Summary
For patients with advanced stage non-small cell lung cancer (NSCLC), treatment strategies have changed significantly due to the introduction of targeted therapies and immunotherapy. In the last few years, we have seen an explosive growth of newly introduced targeted therapies in oncology and this development is expected to continue in the future. Besides primary targetable aberrations, emerging diagnostic biomarkers also include relevant co-occurring mutations and resistance mechanisms involved in disease progression, that have impact on optimal treatment management. To accommodate testing of pending biomarkers, it is necessary to establish routine large-panel next-generation sequencing (NGS) for all patients with advanced stage NSCLC. For cost-effectiveness and accessibility, it is recommended to implement predictive molecular testing using large-panel NGS in a dedicated, centralized expert laboratory within a regional oncology network. The central molecular testing center should host a regional Molecular Tumor Board and function as a hub for interpretation of rare and complex testing results and clinical decision-making.
Keywords: Predictive biomarker testing, Next-generation sequencing, Non-small cell lung cancer
Key messages.
-
•
The number of clinically relevant biomarkers in patients with advanced stage NSCLC has increased rapidly over the past years and this is expected to continue faster and broader;
-
•
Molecular tumor profiling for all clinically relevant biomarkers in patients with advanced stage NSCLC, including biomarkers for which targeted therapies are available through clinical trials, off-label use, or compassionate use programs, should be performed;
-
•
It is necessary to implement routine large-panel next-generation sequencing (NGS) for all patients with advanced stage NSCLC to enable testing of all clinically relevant biomarkers, both for biomarkers relevant today as well as biomarkers relevant in the (near) future;
-
•
Predictive molecular testing using large-panel NGS should be centralized in a dedicated, centralized expert laboratory within a regional oncology network, for cost-effectiveness and to facilitate equal access to biomarker testing to all patients;
-
•
Within a regional oncology network, the central institution performing molecular testing should also host a regional Molecular Tumor Board.
Introduction
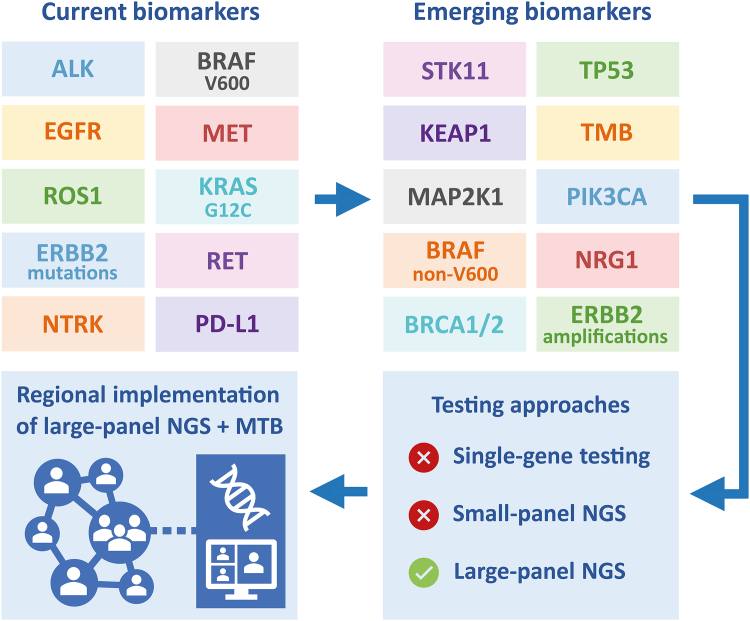
Optimal treatment of patients with advanced stage non-small cell lung cancer (NSCLC) requires appropriate molecular testing of predictive biomarkers, as it provides information that is essential for establishing the appropriate treatment option for each patient. In the corresponding review of this Clinical Series on lung cancer, we describe the current application of predictive molecular testing in patients with advanced stage NSCLC from a European perspective, in relation to European Medicines Agency (EMA)-approved targeted therapies.1 The review paper provides an overview of EMA-approved targeted therapies in patients with NSCLC and presents the current availability of targeted therapies and guidelines on predictive biomarker testing in patients with NSCLC in eleven European countries. In the current viewpoint paper, we look beyond the current guidelines and provide our perspective of what is required with regard to biomarker testing in the (near) future to be able to provide optimal care to patients with advanced stage NSCLC. Future perspectives of predictive biomarker testing are discussed and recommendations are given with regard to the application of molecular testing for treatment decision-making in advanced stage NSCLC. Fig. 1 provides a simplified illustration of the authors’ perspective described in this viewpoint.
Fig. 1.
Recommendations for molecular biomarker testing in metastatic NSCLC. TMB, tumor-mutational burden; NGS, next-generation sequencing; MTB, Molecular Tumor Board.
Molecular biomarkers
For patients with histologically or cytologically confirmed metastatic non-squamous NSCLC, the current Clinical Practice Guidelines of the European Society for Medical Oncology (ESMO) for oncogene-addicted advanced-stage NSCLC (2023) recommend to perform molecular testing for the following predictive biomarkers: EGFR, KRAS (G12C), BRAF (V600), ERBB2, and MET (exon 14 skipping) mutations, MET amplifications, ALK, ROS1, RET, and NTRK1/2/3 fusions, and PD-L1 expression.2 These ESMO guidelines are in line with the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology (2022) and the American Society of Clinical Oncology (ASCO) guidelines (2022).3,4 As of July 2023, for all these biomarkers, except for ERBB2, at least one EMA-approved targeted therapy is available.1 However, beyond predictive markers for EMA-approved therapies, drugs are available through clinical trials, off-label use, or compassionate use programs, which require additional molecular testing. There are several emerging biomarkers that affect optimal treatment decision-making due to their predictive and/or prognostic value (for some examples see Table 1). In the paragraphs below, these biomarkers are described in the context of newly diagnosed advanced stage NSCLC (paragraphs 2.1, 2.2, and 2.3) and advanced stage NSCLC after progression on targeted therapy (paragraph 2.4).
Table 1.
Examples of emerging molecular biomarkers and their expected/potential relevance.
| Emerging predictive biomarkers in non-squamous NSCLC | |
|---|---|
| Biomarker | Relevance |
| ERBB2 mutation | Predictive value for response to HER2-targeted therapy (e.g., trastuzumab deruxtecan) |
| ERBB2 amplification | Predictive value for response to HER2-targeted therapy (e.g., ado-trastuzumab) |
| NRG1 fusion | Predictive value for response to NRG1-targeted therapy |
| MAP2K1 mutation | Predictive value for response to MEK inhibitors |
| BRAF non-V600 mutation | Predictive value for response to BRAF/MEK inhibitors |
| TP53 mutation + KRAS mutation | Predictive value for response to immunotherapy |
| TP53 mutation + EGFR mutation | Predictive value for response to ramucirumab + erlotinib |
| High TMB + KRAS mutation | Predictive value for response to immunotherapy |
| Emerging biomarkers associated with primary resistance to targeted therapy | |
|---|---|
| Biomarker | Relevance |
| PIK3CA mutation + EGFR mutation | Negative prognostic value for response to EGFR TKI |
| KEAP1 mutation + KRAS mutation | Negative predictive value for response to KRASG12C-inhibitor treatment |
| CDKN2A mutation + KRAS mutation | Negative predictive value for response to KRASG12C-inhibitor treatment |
| SMARCA4 mutation + KRAS mutation | Negative predictive value for response to KRASG12C-inhibitor treatment |
| RB1 mutation + TP53 mutation (in EGFR-mutated NSCLC) | Prognostic value for later neuroendocrine trans-differentiation of tumor |
| Emerging biomarkers associated with primary resistance to non-targeted therapy | |
|---|---|
| Biomarker | Relevance |
| STK11 mutation | Negative predictive value for response to immunotherapy treatment |
| KEAP1 mutation | Negative predictive value for response to immunotherapy treatment |
| EGFR driver mutation | Negative predictive value for response to immunotherapy treatment |
| ALK fusion | Negative predictive value for response to immunotherapy treatment |
| KRAS driver mutation | Predictive value for response to immunotherapy treatment |
| BRAF driver mutation | Predictive value for response to immunotherapy treatment |
| NOTCH1/2/3 mutation | Predictive value for response to immunotherapy treatment |
| Emerging biomarkers associated with acquired resistance to targeted therapy | |
| MET amplification (as resistance mechanism to EGFR TKI treatment) | Potentially targetable with MET-targeted therapy |
| CDK6 amplification (as resistance mechanism to EGFR TKI treatment) | Potentially targetable with CDK6-targeted therapy |
| RET fusion (as resistance mechanism to EGFR TKI treatment) | Potentially targetable with RET-targeted therapy |
| ALK mutation (as resistance mechanism to ALK inhibitor treatment) | Potentially targetable with alternative ALK-targeted therapy |
TMB, tumor-mutational burden; TKI, tyrosine kinase inhibitor; NSCLC, non-small cell lung cancer.
Molecular biomarkers for targeted therapy in trial setting
Molecular biomarkers for which there are ongoing clinical trials include NRG1 fusions (NCT02912949), MAP2K1 mutations (NCT04534283), ERBB2 mutations (NCT05048797, NCT02716116), ERBB2 amplifications (NCT04886804, NCT04143711), high c-MET expression (NCT03539536), BRAF non-V600 mutations (NCT04488003, NCT03843775), and BRCA1/2 mutations (NCT03845296).
With regard to ERBB2 mutations, the phase II DESTINY-Lung01 trial demonstrated the efficacy of trastuzumab deruxtecan in (predominantly) pretreated patients with ERBB2-mutated advanced stage NSCLC, with an overall response rate (ORR) of 55% (50 out of 91 patients) and a median progression-free survival (PFS) of 8·2 months.5 As of July 2023, its indication for therapeutic use in patients with advanced stage NSCLC has already been approved by the United Status Food and Drug Administration (FDA), but not by the EMA.
At present, evidence of clinical benefit of targeted therapy treatment in patients with an ERBB2 amplification, NRG1 fusion, MAP2K1 mutation, BRAF non-V600 mutation, or BRCA1/2 mutation is still limited.6, 7, 8, 9, 10 Nevertheless, patients with driver mutations without available EMA-approved targeted therapies, may still benefit from treatment with targeted therapy in a trial setting. As trial availability differs between countries and regions, it is recommended to perform molecular testing for those biomarkers for which clinical trials are ongoing within the region or country of the treating hospital. There are ongoing precision oncology trials in which FDA- and/or EMA-approved therapies are available outside of their approved indication label, including the Drug Rediscovery Protocol (DRUP), the MyPathway study, and the I-PREDICT trial (Investigation of Profile-Related Evidence Determining Individualized Cancer Therapy).11 DNA- and RNA-based large panel testing together would enable analysis of any biomarker for which targeted treatment, EMA-approved or experimental, is available. Also, large panels can easily be expanded to cover novel markers when new exploratory targeted treatments reach clinical trials. Already in 2020, the ESMO Precision Medicine Working Group recommended multigene sequencing in clinical research centers for many of the biomarkers stated in this paragraph.12
Biomarkers associated with primary resistance to targeted therapy
Some biomarkers in NSCLC may co-occur with therapeutically targetable biomarkers and carry prognostic and/or predictive value for treatment with targeted therapy, while not directly interacting with the targeted protein(s) (Table 1). For example, patients with therapeutically targetable EGFR-mutated NSCLC with co-occurring PIK3CA mutation have worse survival outcomes compared to patients with EGFR-mutated NSCLC without PIK3CA mutation.13 In this example, PIK3CA mutation status provides information to the patient and treating physician on the expected efficacy of a specific targeted therapy treatment and may warrant more frequent patient check-ups. Besides PIK3CA mutations, prognostic biomarkers include SMARCA4 co-mutations in KRASG12C-mutated NSCLC treated with sotorasib or adagrasib and TP53 co-mutations in ALK fusion-positive NSCLC treated with ALK inhibitors.14,15 Additionally, the simultaneous co-occurrence of TP53 and RB1 mutations in patients with EGFR-mutated NSCLC is associated with trans-differentiation of the tumor to a neuroendocrine phenotype.16
Other co-occurring mutations carry predictive value for alternative targeted treatment options. In patients with therapeutically targetable EGFR-mutated NSCLC treated with osimertinib, the co-occurrence of a (pathogenic) TP53 mutation has been associated with significantly shorter time to treatment discontinuation (TTD) when compared to patients with EGFR-mutated NSCLC with wildtype TP53.17 In patients with a common EGFR driver mutation (i.e., exon 19 deletion or L858R), the phase III RELAY study demonstrated improved clinical outcome of ramucirumab plus erlotinib treatment in patients with a co-occurring TP53 mutation at baseline, when compared to placebo plus erlotinib treatment.18 Ongoing clinical trials compare treatment outcome of ramucirumab plus erlotinib treatment versus mono-osimertinib treatment (REVOL858R), and ramucirumab plus osimertinib versus mono-osimertinib treatment (TORG1833, RAMOSE, NCT03909334).19, 20, 21 Due to these clinical implications, TP53 mutation status has already become part of routine molecular testing in patients with advanced stage NSCLC in many molecular laboratories.
Biomarkers associated with primary resistance to non-targeted therapy
At present, the expression of programmed death-ligand 1 (PD-L1) in NSCLC is used as the main predictive biomarker for therapy with immune checkpoint inhibitors (ICI). In recent years, it has become apparent that the occurrence of certain driver mutations in patients with NSCLC is associated with treatment efficacy of immunotherapy. For example, in patients with EGFR-mutated or ALK fusion-positive advanced stage NSCLC, treatment with (mono-)immunotherapy yields limited clinical efficacy, while patients with KRAS mutations commonly experience clinical benefit from either chemo-immunotherapy or immunotherapy alone.22 However, besides these driver mutations, co-occurring mutations in STK11, KEAP1, and/or SMARCA4 also provide a negative predictive value for ICI treatment (Table 1). For example, the presence of a co-occurring STK11 and/or KEAP1 mutation has been described to carry a negative predictive value for combined chemo-immunotherapy treatment in patients with KRASG12C-mutated NSCLC.23 Meanwhile, KEAP1 but not STK11 co-occurring mutations in patients with KRASG12C-mutated NSCLC have been associated with reduced treatment efficacy of sotorasib.24 Therefore, the presence of STK11, KEAP1, and/or SMARCA4 mutations may support alternative treatment options in selected patients. Additionally, low tumor-mutational burden (TMB) has been described to have negative predictive value for mono-immunotherapy treatment in patients with advanced stage PD-L1-positive NSCLC, regardless of STK11 and KEAP1 mutation status.25 Therefore we recommend that, besides TP53, these four biomarkers (TMB, STK11, KEAP1, SMARCA4) should be part of routine molecular testing in patients with advanced stage NSCLC due to the clinical consequences on treatment response. On the other hand, NOTCH1/2/3 mutations are associated with increased efficacy of immunotherapy in patients with advanced stage NSCLC.26
Biomarkers associated with acquired resistance to targeted therapy
Both type of primary driver aberration, as well as the prescribed drug have impact on the resulting patterns of resistance at disease progression. Resistance mechanisms may involve target-specific or other mechanisms, either within the originally affected or an alternative pathway, and even transformations to other histological subtypes are reported.27 For example, previously described resistance mechanisms to osimertinib treatment in patients with EGFR-mutated NSCLC include EGFR resistance mutations, MET amplification, amplification of cell cycle genes (e.g., CDK4), gene fusions (most commonly RET fusions), and DNA amplification of EGFR.28 Although some aberrations have been described as a resistance mechanism to different targeted therapies or molecular drivers (i.e., overlapping/common resistance mechanisms), distinct resistance mechanisms to specific therapies or drivers are also reported.27,29 For example, patients with ALK fusion-positive NSCLC treated with alectinib may develop ALK resistance mutations. Various resistant ALK mutations have been reported, with varying responses to the more than five other available ALK-inhibitors.30 Therefore, it is recommended to perform molecular testing for specific resistance mechanisms that can be expected to occur at disease progression.29
Molecular testing approaches
Most of the described biomarkers in sections 2.1, 2.2, 2.3 and 2.4, 2.1, 2.2, 2.3 and 2.4, 2.1, 2.2, 2.3 and 2.4, 2.1, 2.2, 2.3 and 2.4 are currently not included in the ESMO guidelines or national guidelines, but should be considered for inclusion in the next versions of national and European guidelines for molecular testing in NSCLC. To ensure equal access for all patients with NSCLC to any available drug, either EMA-approved or experimental, molecular testing should cover all relevant biomarkers. Restricting testing to only biomarkers in current guidelines would not offer all possible treatment options to each patient, and simultaneously limits experimental data to improve treatment of patients with NSCLC. In the United States, similar recommendations of comprehensive predictive biomarker testing in solid tumors have been made in an ASCO Provisional Clinical Opinion (2022).31
The currently available targeted therapies for patients diagnosed with advanced stage NSCLC already warrant the use of upfront next-generation sequencing (NGS) analysis with large panels rather than (sequential) single-gene testing. Model-based studies estimating the costs, positive test rates of clinically relevant molecular aberrations, and the mean time to appropriate treatment initiation using either NGS or single-gene testing approaches have demonstrated the benefits of upfront NGS testing. Using Europe-based cost estimates, Stenzinger et al. (2023), estimated cost differences favoring NGS compared to sequential single-gene testing approaches, with the largest difference in cost difference found in molecular testing of non-squamous NSCLC.32 Similar conclusions were drawn by modeling studies assessing the cost-effectiveness of NGS for patients with advanced stage NSCLC in the Netherlands and Spain.33,34 In North America, comparable studies have also favoured NGS to sequential single-gene testing in NSCLC.35,36
There are numerous commercial NGS panels for targeted sequencing of cancer samples, with differences in enrichment technique and panel comprehensiveness.37 Large NGS panels (>300 genes) offer a higher number of potentially clinically relevant markers, including single nucleotide polymorphisms (SNP), copy number variation (CNV), homologous recombination deficiency (HRD) signatures, microsatellite instability (MSI), rearrangements, and tumor-mutational burden (TMB). Examples are the TruSight Oncology 500 NGS panel (523 clinically relevant genes), FoundationOne CDx panel (324 relevant genes) and Agilent SureSelect Cancer CGP assay (679 relevant genes).38,39 Many, if not all, emerging biomarkers such as TP53, KEAP1, STK11, MAP2K1, PIK3CA, MET amplification, and TMB are included in these panels. In addition to large-panel NGS, whole-exome sequencing (WES) and whole-genome sequencing (WGS) are considered as even broader panels.40 However, today, WES and WGS require frozen or fresh tissue and cannot be performed on formalin-fixed paraffin-embedded (FFPE) samples, these global approaches are not yet suitable for analysis in routine care.
Our expert opinion is that predictive biomarker testing techniques should continue its shift towards large-panel testing, thereby enabling testing for all currently targetable molecular aberrations, while simultaneously facilitating molecular testing for emerging (predictive and/or prognostic) biomarkers for both EMA-approved and experimental therapies and consequently facilitate equal access to available systemic therapies for all patients.
Importantly, while large-panel NGS is essential for optimal treatment decision-making, tissue material of sufficient quantity and quality is not always available. For such cases, if re-biopsy is not favorable, single-gene tests or limited gene panels can be considered. Moreover, single-gene tests can also be of use as an orthogonal test method in case of doubtful NGS results. However, healthcare professionals must be aware that single-gene tests or limited gene panels may not provide all the molecular information needed for optimal treatment decision-making.
Centralization of molecular testing
Despite the recommendation of the ESMO Precision Medicine Working Group in 2020 to implement multigene sequencing in the current clinical setting12 and (near) future molecular testing requiring larger NGS panels to cover all upcoming predictive markers associated with new targeted therapies, today molecular testing is mostly decentralized and very heterogeneous among European countries as well as between laboratories within the same countries.41, 42, 43 In a low-volume throughput setting, large-panel NGS is expensive due to the relatively high costs of the sequencing machine acquisition, maintenance, and operational costs (e.g., sample preparation, expert personnel, data interpretation and processing). Therefore, optimal cost-effectiveness of large-panel NGS requires high-volume sample testing. Centralized molecular testing within a region, province, or metropolitan area, allows for referred, comprehensive predictive biomarker testing for all patients within its borders, due to optimal utilization of equipment and personnel. It prevents the necessity for individual institutions to perform molecular testing in-house, for whom large-panel testing is not cost-effective and would therefore revert to (sequential) small/medium-panel NGS or single-gene testing. Although turn-around-time might increase due to transport of tissue samples to the central laboratory, a centralized approach can simultaneously contribute to improving the turn-around time by minimization of waiting times for molecular tests due to sample batching as well as obtaining the complete molecular profile using a single large NGS panel assay. On the other hand, the turn-around-time of (central) molecular testing is defined by time-to-treatment and thus opportunities for improvement are larger volumes, efficient NGS workflows, robotization of workflows, optimizing data-flow and defining regional standard-operation-procedures for these workflows as well as logistics, also beyond the molecular testing procedure itself. The use of the same large panels is also suitable for predictive/prognostic testing in other malignancies, as well as diagnostic molecular testing, which is becoming a more common practice with incorporation of molecular characteristics in classifications of the World Health Organization (https://www.iarc.who.int/). Indeed, defining molecular characteristics of some thoracic tumor entities such as NUT midline carcinoma and SMARCA4-deficient undifferentiated carcinoma are already covered in many large NGS-panels, as well as other aberrations associated with metastatic tumors that may be confused with NSCLC. The use of a workflow for all oncologic molecular diagnostics practices using a single NGS large panel is cost-effective and can improve turn-around time and quality control. Additionally, the complexity of large-panel sequencing (and WES/WGS) analysis, interpretation of complex molecular results, differentiation between pathogenic, germline-associated and clinical non-relevant aberrations, and translation of molecular findings towards advice for appropriate treatment strategies, requires not only centralization but also embedding in a center using dedicated and innovative molecular approaches in precision medicine in malignancies other than NSCLC, in human genetics, bioinformatics, and artificial intelligence. A regionally centralized approach for comprehensive predictive biomarker testing in NSCLC has previously been described in certain (regions within) European countries, including the Netherlands, France, Wales, and the United Kingdom.44, 45, 46, 47, 48 From a broader perspective, the use of regional healthcare networks for the implementation of precision medicine – including genomic testing – has also been described in other European countries, including Germany, Italy, and Sweden.48 This centralization of molecular testing can be incorporated within existing regional oncology networks, wherein hospitals collaborate to provide care for patients with NSCLC. These dedicated regional testing centers are recommended to host Molecular Tumor Boards (MTBs) meant to discuss and provide treatment recommendation for patients with complex or rare molecular test results. Regional healthcare professionals should be able to participate and enroll patients for these discussions, whenever in-house expertise or therapy options are insufficient for optimal treatment of a patient with NSCLC. In the future, with the integration of more complex plasma-derived circulating tumor DNA NGS analysis into routine care and the ever-increasing number of clinically relevant molecular aberrations, a central role of MTBs remains essential in treatment-decision-making. Using a centralized approach, testing centers can develop and upkeep the required expertise in molecular diagnostics and targeted treatment outcome, while providing optimal treatment recommendations to all participating members in the regional network when requested. This approach also promotes regional collaboration, educational development, and (clinical) research, with possibilities for regional MTBs to form national research databases.
Conclusion
Despite the comprehensive set of currently recommended predictive biomarkers, the number of clinically relevant molecular aberrations is expected to keep growing in the near future, not only in patients with advanced stage NSCLC, but also in early-stage NSCLC49 and other malignancies. The use of large DNA- and RNA-based NGS panels covering all predictive and diagnostic markers of (nearly) all malignancies is required for workflow-efficiency, practical feasibility, and turn-around time. From the perspective of cost-effectiveness, concentrating knowledge of (rapidly changing) technologies, quality assurance of testing, expertise on interpretation of molecular test results, and assistance in optimal treatment-decision making, a regionally centralized approach for predictive biomarker testing is highly recommended. Centralized biomarker testing for patients with NSCLC can be particularly most feasible if incorporated in the structure of existing regional oncological care networks and national regulations. It is recommended that the molecular testing center within each oncology network hosts a regional MTB, thereby adopting an assisting role, with continuously expanding expertise, in the interpretation of test results and clinical-decision making for those patients with rare or complex molecular aberrations for which there is insufficient in-house expertise or limited treatment options. It is essential that all healthcare professionals within an oncology network are able to participate and request advice on treatment and testing for their patients in the regional MTB.
Contributors
VdJ contributed to the conceptualization, data curation, methodology, visualization, writing - original draft, writing - review & editing; WT contributed to writing - original draft, writing - review & editing; AB, JB, LB, RB, MGOF, LH, MH, PH, AJ, LvK, IK, JCM, KM, SP, AR, and JW contributed to the investigation, writing - review & editing; ES and AvdW contributed to the conceptualization, methodology, supervision, writing - original draft, writing - review & editing.
Declaration of interests
WT has received consulting fees from Merck Sharp and Dohme, Bristol-Myers Squibb, and Altana (fees to institution), is board member of Dutch Society of Pathology and member of Council for Research and Innovation of the Federation of Medical Specialists (FMS); AB has received consulting fees from Sanofi (OncoCollective advisory board), payments or honoraria from Roche (oral presentation), support for attending ASCO 2023 from Pfizer; JB has received research grants from Amgen and Bristol-Myers Squibb, lecture honoraria from AstraZeneca, Merck Sharp and Dohme, Roche, Pfizer, Bristol-Myers Squibb, Boehringer Ingelheim, Novartis, GSK, Eli Lilly, Amgen, and Sanofi, and support for attending meetings and/or travel from Amgen; LB has received grants or contract from Takeda, Roche, AstraZeneca, and Bristol-Myers Squibb, payments or honoraria from Invitae, Eli Lilly, AstraZeneca, Roche, Merck Sharp and Dohme, Merck, Bristol-Myers Squibb, Pfizer, Novartis, Takeda, and Janssen, has participated in data safety monitoring boards or advisory boards for Invitae, Eli Lilly, AstraZeneca, Roche, Merck Sharp and Dohme, Merck, Bristol-Myers Squibb, Pfizer, Novartis, Takeda, and Janssen, is Int. Secretary of the Austrian Society of Pathology, PPS Membership and Awards Committee, and is Member of the Mesothelioma Committee of IASLC; RB has received payments or honoraria for lectures and advisory boards for Abbvie, Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer-Ingelheim, Illumina, Janssen, Lilly, Merck-Serono, Merck Sharp and Dohme, Novartis, Qiagen, Pfizer, Roche, and Targos MP Inc., has received payments for expert testimony from Merck Sharp and Dohme, is co-founder and co-owner of Gnothis Inc (SE), Timer Therapeutics Inc (GE); MGOF has received fees for advisory boards participation, meetings, or as invited speaker, from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Lilly, Janssen, Merck Sharp and Dohme, Pfizer, Roche Farmacêutica Química Lda, and Takeda; MH has received payments or honoraria from Merck Sharp and Dohme, Roche, Lilly, AstraZeneca, and Takeda; PH has received consulting fees from AstraZeneca, Roche, Abbvie, Qiagen, and Pfizer, has received payments or honoraria from Thermofisher Scientist, Novartis, Janssen, AstraZeneca, and Biocartis, has received support for attending meetings and/or travel from Thermofisher Scientist, Novartis, Janssen, AstraZeneca, and Biocartis, has participated on a Data Safety Monitoring Board or Advisory Board for Thermofisher Scientist, Novartis, Janssen, AstraZeneca, Biocartis, Bristol-Myers Squibb, Sanofi, Roche, and Abbvie; LvK has received institutional grants or contract from Amgen, AstraZeneca, Bayer, Janssen-Cilag, Merck, Roche, and Servier, has received institutional payments or honoraria from Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, Eli Lilly, Novartis, Pfizer, and Roche, has received institutional support for attending meeting and/or travel from Roche, has participated on a Data Safety Monitoring Board or Advisory Board from Janssen-Cilag, Merck, and Roche, has a leadership or fiduciary role in the Commission Personalized Medicine – Belgium (unpaid), has stock or stock options in Cyclomics (institutional/personal); JCM has received payments or honoraria from Lilly Portugal, Novartis Portugal, Roche Portugal, Fresco Produções, Janssen-Cilag Portugal, Pierre Fabre Portugal, and Servier Portugal; has participated on a Data Safety Monitoring Board or Advisory Board of Roche Portugal; KM has received payments or honoraria from Takeda, Janssen, Pfizer, Merck Sharp and Dohme, Bristol-Myers Squibb, Roche, Amgen, Novartis, and Eli Lilly, has participated on a Data Safety Monitoring Board or Advisory Board of BMS, Takeda, AstraZeneca, Amgen, Roche, Boehringer Ingelheim, and Janssen; SP has received consulting fees (personal fees) from Amgen, AstraZeneca, Bayer, Blueprint, Bristol-Myers Squibb, Boehringer Ingelheim, Daiichi Sankyo, GSK, Guardant Health, Incyte, Janssen, Lilly, Merck Serono, Merck Sharp and Dohme, Novartis, Roche, Takeda, Pfizer, Seattle Genetics, Turning Point Therapeutics, and EQRx, has received payments or honoraria (personal fees) from AstraZeneca, Bayer, Guardant Health, Janssen, Merck Serono, Roche, and Takeda, has received payments for expert testimony (personal fees) from Roche, and Merck Serono, has received reimbursement of travel expenses from Janssen, and Roche, has participated on a Data Safety Monitoring Board or Advisory Board as per consulting fees, has a leadership role or fiduciary role (all unpaid) in the British Thoracic Oncology Group, ALK Positive UK, Lung Cancer Europe, Ruth Strauss Foundation, Mesothelioma Applied Research Foundation, and ETOP-IBCSG Partners Foundation Board; AR has received consulting fees for participation on advisory boards of AstraZeneca, Bristol-Myers Squibb, Eli Lilly, Amgen, Merck Sharp and Dohme, Gilead, and Sanofi, has received honoraria for lectures and presentations from AstraZeneca, Bristol-Myers Squibb, Eli Lilly, Amgen, Merck Sharp and Dohme, Gilead, Roche, Merck-Serono, has received support for attendings meetings and travel from Sanofi, Gilead, Roche, and Bristol-Myers Squibb; JW has received payments or honoraria, has received support for attendings meetings and travel, and has participated in a Data Safety Monitoring Board or Advisory Board, from Amgen, AstraZeneca, Bayer, Blueprint, BMS, Boehringer-Ingelheim, Chugai, Daiichi Sankyo, Janssen, Lilly, Loxo, Merck, Mirati, MSD, Novartis, Nuvalent, Pfizer, Pierre-Fabre, Roche, Seattle Genetics, Takeda, and Turning Point; ES has received unrestricted grants (all paid to UMCG institution) from Abbott, Biocartis, AstraZeneca, Invitae/Archer, Bayer, Bio-Rad, Roche, Agena Bioscience, CC Diagnostics, MSD/MERCK, and Boehringer Ingelheim, has received consulting fees (all paid to UMCG institution) from MSD/Merck, AstraZeneca, Roche, Novartis, Bayer, BMS, Lilly, Amgen, Illumina, Agena Bioscience, CC Diagnostics, Janssen Cilag (Johnson & Johnson), Astellas Pharma, GSK, Sinnovisionlab, and Sysmex, has received payments or honoraria (all paid to UMCG institution) from Bio-Rad, Seracare, Roche, Biocartis, Lilly, Agena Bioscience, and Illumina, has received support for attending meetings and/or travel from BioRad, Biocartis, Ageno Sciences, and Illumina, is a board member for the Dutch Society of Pathology (unpaid), European Society of Pathology (unpaid), European Liquid Biopsy Society (unpaid), is a secretary/member of the advisory committee for assessment of molecular diagnostics (cieBOD) (honoraria paid to UMCG institution), is committee member of national guideline advisory (honoraria paid to UMCG institution); AvdW, has received grants or contracts from AstraZeneca, Boehringer-Ingelheim, Pfizer, Roche, and Takeda, has received consulting fees from AstraZeneca, Janssen, Lilly, Roche, and Takeda, has received payments or honoraria from AstraZeneca, BMS, Lilly, Pfizer, and Roche, has a leadership or fiduciary role in the oncology section NVALT, guideline committee NSCLC and CUP, dure geneesmiddelen committee NVALT and FMS. All other authors declare no competing interests.
Contributor Information
Ed Schuuring, Email: e.schuuring@umcg.nl.
Anthonie J. van der Wekken, Email: a.j.van.der.wekken@umcg.nl.
References
- 1.de Jager V.D., Timens W., Bayle A., et al. Developments in predictive biomarker testing and targeted therapy in advanced stage non-small cell lung cancer and their application across European countries. Lancet Reg Health Eur. 2024;38:100838. doi: 10.1016/j.lanepe.2024.100838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriks L.E., Kerr K.M., Menis J., et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34(4):339–357. doi: 10.1016/j.annonc.2022.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Ettinger D.S., Wood D.E., Aisner D.L., et al. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2022;20(5):497–530. doi: 10.6004/jnccn.2022.0025. [DOI] [PubMed] [Google Scholar]
- 4.Singh N., Temin S., Baker S., Jr., et al. Therapy for stage IV non-small-cell lung cancer with driver alterations: ASCO living guideline. J Clin Oncol. 2022;40(28):3310–3322. doi: 10.1200/JCO.22.00824. [DOI] [PubMed] [Google Scholar]
- 5.Li B.T., Smit E.F., Goto Y., et al. Trastuzumab deruxtecan in HER2-mutant non-small-cell lung cancer. N Engl J Med. 2022;386(3):241–251. doi: 10.1056/NEJMoa2112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganti A.K., Rothe M., Mangat P.K., et al. Pertuzumab plus trastuzumab in patients with lung cancer with ERBB2 mutation or amplification: results from the targeted agent and profiling utilization registry study. JCO Precis Oncol. 2023;7 doi: 10.1200/PO.23.00041. [DOI] [PubMed] [Google Scholar]
- 7.Cheng M.L., Lee J.K., Kumar R., et al. Response to MEK inhibitor therapy in MAP2K1 (MEK1) K57N non-small-cell lung cancer and genomic landscape of MAP2K1 mutations in non-small-cell lung cancer. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00382. [DOI] [PubMed] [Google Scholar]
- 8.Laskin J., Liu S.V., Tolba K., et al. NRG1 fusion-driven tumors: biology, detection, and the therapeutic role of afatinib and other ErbB-targeting agents. Ann Oncol. 2020;31(12):1693–1703. doi: 10.1016/j.annonc.2020.08.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dankner M., Wang Y., Fazelzad R., et al. Clinical activity of mitogen-activated protein kinase-targeted therapies in patients with non-V600 BRAF-mutant tumors. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z., Wang K., Zhao L., Gong L. BRCA2 mutation in advanced lung squamous cell carcinoma treated with Olaparib and a PD-1 inhibitor: a case report. Front Oncol. 2023;13 doi: 10.3389/fonc.2023.1190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song I.W., Vo H.H., Chen Y.S., et al. Precision oncology: evolving clinical trials across tumor types. Cancers. 2023;15(7) doi: 10.3390/cancers15071967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosele F., Remon J., Mateo J., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Qiu X., Wang Y., Liu F., et al. Survival and prognosis analyses of concurrent PIK3CA mutations in EGFR mutant non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. Am J Cancer Res. 2021;11(6):3189–3200. [PMC free article] [PubMed] [Google Scholar]
- 14.Kron A., Alidousty C., Scheffler M., et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol. 2018;29(10):2068–2075. doi: 10.1093/annonc/mdy333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang B., Hu L., Dong D., et al. TP53 or CDKN2A/B covariation in ALK/RET/ROS1-rearranged NSCLC is associated with a high TMB, tumor immunosuppressive microenvironment and poor prognosis. J Cancer Res Clin Oncol. 2023;149(12):10041–10052. doi: 10.1007/s00432-023-04924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Offin M., Chan J.M., Tenet M., et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol. 2019;14(10):1784–1793. doi: 10.1016/j.jtho.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Lin G., Zhuo M., et al. Next-generation sequencing based mutation profiling reveals heterogeneity of clinical response and resistance to osimertinib. Lung Cancer. 2020;141:114–118. doi: 10.1016/j.lungcan.2019.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishio M., Paz-Ares L., Reck M., et al. RELAY, ramucirumab plus erlotinib (RAM+ERL) in untreated metastatic EGFR-mutant NSCLC (EGFR+ NSCLC): association between TP53 status and clinical outcome. Clin Lung Cancer. 2023;24(5):415–428. doi: 10.1016/j.cllc.2023.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Haratake N., Hayashi H., Shimokawa M., et al. Phase III clinical trial for the combination of erlotinib plus ramucirumab compared with osimertinib in previously untreated advanced or recurrent non-small cell lung cancer positive for the L858R mutation of EGFR: REVOL858R (WJOG14420L) Clin Lung Cancer. 2022;23(3):E257–E263. doi: 10.1016/j.cllc.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Nakahara Y., Kato T., Isomura R., et al. A multicenter, open label, randomized phase II study of osimertinib plus ramucirumab versus osimertinib alone as initial chemotherapy for EGFR mutation positive non-squamous non -small cell lung cancer: TORG1833. J Clin Oncol. 2019;37(15) [Google Scholar]
- 21.Saltos A., Baik C., Sanborn R.E., et al. RAMOSE: an open-label randomized phase II study of osimertinib with or without ramucirumab in TKI-naive EGFR-mutant metastatic NSCLC. J Thorac Oncol. 2021;16(3):S614. [Google Scholar]
- 22.Seegobin K., Majeed U., Wiest N., Manochakian R., Lou Y., Zhao Y. Immunotherapy in non-small cell lung cancer with actionable mutations other than EGFR. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.750657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West H.J., McCleland M., Cappuzzo F., et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: subgroup results from the phase III IMpower150 trial. J Immunother Cancer. 2022;10(2) doi: 10.1136/jitc-2021-003027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thummalapalli R., Bernstein E., Herzberg B., et al. Clinical and genomic features of response and toxicity to sotorasib in a real-world cohort of patients with advanced KRAS G12C-mutant non-small cell lung cancer. JCO Precis Oncol. 2023;7 doi: 10.1200/PO.23.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mok T.S.K., Lopes G., Cho B.C., et al. Associations of tissue tumor mutational burden and mutational status with clinical outcomes in KEYNOTE-042: pembrolizumab versus chemotherapy for advanced PD-L1-positive NSCLC. Ann Oncol. 2023;34(4):377–388. doi: 10.1016/j.annonc.2023.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Mazzotta M., Filetti M., Occhipinti M., et al. Efficacy of immunotherapy in lung cancer with co-occurring mutations in NOTCH and homologous repair genes. J Immunother Cancer. 2020;8(2) doi: 10.1136/jitc-2020-000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Xing Y., Li B., Li X., Liu B., Wang Y. Molecular pathways, resistance mechanisms and targeted interventions in non-small-cell lung cancer. Mol Biomed. 2022;3(1):42. doi: 10.1186/s43556-022-00107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laface C., Maselli F.M., Santoro A.N., et al. The resistance to EGFR-TKIs in non-small cell lung cancer: from molecular mechanisms to clinical application of new therapeutic strategies. Pharmaceutics. 2023;15(6) doi: 10.3390/pharmaceutics15061604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koopman B., van der Wekken A.J., Ter Elst A., et al. Relevance and effectiveness of molecular tumor board recommendations for patients with non-small-cell lung cancer with rare or complex mutational profiles. JCO Precis Oncol. 2020;4:393–410. doi: 10.1200/PO.20.00008. [DOI] [PubMed] [Google Scholar]
- 30.Koopman B., Groen H.J.M., Schuuring E., et al. Actionability of on-target ALK resistance mutations in patients with non-small cell lung cancer: local experience and review of the literature. Clin Lung Cancer. 2022;23(2):e104–e115. doi: 10.1016/j.cllc.2021.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Chakravarty D., Johnson A., Sklar J., et al. Somatic genomic testing in patients with metastatic or advanced cancer: ASCO provisional clinical opinion. J Clin Oncol. 2022;40(11):1231–1258. doi: 10.1200/JCO.21.02767. [DOI] [PubMed] [Google Scholar]
- 32.Stenzinger A., Cuffel B., Paracha N., et al. Supporting biomarker-driven therapies in oncology: a genomic testing cost calculator. Oncol. 2023;28(5):e242–e253. doi: 10.1093/oncolo/oyad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff H.B., Steeghs E.M.P., Mfumbilwa Z.A., et al. Cost-effectiveness of parallel versus sequential testing of genetic aberrations for stage IV non-small-cell lung cancer in The Netherlands. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arriola E., Bernabe R., Campelo R.G., et al. Cost-effectiveness of next-generation sequencing versus single-gene testing for the molecular diagnosis of patients with metastatic non-small-cell lung cancer from the perspective of Spanish reference centers. JCO Precis Oncol. 2023;7 doi: 10.1200/PO.22.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou D., Ye W., Hess L.M., et al. Diagnostic value and cost-effectiveness of next-generation sequencing-based testing for treatment of patients with advanced/metastatic non-squamous non-small-cell lung cancer in the United States. J Mol Diagn. 2022;24(8):901–914. doi: 10.1016/j.jmoldx.2022.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Sheffield B.S., Eaton K., Emond B., et al. Cost savings of expedited care with upfront next-generation sequencing testing versus single-gene testing among patients with metastatic non-small cell lung cancer based on current Canadian practices. Curr Oncol. 2023;30(2):2348–2365. doi: 10.3390/curroncol30020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ionescu D.N., Stockley T.L., Banerji S., et al. Consensus recommendations to optimize testing for new targetable alterations in non-small cell lung cancer. Curr Oncol. 2022;29(7):4981–4997. doi: 10.3390/curroncol29070396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroeze L.I., de Voer R.M., Kamping E.J., et al. Evaluation of a hybrid capture-based pan-cancer panel for analysis of treatment stratifying oncogenic aberrations and processes. J Mol Diagn. 2020;22(6):757–769. doi: 10.1016/j.jmoldx.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Milbury C.A., Creeden J., Yip W.K., et al. Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One. 2022;17(3) doi: 10.1371/journal.pone.0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samsom K.G., Schipper L.J., Roepman P., et al. Feasibility of whole-genome sequencing-based tumor diagnostics in routine pathology practice. J Pathol. 2022;258(2):179–188. doi: 10.1002/path.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Normanno N., Apostolidis K., Wolf A., et al. Access and quality of biomarker testing for precision oncology in Europe. Eur J Cancer. 2022;176:70–77. doi: 10.1016/j.ejca.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Hofman P., Calabrese F., Kern I., et al. Real-world EGFR testing practices for non-small-cell lung cancer by thoracic pathology laboratories across Europe. ESMO Open. 2023;8(5) doi: 10.1016/j.esmoop.2023.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayle A., Bonastre J., Chaltiel D., et al. ESMO study on the availability and accessibility of biomolecular technologies in oncology in Europe. Ann Oncol. 2023;34(10):934–945. doi: 10.1016/j.annonc.2023.06.011. [DOI] [PubMed] [Google Scholar]
- 44.de Jager V.D., Cajiao Garcia B.N., ter Elst A., et al. Optimalisatie van de zorgketen moleculaire diagnostiek bij niet-kleincellig longcarcinoom in Noord-Nederland. Ned Tijdschr Oncol. 2023;20:70–76. [Google Scholar]
- 45.Levy Y. Genomic medicine 2025: France in the race for precision medicine. Lancet. 2016;388(10062):2872. doi: 10.1016/S0140-6736(16)32467-9. [DOI] [PubMed] [Google Scholar]
- 46.Cox S., Powell C., Morgan S. Implementing genomic testing for lung cancer into routine clinical practice - the Welsh experience. Clin Oncol. 2022;34(11):716–723. doi: 10.1016/j.clon.2022.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Taniere P., Nicholson A.G., Gosney J.R., et al. Landscape of cancer biomarker testing in England following genomic services reconfiguration: insights from a nationwide pathologist survey. J Clin Pathol. 2023 doi: 10.1136/jcp-2023-208890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenzinger A., Moltzen E.K., Winkler E., et al. Implementation of precision medicine in healthcare-A European perspective. J Intern Med. 2023;294(4):437–454. doi: 10.1111/joim.13698. [DOI] [PubMed] [Google Scholar]
- 49.Houda I., Dickhoff C., Uyl-de Groot C.A., et al. Challenges and controversies in resectable non-small cell lung cancer: a clinician’s perspective. Lancet Reg Health Eur. 2024;38:100841. doi: 10.1016/j.lanepe.2024.100841. [DOI] [PMC free article] [PubMed] [Google Scholar]