Abstract
As part of a cDNA library screen for clones that induce transformation of NIH 3T3 fibroblasts, we have isolated a cDNA encoding the murine homolog of the guanine nucleotide exchange factor RasGRP. A point mutation predicted to prevent interaction with Ras abolished the ability of murine RasGRP (mRasGRP) to transform fibroblasts and to activate mitogen-activated protein kinases (MAP kinases). MAP kinase activation via mRasGRP was enhanced by coexpression of H-, K-, and N-Ras and was partially suppressed by coexpression of dominant negative forms of H- and K-Ras. The C terminus of mRasGRP contains a pair of EF hands and a C1 domain which is very similar to the phorbol ester- and diacylglycerol-binding C1 domains of protein kinase Cs. The EF hands could be deleted without affecting the ability of mRasGRP to transform NIH 3T3 cells. In contrast, deletion of the C1 domain or an adjacent cluster of basic amino acids eliminated the transforming activity of mRasGRP. Transformation and MAP kinase activation via mRasGRP were restored if the deleted C1 domain was replaced either by a membrane-localizing prenylation signal or by a diacylglycerol- and phorbol ester-binding C1 domain of protein kinase C. The transforming activity of mRasGRP could be regulated by phorbol ester when serum concentrations were low, and this effect of phorbol ester was dependent on the C1 domain of mRasGRP. The C1 domain could also confer phorbol myristate acetate-regulated transforming activity on a prenylation-defective mutant of K-Ras. The C1 domain mediated the translocation of mRasGRP to cell membranes in response to either phorbol ester or serum stimulation. These results suggest that the primary mechanism of activation of mRasGRP in fibroblasts is through its recruitment to diacylglycerol-enriched membranes. mRasGRP is expressed in lymphoid tissues and the brain, as well as in some lymphoid cell lines. In these cells, RasGRP has the potential to serve as a direct link between receptors which stimulate diacylglycerol-generating phospholipase Cs and the activation of Ras.
The Ras family of small GTPases (5) is comprised of the classical Ras GTPases (H-Ras, N-Ras, and K-Ras, henceforth collectively referred to as Ras) as well as a more divergent group of Ras-related proteins (TC21, R-Ras, R-Ras3, and the Rals and Raps). By oscillating between GDP-bound forms, which are inert, and GTP-bound forms, which are able to bind and activate multiple effector proteins, these GTPases can serve as switches for the propagation and divergence of signalling pathways. The critical role of Ras in communicating signals from cell surface receptors to intracellular kinases has now been established. Ras is rapidly converted to its GTP-bound form in response to the ligation of many different types of signalling receptors on the cell surface (18). Once GTP bound, Ras can bind to the Raf kinase, bringing it up to the membrane where it can be activated by tyrosine and/or serine phosphorylation by kinases in membrane-localized signalling complexes (46). Raf then initiates a kinase cascade through MEK, ERK1, and ERK2 (56), leading to the phosphorylation and activation of transcription factors such as Elk-1 (61). GTP-bound Ras also binds to and activates phosphatidylinositol 3-kinase, which may lead to the activation of GTPases of the Rho family (57), and binds to and activates RalGDS, thus potentially leading to the activation of Ral GTPases (24). The Ras-related proteins TC21, R-Ras, R-Ras3, and Ral have been relatively neglected, but the available evidence implicates them as collaborators of Ras in signal transduction, and, as suggested for Ral, they may be downstream effectors of Ras which serve to propagate and diversify signalling via GTPase cascades (5, 24, 29). The Rap GTPases are distinct in that they seem to act as repressors of Ras function, possibly by competing with Ras for Raf binding (32).
Like other small GTPases, the Ras family members are themselves controlled by proteins which modulate their nucleotide binding state or their intrinsic GTPase activity. Ras activation appears to be determined largely by specific guanine nucleotide exchange factors (GEFs), which promote the release of GDP from Ras and thus facilitate their conversion to the GTP-bound state (4, 23, 55). A common mechanism of activation of these GEFs is via their translocation to membranes, which is assumed to work simply by bringing the GEF into contact with membrane-bound GTPase targets. Several mammalian GEFs capable of activating Ras or Ras-related GTPases have been identified so far. All of these GEFs have a common GEF domain, which houses their guanine nucleotide exchange activities. Sos1 and Sos2 are a pair of very closely related GEFs which act only on Ras (11). The Sos GEFs are drawn to the membrane in response to tyrosine kinase activation by their interaction with adapter proteins, e.g., Grb2, which binds to phosphorylated tyrosines via its SH2 domain and binds to a proline cluster on Sos via its SH3 domains (18). RasGRF-1 and RasGRF-2 comprise a second class of GEFs which are related to Sos both within the GEF domain and in the possession of PH and DH domains (9, 21, 59). Like the two forms of Sos, both RasGRFs act on Ras. In addition, RasGRF-1 can serve as a GEF for R-Ras (28). The RasGRFs appear to be specialized for activating Ras in response to calcium signalling, via their calmodulin-binding IQ motifs (21, 22). RGL, RLF, and RalGDS are a third group of GEFs, which are homologous to Sos in the GEF catalytic domain but are otherwise quite different in sequence. RalGDS is a GEF specific for the Ral GTPases (1) and is activated by binding to Ras, thus directly coupling Ras activation and Ral activation (24). C3G is a GEF for R-Ras and Rap (27, 28). Its similarity to Sos and the other GEFs is also confined to the GEF domain. It forms complexes with the adapter protein Crk and may thereby be activated in response to tyrosine kinase-coupled receptors.
In lymphocytes, the classical Ras GTPases are activated following ligation of the T-cell receptor or B-cell receptor, as well as by receptors for some cytokines and costimulators (18). All lymphocytes probably express Sos, and this GEF has been implicated in Ras activation occurring downstream of T-cell and B-cell receptors (31, 39, 58, 60). However, in at least some circumstances, T-cell receptor signalling that leads to Ras activation does not seem to result in recruitment of Sos to receptor complexes (8, 49). RasGRF-1 is not expressed by lymphocytes. The expression of RasGRF-2 is much more widespread (21), although it has not yet been determined if this GEF is expressed by lymphocytes. Thus, it is not clear whether the known GEFs are able to mediate all Ras activation events in lymphocytes or whether additional Ras-specific GEFs are required.
In a general attempt to identify additional participants in signalling pathways leading to cell proliferation, we have screened cDNA libraries for clones whose expression causes loss of contact inhibition and morphological transformation of fibroblast cell lines (63). Most of the clones isolated in these screens have been known or novel upstream activators or downstream effectors of Ras or Rho family GTPases. This paper describes the selection from a murine T-cell line of a strongly transforming cDNA which encodes RasGRP, a Ras-specific GEF recently identified via a similar screen for transforming cDNAs derived from rat brain (19). We show that the ability of murine RasGRP (mRasGRP) to transform fibroblasts and activate mitogen-activated protein kinases (MAP kinases) is dependent on its GEF and REM domains, with the C1 domain playing an essential role in mediating the translocation of mRasGRP to cell membranes.
MATERIALS AND METHODS
Cell lines.
T28 cells (53) and BOSC 23 retroviral packaging cells (52) (derived from the human epithelial cell line 293T) were cultured in Dulbecco’s modified Eagle’s medium (DME) supplemented with 10% fetal calf serum. NIH 3T3 cells were obtained from the American Type Culture Collection and cultured at low density in DME containing 9% calf serum.
cDNA library synthesis, viral transmission, and screening.
The vectors and methodology for the construction of cDNA libraries, their conversion to retroviral form, infection of NIH 3T3 cells, selection of transformed cell clones, recovery of proviral cDNAs, and secondary screening for transforming cDNA clones have been described previously (63). One important modification used in the cloning of the CXR-CT cDNA was the use of BOSC 23 packaging cells for production of retroviral libraries, as these cells produce much higher library titers than do the NIH 3T3-derived packaging lines that had been used previously. Transfection of BOSC 23 cells was via calcium phosphate precipitates, as described previously (52). Details relevant to the screening of the T28 library are given below.
Total mRNA from exponentially growing T28 cells was used as a template for cDNA synthesis with random hexamer primers. The cDNA was ligated into pCTV1B, yielding 3.5 × 106 cDNA clones. An estimated 106 retroviruses were produced from this library by transient transfection into BOSC 23 cells and were used to infect 106 NIH 3T3 cells plated at very low density. After 2 weeks of culture, transformed cell foci were picked, and proviral cDNAs were recovered by PCR, using a mixture of Taq polymerase and Pfu polymerase to reduce the frequency of rearrangements and mutations in the PCR products (3), and recloned into pCTV3. After conversion to retrovirus, these clones were tested for transforming activity by infecting NIH 3T3 cells. Positive clones were recloned by purification of the cDNA insert and insertion into pCTV3 or related retroviral vectors and were retested for transforming activity. This ensures that transformation is caused by the cDNA itself, rather than by a contaminating plasmid or a rearranged retroviral vector.
Cloning of 5′- and 3′-extended cDNAs and sequence analysis and comparison.
cDNAs which overlapped with the original CXR-CT cDNA but extended further 3′ or 5′ were isolated from the T28 cell cDNA library by PCR amplification with a CXR-specific primer plus vector-specific primers.
cDNA sequences were determined with ABI 373 and 377 DNA sequencers by using FS Taq Dye Terminators. Continuous sequences were determined for both strands. Database comparisons were performed with BlastX. Sequence similarities of CXR and other sequences were determined with the Bestfit and Pileup programs of the Genetics Computer Group (Madison, Wis.).
Construction of mutant forms of mRasGRP cDNAs.
All deletion mutants were made by fusing natural or PCR-generated restriction enzyme sites to start codons, stop codons, green fluorescence protein (GFP) (derived from pEGFP-C1; Clontech), and/or hemagglutinin (HA) epitope tags, provided by pCTV derivative vectors.
The R271E point mutation in the GEF domain (CXR-GEFμ) was made by the fusion of two PCR fragments, introducing the base changes at the site of fusion. The resulting encoded peptide sequence is PTPQLEAEVFIK, compared to the normal sequence of PTPQLRAEVFIK. The deletion of the EF hands (CXR-EFHΔ) was made by replacing the fragment of the CXR-CT cDNA lying between the HindIII and StuI sites with a synthetic oligonucleotide hybrid encoding the appropriate amino acids. The mutation affecting the proline cluster (CXR-Proμ) was made by replacing the fragment of the CXR-CT cDNA lying between the SstI and AccI sites with a synthetic oligonucleotide hybrid encoding the appropriate amino acids. The encoded peptide sequence of this mutant is RAQGLTGSKGGVVV; the normal sequence is RAPPLTPSKPPVVV. The deletion of the REM domain (CXR-REMΔ) was made by replacing the fragment of the CXR-CT cDNA lying between the MscI and BglII sites with a synthetic oligonucleotide hybrid encoding the appropriate amino acids. The prenylation signal was made by inserting an oligonucleotide hybrid downstream of an HA tag in a pCTV retroviral vector. The resulting prenylation vector encodes the following amino acids from the C terminus of K-Ras: xzGSRKHKEKMSKDGKKKKKKSKTKCVIM#, where x is the site of insertion of cDNA, z is the HA epitope tag, and # is the stop codon. mRasGRP cDNAs were then inserted into this vector, resulting in in-frame fusion of the cDNA to the HA tag and prenylation signal.
The second PKC-δ C1 domain and the mRasGRP C1 domain were generated by PCR amplification from the T28 library and from the CXR-CT cDNA, respectively, using primers which modify the N and C termini of the encoded peptides. The encoded peptide of the mRasGRP C1 domain is STFPHNF.....LVVFECKKRIKPT (see Fig. 2 for complete sequences of C1 domains). The murine PKC-δ2 C1 domain is stMPHRF.....KVANLCkkrikpt, with the uppercase letters indicating the PKC-δ2 C1 domain sequence and the lowercase letters indicating sequence identical to those at equivalent positions in the mRasGRP C1 domain sequence. The PCR-generated C1 domains were inserted into an mRasGRP cDNA, such that their N termini are fused to amino acid 539 of Cxr, and their C termini are fused to an HA tag and a stop codon. Thus, the PKC-δ2 C1 domain-encoding (CXR-CΔ1/PKC) and mRasGRP C1 domain-encoding (CXR-CΔ1) cDNAs are completely identical except for the C1 domain-encoding sequences themselves.
FIG. 2.
(A) Sequence of the mRasGRP peptide. The peptide sequence is derived from the continual open reading frame obtained from the combined sequences of the CXR-CT cDNA and four overlapping cDNAs isolated from the T28 cell library. The REM and GEF domains are indicated by boldface, while the EF hands and C1 domain are indicated by the solid and dotted underline, respectively. Potential MAP kinase phosphorylation sites (16) are underlined. The potential amphipathic α-helix is double underlined, with bars under the aliphatic residues at seventh positions in the sequence. The arrows indicate the boundaries of the peptides encoded by the CT, FL, and XFL forms of mRasGRP and the various C-terminal deletions. The open and closed circles indicate positions affected by point mutations in CXR-GEFμ and CXR-Proμ, respectively. (B) Comparison of C1 domain sequences. The C1 domain of mRasGRP is aligned with C1 domains that bind phorbol ester (the second C1 domains of PKC-α, PKC-δ, and PKC-θ and the single C1 domain of n-chimerin) and those that do not bind phorbol ester (the single C1 domains of PKC-ζ, Raf, and Vav). Columns of histidines and cysteines involved in zinc binding are marked with asterisks. Residues predicted to be capable of participating in phorbol ester binding have solid highlighting.
All HA tag and GFP fusions, mutations, and PCR-generated DNA fragments were sequenced to confirm specific mutations and reading frame preservation and to ensure that no secondary mutations had occurred.
Construction of K-Ras mutants.
The starting cDNA was an activated human K-Ras clone. This was fused at its N terminus to GFP, making a cDNA that was as fully transforming as was the unmodified K-Ras. The prenylation site was mutated by replacing sequence downstream of a BstBI site with a synthetic oligonucleotide and then ligating into an HA-tagging vector, resulting in a cDNA encoding K-Ras with the C-terminal sequence RKHKEKMSKDGs(HA tag)stop, where the uppercase letters indicate natural K-Ras sequence. The C1 domain of RasGRP was then attached to this modified K-Ras, resulting in the C-terminal sequence RKHKEKMSKDGs(C1 domain)(HA tag)stop. All three forms of K-Ras were expressed at high levels, as determined by flow cytometry.
Detection of expression of HA-tagged or GFP-tagged mRasGRP or K-Ras constructs.
Tagged, mutated, and other variants of mRasGRP and control cDNAs in CTV retroviral vectors were converted to retroviral form by transfection into BOSC 23 cells, and then viral supernatants were used to infect NIH 3T3 cells. All mRasGRP constructs were tested for protein expression and structural integrity by Western blot analysis of infected NIH 3T3 cells, using anti-HA monoclonal antibody HA.11 (BABCo) or anti-GFP (Clontech) as a primary antibody and peroxidase-conjugated polyclonal anti-mouse immunoglobulin G (Jackson Laboratories) as a secondary antibody, and detection by chemiluminescence with the ECL system (Amersham). The range of expression in cell populations was determined for GFP-tagged constructs with a FACScan flow cytometer (Becton Dickinson).
Fluorescence analysis of mRasGRP localization.
NIH 3T3 cells infected with retroviral vectors expressing GFP-tagged forms of mRasGRP were treated as indicated in the figure legends. The cells were then fixed in 4% paraformaldehyde. For colocalization of GFP-tagged forms of mRasGRP and the endoplasmic reticulum-specific protein BiP, the cells were permeabilized with 0.1% Triton X-100, blocked with 2% bovine serum albumin, and incubated with the anti-BiP monoclonal antibody SPA-827 (StressGen Biotechnologies, Inc., Victoria, British Columbia, Canada), and then with Texas red-conjugated secondary antibody. Photomicrographs were taken under UV illumination, using a Zeiss fluorescent microscope.
Quantification of transformation efficiency.
NIH 3T3 cells expressing GFP-tagged derivatives of mRasGRP were plated at very low densities to allow individual colonies to form and were maintained in high-serum medium for 13 days. At that time, colonies were scored as transformed if they showed an obvious absence of contact inhibition and high refractility. The proportion of transformed colonies was normalized for the proportion expressing mRasGRP, as previously determined by flow cytometry. For mRasGRP derivatives which caused transformation, the number of colonies scored was relatively low (20 to 40) because they had to be completely separated to be distinguishable. For derivatives with very low or nil transformation efficiencies, up to 1,000 colonies could be scored, because rare transformed colonies (or their complete absence) could be individually scored despite being surrounded by and merged with a high density of nontransformed colonies (which were countable by extrapolation to very-low-density plates in a serial dilution).
Elk-1 activation assays.
The indicated cDNAs were inserted into the pAX142 (15) or pZIPNeo (10) expression vector and cotransfected into NIH 3T3 cells, along with pGal4-Elk-1, which expresses a fusion of the Gal4 DNA-binding domain and the transactivation domain and MAP kinase sites of Elk-1, and with pGal4-LUC, which contains a luciferase reporter driven by a minimal promoter and tandem Gal-4-binding sites (29). The cells were switched to DME containing 0.5% fetal bovine serum at 34 h posttransfection, and 14 h later the cells were lysed and assayed for luciferase activity as described previously (29).
ERK1 and ERK2 activation assays.
NIH 3T3 cells stably transduced with the indicated retroviral vectors were maintained in DME containing 9% calf serum or switched to serum-free DME for 2 h and then treated as indicated. BOSC 23 cells were transiently transfected with the indicated cDNAs in pAX142, using the calcium phosphate procedure (52). At 32 h after transfection, the BOSC 23 cells were switched to serum-free RPMI medium containing 4 μg of insulin per ml and 5 ng of sodium selenite per ml for 19 h and then were stimulated for 15 min with 10% fetal calf serum as indicated. Total cell lysates were prepared and analyzed for activation of ERK1 and ERK2 by Western blotting with a polyclonal antibody specific to ERK1 and ERK2 phosphorylated at Tyr204, using the procedures specified by New England Biolabs.
Hybridization analysis of RNA.
Total cellular RNA was separated on 5% formaldehyde agarose gels and transferred to a Hybond N+ nylon membrane (Amersham). Hybridization and high-stringency washing were performed as described previously (20). The probe was a 1,250-bp fragment of the CXR-CT cDNA which encompasses the region encoding amino acids 49 to 448.
RESULTS
Selection of a cDNA clone which causes Ras-like transformation of NIH 3T3 cells.
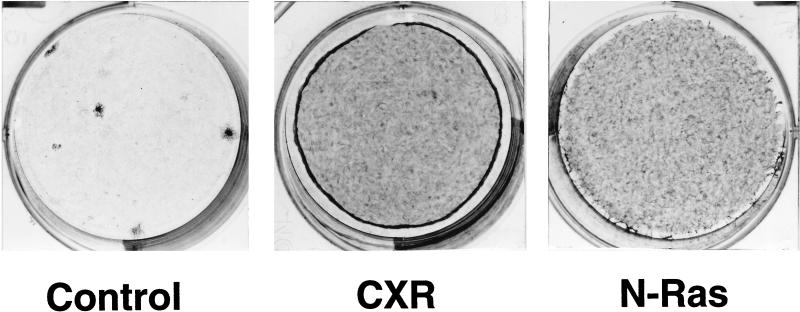
To identify novel Ras activators expressed by T lymphocytes, we screened for cDNAs capable of transforming NIH 3T3 cells within a large cDNA library made from a murine T-cell hybridoma, T28. This cell line has functional signalling from CD3 and can also be activated by cotreatment with phorbol ester and calcium ionophores (36a). The cDNA library was made in plasmid form in the pCTV1 retroviral vector, converted to retroviral form by transfection into the packaging cell line BOSC 23, and then transferred to NIH 3T3 cells by infection. Among the approximately 25 transformed cell foci arising in the infected NIH 3T3 cultures was one which had the refractile, swirling morphology characteristic of Ras-induced transformation. This cell clone was picked and expanded, and the single proviral cDNA within it was recovered by PCR and recloned into the pCTV3 retroviral vector to produce clone pCXR-CT. Infection of NIH 3T3 cells with retrovirus derived from pCXR-CT resulted in massive transformation of the cell culture (Fig. 1), with essentially every infected cell in the culture becoming refractile. The cell cultures continued to proliferate beyond confluence, eventually peeling off the culture surface in large masses. The onset of obvious morphological transformation occurred about 2 days later than that induced by expression of activated mutants of N-Ras or K-Ras.
FIG. 1.
Transforming activity of the CXR-CT cDNA clone encoding truncated mRasGRP. NIH 3T3 cells were plated at low density and infected with CTV83B, which is an empty retroviral vector (Control); with retroviral vector CTV82, carrying the CXR-CT cDNA (CXR); or with retroviral vector CTV80, carrying a cDNA encoding N-Ras activated by a Q61K mutation (N-Ras). After 8 days (5 days postconfluence), the cell cultures were stained with methylene blue.
The CXR-CT cDNA encoded a protein of 570 amino acids (Fig. 2). It lacked a stop codon and thus represented an artificial C-terminal truncation arising from synthesis of an incomplete cDNA. Three overlapping cDNAs extending further at the 3′ end and two extending further at the 5′ end were subsequently isolated from the T28 library. In combination, the original cDNA and the 5′- and 3′-extended cDNAs contain a complete open reading frame encoding a protein of 795 amino acids (Fig. 2A). The C-terminally extended form (FL) (Fig. 2A) or the complete protein (XFL) (Fig. 2A) caused transformation of NIH 3T3 cells with an efficiency similar to that of CXR-CT (Fig. 3). Thus, the N-terminal and C-terminal regions are not required for and do not suppress transformation.
FIG. 3.
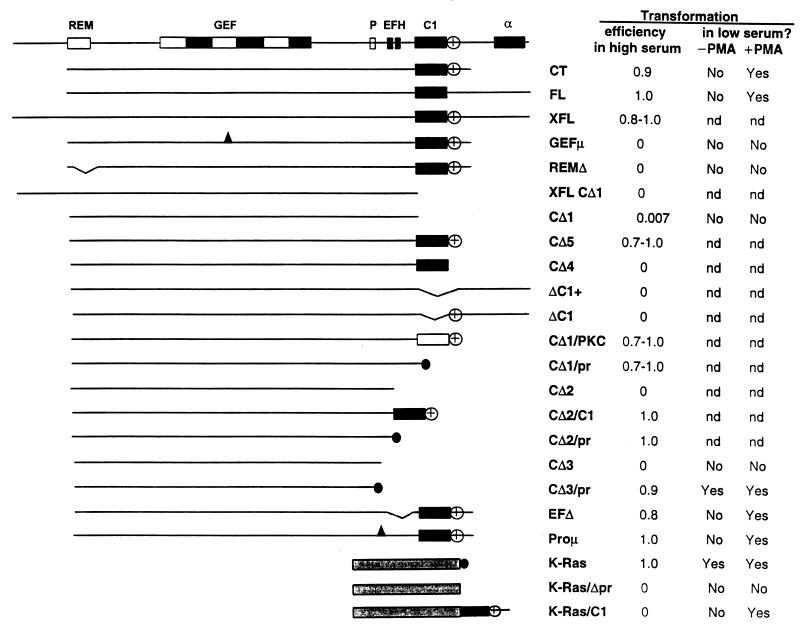
Mutational analysis of regions of mRasGRP required for transformation of NIH 3T3 cells. The domain structure of mRasGRP is illustrated in the upper diagram (REM, Ras exchanger motif; GEF, region of homology to the GEF domain of Sos; P, proline cluster; EFH, each box represents one EF hand motif; C1, C1 domain; α, putative α-helical segment). Deletion and other mutant constructs are depicted in the other diagrams. All constructs are GFP tagged at N termini and/or HA tagged at C termini (or between the mRasGRP sequence and the prenylation signal). The prenylation signal is symbolized by the black circle, and sites of point mutations are symbolized by the black triangles. Exact boundaries of domains and predicted translation products of the deletion and mutant constructs are indicated in Fig. 2, as are positions of point mutations. The open box in construct CΔ1/PKC represents the second C1 domain of PKC-δ. The stippled box represents the region of K-Ras N terminal to its prenylation signal. Transformation efficiency in high-serum medium is the proportion of isolated colonies expressing N-terminally GFP-tagged, retrovirally transduced mRasGRP constructs which were morphologically transformed and not contact inhibited after 13 days in culture in medium with 9% calf serum. The spontaneous rate of transformation was less than 1 focus/105 cells. Transformation in low-serum medium was assessed after 6 days of culture with daily feedings of medium containing 0.5% fetal bovine serum and 4 μg of insulin per ml, with or without 10 ng of PMA per ml.
The protein encoded by the CXR cDNA is 98% identical in amino acid sequence to rat RasGRP, a new member of the Sos family of GEFs for Ras GTPases that was described after the submission of this paper (19). Therefore, we now refer to the protein encoded by our cDNAs as mRasGRP, while using the CXR designation when referring to the cDNAs themselves.
The GEF domain of mRasGRP is essential for its transforming activity.
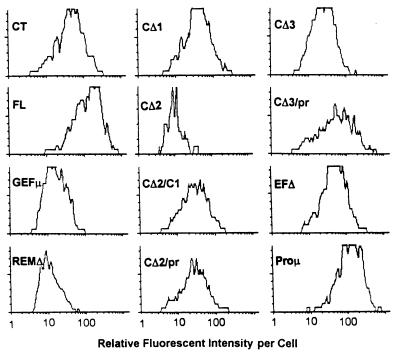
Amino acids 201 to 371 of mRasGRP are clearly homologous to the GEF domains of Sos and related exchange factors which are known or assumed to act on one or more members of the Ras family of GTPases (4, 23, 55). Among mammalian GEFs, this region of RasGRP is most similar to RasGRF and C3G and somewhat less similar to Rgl, RalGDS, and Sos1 (determined by using pairwise similarity scores derived by the Genetics Computer Group Pileup program). Similarities to all GEFs are particularly strong within the conserved SCR-1, -2, and -3 boxes within the GEF domain (4). The first residue in the SCR-2 box is invariably arginine in all Sos family members, and mutation of this residue to glutamate in the Saccharomyces cerevisiae CDC25 GEF eliminates its function, apparently by preventing interaction with Ras while otherwise retaining the structure and catalytic competence of the GEF domain (51). An equivalent arginine-to-glutamate mutation at position 271 in mRasGRP eliminates its ability to transform NIH 3T3 cells (Fig. 3, GEFμ). The expression of GEFμ was somewhat lower on average than the expression of CXR-CT (Fig. 4). However, about 10% of the GEFμ cells (none of which were transformed) expressed amounts of mRasGRP protein equivalent to or greater than the levels expressed by 50% of the CXR-CT cells (of which at least 80% were transformed). Therefore, in cells expressing equivalent levels of protein, the point mutation in the GEF domain eliminated the ability of mRasGRP to induce transformation.
FIG. 4.
Expression levels of mRasGRP in retrovirally transduced NIH 3T3 cells. mRasGRP constructs were tagged by fusion of GFP to the CXR-CT N terminus. Expression levels were determined by flow cytometry just prior to plating of cells for the transformation assays shown in Fig. 3. The histograms show the distribution of fluorescence values in the population of cells expressing the indicated GFP-tagged mRasGRP construct, after gating out of cells with fluorescence values not above those of control, non-GFP-expressing cells. Fluorescence intensity is on a log scale.
All members of the Sos family have an additional sequence homology (37), which has been termed a Ras exchanger motif or REM box (21), at a variable distance N terminal to the GEF domain. mRasGRP has a REM box near its N terminus, although the middle portion of the mRasGRP REM is markedly variant from those of other GEFs, particularly by having a five-amino-acid insertion. A deletion which precisely removes the REM eliminated mRasGRP’s ability to transform NIH 3T3 cells (Fig. 3, REMΔ).
A C1 domain within mRasGRP is required for its transforming activity.
A unique feature of the C-terminal region of mRasGRP is the presence of a C1 domain (33) very similar to those found in classical and novel protein kinase Cs (PKCs). In most of these PKCs (and in some other proteins, such as n-chimerin), the C1 domains serve as binding sites for diacylglycerol or phorbol ester and act as positive regulatory domains by mediating the translocation of the C1 domain-containing protein to membranes enriched in diacylglycerol or phorbol ester (47, 48). In addition to the histidines and cysteines required for zinc coordination, the C1 domain of mRasGRP contains appropriate residues at the 11 positions that participate in forming a phorbol ester-binding pocket in the second C1 domain of PKC-δ (64) (Fig. 2B). C1 domains which do not bind phorbol ester, e.g., those in Raf, Vav, or the atypical PKC-ζ, have inappropriate residues or gaps in at least three of these positions. Thus, the C1 domain of mRasGRP is predicted to bind phorbol ester and diacylglycerol and by implication is expected to regulate the function of mRasGRP by mediating its binding to membranes enriched in the second messenger diacylglycerol.
All deletions which removed the C1 domain from mRasGRP eliminated transforming activity (Fig. 3, CΔ1, XFL-CΔ1, ΔC1+, ΔC1, CΔ2, and CΔ3). A deletion that left the C1 domain intact but excised a cluster of basic amino acids (KKRIK) found immediately C terminal to the C1 domain also resulted in loss of transforming activity (compare CΔ4 and CΔ5 in Fig. 3). Conversely, removal of the C1 domain with retention of the basic cluster also eliminated transforming activity (Fig. 3, ΔC1). The basic cluster may serve to increase the affinity of the C1 domain for membranes, by interacting with negatively charged phospholipid head groups.
The transforming activity of mRasGRP with the C1 domain deleted could be restored by attaching to it either its own C1 domain or the second C1 domain of murine PKC-δ, along with the basic cluster (Fig. 3, CΔ1/PKC). Therefore, the C1 domain of mRasGRP is essential for transformation by mRasGRP, and it is functionally equivalent in this assay to a bona fide phorbol ester- and diacylglycerol-binding C1 domain. The absence of the C1 domain was also fully compensated for by adding to mRasGRP a C-terminal signal for membrane localization, provided by the prenylation signal of K-Ras (Fig. 3, CΔ1/pr, CΔ2/pr, or CΔ3/pr). Expression of these prenylated forms of mRasGRP induced transformation within 2 days, i.e., as rapidly as did expression of activated mutants of K-Ras or N-Ras.
The immediate N terminus of mRasGRP has a glycine at position 2 and a lysine and an arginine at positions 6 and 8, respectively. This makes it a potential target for N-terminal myristoylation (36), which could provide an alternative means of membrane localization. However, a form of mRasGRP that contains this N terminus but lacks the C1 domain is nontransforming (Fig. 3, XFL-CΔ1), indicating that if N-terminal myristoylation occurs, it cannot substitute on its own for the function of the C1 domain.
EF hands and a potential SH3 domain-binding site are not needed for transformation via mRasGRP.
The region between the N-terminal GEF domain and the C-terminal C1 domain contains two features which could potentially contribute to the regulation of mRasGRP. The first is a pair of EF hands (Fig. 2), compact domains which serve as sensors of calcium concentration changes in the cell (34). EF hands typically function in pairs, making cooperative changes in conformation following calcium binding which create hydrophobic surfaces that can participate in intra- or interprotein interactions. The EF hand pair of mRasGRP thus has the potential to serve as a calcium-responsive element that could alter interdomain interactions within mRasGRP or alter interactions of mRasGRP with other proteins or with membranes. However, deletion of the EF hand pair (Fig. 3, EFΔ) or point mutations in the putative calcium-binding residues of the second EF hand (data not shown) had no effect on transformation activity. The second feature is a proline cluster (Fig. 2) which could serve as a binding site for SH3 domains (RapPLtPsKppvvv, where the uppercase letters indicate residues that could participate in SH3 binding [44]). A cluster of point mutations which eliminate the potential for SH3 binding (Fig. 3, Proμ) in this sequence had no effect on transformation activity. Double mutants of mRasGRP lacking both the EF hand pair and the proline cluster were also fully active in transformation assays (data not shown). Therefore, the EF hands and the proline cluster, either singly or collectively, have no discernible influence on the function of mRasGRP in the transformation assay.
Localization of mRasGRP to cell membranes in response to either serum or phorbol ester.
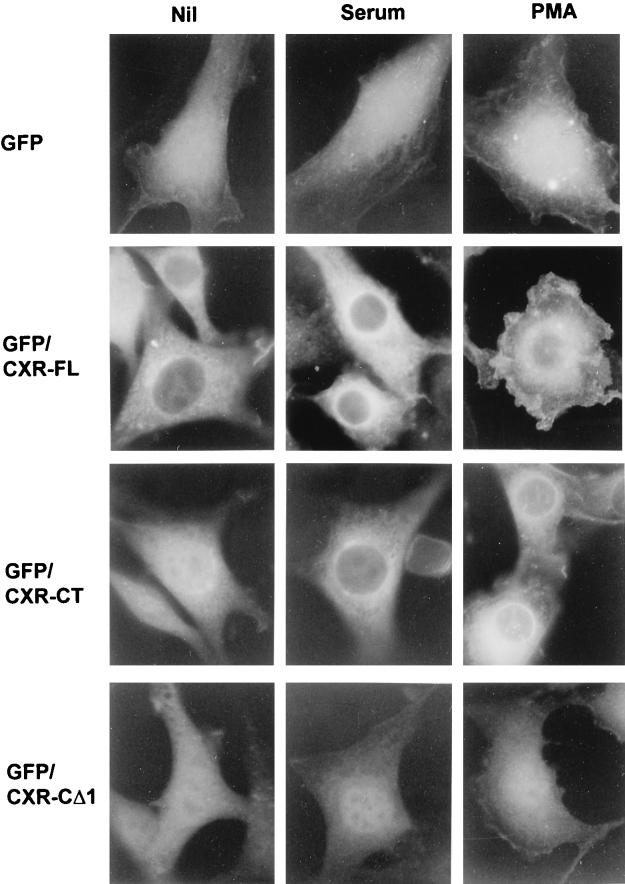
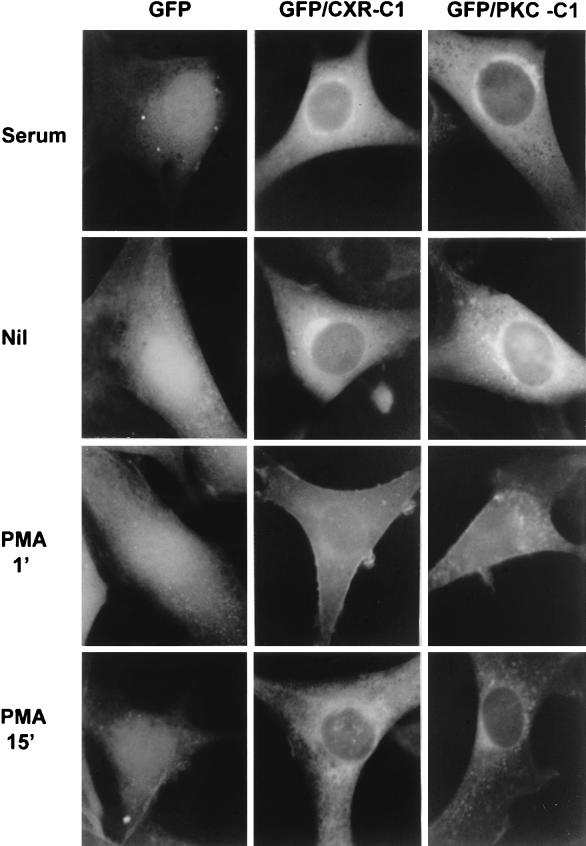
In serum-starved NIH 3T3 cells, the CXR-CT form of mRasGRP is distributed relatively uniformly throughout the cell, including the nucleus (Fig. 5). After a 15-min stimulation with serum or the phorbol ester phorbol 12-myristate 13-acetate (PMA), CXR-CT became predominantly localized to cellular structures concentrated around the nucleus and in a punctate network spreading further out into the cell periphery (Fig. 5). Translocation of CXR-CT was induced by PMA at concentrations as low as 0.5 ng/ml. Most of the mRasGRP in serum- or PMA-stimulated cells was in the endoplasmic reticulum, as determined by its colocalization with the endoplasmic reticulum-specific protein BiP (data not shown). The occurrence of mRasGRP in BiP-negative structures around and to one side of the nucleus probably represents localization to the nuclear envelope and Golgi. An equivalent concentration of some PKC isoforms in the endoplasmic reticulum and Golgi is seen in PMA-stimulated fibroblasts (26).
FIG. 5.
Translocation of mRasGRP in response to serum or PMA stimulation. NIH 3T3 cells stably expressing the indicated forms of N-terminally GFP-tagged mRasGRP via retroviral infection were serum starved in DME for 4 h. The medium was then replaced with DME (Nil), DME containing 10% calf serum (serum), or DME containing 50 ng of PMA per ml (PMA). The cells were fixed 15 min later, UV illuminated, and photographed.
The translocation of CXR-CT to membranes in response to PMA or serum was mediated by the C1 domain, because precise deletion of the C1 domain resulted in a diffuse distribution of mRasGRP throughout the cell, with no apparent concentration in cell membranes and no relocalization being induced by either PMA or serum (Fig. 5, CXR-CΔ1).
The CXR-FL form of mRasGRP was also localized predominantly to internal membranes in serum-stimulated cells, but in this case serum withdrawal did not result in appreciable delocalization (Fig. 5). In contrast to the case for CXR-CT, PMA treatment induced translocation of a considerable portion of CXR-FL to the cell periphery (Fig. 5). This appeared to be specific localization at or closely beneath the plasma membrane, because it was equivalent to the distribution of prenylated GFP in PMA-stimulated cells (not shown). Stable targeting to the cell periphery in response to PMA was dependent on the C1 domain but also required the presence of both the N-terminal and C-terminal regions of mRasGRP (data not shown).
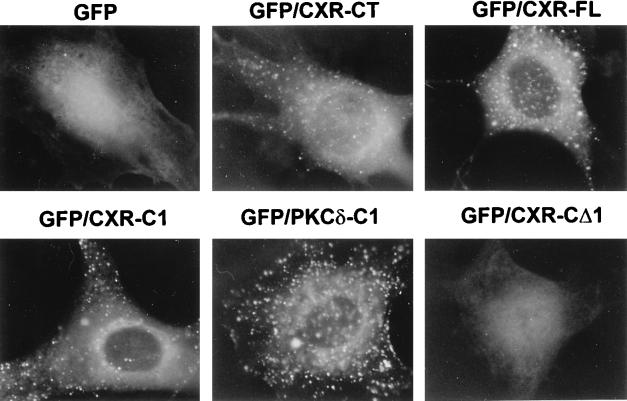
The C1 domain plus basic cluster of mRasGRP had a distribution in serum-stimulated cells that was very similar to that of CXR-CT or CXR-FL and essentially identical to that of the second C1 domain of PKC-δ plus the basic cluster (Fig. 6). However, serum deprivation for several hours did not result in a significant delocalization of either of these C1 domains. The ability of CXR-CT to delocalize upon serum deprivation and the failure of the isolated C1 domains to do so was not a result of the cells being transformed or not, because when the GFP-tagged C1 domains were coexpressed with untagged CXR-CT (resulting in cell transformation), the isolated C1 domain still did not delocalize when the cells were deprived of serum. Therefore, CXR-CT delocalization following serum starvation may be an active process which is promoted by a part of the protein N-terminal to the C1 domain and which is suppressed by the C-terminal region found in CXR-FL.
FIG. 6.
Translocation of C1 domains in response to serum or PMA stimulation. NIH 3T3 cells were infected with retroviral vectors expressing GFP alone (GFP) or fusions of GFP to the C termini of isolated C1 domains of mRasGRP (GFP/CXR-C1) or PKC-δ (GFP/PKC-C1). Cells were serum stimulated, serum deprived for 3.5 hours (Nil), or serum deprived and then stimulated with 50 ng of PMA per ml for 1 or 15 min. The cells were then fixed and photographed under UV illumination.
The localization of the C1 domains was dynamically regulated by PMA. One minute after PMA stimulation, a portion of the C1 domains of mRasGRP and PKC-δ translocated away from internal membranes and became distinctively concentrated at the plasma membrane (Fig. 6). After that, the C1 domains gradually returned to internal membranes, such that at 15 min after the addition of PMA, the distribution of the C1 domain was identical to that in nonstimulated or serum-stimulated cells. Migration of the C1 domains was presumably driven by the initial accumulation of PMA in the plasma membrane, followed by diffusion of PMA to internal membranes. When the basic cluster was removed, the C1 domain of mRasGRP was still predominantly localized to internal membranes, and it became partially delocalized immediately following PMA treatment, but it was not as distinctively attracted to the plasma membrane as was the C1 domain including the basic cluster (data not shown). These results indicate that the basic cluster is not essential for membrane localization of the C1 domain but that it may play a role in specifically stabilizing interactions between the C1 domain and the plasma membrane.
CXR-CT and CXR-FL did not show the same rapid and transient translocation to the plasma membrane following PMA treatment that was seen for the isolated C1 domain plus basic cluster. Translocation of CXR-FL from internal membranes to the plasma membrane was maximal only after 15 min of PMA stimulation. This may in part be due to reduced mobility of the full-length protein relative to the small C1 domains. It is also possible that the migration of the full-length protein needs to be assisted by PKC-dependent processes, such as the changes in cell shape that start to occur after about 5 min of PMA stimulation. In contrast, the migration of the C1 domains is probably a direct effect of PMA, because it occurs very rapidly, well before there are any discernible effects of PMA stimulation on cell structure.
To test the ability of the mRasGRP C1 domain to bind diacylglycerol, NIH 3T3 cells were treated with exogenous phosphatidylcholine-specific phospholipase C (PC-PLC). This enzyme generates large quantities of diacylglycerol at the plasma membrane, which then rapidly redistributes to internal structures in the cell (38). PC-PLC treatment had no discernible effect on the distribution of either GFP alone or CXR-CΔ1, the form of mRasGRP that lacks the C1 domain (Fig. 7). In contrast, after 45 min of PC-PLC treatment, there was an intense relocalization of the C1 domains of mRasGRP and PKC-δ, as well as the C1 domain-containing forms of mRasGRP, CXR-CT, and CXR-FL, to multiple small spherical structures within the cell (Fig. 7). These structures closely resemble the lipid droplets that are intensely labelled when fibroblasts are treated with fluorescently tagged phosphatidic acid, which is metabolized to diacylglycerol and then triacylglycerol once inside the cell (50). The PC-PLC treatment apparently results in the migration of large quantities of diacylglycerol from the plasma membrane into the lipid droplets, as indicated by the ability of the C1 domain of PKC-δ to bind to droplets only following PC-PLC treatment. The concentration of the mRasGRP C1 domain in the same droplets under the same conditions and with the same kinetics indicates that the mRasGRP C1 domain could be equally capable of binding diacylglycerol. It is also possible that the C1 domain of mRasGRP is binding another lipid that is generated in response to PC-PLC treatment and colocalizes with diacylglycerol in these structures. However, the major products of diacylglycerol metabolism, such as phosphatidylcholine, tri- and mono-acylglycerols, and fatty acids (14, 50), are not expected to bind the C1 domain of mRasGRP, given the size and hydrophobicity of the residues predicted to compose its ligand-binding pocket (33, 64). PC-PLC had no obvious effect on the distribution of mRasGRP or the C1 domains during the first 15 min of treatment, even though activation of ERK1 and ERK2 (presumably via diacylglycerol-mediated activation of PKCs) was occurring during this time. This indicates that the amount of diacylglycerol that could be generated and maintained at the plasma membrane by PC-PLC treatment was not sufficient to draw the C1 domains away from their normal sites of localization in internal membranes.
FIG. 7.
Phospholipase C-induced localization of mRasGRP and isolated C1 domains. NIH 3T3 cells infected with retroviral vectors expressing GFP alone, fusions of GFP to the isolated C1 domains of mRasGRP (GFP-CXR-C1) or PKC-δ (GFP/PKCδ-C1), or GFP fusions of the indicated forms of mRasGRP were serum deprived for 3.5 h in DME, and then Bacillus cereus PC-PLC (Sigma) was added to the medium at 4 U/ml and the cells were cultured at 37°C for 45 min. The cells were then fixed and photographed under UV illumination.
In total, the localization results with mRasGRP and its isolated domains demonstrate that the C1 domain is the primary mediator of mRasGRP translocation to membranes. This process can be directly regulated by PMA and potentially also by the activation of phospholipases that generate diacylglycerol. The specific and stable targeting of the full-length form of mRasGRP to the plasma membrane following PMA stimulation indicates that interaction of mRasGRP with Ras family GTPases may be subject to more complex regulation, with the C1 domain serving as a PMA-responsive membrane-targeting element and the combined action of the basic cluster and N-terminal and C-terminal regions serving to retain mRasGRP specifically in the plasma membrane.
mRasGRP expression induces MAP kinase activation.
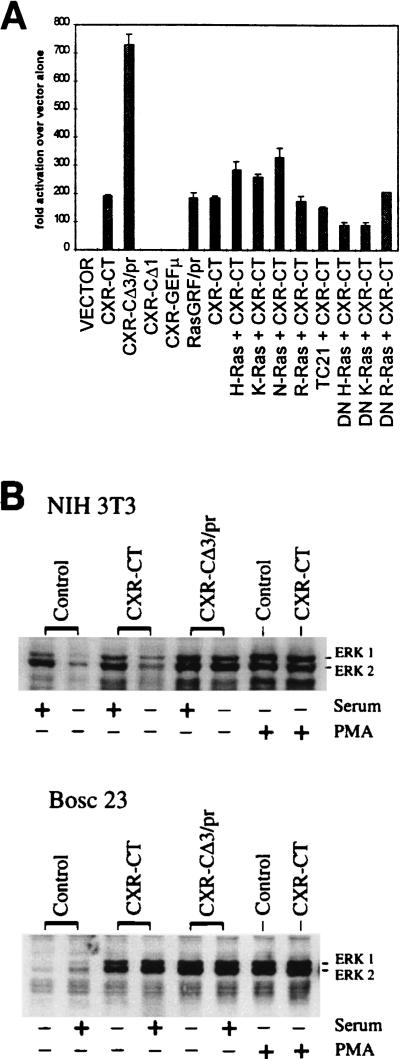
The transcription factor Elk-1 is activated by C-terminal phosphorylation via the MAP kinases ERK-1 and ERK-2, as well as by the related MAP kinase JNK (61). These MAP kinases are themselves indirectly activated in fibroblasts by GTP-loaded Ras, in the case of the ERKs via a kinase cascade from Raf to MEK kinases (18). We have used an assay based on MAP kinase-dependent stimulation of transactivation of a reporter gene via the C-terminal domain of Elk-1 (29) to indirectly measure mRasGRP activity and to determine the ability of mRasGRP to act in conjunction with specific Ras family members. Expression of mRasGRP caused a very high level of MAP kinase activation in NIH 3T3 cells cultured in low-serum medium, equivalent to that attained by expression of a prenylated and thus hyperactivated form of RasGRF-1 (Fig. 8A). A prenylated form of mRasGRP (CXR-CΔ3/pr) induced fourfold-higher levels of MAP kinase activation, while the point mutation in the GEF domain and the deletion of the C1 domain eliminated the ability of mRasGRP to activate MAP kinases (Fig. 8A). MAP kinase activation by mRasGRP was increased by coexpression with normal forms of either H-Ras, K-Ras, or N-Ras but not by coexpression with the Ras-related GTPase R-Ras or TC21 (Fig. 8A). Expression of these GTPases on their own had only very minor effects on MAP kinase activation (data not shown). Coexpression of dominant negative forms of H-Ras and K-Ras (which bind to GEFs but are not substrates for guanine nucleotide exchange) partially suppressed MAP kinase activation via mRasGRP, while dominant negative R-Ras had no effect (Fig. 8A). The dominant negative forms of H-Ras and K-Ras did not inhibit MAP kinase activation via expression of an activated form of Raf-1 (data not shown), indicating that they were acting specifically on the process of Ras activation rather than at a later point in the transduction of activating signals from mRasGRP to MAP kinases. mRasGRP also strongly induced expression from NF-κB-, Ets/AP1-, and cyclin D-responsive promoters, each of which is inducible via Ras activation (data not shown). For all three of these Ras-responsive reporter systems, mRasGRP stimulated expression equivalently to prenylated RasGRF, while prenylated mRasGRP was considerably more effective.
FIG. 8.
Induction of MAP kinase activation and ERK1 and -2 phosphorylation by expression of mRasGRP. (A) NIH 3T3 cells were cotransfected with Gal4-LUC and Gal4-Elk-1, along with expression vectors expressing mRasGRP cDNAs, prenylated RasGRF-1, and normal or dominant negative forms of Ras family GTPases. The data shown are representative of those from three separate experiments, with data for each point determined in triplicate in each experiment. (B) NIH 3T3 cells were stably expressing the indicated mRasGRP cDNAs by retroviral infection. BOSC 23 cells were transiently transfected with the indicated cDNAs. Cells were stimulated with serum as described in Materials and Methods and then analyzed for ERK1 and ERK2 phosphorylation by Western blotting. Equivalent loading of protein was checked by Coomassie blue staining and by reprobing the blot with an anti-ERK1 and -2 antibody that was not phosphorylation specific.
Detection of tyrosine 204 phosphorylation via a phospho-specific antibody (New England Biolabs) was used to assess the activation state of the MAP kinases ERK1 and ERK2 in NIH 3T3 cells stably expressing mRasGRP. Expression of mRasGRP (CXR-CTR) resulted in only a minor increase in ERK1 and ERK2 phosphorylation when cells were cultured in high-serum medium, while expression of prenylated mRasGRP (CΔ3/pr) had a greater effect (Fig. 8B). When the cells were withdrawn from serum for 2.5 h, ERK1 and ERK2 phosphorylation did not decline at all in the CΔ3/pr-expressing cells, while it did decline in the CXR-CT-expressing cells, although not down to the levels seen in control cells. The level of ERK1 and ERK2 phosphorylation induced by prenylated mRasGRP in the presence or absence of serum was equivalent to that induced by the expression of a transforming, GTPase-defective form of K-Ras (data not shown).
In BOSC 23 cells, a derivative of the 293T human epithelial cell line, transient expression of either CXR-CT or CXR-CΔ3/pr resulted in a considerable increase in ERK1 and ERK2 phosphorylation in either the presence or absence of serum (Fig. 8B). The greater ability of mRasGRP to activate ERK1 and ERK2 in the absence of serum in BOSC cells may be due to its higher level of expression in these cells than in stably infected NIH 3T3 cells or to a difference in regulation of mRasGRP in these two distinct cell types.
PMA treatment on its own resulted in a large increase in ERK1 and -2 phosphorylation, presumably due to PKC stimulation. There was no further discernible increase in PMA-induced ERK1 and -2 phosphorylation resulting from expression of either C1 domain-containing or prenylated forms of mRasGRP (Fig. 8B) or from expression of activated N-Ras (not shown).
The C1 domain of mRasGRP mediates PMA-dependent transformation in low concentrations of serum.
When the amount of serum in the culture medium was reduced from the normal concentration of 9% down to 0.5%, NIH 3T3 cells expressing CXR-FL or CXR-CT lost their transformed appearance and stopped proliferating. This reversion of the transformed phenotype allowed us to determine if PMA could regulate the ability of mRasGRP to induce cell transformation. Addition of PMA to the low-serum medium caused CXR-CT- and CXR-FL-expressing cells to regain their transformed appearance within 1 day and also enabled them to proliferate beyond confluence (Fig. 3). Control cells grew to higher density in the low-serum medium when it was supplemented with 10 ng of PMA per ml, but they were not morphologically transformed and were fully contact inhibited. Morphological transformation of mRasGRP-expressing cells in low-serum medium was induced by PMA concentrations as low as 0.1 ng/ml. PMA-dependent transformation in low-serum medium was also observed with cells expressing the Proμ or EFΔ form of mRasGRP (Fig. 3). The CΔ1 form lacking the C1 domain as well as the more extensive CXR-CΔ3 C-terminal deletion and the REMΔ and GEFμ mutants did not cause morphological transformation in the presence of PMA concentrations as high as 300 ng/ml. In contrast, cells expressing the prenylated form of mRasGRP (Fig. 3, CΔ3/pr) were transformed in low-serum medium in the presence or absence of PMA, as were cells expressing GTPase-defective K-Ras (Fig. 3). These results demonstrate that PMA can regulate the ability of mRasGRP to induce transformation when the serum concentration is low. This effect of PMA requires the C1 domain of mRasGRP but not its EF hands or proline cluster, and the requirement for PMA or the C1 domain is abrogated by constitutive plasma membrane localization of mRasGRP.
When the prenylation signal of the GTPase-defective K-Ras was replaced with the C1 domain of mRasGRP, it was no longer able to transform cells when they were grown in high-serum medium. However, the C1 domain-containing form of K-Ras was transforming when the cells were cultured in PMA and low-serum medium (Fig. 3). Removal of the C1 domain and its replacement by a prenylation signal restored the ability of K-Ras to induce PMA- and serum-independent transformation (data not shown). Therefore, the C1 domain of mRasGRP by itself could confer PMA responsiveness on a protein whose transforming activity normally is PMA independent and whose activity normally requires constitutive localization to the plasma membrane. The inability of the C1 domain-containing K-Ras to transform in the absence of PMA may be due to almost all of it being localized to internal membranes, and thus depleted from the plasma membrane, under these conditions.
mRasGRP can induce transformation via a C1 domain-independent mechanism.
None of the cells expressing the CΔ1 form of mRasGRP were detectably transformed when the cells were continually passaged below confluence or in cultures that had been maintained as monolayers for up to 10 days. Beyond that time, highly transformed cells began to gradually appear, until after 28 days they formed the majority of cells in the culture. This accumulation represented both overgrowth of the monolayer by transformed cells and the transformation of previously nontransformed cells. Conversion of the majority of cells in an isolated colony to a highly transformed phenotype could occur over a few days. This conversion presumably reflects de novo activation of mRasGRP, because there is no enhanced activation of MAP kinases in the CΔ1-expressing cells prior to their conversion to a transformed phenotype (Fig. 8A). Amounts of CΔ1 protein were equivalent among both nontransformed and transformed cells (data not shown), indicating that transformation did not result from unusually high levels of CXR-CΔ1 expression in the subset of cells that became transformed or that transformation was caused by a shift to higher levels of expression of CΔ1.
The CΔ1 form of mRasGRP was found distributed throughout the cytoplasm and nucleus in both the untransformed cells and the transformed variants that arose after prolonged culture, and this delocalized distribution was unaffected by serum or PMA stimulations. When cultured in low-serum medium, the two types of CΔ1-expressing cells were unresponsive to PMA, i.e., the untransformed cells remained untransformed, and the transformed variants remained transformed, irrespective of whether PMA was added to the medium. This extends our previous observations that only C1 domain-containing forms of mRasGRP are responsive to PMA in the low-serum transformation assay. The accelerating spontaneous transformation that was observed with CΔ1-expressing cells did not occur in cells expressing CΔ2 or CΔ3, which lack the EF hands as well as the C1 domain, or in cells expressing the REMΔ or GEFμ form of mRasGRP. In combination, these results indicate that activation of mRasGRP can occur via a mechanism that does not involve the C1 domain and is not regulated by PMA but that does require the GEF functions and possibly the EF hands or adjacent structures.
Expression pattern of mRasGRP.
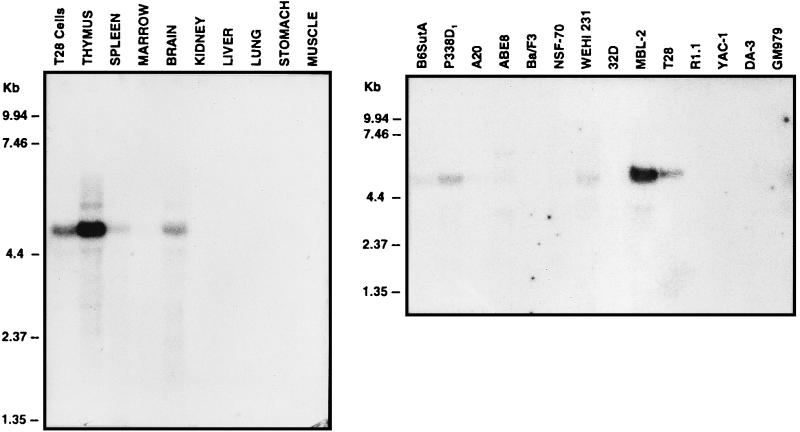
Northern analysis of total RNA identified a predominant mRasGRP transcript of about 4.8 kb in T28 cells (Fig. 9). The same mRNA species was abundant in thymus, at moderate levels in brain, and at lower levels in spleen and bone marrow. No RasGRP transcripts were detectable in kidney, lung, stomach, and skeletal muscle (Fig. 9) or in heart or testis (data not shown). In addition to the T28 T-cell hybridoma, the RasGRP mRNA was detected in the MBL-2 T-cell line; the B-cell lines WEHI 231, ABE8, and A20; the primitive hemopoietic line B6SutA; and P388D1, a cell line which was derived from a B lymphoma but subsequently underwent monocytic differentiation (Fig. 9). RasGRP transcripts were not detected in the R1.1 or YAC-1 T-cell line, the NSF-70 or Ba/F3 B-cell line, the 32D or DA-3 myeloid cell line, or the GM979 erythroid cell line (Fig. 9). RasGRP transcripts were also not detectable in the murine fibroblast cell line NIH 3T3 or C3H10T1/2 or in the breast-derived cell line C127 (data not shown). Overall, this suggests that RasGRP is expressed in some but not all murine lymphoid cell types and possibly also in uncommitted hemopoietic precursors, as represented by the B6SutA cell line.
FIG. 9.
Expression of mRasGRP in murine tissues and hemopoietic cell lines. Northern blots of total RNAs from the indicated tissues of a 6-week-old C57BL/6J mouse or murine hemopoietic cell lines were probed with the CXR-CT cDNA. Marker sizes are indicated.
DISCUSSION
The GEF function of RasGRP.
Purified rat RasGRP catalyzes guanine nucleotide exchange on H-Ras but not on R-Ras (19). Our results obtained by using coexpression of mRasGRP with various Ras family members indicate that this specificity is retained in vivo and that mRasGRP acts on K-Ras and N-Ras as well as H-Ras. The failure of the catalytic domain mutant of mRasGRP to induce transformation suggests that the in vivo function of RasGRP requires direct activation of Ras GTPases via guanine nucleotide exchange. The ability of the REM-plus-GEF region of mRasGRP to induce transformation when prenylated indicates that GEF activity is probably the only function of RasGRP required for transformation, as the only known function of the REM box is to increase the efficiency of GEF catalysis in vitro (37). It remains possible that beyond serving as a Ras-specific GEF, RasGRP has an additional biological function that is not apparent via fibroblast transformation assays.
Regulation of membrane localization of RasGRP by serum, PMA, and diacylglycerol.
RasGRP is found predominantly in internal cell membranes in serum-stimulated fibroblasts. This is dependent on its C1 domain, as demonstrated by the equivalent localization pattern of the isolated C1 domain of mRasGRP and the absence of membrane localization of the mRasGRP mutant lacking the C1 domain. The C1 domain of RasGRP is very similar in sequence to the C1 domains of PKCs and n-chimerin, which bind phorbol esters and diacylglycerol. This suggests that membrane localization of RasGRP could be driven by binding of its C1 domain to phorbol ester or diacylglycerol. The best evidence for this mechanism of membrane localization is the complete equivalence of the C1 domains of mRasGRP and PKC-δ in their abilities to restore serum- and PMA-dependent transformation competence to mRasGRP deletion mutants, their localization in serum-stimulated cells, their transient translocation to the plasma membrane in response to PMA, and their concentration in lipid droplets following PC-PLC treatment.
The rapidity of the migration of both C1 domains to the plasma membrane following addition of PMA to the medium implies that PMA is a direct, high-affinity ligand for the RasGRP C1 domain, as it is for the PKC-δ C1 domain. Purified C1 domain of rat RasGRP binds to dibutyryl phorbol ester (19), although the affinity of this interaction was not established. It is also plausible to conclude that diacylglycerol is a physiological ligand for the C1 domain of RasGRP. The cellular distribution of the C1 domains is what is expected if their location is dictated by the distribution of diacylglycerol, as almost all of this lipid is found in internal membranes. Serum stimulation, PC-PLC treatment, or the addition of short-chain diacylglycerols did not drive translocation of the mRasGRP C1 domain to the plasma membrane, but this cannot be taken as evidence that diacylglycerol is not a ligand for the RasGRP C1 domain, because the PKC-δ C1 domain also failed to translocate in response to these stimuli. Transient increases in diacylglycerol concentrations at the plasma membrane produced by serum stimulation, treatment with PC-PLC, or short-chain diacylglycerol addition may be sufficient to induce translocation to the plasma membrane of C1 domains that are free in the cytosol (48) but might not be able to attract a significant portion of C1 domains that are already bound to diacylglycerol in internal membranes. In contrast, PMA could draw C1 domains away from internal membranes, because its higher affinity would allow it to out-compete diacylglycerol in internal membranes for C1 domain binding. PC-PLC treatment could cause translocation of the C1 domains to lipid droplets if diacylglycerol is continually generated at the plasma membrane but then rapidly transported to lipid droplets (50). The site of the highest and most stable diacylglycerol concentrations, and thus the site of translocation of the C1 domains, would therefore be the lipid droplets rather than the plasma membrane.
The observed distribution of RasGRP within NIH 3T3 cells raises the question of where it normally functions. A small but functionally significant portion of RasGRP could be transiently recruited to the plasma membrane during the very brief period of diacylglycerol accumulation there following serum stimulation (42, 48) and thus could be transiently brought in contact with plasma membrane-localized Ras GTPases. It should be noted that immunofluorescence studies of Sos1 and RasGRF-2 in serum-stimulated or ionophore-stimulated cells show predominant cytoplasmic distribution, with no detectable concentration at the plasma membrane (2, 21), despite the evidence that these stimuli cause translocation of these GEFs to particulate fractions of the cell and stimulate GTP loading of Ras. Therefore, it may generally be the case that translocation signals are effective even if they are able to colocalize only a minor portion of a GEF with its GTPase targets. The localization of a considerable portion of CXR-FL at the plasma membranes of PMA-stimulated cells suggests that this extended form of mRasGRP is indeed specialized to some extent to being targeted to the plasma membrane. This is comparable to the way that some but not all isoforms of PKC are targeted to and held at the plasma membrane following PMA stimulation (26). This retention function is evidently not needed for mRasGRP function when it is overexpressed in NIH 3T3 cells, but it may make a significant contribution to the process of RasGRP activation under more physiological conditions, e.g., increasing the number of encounters with Ras after a RasGRP molecule has been attracted to the plasma membrane by local and transient bursts of diacylglycerol production.
Is membrane localization sufficient to fully activate RasGRP? The ability of the prenylated form of RasGRP to maximally activate MAP kinases in the absence of serum suggests that this is the case. We were not able to show that PMA-induced translocation could fully activate mRasGRP, due to the ability of PMA treatment to saturate ERK activation via RasGRP-independent mechanisms. However, the ability of RasGRP to stimulate GTP loading of Ras in vivo is considerably increased by PMA treatment (19). PMA also substituted for serum in enabling mRasGRP or rat RasGRP (19) expression to induce cell transformation. Given the nature of the transformation assay, it is not possible to determine whether this represents a direct effect of PMA on RasGRP or a synergy between RasGRP and PKC signalling. However, the observation that all prenylated forms of RasGRP and the sporadic transformants arising via expression of CΔ1 did not require PMA for transformation while all of the C1 domain-containing forms of mRasGRP did is most simply explained by assuming that PMA was acting directly on the C1 domain of RasGRP.
Although the C1 domain and membrane localization appear to have dominant roles in activating RasGRP in NIH 3T3 cells, there may be additional modes of regulation of RasGRP which are operative only when RasGRP is expressed at physiological levels in lymphocytes. The activation of both Sos and RasGRF has recently been shown to involve complex interactions and cooperativity between multiple domains, with membrane localization being required but not always fully sufficient for activation (6, 7, 12, 25, 54, 62).
Potential roles of RasGRP in lymphocytes.
Ras activation can be induced in lymphocytes by tyrosine kinase-coupled receptors for antigen, cytokines, or costimulators, by G protein-coupled receptors, and by phorbol ester treatment (8, 18, 45). Given that most of these trigger or mimic diacylglycerol production, there is considerable potential for RasGRP to participate in the diverse processes that activate Ras in lymphocytes.
For both B-cell and T-cell antigen receptors, Ras activation seems to be at least partially dependent on tyrosine kinases, and the stimulation of these receptors can lead to the formation of complexes of Shc, Grb2, and/or Sos (31, 39, 58, 60), suggesting that Ras activation can occur through a mechanism similar to the relatively well-characterized recruitment of Sos to receptor tyrosine kinases such as epidermal growth factor receptor. However, Sos has not yet been detected in antigen receptor complexes (8, 49). This leaves open the possibility that a GEF other than Sos is responsible for at least some of the Ras activation stimulated by antigen receptors. Part of the Ras activation stimulated by the T-cell receptor may also be due to Ras GAP inhibition rather than GEF activation (17, 35). Lymphocytes have a Ras activation pathway that is initiated by phorbol ester treatment, and this is only partially or not at all dependent on tyrosine kinase activity (17, 35). Most of this phorbol ester-stimulated activation of Ras seems to be mediated by PKCs (35), but it is certainly possible that RasGRP could also contribute, perhaps with a dependence on concurrent PKC activity.
Many G protein-coupled receptors are potent activators of both PLCs and Ras (45). Ras activation via some G protein-coupled receptors seems to be at least partially dependent on a tyrosine kinase(s) and may occur via Shc-Grb2-Sos complex formation (13, 41) or via RasGRF (43). None of these studies have been conducted with cells which express RasGRP, and so it is presently an open question whether RasGRP plays a role in G protein-coupled receptor signalling in lymphocytes.
In some B-cell types, CD40 ligation results in transient Ras and/or Erk activation (30, 40), and this can be partially suppressed by calphostin C (30), which disrupts the ability of C1 domains to bind to diacylglycerol. Since Ras activation via CD40 is not suppressed by specific inhibitors of PKCs (40), RasGRP is a good candidate for mediating this diacylglycerol-dependent route to Ras activation.
ACKNOWLEDGMENTS
We thank Rosemary Cornell for very helpful discussions on lipid metabolism and localization and William Pear for provision of BOSC 23 cells and advice on using them.
This work was supported by grants from the National Cancer Institute of Canada, the Medical Research Council of Canada, the British Columbia Health Research Foundation, and the Leukemia Research Fund to R.J.K. and by grants from the National Institutes of Health (CA42978, CA55008, and CA63071) to J.C.D. I.P.W. is a Research Fellow of the National Cancer Institute of Canada supported by funds provided by the Terry Fox Run, and L.A.P. was supported by the J. M. Warren Studentship award provided by the British Columbia Cancer Foundation.
REFERENCES
- 1.Albright C F, Giddings B W, Liu J, Vito M, Weinberg R A. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J. 1993;12:339–347. doi: 10.1002/j.1460-2075.1993.tb05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronheim A, Engelberg D, Li N, Al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 3.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc Natl Acad Sci USA. 1993;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M, Der C J. Emerging concepts in the Ras superfamily of GTP-binding proteins. FASEB J. 1993;7:750–759. doi: 10.1096/fasebj.7.9.8330683. [DOI] [PubMed] [Google Scholar]
- 6.Buchsbaum R, Telliez J-P, Goonesekera S, Feig L A. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne J L, Paterson H F, Marshall C J. p21 Ras activation by the guanine nucleotide exchange factor Sos requires the Sos/Grb2 interaction and a second ligand-dependent signal involving the Sos N-terminus. Oncogene. 1996;13:2005–2065. [PubMed] [Google Scholar]
- 8.Cantrell D. T cell antigen receptor signal transduction pathways. Annu Rev Immunol. 1996;14:259–274. doi: 10.1146/annurev.immunol.14.1.259. [DOI] [PubMed] [Google Scholar]
- 9.Cen H, Papageorge G, Vass W C, Zhang K, Lowy D R. Regulated and constitutive activity by CDC25Mm (GRF), a Ras-specific exchange factor. Mol Cell Biol. 1993;13:7718–7724. doi: 10.1128/mcb.13.12.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepko C L, Roberts B E, Mulligan R C. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984;84:1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- 11.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to Grb2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 12.Chen R-H, Corbalan-Garcia S, Bar-Sagi D. The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J. 1997;16:1351–1359. doi: 10.1093/emboj/16.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y-H, Grall D, Salcini A E, Pelicci P G, Pouysségur J, Van Obberghen-Schilling E. Shc adaptor proteins are key transducers of mitogenic signaling mediated by the G protein-coupled thrombin receptor. EMBO J. 1996;15:1037–1044. [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman R A, Zeisel S H. Diacylglycerol metabolism in cellular membranes. In: Gross R W, editor. Advances in lipidology. Greenwich, Conn: JAI Press; 1996. pp. 337–366. [Google Scholar]
- 15.Craig W, Kay R, Cutler R L, Lansdorp P M. Expression of Thy-1 on human hematopoietic progenitor cells. J Exp Med. 1993;177:1331–1342. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis R J. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 17.Downward J, Graves J D, Warne P H, Rayter S, Cantrell D A. Stimulation of p21ras upon T-cell activation. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- 18.Downward J. Control of Ras activation. Cancer Surveys. 1996;27:87–100. [PubMed] [Google Scholar]
- 19.Ebinu J O, Bottorff D A, Chan E Y W, Stang S L, Dunn R J, Stone J C. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 20.Engler-Blum G, Meier M, Frank J, Muller G A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 21.Fam N P, Fan W-T, Wang Z, Zhang L-J, Chen H, Moran M F. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol Cell Biol. 1997;17:1396–1406. doi: 10.1128/mcb.17.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnsworth C L, Freshney N W, Rosen L B, Ghosh A, Greenberg M E, Feig L A. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 23.Feig L A. Guanine nucleotide exchange factors: a family of positive regulators of Ras and related GTPases. Curr Opin Cell Biol. 1994;6:204–211. doi: 10.1016/0955-0674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 24.Feig L A, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 25.Freshney N W, Goonesekera S D, Feig L A. Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett. 1997;407:111–115. doi: 10.1016/s0014-5793(97)00309-8. [DOI] [PubMed] [Google Scholar]
- 26.Goodnight J, Mischak H, Kolch W, Mushinski J F. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH 3T3 fibroblasts. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- 27.Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, Kurata T, Matsuda M. Identification of Rap1 as a target for the Crk SH3 domain binding guanine nucleotide-releasing factor C3G. Mol Cell Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotoh T, Niino Y, Tokuda M, Hatase O, Nakamura S, Matsuda M, Hattori S. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J Biol Chem. 1997;272:18602–18607. doi: 10.1074/jbc.272.30.18602. [DOI] [PubMed] [Google Scholar]
- 29.Graham S M, Vojtek A B, Huff S Y, Cox A D, Clark G J, Cooper J A, Der C J. TC21 causes transformation by Raf-independent signaling pathways. Mol Cell Biol. 1996;16:6132–6140. doi: 10.1128/mcb.16.11.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulbins E, Brenner B, Schlottmann K, Koppenhoefer U, Linderkamp O, Coggeshall K M, Lang F. Activation of the Ras signaling pathway by the CD40 receptor. J Immunol. 1996;157:2844–2850. [PubMed] [Google Scholar]
- 31.Harmer S L, DeFranco A L. Shc contains two Grb2 binding sites needed for efficient formation of complexes with Sos in B lymphocytes. Mol Cell Biol. 1997;17:4087–4095. doi: 10.1128/mcb.17.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu C D, Kariya K, Kotani G, Shirouzu M, Yokohama S, Kataoka T. Co-association of Rap1A and Ha-Ras with Raf-1 N-terminal region interferes with Ras-dependent activation of Raf-1. J Biol Chem. 1997;272:11702–11705. doi: 10.1074/jbc.272.18.11702. [DOI] [PubMed] [Google Scholar]
- 33.Hurley J H, Newton A C, Parker P J, Blumberg P M, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikura I. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- 35.Izquierdo M, Downward J, Graves J D, Cantrell D A. Role of protein kinase C in T-cell antigen receptor regulation of p21ras: evidence that two p21ras regulatory pathways coexist in T cells. Mol Cell Biol. 1992;12:3305–3312. doi: 10.1128/mcb.12.7.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan J M, Mardon G, Bishop J M, Varmus H E. The first seven amino acids encoded by the v-src oncogene act as a myristylation signal: lysine 7 is a critical determinant. Mol Cell Biol. 1988;8:2435–2441. doi: 10.1128/mcb.8.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Kay, R. J. Unpublished data.
- 37.Lai C-C, Boguski M, Broek D, Powers S. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol Cell Biol. 1993;13:1345–1352. doi: 10.1128/mcb.13.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larrodera P, Cornet M E, Diaz-Meco M T, Lopez-Barahona M, Diaz-Laviada I, Guddal P H, Johansen T, Moscat J. Phospholipase C-mediated hydrolysis of phosphatidylcholine is an important step in PDGF-stimulated DNA synthesis. Cell. 1990;61:1113–1120. doi: 10.1016/0092-8674(90)90074-o. [DOI] [PubMed] [Google Scholar]
- 39.Li B-Q, Subleski M, Fusaki N, Yamamoto T, Copeland T, Princler G L, Kung H-F, Kamata T. Catalytic activity of the mouse guanine nucleotide exchanger mSos is activated by Fyn tyrosine kinase and the T-cell antigen receptor in T cells. Proc Natl Acad Sci USA. 1996;93:1001–1005. doi: 10.1073/pnas.93.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y-Y, Baccam M, Waters S B, Pressin J E, Bishop G A, Koretzky G A. CD40 ligation results in protein kinase C-independent activation of ERK and JNK in resting murine splenic B cells. J Immunol. 1996;157:1440–1447. [PubMed] [Google Scholar]
- 41.Lopez-Ilasaca M, Crespo P, Pellici P G, Gutkind J S, Wetzker R. Linkage of G protein-coupled receptors to the MAPK signaling pathway through PI-3 kinase γ. Science. 1997;275:394–397. doi: 10.1126/science.275.5298.394. [DOI] [PubMed] [Google Scholar]
- 42.Martin T F J, Hsieh K-P, Porter B W. The sustained phase of hormone-stimulated diacylglycerol accumulation does not activate protein kinase C in GH3 cells. J Biol Chem. 1990;265:7623–7631. [PubMed] [Google Scholar]
- 43.Mattingly R R, Macara I G. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein βγ subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 44.Mayer B J, Eck M J. SH3 domains: minding your p’s and q’s. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 45.Moolenaar W H, Kranenburg O, Postuma F R, Zondag G C M. Lysophosphatidic acid: G-protein signalling and cellular responses. Curr Opin Cell Biol. 1997;9:168–173. doi: 10.1016/s0955-0674(97)80059-2. [DOI] [PubMed] [Google Scholar]
- 46.Morrison D K, Cutler R E. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 47.Newton A C. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 48.Oancea E, Teruel M N, Quest A F G, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signalling in living cells. J Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osman N, Lucas S C, Turner H, Cantrell D. A comparison of the interaction of Shc and the tyrosine kinase ZAP-70 with the T cell antigen receptor ζ chain tyrosine-based activation motif. J Biol Chem. 1995;279:13981–13986. doi: 10.1074/jbc.270.23.13981. [DOI] [PubMed] [Google Scholar]
- 50.Pagano R E, Longmuir K J, Martin O C. Intracellular translocation and metabolism of a fluorescent phosphatidic acid analogue in cultured fibroblasts. J Biol Chem. 1983;258:2034–2040. [PubMed] [Google Scholar]
- 51.Park W, Mosteller R D, Broek D. Amino acid residues in the CDC25 guanine nucleotide exchange factor critical for interaction with Ras. Mol Cell Biol. 1994;14:8117–8122. doi: 10.1128/mcb.14.12.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pyszniak A M, Welder C A, Takei F. Cell surface distribution of high-avidity LFA-1 detected by soluble ICAM-1-coated microspheres. J Immunol. 1994;152:5241–5249. [PubMed] [Google Scholar]
- 54.Qian X, Vass W C, Papageorge A G, Anborgh P H, Lowy D R. N-terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol Cell Biol. 1998;18:771–778. doi: 10.1128/mcb.18.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quilliam L A, Khosravi-Far R, Huff S Y, Der C J. Guanine nucleotide exchange factors: activators of the Ras superfamily of proteins. Bioessays. 1995;17:395–404. doi: 10.1002/bies.950170507. [DOI] [PubMed] [Google Scholar]
- 56.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphatidylinositol 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 58.Saxton T M, van Oostveen I, Bowtell D, Aebersold R, Gold M R. B cell antigen receptor cross-linking induces phosphorylation of the p21ras oncoprotein activators SHC and mSos1 as well as assembly of complexes containing SHC, GRB-2, mSos1, and a 145-kDa tyrosine-phosphorylated protein. J Immunol. 1994;153:623–636. [PubMed] [Google Scholar]
- 59.Shou C, Farnsworth C L, Neel B G, Feig L A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992;258:351–358. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- 60.Smit L, van der Horst G, Borst J. Sos, Vav, and C3G participate in B cell receptor-induced signaling pathways and differentially associate with Shc-Grb2, Crk, and Crk-L adaptors. J Biol Chem. 1996;271:8564–8569. doi: 10.1074/jbc.271.15.8564. [DOI] [PubMed] [Google Scholar]
- 61.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 62.Wang W, Fisher E M C, Jia Q, Dunn J M, Porfiri E, Downward J, Egan S E. The Grb2 binding domain of mSos1 is not required for downstream signal transduction. Nat Genet. 1995;10:294–300. doi: 10.1038/ng0795-294. [DOI] [PubMed] [Google Scholar]
- 63.Whitehead I, Kirk H, Kay R. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang G, Kazanietz M G, Blumberg P M, Hurley J H. Crystal structure of the Cys2 activator-binding domain of protein kinase Cδ in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]