concentration within the LSC region and IGS were similar to patterns observed in other angiosperms chloroplast genome32–34. The analysis of codon usage revealed that all but one of the codons with RSCU values exceeding 1 ended in either A or T (Table S1). This corresponds with research results indicating a preference for A/T-ending codons among synonymous codons, as it offers advantages in codon optimization during gene expression compared to codons ending in C/G35. Codon usage patterns are fundamentally dependent on the gene sequences, thereby mirroring nucleotide substitutions that lead to variations of gene, protein structure, and function. Consequently, by understanding codon usage, it was possible to gain a foundational framework for insights into the history of W. arrhiza in the context of genome evolution36.
The analysis of genome information for the Lemnoideae subfamily, including W. arrhiza, was conducted to identify the various changes that occurred during evolution. First, diversity within the Lemnoideae subfamily, including W. arrhiza, was analyzed at the nucleotide level, and a total of 12 hotspots were detected. There were several peaks in the data with significantly high Pi values, such as trnS(GCU)-trnG(GCC) (0.21294)37, rps16-trnQ(UUG) (0.18500)38, and ndhF-rpl32 (0.19456)39, indicating a substantial nucleotide variability in other angiosperm plants. One can surmise that these genes are inclined to undergo rapid nucleotide substitutions, suggesting at their applicability as molecular markers for phylogenetic analysis and species identification. Subsequently, within the Lemnoideae, it was confirmed that W. arrhiza had the longest chloroplast genome length among its fellow species (Table S5). Various elements influencing chloroplast length have been reported as the contraction and expansion of the IR region, gene insertions, deletions, duplications, inversions, and alterations in introns40–43. In the case of W. arrhiza, the JLB and JSA boundaries were positioned similarly to most other Lemnoideae species. There was no noticeable expansion or contraction in the IR region, which was 31,872 bp in size. This falls within the average IR range of 31,223 to 31,930 bp observed in other species (Fig. 7 and Table S5). To explore whether there were additional alterations, such as gene rearrangements or inversions, a synteny analysis of the Lemnoideae chloroplast genome was performed utilizing the MAUVE program44. The seven Lemnoideae species visually demonstrated a highly similar gene position pattern, suggesting a significant level of collinearity (Fig. S1). The events such as the deletion of ycf68 and ycf15, resulted in changes to the types and number of genes (Fig. 4). However, they could not be considered to have had a significant impact on the length of the whole chloroplast genome. On the other hand, when comparing the lengths of genes, CDS, introns, and intergenic regions, there was a noticeable increase in the total intron length (Fig. 3). This suggests that intron evolution may have occurred in the W. arrhiza chloroplast genome. As a result, significant alterations were identified in the intron analysis of petB and rpl16 showing from 642–701 bp to 1400 bp, and from 411 to 1983 bp, respectively (Tables S2, S3). While the exact reasons for the insertion of these introns require additional exploration, it implied the significance of both the petB and rpl16 genes. From this, it can be inferred that changes in introns impacted the chloroplast genome length of W. arrhiza, resulting in it possessing the longest genome length among Lemnoideae species. Additionally, the alterations in introns were commonly detected in various angiosperms, indicating their potential utility as molecular markers for conducting phylogenetic studies and identifying different species45–49.
The chloroplast, which contains all of this information, was used to explore the phylogenetic history of the species. The phylogenetic tree was partitioned into three sections: W. arrhiza—Spirodela polyrhiza, Colocasia esculenta—Symplocarpus renifolius, and the outgroup (Fig. 8, Table S4). In the W. arrhiza—Spirodela polyrhiza section, the majority of posterior values were 1.0, providing a strong representation of the Lemnoideae subfamily lineage. Specifically, the relationship between W. arrhiza and W. globosa is affirmed with a posterior value of 1, signifying that they are sister species. W. brasiliensis showed unclear taxonomic results that did not align with the same genus, Wolffia, as observed in previous research8,9,23. Nevertheless, through conducting a phylogenetic classification of the Lemnoideae subfamily, which incorporated W. arrhiza, valuable insights into the evolutionary dynamics affecting populations and species were gained.
Conclusion
In this study, an analysis of the complete chloroplast genome of W. arrhiza is provided. The chloroplast genome of W. arrhiza exhibits resemblances in terms of size, structure, gene composition, GC content, and codon preferences when compared to the typical characteristics observed in land plants and angiosperms. The comparison of Lemnoideae species with W. arrhiza also demonstrates a significant level of conservation in the chloroplast genome. It also offers details about genes containing nucleotide or intron variations that can be used as molecular markers in the species classification and evolutionary research. The phylogenetic analysis verified that W. arrhiza is closely related as a sister species to W. globosa. In summary, the characterization of chloroplast genomic data for W. arrhiza has provided insights and enriched understanding of the phylogeny of the challenging-to-classify Lemnoideae subfamily using traditional methods.
Materials and methods
Plant materials, DNA extraction, and sequencing
The samples of W. arrhiza were obtained from the Rutgers Duckweed Stock Cooperative (RDSC) located at Rutgers University in New Jersey (http://www.ruduckweed.org/, ruduckweed@gmail.com). The plant collection and use were in accordance with all the relevant guidelines. Among several strains of W. arrhiza, voucher number of 7193 was used, which was collected in Masaka, Uganda, located at 0°19′36.3″ S 31°45′13.5″ E. Total genomic DNA was isolated from the whole plant by the modified cetyltrimethylammonium bromide (CTAB) method and quantified by Nanodrop spectrophotometer (Thermo Fisher Scientific, ND-1000) and Qubit Fluorometer (Invitrogen, Thermo Fisher Scientific, Qubit 4). The paired-end libraries were constructed using the MGI Eazy FS DNA Library Prep Kit, with an insert length of 350 bp. Sequencing was performed using reads of 150 bp on the MGI DNBSEQ-G50 second-generation sequencing platform, resulting in a total of approximately 37.82 GB of raw reads being generated.
Chloroplast genome assembly and annotations
GetOrganelle v1.7.5.350 was utilized to assemble the chloroplast genome using following command (get_organelle_from_reads.py -1 left.fq -2 right.fq -k 21,45,65,85,105 -t 3 -o result_folder -F embplant_pt), and annotation was done with GeSeq51. The circular maps for newly sequenced plastomes were generated using the OGDRAW v1.3.152.
Repeat sequences analysis
The web application MISA53 was utilized to detect microsatellites, known as simple sequence repeats (SSRs). The minimum number of repetitions were set as follows: 10 repeat units for mononucleotide SSRs, 6 repeat units for dinucleotide SSRs, and 5 repeat units for tri-, tetra-, penta-, and hexanucleotide SSRs. Furthermore, the web application REPuter54 was used to identify long repeats. This process encompassed detecting forward, reverse, complement, and palindromic repeats with a minimum repeat size set at 30 bp and a Hamming distance of three.
Codon usage
The chloroplast genome's Protein-Coding Genes (PCGs) and their protein coding sequences (CDS) were extracted from the genbank file using Python's SeqIO object. Following that, the Relative Synonymous Codon Usage (RSCU) was calculated using codonW v1.4.455. Synonymous codons refer to codons encoding the same amino acid, and RSCU assesses the relative frequency of such synonymous codons. An RSCU value greater than 1.00 signifies a relatively higher frequency of codon usage, while values less than 1.00 indicate the opposite.
Chloroplast genome comparison
The tool mVISTA56, with the Shuffle-LAGAN mode, was used to identify variations of whole chloroplast genome sequences using Wolffia australiana (MN912638.1, strain 8730) as the reference. It visually represents the similarities and differences among seven species, namely Wolffia arrhiza (OQ079152.1), Wolffia globosa (MN881100.1), Wolffia australiana (NC_015899.1, strain 7733), Wolffia brasiliensis (MN850406.1), Wolffiella lingulata (NC_015894.1), Lemna minor (DQ400350.1), and Spirodela polyrhiza (NC_015891.1). This entailed utilizing reference information as annotation notes to gain a deeper comprehension of the observed patterns. Additionally, the genome sequences of seven chloroplasts were aligned using the multiple sequence alignment program, MAFFT v7.52057. In this alignment, nucleotide diversity (π, Pi) was calculated using DnaSP (DNA Sequences Polymorphism) v6.12.0358, employing a window length of 600 bp and a step size of 200 bp. It was also utilized for the calling of SNPs and InDels. The visualization and comparison of the genes located at the junctions of chloroplast genomes in the seven species were carried out using the web application IRscope59.
Phylogenetic analysis
The phylogenetic analysis encompassed a total of 14 species from the Araceae family, which included W. arrhiza. To facilitate the comparative analysis, Zea mays was chosen as the outgroup. The chloroplast genome information was acquired from GenBank, and Python's SeqIO object was employed for parsing and extracting the protein-coding genes and their corresponding protein sequences, which were shared among 15 species. Afterward, the alignment process was conducted using PRANK60, and to select the most suitable amino acid substitution model for the alignment data, ProtTest61 was employed. The Beast v1.10.4 software62 performed Bayesian-based evolutionary analysis, with the CpREV + G + I substitution model, Yule model for prior tree, and the uncorrelated relaxed clock, widely regarded as the most suitable model for datasets at the species level63. The Markov Chain Monte Carlo (MCMC) was also set for 50,000,000 generations. Following that, a maximum credibility tree was constructed using TreeAnnotator v1.10.4, discarding the initial 10% of trees as burn-in using Tracer v1.7.1. The phylogenetic tree was then created using FigTree v1.4.4.
Supplementary Information
Acknowledgements
We would like to express our thanks to the professors in the Division of Bio & Medical Big Data Department at Gyeongsang National University for their priceless advice and assistance during this research project.
Author contributions
H.P. and Y.J.K.conceptualized and designed this research project. H.P. and J.H.P. collected samples and conducted DNA extraction and sequencing. H.P. assembled, annotated, and analyzed the sequencing data, and wrote the manuscript. All authors read and approved the manuscript and agreed to be accountable for all aspects of the work.
Funding
This work was supported by a grant from the "Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01627602)" from the Rural Development Administration, Republic of Korea and was supported by Learning & Academic research institution for Master’s·PhD students, and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (RS-2023-00301974).
Data availability
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under the Accession No. OQ079152.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-56394-7.
References
- 1.Landolt, E. Biosystematic investigations in the family of duckweeds (Lemnaceae). II: The family of Lemnaceae: a monographic study. 1. Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Rübel, Zürich (1986).
- 2.Landolt, E. Taxonomy and ecology of the section Wolffia of the genus Wolffia (Lemnaceae). Ber. Geobot. Inst. ETH, Stiftung Rübel, Zürich60, 137–151 (1994).
- 3.Xu Y, et al. Species distribution, genetic diversity and barcoding in the duckweed family (Lemnaceae) Hydrobiologia. 2015;743:75–87. doi: 10.1007/s10750-014-2014-2. [DOI] [Google Scholar]
- 4.Cheng JJ, Stomp AM. Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean-Soil Air Water. 2009;37:17–26. doi: 10.1002/clen.200800210. [DOI] [Google Scholar]
- 5.Bog M, et al. A taxonomic revision of Lemna sect: Uninerves (Lemnaceae) Taxon. 2020;69:56–66. doi: 10.1002/tax.12188. [DOI] [Google Scholar]
- 6.Bog M, et al. Key to the determination of taxa of Lemnaceae: An update. Nordic J. Botany. 2020 doi: 10.1111/njb.02658. [DOI] [Google Scholar]
- 7.Les DH. Aquatic Monocotyledons of North America: Ecology, Life History, and Systematics. CRC Press; 2020. [Google Scholar]
- 8.Les DH, et al. Phylogeny and systematics of Lemnaceae, the duckweed family. Syst. Botany. 2002;27:221–240. [Google Scholar]
- 9.Tippery N, et al. Evaluation of phylogenetic relationships in Lemnaceae using nuclear ribosomal data. Plant Biol. 2015;17:50–58. doi: 10.1111/plb.12203. [DOI] [PubMed] [Google Scholar]
- 10.Les DH, et al. Systematics of the Lemnaceae (duckweeds): Inferences from micromolecular and morphological data. Plant Syst. Evol. 1997;204:161–177. doi: 10.1007/BF00989203. [DOI] [Google Scholar]
- 11.Pan, S. & Chen, S. Morphology of Wolffia arrhiza: a scanning electron microscopic study. Botanical bulletin of Academia Sinica. New series (1979).
- 12.Appenroth KJ, et al. Telling duckweed apart: Genotyping technologies for the Lemnaceae. Chin. J. Appl. Environ. Biol. 2013;19:1–10. doi: 10.3724/SP.J.1145.2013.00001. [DOI] [Google Scholar]
- 13.Yang J, et al. Frond architecture of the rootless duckweed Wolffia globosa. BMC Plant Biol. 2021;21:1–10. doi: 10.1186/s12870-021-03165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iqbal, S. Duckweed aquaculture. Potentials, possibilities and limitations for combined wastewater treatment and animal feed production in developing countries. SAn-DEC Report (1999).
- 15.Skillicorn P, et al. Duckweed Aquaculture: A New Aquatic Farming System for Developing Countries. World Bank; 1993. [Google Scholar]
- 16.Ansal M, et al. Duckweed based bio-remediation of village ponds: An ecologically and economically viable integrated approach for rural development through aquaculture. Livestock Res. Rural Dev. 2010;22:129. [Google Scholar]
- 17.Jorgensen SE, Fath BD. Encyclopedia of Ecology. Elsevier; 2008. [Google Scholar]
- 18.Fujita M, et al. Nutrient removal and starch production through cultivation of Wolffia arrhiza. J. Biosci. Bioeng. 1999;87:194–198. doi: 10.1016/S1389-1723(99)89012-4. [DOI] [PubMed] [Google Scholar]
- 19.Czerpak R, Szamrej I. The effect of β-estradiol and corticosteroids on chlorophylls and carotenoids content in Wolffia arrhiza (L.) Wimm.(Lemnaceae growing in municipal Bialystok tap Tap T water. Pol. J. Environ. Stud. 2003;12:677. [Google Scholar]
- 20.Körner S, et al. The capacity of duckweed to treat wastewater: ecological considerations for a sound design. J. Environ. Qual. 2003;32:1583–1590. doi: 10.2134/jeq2003.1583. [DOI] [PubMed] [Google Scholar]
- 21.Bhanthumnavin K, Mcgarry MG. Wolffia arrhiza as a possible source of inexpensive protein. Nature. 1971;232:495–495. doi: 10.1038/232495a0. [DOI] [PubMed] [Google Scholar]
- 22.Chareontesprasit, N. & Jiwyam, W. An evaluation of Wolffia meal (Wolffia arrhiza) in replacing soybean meal in some formulated rations of Nile tilapia (Oreochromis niloticus L.). (2001).
- 23.Wang W, et al. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010;10:1–11. doi: 10.1186/1471-2229-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borisjuk N, et al. Assessment, validation and deployment strategy of a two-barcode protocol for facile genotyping of duckweed species. Plant Biol. 2015;17:42–49. doi: 10.1111/plb.12229. [DOI] [PubMed] [Google Scholar]
- 25.Bog M, et al. Duckweed (Lemnaceae): Its molecular taxonomy. Front. Sustain. Food Syst. 2019;3:117. doi: 10.3389/fsufs.2019.00117. [DOI] [Google Scholar]
- 26.Palmer, J. D. et al. Chloroplast DNA variation and plant phylogeny. Ann. Missouri Botanical Garden, 1180–1206 (1988).
- 27.Li X, et al. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 28.Nauheimer L, et al. Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol. 2012;195:938–950. doi: 10.1111/j.1469-8137.2012.04220.x. [DOI] [PubMed] [Google Scholar]
- 29.Les, D. H. & Crawford, D. J. Landoltia (Lemnaceae), a new genus of duckweeds. Novon, 530–533 (1999).
- 30.Mardanov AV, et al. Complete sequence of the duckweed (Lemna minor) chloroplast genome: Structural organization and phylogenetic relationships to other angiosperms. J. Mol. Evol. 2008;66:555–564. doi: 10.1007/s00239-008-9091-7. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y-Y, et al. Chloroplast genomes of two species of Cypripedium: Expanded genome size and proliferation of AT-biased repeat sequences. Front. Plant Sci. 2021;12:609729. doi: 10.3389/fpls.2021.609729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton BR, Clegg MT. Neighboring base composition is strongly correlated with base substitution bias in a region of the chloroplast genome. J. Mol. Evol. 1995;41:597–603. doi: 10.1007/BF00175818. [DOI] [PubMed] [Google Scholar]
- 33.Morton, B. R. The influence of neighboring base composition on substitutions in plant chloroplast coding sequences. (1997).
- 34.Zhang R-S, et al. A high level of chloroplast genome sequence variability in the Sawtooth Oak Quercus acutissima. Int. J. Biol. Macromol. 2020;152:340–348. doi: 10.1016/j.ijbiomac.2020.02.201. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, et al. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ. 2020;8:e8251. doi: 10.7717/peerj.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wicke S, et al. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol. Biol. Evol. 2014;31:529–545. doi: 10.1093/molbev/mst261. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, et al. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022;22:1–21. doi: 10.1186/s12870-022-03529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y, et al. Development of chloroplast genomic resources for Oryza species discrimination. Front. Plant Sci. 2017;8:1854. doi: 10.3389/fpls.2017.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw J, et al. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- 40.Tsudzuki J, et al. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: Nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol. Gener. Genet. MGG. 1992;232:206–214. doi: 10.1007/BF00279998. [DOI] [PubMed] [Google Scholar]
- 41.Lin C-P, et al. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol. 2012;4:374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White E. Chloroplast DNA in Pinus monticola: 1 Physical map. Theor. Appl. Genet. 1990;79:119–124. doi: 10.1007/BF00223797. [DOI] [PubMed] [Google Scholar]
- 43.Li C, et al. Initial characterization of the chloroplast genome of Vicia sepium, an important wild resource plant, and related inferences about its evolution. Front. Genet. 2020;11:73. doi: 10.3389/fgene.2020.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling AC, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butterworth CA, et al. Molecular systematics of tribe Cacteae (Cactaceae: Cactoideae): A phylogeny based on rpl16 intron sequence variation. Syst. Botany. 2002;27:257–270. [Google Scholar]
- 46.Kelchner SA, Clark LG. Molecular evolution and phylogenetic utility of the chloroplast rpl16 intron in Chusquea and the Bambusoideae (poaceae) Mol. Phylogen. Evol. 1997;8:385–397. doi: 10.1006/mpev.1997.0432. [DOI] [PubMed] [Google Scholar]
- 47.Downie SR, et al. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: Towards a suprageneric classification of subfamily Apioideae. Am. J. Botany. 2000;87:273–292. doi: 10.2307/2656915. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W. Phylogeny of the grass family (Poaceae) from rpl16 intron sequence data. Mol. Phylogen. Evol. 2000;15:135–146. doi: 10.1006/mpev.1999.0729. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda T, et al. Molecular phylogeny of the genus Asparagus (Asparagaceae) inferred from plastid petB intron and petD–rpoA intergenic spacer sequences. Plant Species Biol. 2005;20:121–132. doi: 10.1111/j.1442-1984.2005.00131.x. [DOI] [Google Scholar]
- 50.Jin J-J, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:1–31. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tillich M, et al. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greiner S, et al. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beier S, et al. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurtz S, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peden, J. F. Analysis of codon usage. (2000).
- 56.Frazer KA, et al. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rozas J, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 59.Amiryousefi A, et al. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 60.Löytynoja, A. Phylogeny-aware alignment with PRANK. Multiple sequence alignment methods, 155–170 (2014). [DOI] [PubMed]
- 61.Darriba D, et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drummond AJ, et al. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under the Accession No. OQ079152.