Abstract
The roles of protein dimerization and double-stranded RNA (dsRNA) binding in the biochemical and cellular activities of PKR, the dsRNA-dependent protein kinase, were investigated. We have previously shown that both properties of the protein are mediated by the same domain. Here we show that dimerization is mediated by hydrophobic residues present on one side of an amphipathic α-helical structure within this domain. Appropriate substitution mutations of residues on that side produced mutants with increased or decreased dimerization activities. Using these mutants, we demonstrated that dimerization is not essential for dsRNA binding. However, enhancing dimerization artificially, by providing an extraneous dimerization domain, increased dsRNA binding of both wild-type and mutant proteins. In vitro, the dimerization-defective mutants could not be activated by dsRNA but were activated normally by heparin. In Saccharomyces cerevisiae, unlike wild-type PKR, these mutants could not inhibit cell growth and the dsRNA-binding domain of the dimerization-defective mutants could not prevent the antigrowth effect of wild-type PKR. These results demonstrate the biological importance of the dimerization properties of PKR.
The double-stranded RNA (dsRNA)-dependent protein kinase PKR is an interferon (IFN)-inducible protein (20, 41). Although it is present in most mammalian cells at a low constitutive level, PKR is induced at the transcriptional level by treatment of the cells with IFNs. The PKR protein is enzymatically inactive unless it is activated by binding to dsRNA; polyanionic molecules such as heparin have also been shown to activate PKR (21). In the presence of the activator, PKR undergoes autophosphorylation and renders itself active (20, 34). Activated PKR is able to phosphorylate the eukaryotic translation initiation factor eIF2α at serine 51, which leads to a general block in protein synthesis (9, 19, 56). dsRNA produced during replication of many viruses triggers PKR activation. The resulting block in protein synthesis is detrimental to virus replication, and many viruses have therefore evolved strategies to inhibit PKR activation (24, 25, 58). These include production of other dsRNA-binding proteins (22, 61), production of decoy substrates with structural similarity to eIF2α (12), production of inhibitory RNAs (28, 46), sequestration of PKR (14), degradation of PKR (4), and induction of cellular inhibitors of PKR (33). In addition to the well-characterized role in IFN-mediated antiviral pathways, PKR has been implicated in several important cellular processes such as signal transduction (31, 36, 37, 60, 62, 65), cell growth (2, 29, 43), apoptosis (13, 32), and differentiation (23, 53). PKR’s role in IFN-γ and dsRNA-mediated signaling has become clear from the studies with PKR-knockout mice (31). Overexpression of enzymatically inactive PKR has been shown to lead to oncogenic transformation of NIH 3T3 cells (29, 42).
We have been investigating the mechanism of activation of PKR by dsRNA and heparin. In this context, we have identified the dsRNA-binding domain (DRBD) of the protein to reside within its amino-terminal 170 residues (49), as have others (8, 15, 18, 26, 39). The DRBD is required for activation of the enzyme by dsRNA but is dispensable for activation by heparin (51). This domain contains two copies of a conserved motif present in many dsRNA-binding proteins (59). The carboxyl-terminal part of this motif can form an α helix, as shown by nuclear magnetic resonance (NMR) analysis (5, 27), and mutations of positively charged residues within this helix affect dsRNA binding (18, 38, 40, 52). PKR is a dimeric protein, and we have shown that the dimerization process is also mediated by the DRBD (50, 52). The two properties, dimerization and dsRNA binding, are, however, genetically dissociable. We generated several mutants which lost their dsRNA-binding ability but still dimerized. Thus, dsRNA binding is not required for PKR dimerization.
In this study, we demonstrated that the converse is also true: dimerization of DRBD is not required for dsRNA binding. Using site-directed mutagenesis, we established that hydrophobic residues, present on one side of the amphipathic helix in the DRBD, mediate dimerization. The dimerization-defective PKR proteins could not be activated by dsRNA but were activated by heparin. These mutants were also ineffective in inhibiting the growth of the yeast Saccharomyces cerevisiae. Thus, DRBD-mediated dimerization of PKR is required for its biochemical activation and cellular actions.
MATERIALS AND METHODS
Site-directed mutagenesis.
The point mutants were generated by oligonucleotide-directed mutagenesis of p68/BS, using the Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad). To generate these mutants, the oligonucleotides K60A (5′-GGTAGATCAGCGAAGGAAGCA-3′), A63E (5′-CAAAGAAGGAAGACAAAAATGCCGC-3′), A66E (5′-GCAAAAAATGACGCAGCCAAATTAGC-3′), A67E (5′-GCAAAAAATGCCGACGCCAAATTAGC-3′), L70E (5′-GCCGCAGCCAAAGAAGCTGTTGAGATAC-3′), L74E (5′-GCTGTTGAGGAACTTAATAAGG-3′), and L75E (5′-GCTGTTGAGATAGATAATAAGGAAAAG-3′) were used.
Plasmids and yeast strains.
The full-length and DRBD portions of point mutants were subcloned into the yeast vector pYES2 (Invitrogen) for analyzing growth regulatory effects. The S. cerevisiae strain used for this purpose was INVSc1 (MATα his3-D1 leu2 trp1-289 ura3-52; Invitrogen). For the rescue of growth inhibition by various DRBDs, wild-type (wt) PKR was subcloned into pRS314 (57), which has a centromeric sequence. For the analysis of in vivo interactions in yeast, the mutants were subcloned into the pGBT9 vector (Clontech). The K296R mutant was subcloned into yeast vectors pGBT9 and pGAD424 (Clontech) to be expressed as GAL4 DNA-binding domain (DBD) and activation domain (AD) hybrids, respectively. The interactions were measured in strain HF7c (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112, canr gal4-542 gal80-538 URA3::GAL1-lacZ; Clontech). To generate the plasmids encoding fusion proteins, the GAL4 DBD fragment from pSG424 (55) was subcloned in frame with the DRBD mutants in pBSII KS+ (Stratagene). This results in the fusion of amino acids 1 to 147 of the GAL4 protein to the amino terminus of the DRBD of PKR.
Chemical cross-linking with dimethylsuberimidate.
The DRBD proteins and the GAL4-DRBD fusion proteins were expressed in bacteria and purified as described elsewhere (50). The purified DRBD (wild type and mutant) proteins were dialyzed against 2,000 volumes of buffer (20 mM HEPES [pH 7.5], 10% glycerol) at 4°C for 17 h. Then 4 μg of proteins was cross-linked in 200 μl with 1 mM dimethylsuberimidate in cross-linking buffer (10 mM HEPES [pH 8.0], 100 mM NaCl) at 25°C for 2 h; 10-μl aliquots were removed at times indicated, and the reaction was stopped by adding 1 M glycine to a concentration of 100 mM. Protein was then denatured by boiling in Laemmli buffer for 2 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel followed by Western blot analysis using a polyclonal antibody raised against bacterially produced DRBD.
In vivo interaction assay in COS-1 cells.
The DRBD portions of the point mutants were amplified by PCR as described previously (52) and subcloned into pSG424. The DRBD of each point mutant was subcloned into pSG424 as a GAL4 DBD fusion protein and tested for interaction with a VP16 AD fusion of wt DRBD. The GAL4-wt DRBD and VP16-wt DRBD fusions were as described elsewhere (50, 52). COS-1 cells were transfected with 200 ng of each of the four (two test plasmids encoding proteins to be tested for interaction, the reporter plasmid pG5Luc, and plasmid pRSV-β-galactosidase to normalize transfection efficiency) plasmid DNAs by the Lipofectamine (GIBCO BRL) procedure. Cells were harvested 48 h after transfection and assayed for luciferase activity after normalization for transfection efficiency by measuring β-galactosidase activity.
Western blot analysis.
Western blot analysis was performed either with a polyclonal antibody against DRBD (48) or with an anti-PKR monoclonal antibody (MAb) (Ribogene), using enhanced chemiluminescence reagents from Amersham.
dsRNA-binding assay.
The in vitro-translated, 35S-labeled proteins were synthesized by using the Promega TNT T7 coupled reticulocyte lysate system. dsRNA-binding activity was measured by poly(I-C)-agarose binding assay performed with 35S-labeled proteins (49). Aliquots of 4 μl of translation products diluted with 25 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 0.3 M NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% Nonidet P-40, 10% glycerol) were mixed with 25 μl of poly(I-C)-agarose beads and incubated at 30°C for 30 min with intermittent shaking. The beads were then washed with 500 μl of binding buffer four times. The proteins bound to beads after washing were analyzed by SDS-PAGE followed by fluorography.
PKR activity assay.
The kinase activity assay of in vitro-translated wt PKR and mutant proteins was performed as described previously (51). In vitro-translated 35S-labeled proteins (6 μl) were incubated with 1 μl of antiserum in 200 μl of high-salt buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 400 mM NaCl, 1 mM EDTA, 0.2 mg of aprotinin per ml, 20% glycerol) at 4°C for 1 h on a rotating wheel; 10 μl of protein A-Sepharose slurry was added, and incubation continued for an additional hour. The protein A-Sepharose beads were washed four times in 500 μl of high-salt buffer and two times in activity buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 10 μg of aprotinin per ml, 0.1 mM PMSF, 5% glycerol). The PKR assay was performed in activity buffer containing 500 ng of purified eIF2, 0.1 mM ATP, and 10 μCi of [γ-32P]ATP at 30°C for 10 min. Poly(I-C) (2 μg/ml) or heparin (10 U/ml) was used as the enzyme activator. Labeled proteins were then analyzed by SDS-PAGE on a 12% gel. Autoradiography was performed at −80°C with intensifying screens.
In vitro protein-protein interaction assay.
The proteins were in vitro translated from 2 μg of plasmid DNA by using a coupled rabbit reticulocyte in vitro translation kit (Promega). After translation, equal quantities of the reticulocyte extracts containing the two proteins to be tested for interaction were mixed; 4 μl of the mix was incubated with 10 μl of anti-Flag MAb-resin (Kodak) at 30°C for 30 min in immunoprecipitation buffer (100 mM NaCl, 20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 1 mM DTT, 10 μg of aprotinin per ml, 0.1 mM PMSF, 10 U of heparin per ml, 20% glycerol). After binding, the beads were washed six times with 500 μl of immunoprecipitation buffer. The washed beads were then boiled in 2× Laemmli buffer (150 mM Tris-HCl [pH 6.8], 5% SDS, 5% β-mercaptoethanol, 20% glycerol) for 2 min and analyzed by SDS-PAGE on a 12% gel. Fluorography was performed at −80°C with intensifying screens.
In vivo interaction assay of yeast.
The interaction between K296R mutant and the A63E, A67E, and L75E mutants in vivo in yeast was measured by activation of the lacZ reporter constructs as detected by β-galactosidase assays. Yeast strain HF7C was transformed with the appropriate plasmids encoding the K296R-GAL4 DBD fusion protein and dimerization-defective–AD hybrid proteins. The colonies were selected on synthetic medium lacking l-leucine and l-tryptophan at 30°C for 3 days, and β-galactosidase activity in extracts prepared from liquid cultures was determined. Cultures were grown in 5 ml of synthetic medium lacking l-leucine and l-tryptophan to an optical density at 600 nm (OD600) of 1.0 to 1.5. Cells were harvested and disrupted by vortexing vigorously in 0.1 M Tris-HCl (pH 8.0) containing glass beads plus 1 mM DTT, 20% glycerol, and 2 mM PMSF. The cell extract was used to determine β-galactosidase activity by using a luminescent assay kit from Tropix.
Growth rate analysis.
The expression plasmids encoding various mutant and wt PKR proteins were introduced into yeast strain INVSc1 by the lithium acetate method (8). Transformed yeast strains were grown to an OD600 of about 1.5 in synthetic medium containing 0.1% glucose and lacking uracil or, for the rescue experiment, lacking uracil and tryptophan. The cultures were then harvested and washed with synthetic medium containing 2% galactose. The cultures were then diluted to OD600 of 0.4 in synthetic medium containing 2% galactose. At various time points, cell growth was monitored by measuring the OD600.
RESULTS
Dimerization is mediated by the hydrophobic side of the amphipathic α helix.
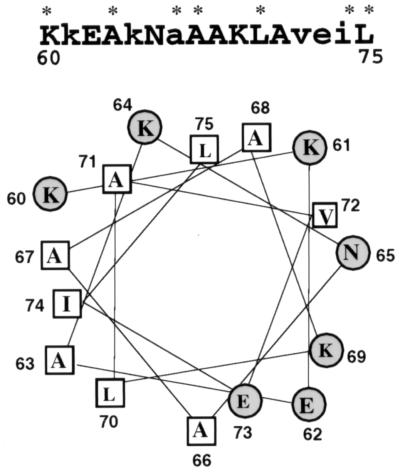
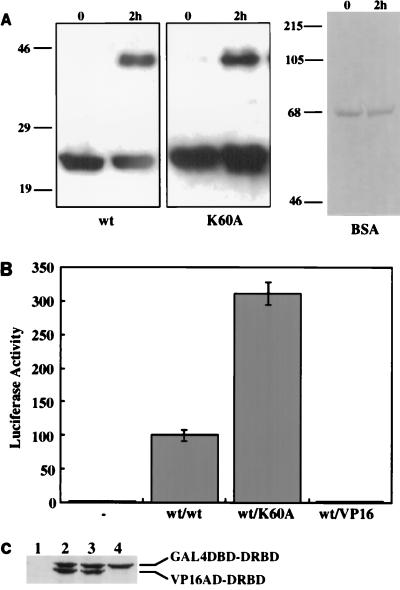
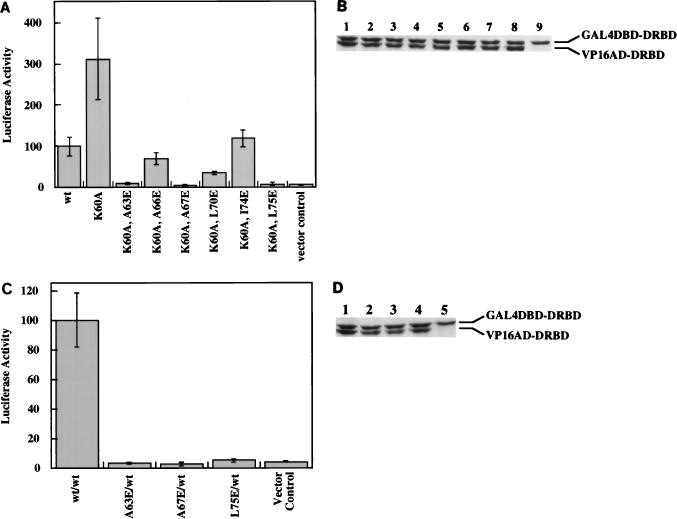
The DRBD of PKR contains two copies of a 65-residue motif found in many dsRNA-binding proteins. The most conserved region of this motif from RNase III and Staufen proteins forms an α helix in solution, as shown by NMR analysis (5, 27). The corresponding region in the first motif of PKR encompasses the residues 60 through 75. A helical wheel analysis of this region showed that it is an amphipathic helix (Fig. 1). Charged residues, mostly basic, are primarily on one side of the helix, whereas hydrophobic residues are located on the other side. Since the hydrophobic sides of amphipathic helices present in other unrelated proteins have been shown to mediate dimerization (44), we wanted to examine whether the same is true for the DRBD of PKR. Specific residues, marked by asterisks in Fig. 1, were targeted for site-directed mutagenesis for this purpose. They included residues that are conserved in this motif in several proteins and two nonconserved hydrophobic residues. Effects of these mutations on the dimerization property of DRBD were monitored by several assays described previously (50, 52). Mutation of K60 to A has been shown to destroy the dsRNA-binding ability of DRBD (40, 52). This mutation, however, did not affect the protein’s dimerization ability as measured by chemical cross-linking of the purified mutant protein expressed in bacteria (Fig. 2A). The wt and K60A DRBD proteins could be cross-linked as a dimer. As a negative control, we used bovine serum albumin, which did not cross-link with similar treatment and is known to be a monomeric protein. In a different assay (the mammalian two-hybrid assay), the K60A mutant was in fact three times more efficient in dimerization than the wt protein (Fig. 2B). Western blot analysis confirmed that the mutant and wt proteins were expressed at comparable levels (Fig. 2C). The observed enhanced dimerization activity of the K60A mutant supports our hypothesis since this mutation extended the hydrophobic side of the α helix (Fig. 1). To further test our hypothesis, we generated six other mutants in which hydrophobic residues on the putative dimerization face of the α helix were individually replaced with the charged residue E. Since we anticipated a lowering of the dimerization activity by these specific mutations, they were introduced in the background of K60A, which dimerized better than the wt protein, to create a more demanding situation. As shown in Fig. 3A, each of these mutations strongly reduced the dimerization activity of the proteins, and three, at residues 63, 67, and 75, entirely abrogated the activity. All of these residues are conserved in the various dsRNA-binding proteins (Fig. 1). These three mutations were selected for further detailed investigation. As expected, the same mutations, when tested in the background of the wt protein instead of K60A, were equally crippling (Fig. 3C). Western blot analysis ascertained that all proteins were expressed at comparable levels in transfected cells (Fig. 3B and D). These results strongly suggest that, as hypothesized, the hydrophobic side of the α helix mediates dimerization of the protein.
FIG. 1.
Helical wheel projection of residues 60 to 75 within the DRBD of PKR. Secondary structure predictions were performed with the program Protean. The hydrophobic residues are indicated in rectangles, and the charged residues are indicated in circles. The primary sequence of this region is shown at the top. The residues conserved within several dsRNA-binding proteins are capitalized, and the residues targeted by site-directed mutagenesis are indicated with asterisks.
FIG. 2.
(A) Chemical cross-linking of K60A DRBD. The proteins were expressed as hexahistidine-tagged fusion proteins from pET15b (Novagen) and purified on Ni-Sepharose. Purified DRBDs (wt and K60A; 4 μg of each) were cross-linked with 1 mM dimethylsuberimidate in 200 μl of cross-linking buffer (10 mM HEPES [pH 8.0], 100 mM NaCl) at 25°C for 2 h. The reactions were stopped by adding 1 M glycine to a concentration of 100 mM. Proteins were analyzed by SDS-PAGE on a 12% gel. Western blot analysis was performed with a polyclonal anti-DRBD antibody. As a negative control for cross-linking, 4 μg of pure bovine serum albumin (BSA) was cross-linked under identical conditions, and the lack of cross-linking was confirmed by SDS-PAGE on an 8% gel followed by Coomassie blue staining. The positions of the molecular mass markers are indicated in kilodaltons on the left. (B) Dimerization activity of K60A DRBD in vivo. COS-1 cells were transfected with 200 ng of each of the four (two test plasmids encoding proteins to be tested, the reporter plasmid pG5Luc, and plasmid pRSV-β-galactosidase to normalize transfection efficiency) plasmid DNAs by the Lipofectamine procedure. Cells were harvested 48 h after transfection and assayed for luciferase activity after normalization for transfection efficiency by measuring β-galactosidase activity. The relative luciferase activity obtained is represented on the y axis. The proteins assayed for interaction with wt DRBD are indicated below the bars. wt/VP16, negative control (relative luciferase activity obtained with wt DRBD-GAL4 and VP16 vector alone); wt/wt, positive control (luciferase activity obtained with wt DRBD-VP16 and wt DRBD-GAL4 as a percentage of the activity obtained with wt/wt, considered 100%); −, the activity of cells transfected with VP16 and GAL4 vectors alone. Each experiment was repeated six times, and the averages of individual values with standard error bars are presented. (C) Western blot analysis. COS-1 cell extracts were examined by Western blot analysis using a MAb against PKR (Ribogene); 100 μg of total protein was loaded in each lane. Lane 1, vectors VP16 and GAL4 only; lane 2, wt DRBD-VP16 and wt DRBD-GAL4; lane 3, wt DRBD-VP16 and K60A DRBD-GAL4; lane 4, K60A DRBD-GAL4 and VP16 vector. Positions of the hybrid proteins GAL4 DBD-DRBD and VP16 AD-DRBD are indicated on the right.
FIG. 3.
Dimerization activity of mutants within the hydrophobic face of the α helix. (A) Dimerization activity of DRBD mutants in the K60A background. Specific hydrophobic residues on the hydrophobic side of the α helix in DRBD were mutated to E residues in the K60A background. These mutants were tested for dimerization activity in COS-1 cells with the K60A DRBD as described for Fig. 2B. All double-mutant DRBDs were in the GAL4 vector, and K60A DRBD was in the VP16 vector. Vector control, activity obtained with K60A DRBD-GAL4 and VP16 vector alone. (B) Western blot analysis. COS-1 cell extracts were examined by Western blot analysis using a MAb against PKR (Ribogene); 100 μg of total protein was loaded in each lane. Lane 1, wt DRBD-VP16 and wt DRBD-GAL4; lane 2, K60A DRBD-VP16 and K60A DRBD-GAL4; lane 3, K60A DRBD-VP16 and K60A,A63E DRBD-GAL4; lane 4, K60A DRBD-VP16 and K60A,A66E DRBD-GAL4; lane 5, K60A DRBD-VP16 and K60A,A67E DRBD-GAL4; lane 6, K60A DRBD-VP16 and K60A,L70E DRBD-GAL4; lane 7, K60A DRBD-VP16 and K60A,I74E DRBD-GAL4; lane 8, K60A DRBD-VP16 and K60A,L75E DRBD-GAL4; lane 9, K60A DRBD-GAL4 and VP16 vector alone. Positions of the hybrid proteins GAL4 DBD-DRBD and VP16 AD-DRBD are indicated on the right. (C) In vivo dimerization activity of the three most defective point mutants in the wt DRBD background. Mutants A63E, A67E, and L75E were generated in the wt DRBD background and tested for dimerization activity in COS-1 cells with the wt DRBD as described for Fig. 2B. All mutant DRBDs were in the GAL4 vector, and the wt DRBD was in the VP16 vector. (D) Western blot analysis. COS-1 cell extracts were examined by Western blot analysis using a MAb against PKR (Ribogene); 100 μg of total protein was loaded in each lane. Lane 1, wt DRBD-VP16 and wt DRBD-GAL4; lane 2, wt DRBD-VP16 and A63E DRBD-GAL4; lane 3, wt DRBD-VP16 and A67E DRBD-GAL4; lane 4, wt DRBD-VP16 and L75E DRBD-GAL4; lane 5, wt DRBD-GAL4 and VP16 vector.
In vitro dimerization activity of the mutants.
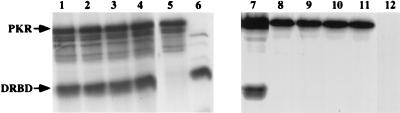
To ascertain that mutants A63E, A67E, and L75E had lost their dimerization activity, an in vitro protein-protein interaction assay was performed. 35S-labeled, Flag-tagged wt PKR and nontagged DRBD proteins were synthesized by in vitro translation. The Flag-tagged wt PKR protein was mixed individually with each DRBD protein, and a coimmunoprecipitation assay was performed with anti-Flag MAb-agarose. The nontagged DRBD proteins can be coimmunoprecipitated with Flag-wt PKR only if they interact with it. As shown in Fig. 4, the wt DRBD bound efficiently to Flag-PKR and could therefore be coimmunoprecipitated with it by anti-Flag MAb-agarose (lane 7). In contrast, A63E DRBD, A67E DRBD, and L75E DRBD (lanes 8 to 10) could not be coimmunoprecipitated with Flag-PKR, confirming that they had lost the ability to interact with wt PKR. The specificity of the coimmunoprecipitation was ascertained by the fact that wt DRBD alone could not be immunoprecipitated with anti-Flag MAb-agarose (lane 12). These results confirmed further that mutations A63E, A67E, and L75E lead to a loss of dimerization activity.
FIG. 4.
In vitro dimerization activity of the mutants. 35S-labeled, Flag epitope-tagged wt PKR and DRBD proteins were in vitro translated in rabbit reticulocyte lysate. Equal amounts of the two proteins to be tested for interaction were mixed and incubated at 30°C for 30 min with anti-Flag MAb-agarose in 50 μl of immunoprecipitation buffer. The proteins remaining bound to agarose beads after extensive washing were analyzed by SDS-PAGE followed by fluorography. Lanes 1 and 7, Flag-tagged wt PKR and wt DRBD; lanes 2 and 8, Flag-tagged wt PKR and A63E DRBD; lanes 3 and 9, Flag-tagged wt PKR and A67E DRBD; lanes 4 and 10, Flag-tagged wt PKR and L75E DRBD; lanes 5 and 11, Flag-tagged wt PKR alone; lane 6 and 12, wt DRBD alone. The positions of PKR and DRBD are indicated by arrows. Lanes 1 to 6 show total proteins from reticulocyte lysate, and lanes 7 to 12 show immunoprecipitated proteins.
Role of dimerization in dsRNA binding.
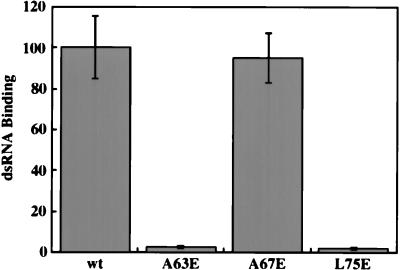
We have previously shown that dimerization of PKR does not require dsRNA binding (50, 52). Here, using the new mutants, we examined the converse, i.e., whether dimerization is absolutely required for dsRNA binding. The results shown in Fig. 5 demonstrated that the two properties are independent. The A67E mutant was as efficient as the wt protein in dsRNA binding, although it was incapable of dimerization. The two other dimerization-defective mutants, A63E and L75E, were defective in dsRNA binding. The mutations in the latter probably altered the structure of this region such that both properties of the protein were independently affected. This possibility is supported by the observation made by McMillan et al. (40) that dsRNA binding is drastically reduced upon replacement of L75 by A. The full-length A63E and L75E mutant proteins showed similar defects in dsRNA binding, and the full-length A67E mutant retained about 70% of its dsRNA-binding activity (data not shown).
FIG. 5.
dsRNA-binding activity of dimerization-negative mutants. The wt DRBD and point mutants were tested for poly(I-C)-agarose binding activity; 4 μl of in vitro-translated 35S-labeled proteins was bound to poly(I-C)-agarose beads. Proteins which remained bound to beads after washing were analyzed by SDS-PAGE followed by fluorography. Equal amounts of the total translation mix were also loaded for all samples. PhosphorImager analysis was done to quantify binding activity. The fraction of bound protein was calculated as radioactivity in the bound protein band/total radioactivity assayed. The dsRNA binding of wt DRBD was considered 100%, and values for other point mutants are presented as a percentage of that value.
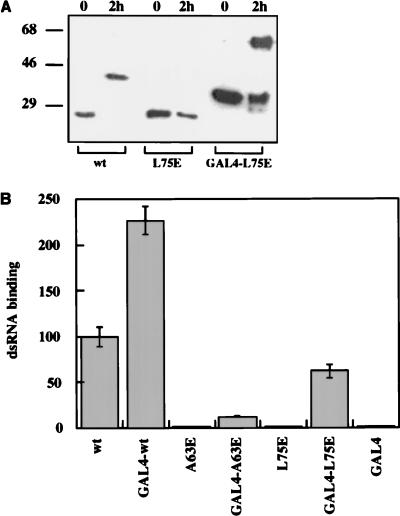
In the next series of experiments, we investigated whether dimerization of DRBD can enhance dsRNA binding, although it is not essential. For this purpose, an exogenous dimerization domain, provided by the DBD of the GAL4 protein, was fused to DRBD. The DBD is known to contain a motif that mediates dimerization of the GAL4 protein (6). The GAL4 dimerization domain was fused to the amino termini of the wt, A63E, and L75E proteins. To confirm the expected dimerization upon the addition of the GAL4 dimerization domain, the different proteins were expressed in Escherichia coli, purified, and tested for cross-linking by dimethylsuberimidate (Fig. 6A). As expected, the wt protein dimerized. The L75E mutant did not dimerize on its own but dimerized efficiently when fused to the GAL4 DBD. These results confirmed by an independent assay that the L75E mutant is indeed defective in dimerization and that the GAL4 dimerization domain functions as expected. The dsRNA-binding properties of the different mutants were examined in assays using in vitro-synthesized radiolabeled proteins (Fig. 6B). The GAL4 dimerization domain by itself did not bind dsRNA, and the wt DRBD bound it efficiently. Both A63E and L75E DRBD proteins were individually incapable of binding dsRNA, whereas the corresponding GAL4 fusion proteins bound dsRNA, albeit inefficiently. Surprisingly, the dsRNA-binding activity of the wt protein was also increased more than twofold upon addition of the GAL4 dimerization domain. These results suggest that dimerization, although not required for dsRNA binding, enhances this property. They also suggest that the A63E and the L75E mutants probably retained an intrinsic ability to bind dsRNA, although this property was not detectable without the artificial enhancement by the addition of the GAL4 DBD.
FIG. 6.
(A) Chemical cross-linking of L75E DRBD and GAL4 DBD-L75E DRBD. The L75E DRBD and GAL4 DBD-L75E DRBD proteins were expressed as hexahistidine-tagged fusion proteins from pET15b (Novagen) and purified on Ni-Sepharose; 4 μg of purified proteins was cross-linked and analyzed as described for Fig. 2A. (B) dsRNA-binding activity of the DRBD mutant proteins. The wt and point mutant DRBDs and their GAL4 DBD hybrids were tested for poly(I-C)-agarose binding activity as described for Fig. 5. The dsRNA binding of wt DRBD was considered 100%, and values for other point mutants are presented as a percentage of that value.
Dimerization and enzyme activity.
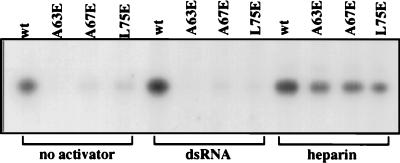
To determine if the DRBD-mediated dimerization of PKR is essential for its enzyme activity, appropriate mutations were introduced to the full-length PKR protein. The different mutant proteins were synthesized in vitro, immunoprecipitated, and tested for the ability to phosphorylate eIF2α (Fig. 7). The wt protein had a constitutive level of activity, but the three dimerization-defective mutants were inactive. Addition of dsRNA activated the wt protein fourfold but had no effect on the mutant proteins. This result was expected for the A63E and L75E mutants since they cannot bind dsRNA. The A67E mutant, however, can bind dsRNA efficiently, although it cannot dimerize. These results strongly indicate that dsRNA cannot activate PKR unless the protein is capable of dimerization. In contrast to the dsRNA activation results, heparin could activate the three mutants and the wt protein about threefold. Thus, the three mutant proteins are capable of activation, and the introduced mutations had not caused gross distortions in the structures of these proteins.
FIG. 7.
eIF2 phosphorylation activities of the point mutants of PKR. In vitro-translated full-length PKR proteins (5 μl) were immunoprecipitated with 1 μl of polyclonal antiserum and protein A-Sepharose. The eIF2α phosphorylation assay was performed with the immunoprecipitated proteins on protein A-Sepharose beads as described in Materials and Methods either in the absence of any activator or in the presence of poly(I-C) (2 μg/ml) or heparin (10 U/ml). Phosphorylated eIF-2 was analyzed by SDS-PAGE followed by autoradiography. The intensities of the bands were quantitated by densitometric scanning.
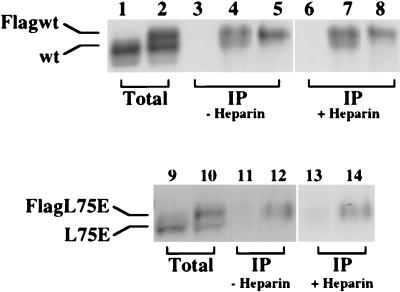
The above results suggested that heparin activation of PKR does not require dimerization of the protein. It remained possible, however, that binding of heparin to PKR caused dimerization of the protein through a different domain. To test this possibility, we used an in vitro PKR-PKR interaction assay. In this assay, a Flag-tagged PKR protein is cotranslated with untagged PKR. They are immunoprecipitated with a Flag antibody, and the presence of the untagged protein in the immunoprecipitate is monitored by gel electrophoresis. As shown in Fig. 8, untagged wt PKR could not be immunoprecipitated with anti-Flag MAb-agarose (lane 3) but coimmunoprecipitated only in the presence of Flag-tagged PKR (lane 4), thus validating the assay. Lane 5, representing Flag-tagged PKR alone immunoprecipitated with anti-Flag MAb-agarose, shows a single band corresponding in position to the Flag-tagged wt PKR. Coimmunoprecipitation in the presence of heparin (lanes 6 to 8) gave results identical to those obtained in the absence of heparin. In contrast, the L75E mutant did not coimmunoprecipitate with the Flag-tagged counterpart either in the presence or in the absence of heparin (lanes 12 and 14), confirming that this mutant is incapable of dimerization. We concluded, therefore, that heparin can activate PKR in its monomeric form whereas dsRNA cannot.
FIG. 8.
Effect of heparin on PKR dimerization. Flag-tagged and nontagged wt and L75E PKR proteins were synthesized by cotranslation in the reticulocyte lysate; 5 μl of in vitro-translated, 35S-labeled proteins were immunoprecipitated with Flag MAb-agarose either in the absence (lanes 3 to 5, 11, and 12) or in the presence (lanes 6 to 8, 13, and 14) of heparin (10 U/ml). The immunoprecipitated proteins were analyzed by SDS-PAGE on an 8% gel followed by fluorography. Lanes 1, 3, and 6, nontagged wt PKR alone; lanes 2, 4, and 7, cotranslated nontagged wt PKR and Flag-tagged wt PKR; lanes 5 and 8, Flag-tagged wt PKR alone; lanes 9, 11, and 13, nontagged L75E PKR alone; lanes 10, 12, and 14, cotranslated nontagged L75E PKR and Flag-tagged L75E PKR. Lanes 1, 2, 9, and 10 represent total proteins; lanes 3 to 8 and 11 to 14 represent immunoprecipitated (IP) proteins. Positions of nontagged and Flag-tagged proteins are as indicated.
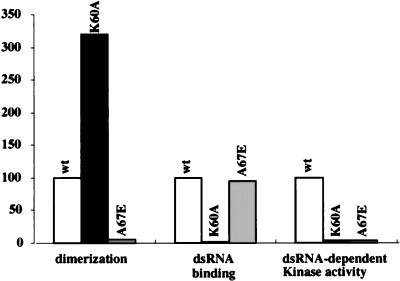
Comparison of the dimerization, dsRNA-binding, and kinase activities of K60A and A67E mutants.
The properties of the wt, K60A, and A67E proteins are summarized in Fig. 9. The dimerization activity presented in this figure was derived from the mammalian two-hybrid assays; similar conclusions were drawn from coimmunoprecipitation of in vitro-translated proteins. The dsRNA-binding values were determined from the poly(I-C)-agarose binding abilities of in vitro-translated proteins. The kinase activity values were from their abilities to phosphorylate eIF2 in the presence of dsRNA. Both mutants were enzymatically inactive, but for different reasons: the K60A mutant could dimerize but not bind dsRNA, whereas the A67E mutant could not dimerize but could bind dsRNA efficiently.
FIG. 9.
Dimerization, dsRNA-binding, and kinase activities of K60A and A67E mutants and the wt protein. Activities of the mutants are presented as percentages of the wt level, considered 100%. Dimerization activities were calculated from mammalian two-hybrid assays using DRBD, dsRNA-binding activities were calculated from the poly(I-C)-agarose binding assays of in vitro-translated DRBD, and kinase activities were calculated from the ability of PKR and its mutants to phosphorylate eIF2. Data from three different sets of experiments in each category were averaged to compile the graph; the primary data used included some derived from Fig. 2, 3, 5, and 7 as well as data not shown.
Dimerization and cellular functions of PKR.
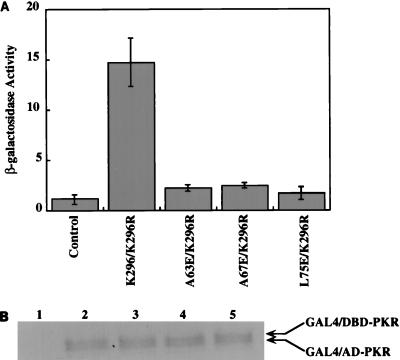
To determine whether the dimerization property of PKR is required for the enzyme’s in vivo functions, we chose the yeast system. It has been shown that wt PKR can inhibit the growth of S. cerevisiae (8, 54), and this growth inhibition phenotype can be rescued by the coexpression of DRBD. We wanted to examine similar properties of the dimerization-defective mutants. However, it was first necessary to confirm that these mutants fail to dimerize in vivo in yeast. A two-hybrid transcriptional activation assay in yeast was set up for this purpose. Because wt PKR is growth inhibitory in yeast, the K296R mutant, which is enzymatically inactive but can dimerize and bind dsRNA, was used in lieu of the wt protein. As shown in Fig. 10A, the K296R mutant interacted with itself strongly, but none of the three mutants showed any interaction with this protein. Thus, the newly identified dimerization-defective mutant PKR proteins were also defective, as expected, in vivo. To ensure that the mutant proteins were expressed in yeast, a Western blot analysis was performed with the yeast extracts. All of the hybrid proteins were found to be expressed at comparable levels (Fig. 10B).
FIG. 10.
(A) Interaction assay of point mutants of PKR in yeast cells. Plasmids expressing the mutant PKR proteins as a GAL4 DBD hybrid and a plasmid expressing K296R PKR as a GAL4 AD hybrid were cotransformed in yeast strain HF7C. The colonies were selected on Leu- and Trp-deficient plates and grown in liquid synthetic medium lacking these two amino acids. After 2 days, the cells were harvested and lysates were prepared for β-galactosidase activity assay. The bars represent averages from three separate experiments. (B) Western blot analysis. Yeast cell extracts were examined by Western blot analysis using a MAb against PKR (Ribogene); 150 μg of total protein was loaded in each lane. Lane 1, pGBT9 vector and pGAD424 vector; lane 2, K296R/pGBT9 and K296R/pGAD424; lane 3, K296R/pGBT9 and A63E/pGAD424; lane 4, K296R/pGBT9 and A67E/pGAD424; lane 5, K296R/pGBT9 and L75E/pGAD424. The arrows represent positions of the hybrid proteins.
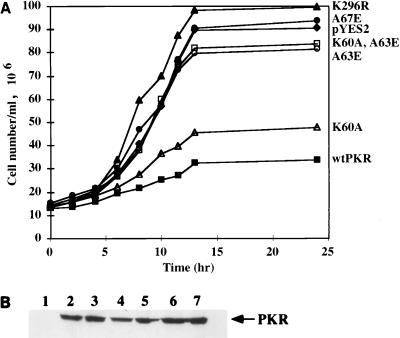
For testing antigrowth effects in yeast, we selected four mutants: K60A, which can dimerize but does not bind dsRNA; A67E, which binds dsRNA but cannot dimerize; A63E, which is defective in both activities; and the double mutant K60A,A63E, which is also devoid of both properties. As reported previously, wt PKR strongly inhibited cell growth but the K296R mutant did not (Fig. 11A). The A67E mutant did not inhibit cell growth, suggesting that the dimerization but not the dsRNA-binding ability of the protein is required for this property. In accord with this conclusion, the K60A mutant, which cannot bind dsRNA but dimerizes very efficiently, inhibited cell growth. However, once the ability to dimerize was eliminated by introducing the A63E mutation in the K60A background, the protein no longer inhibited cell growth. The A63E mutant, which is devoid of both dimerization and dsRNA-binding activities, also was found to be unable to inhibit growth. Western blot analysis with anti-PKR antibody was done to compare the levels of expression of different PKR mutants (Fig. 11B). As expected, the inactive mutants were expressed at slightly higher levels (lanes 2, 5, 6, and 7) than wt PKR (lane 4). Surprisingly, the K60A mutant, although growth inhibiting, was expressed better than wt PKR. This result indicates that K60A is less functional than the wt protein although it still inhibits cell growth. The results presented here demonstrate that the dimerization property of PKR profoundly affects its ability to inhibit yeast cell growth.
FIG. 11.
(A) Growth characteristics of the yeast strains carrying PKR point mutants. Growth of transformed yeast strains containing pYES2 alone (diamonds), wt PKR/pYES2 (closed squares), K296R/pYES2 (closed triangles), K60A/pYES2 (open triangles), A63/pYES2 (open circles), K60A,A63E/pYES2 (open squares), and A67E/pYES2 (closed circles) was assayed in synthetic medium lacking uracil and containing 2% galactose. (B) Western blot analysis. Yeast cell extracts were examined by Western blot analysis using a MAb against PKR; 100 μg of total protein was loaded in each lane. Lane 1, pYES2 vector alone; lane 2, K296R/pYES2; lane 3, K60A/pYES2; lane 4, wt PKR/pYES2; lane 5, A63E/pYES2; lane 6, K60A,A63E/pYES2; lane 7, A67E/pYES2. The arrow represents the position of the PKR protein.
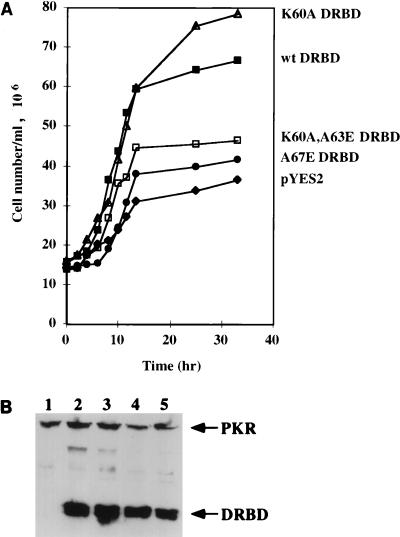
The same general conclusion was confirmed by the rescue assay shown in Fig. 12A. In this assay, yeast cell growth was inhibited by expressing a low level of wt PKR, and the rescue of the slow-growth phenotype by concomitant high-level expression of wt or mutant DRBD was monitored. As expected, wt DRBD was very efficient in rescue, as was the K60A mutant. In contrast, the A67E and K60A,A63E mutant DRBDs were ineffective in rescuing the slow-growth phenotype. A Western blot analysis was performed to ascertain that the mutant proteins were expressed at nearly equal amounts in yeast cells (Fig. 12B). The results presented in Fig. 12 indicate that the in vivo activity of DRBD, as measured by its ability to alleviate PKR’s antigrowth action, requires retention of its dimerization property.
FIG. 12.
(A) Assay for rescue of growth-suppressive phenotype of wt PKR by coexpression of DRBD mutants. Yeast strain INVSc1 expressing wt PKR from a modified pRS314 vector with Trp as the selection marker was grown in synthetic medium containing 2% glucose. The competent cells prepared from this strain were transformed with 1 μg of plasmids encoding wt and mutant DRBDs from pYES2, with Ura as the selection marker. The colonies were selected on medium lacking tryptophan and uracil. Growth of transformed yeast strains containing wt PKR/pRS314+ alone (diamonds) or with wt DRBD/pYES2 (closed squares), K60A DRBD/pYES2 (open triangles), K60A,A63E DRBD/pYES2 (open squares), and A67E DRBD/pYES2 (closed circles) was assayed in synthetic medium lacking tryptophan and uracil and containing 2% galactose. (B) Western blot analysis. Yeast cell extracts were examined by Western blot analysis using a MAb against PKR; 100 μg of total protein was loaded in each lane. Lane 1, wt PKR/pRS314 and pYES2 vector alone; lane 2, wt PKR/pRS314 and K60A DRBD/pYES2; lane 3, wt PKR/pRS314 and wt DRBD/pYES2; lane 4, wt PKR/pRS314 and K60A,A63E DRBD/pYES2; lane 5, wt PKR/pRS314 and A67E DRBD/pYES2. The arrows represent the positions of the PKR and DRBD proteins.
DISCUSSION
PKR is an important cellular enzyme that regulates a diverse array of biological processes. Because its synthesis is induced by IFNs, viral dsRNA activates it, and synthesis of many viral proteins is inhibited as a consequence of its activation, PKR is considered an important component of the antiviral machinery of IFNs. This role of PKR is underscored by the fact many mammalian viruses combat PKR’s action by encoding or inducing the synthesis of different PKR inhibitors (24, 58). Recent studies have made it amply clear, however, that PKR has cellular functions beyond its role in inhibiting viral protein synthesis. Its role in transcriptional signal transduction was revealed by the finding that it can activate NF-κB (30, 31, 36). Observed defects in cytokine signaling pathways in PKR-knockout cells confirmed the pleiotropic nature of its action (31). Its possible involvement in the regulation of cell growth has been suggested by a variety of observations: expression of wt PKR retards the growth of not only mammalian (29) but also yeast (8) cells, and expression of catalytically inactive mutants of PKR or its inhibitor, P58, oncogenically transforms NIH 3T3 cells (1). The underlying mechanisms, however, remain unclear. The crucial substrate that mediates cell growth regulation by the enzyme and its relevant cellular activators remain unidentified. Adding to the complexity are the facts that (i) like other protein kinases, PKR can have multiple substrates and (ii) in addition to dsRNA, polyanionic molecules such as heparin can activate the enzyme. Moreover, PKR can possibly affect the functions of other dsRNA-binding proteins by either acting as a sink of dsRNA or forming heterodimers with them (10). We and others have generated a variety of PKR mutants devoid of a subset of the protein’s properties for the purpose of analyzing PKR’s cellular actions (18, 39, 40, 50–52). This study adds to this endeavor an important set of new mutants that are defective in dimerization. Experiments using the new mutants have allowed us to make several significant conclusions regarding the mechanisms of PKR activation and its action. First, we demonstrated that the hydrophobic side of the amphipathic α helix in the first DRBD of PKR mediates its dimerization. Second, we established that PKR dimerization and dsRNA binding are independent of each other, although the same region of the protein can mediate both processes. Third, our data showed that dimerization is required for activation of PKR by dsRNA but not by heparin. Finally, we demonstrated that the dimerization property of PKR is crucial for its antigrowth effects in yeast.
We have previously reported that PKR is a dimeric molecule and that its dimerization is mediated partly by a domain near the amino terminus that physically overlaps the DRBD (50). This observation was supported by results reported by others (10, 47, 63). The dimerization region of PKR contains two homologous motifs that are involved in dsRNA binding, although residues outside of these motifs also affect this process (18, 48, 52). Parts of similar motifs present in two other dsRNA-binding proteins, RNase III and Staufen, can form α-helical structures, as shown by NMR analyses (5, 27). The corresponding putative α helix in PKR is amphipathic, with one side consisting of mostly hydrophobic residues and the other partially overlapping side consisting of mostly charged residues. Mutation studies from several laboratories have shown that many of these charged residues are required for dsRNA binding (18, 39, 40, 52). In the present study, we tested the complementary function of this α helix in mediating protein-protein interaction. We postulated that the hydrophobic side of the helix mediates this interaction and experimentally tested this hypothesis by introducing appropriate mutations. Substitution of K60 by A extended the hydrophobic surface and, as expected from our hypothesis, enhanced dimerization of the protein. On the other hand, substitution of different hydrophobic residues with glutamic acid reduced protein-protein interactions. The effects were not uniform, however, indicating that all hydrophobic residues on the α helix do not contribute equally to the dimerization property of the protein. Nonetheless, the new mutational data along with published results strongly support the notion that the two sides of the α helix mediate primarily two different functions: the hydrophobic side mediates protein-protein interactions, and the charged side mediates protein-RNA interactions.
The next major issue addressed in this report is the possible interdependence of dsRNA binding and dimerization. Such an interdependence has been demonstrated for RNA binding by the human immunodeficiency virus type 1 Rev protein, which is also an oligomer (45, 64). Previous studies by us and others (47, 50, 52, 63) indicated that dsRNA binding is not essential for dimerization of PKR, although another report presented results to the contrary (10). In this study, our previous conclusion was reinforced by the behavior of the K60A protein, which was known to be defective in dsRNA binding. It dimerized more efficiently than the wt protein. Reciprocally, the A67E mutant, though unable to dimerize, bound dsRNA as efficiently as the wt protein. Because the same region of the protein is involved in both processes, there were several mutations that affected both interactions. The observed properties of the K60A and the A67E mutants, however, firmly support the notion that the dsRNA-binding and dimerization properties of the protein are not interdependent. Further experiments revealed that artificial enhancement of dimerization can enhance the dsRNA-binding ability of the mutants and the wt protein. This observed enhancement can be explained by invoking stabilization of the dsRNA-protein complex as a consequence of the presence of two RNA-binding sites on a dimeric protein, as opposed to only one site on a monomeric protein. The binding of dsRNA by the wt protein is strong per se but was further enhanced by artificially enhanced dimerization of the protein. For the A63E and the L75E mutants, dsRNA binding was too weak to be detected under our experimental conditions, but dimerization brought the two weak binding sites together and produced detectable levels of dsRNA binding. We and others have noted previously that dsRNA binding promotes oligomerization of the protein by virtue of a single dsRNA molecule binding to more than one molecule of protein (35, 48). Thus, the two processes, dimerization and dsRNA binding, are mutually helpful, but neither is absolutely needed for the other.
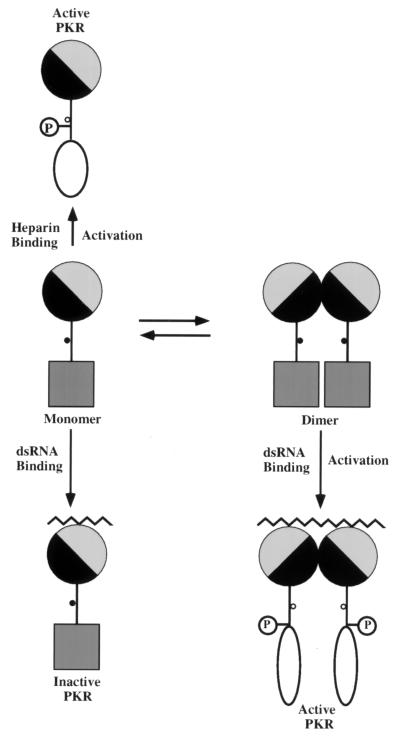
When the new PKR mutants were used for activation analyses, interesting patterns emerged. None of the dimerization-defective mutants could be activated by dsRNA; they also lacked the basal activity of the wt protein. The A67E mutant, although capable of dsRNA binding, could not be activated by dsRNA because of its dimerization defect. Thus, dimerization is absolutely required for activation of the protein by dsRNA. These mutant proteins were, however, capable of activation by heparin. This activation occurred in the monomeric state since even in the presence of heparin, the L75E mutant did not dimerize. The above observations led us to propose a model for PKR activation (Fig. 13). According to this model, there is in cells an equilibrium between monomeric and dimeric inactive PKR. Dimerization is mediated by the hydrophobic side of an amphipathic α helix present near the amino terminus of the protein, although residues present outside this α helix can also influence this process (52). dsRNA binds to the other side of the same α helix of both the monomeric and dimeric molecules, but only the dimeric molecule is activated. The activation process follows conformational changes that can be detected by biophysical measurements (7). It also makes the ATP-binding site accessible, resulting in autophosphorylation of the protein (3, 16). Monomeric proteins, such as the A67E mutant, can bind dsRNA, but the activation process is not completed. Further studies of such a mutant will be required for delineating the exact step in the activation process that is defective. It is conceivable that the conformation change still occurs but the autophosphorylation does not take place because it is an intermolecular process and thus requires dimerization of the protein. If this scenario is true for dsRNA activation, it apparently does not apply to the mode of activation by heparin. Our previous studies demonstrated that heparin could activate PKR mutants that are devoid of the dsRNA-binding and dimerization domain (51). Results presented here demonstrate that the same is true for three monomeric mutants of full-length PKR. Furthermore, unlike dsRNA binding, heparin binding did not cause oligomerization of the protein. These observations suggest that heparin can not only bind monomeric PKR but also activate it by a process that necessarily involves intramolecular phosphorylation. The general conclusion that activations of PKR by dsRNA and heparin involve quite distinct mechanisms is supported by several additional observations reported in the literature. Heparin has been shown to bind to PKR on sites distinct from the DRBD (51), and heparin-activated PKR cannot phosphorylate the K296R mutant, suggesting that heparin activates intramolecular autophosphorylation whereas dsRNA activates intermolecular autophosphorylation (17). Also, heparin-activated PKR probably is phosphorylated on fewer sites than the dsRNA-activated PKR, as judged by its radioactivity and mobility (our unpublished observations).
FIG. 13.
A model for PKR activation. The dimerization and dsRNA-binding domain of the protein is shown as a circle. The hydrophobic side of the α helix within this domain is shaded black, and the charged side is shaded gray. The catalytic domain of the protein is shown as a filled square in the inactive state and as an open oval in the active state. Dots on the stems denote ATP-binding sites, either accessible (open dots) or not accessible (solid dots) to ATP. “P” denotes phosphorylation of the protein, and squiggly lines denote dsRNA. Inactive PKR monomers and dimers exist in equilibrium. The dimerization is mediated by interactions between the hydrophobic sides of the α helix. dsRNA can bind to the charged side of either monomers or dimers, but only the latter leads to a conformational change, ATP binding, intermolecular autophosphorylation, and acquisition of enzyme activity. Heparin, on the other hand, binds to monomers and causes a different conformational change leading to ATP binding, intramolecular autophosphorylation, and enzyme activation.
The last issue addressed in our study was the contribution of the dimerization property of PKR to its cellular antigrowth effects. For this set of in vivo experiments, we chose yeast over mammalian cells for a number of reasons: yeast strains provide a null background with no endogenous PKR, and they are more amenable to experimental manipulations to achieve regulated expression of transfected genes; moreover, effects of PKR and its mutants on yeast cell growth have been studied extensively by several laboratories (8, 11, 54), and thus the properties of our new dimerization-defective mutants could be interpreted in the context of a substantial body of literature. First, we established that the three dimerization-defective mutants were also incapable of interacting with PKR in vivo in yeast strains (Fig. 10). Two of these mutants with different properties were selected for further studies; both A67E and A63E are dimerization defective, but A67E can still bind dsRNA. As observed by others (8, 54), expression of wt PKR inhibited yeast growth severely whereas expression of the enzymatically inactive mutant K296R had no effect (Fig. 11). Curiously, the K60A mutant, which does not bind dsRNA but dimerizes efficiently, also inhibited cell growth appreciably (40, 54). When the A63E mutation was introduced to the K60A background, the growth-inhibiting effect was abolished, indicating that the dimerization property is the crucial determinant. Similarly, the A63E single mutant, which is devoid of both dimerization and dsRNA binding, was not growth suppressive in yeast. The same conclusion was reinforced by analysis of the A67E mutant, which, in contrast to K60A can bind dsRNA but not dimerize. Thus, among the three mutants tested, the growth inhibition phenotype accompanied the dimerization phenotype. It is worth noting in this context that all three new mutants, but not the K296R mutant, are capable of activation by heparin. It therefore appears that the potential to be enzymatically active is not sufficient for the growth inhibition phenotype but the dimerization property of the protein is essential. The same conclusion was supported by the rescue experiment (Fig. 12). Growth inhibition by wt PKR was rescued by wt DRBD, which can both dimerize and bind dsRNA. The K60A DRBD was more efficient than the wt DRBD because it dimerizes better (Fig. 2B). If this dimerization property was destroyed (K60A, A63E, and A67E), the rescue activity was also abolished. Thus, dimerization ability, not dsRNA binding, of the DRBD protein was crucial for the rescue phenotype, a conclusion supported by two earlier studies (40, 54).
After completion of this study, Tan et al. (59a) reported the characterization of a second dimerization domain of PKR located between residues 244 and 296. The existence of such a domain was indicated in our earlier study where we observed that PKR mutants missing the DRBD interacted with wt PKR, albeit weakly (50). Thus, it appears that PKR, like many proteins (59a), has more than one dimerization domain. In the physiologically relevant context of the full-length protein, the two domains may synergize to form a more stable dimer. Given the experimental data from our studies and that of Tan et al., it is also conceivable that initial dimerization through the DRBD is essential for making the internal site available for protein-protein interactions. Such a model can explain the observed properties of the A67E mutant as reported in this paper.
ACKNOWLEDGMENTS
We gratefully acknowledge W. C. Merrick for purified eIF2 and Michael Katze for the polyclonal antiserum against PKR. We thank Paul Stanton and Theresa Rowe for excellent technical assistance, Kurt Runge for plasmid pRS314, and Dorthy Herzberg for editorial assistance.
This work was supported in part by National Institutes of Health grants CA-68782 and CA-62220 to G.C.S. and American Heart Association, North East Ohio chapter, grant 133-BG1A to R.C.P.
REFERENCES
- 1.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber G N, Tomita J, Garfinkel M S, Meurs E, Hovanessian A, Katze M G. Detection of protein kinase homologues and viral RNA-binding domains utilizing polyclonal antiserum prepared against a baculovirus-expressed ds RNA-activated 68,000-Da protein kinase. Virology. 1992;191:670–679. doi: 10.1016/0042-6822(92)90242-h. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff J R, Samuel C E. Mechanism of interferon action: the interferon-induced phosphoprotein P1 possesses a double-stranded RNA-dependent ATP-binding site. J Biol Chem. 1985;260:8237–8239. [PubMed] [Google Scholar]
- 4.Black T L, Safer B, Hovanessian A, Katze M G. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J Virol. 1989;63:2244–2251. doi: 10.1128/jvi.63.5.2244-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bycroft M, Grunert S, Murzin A G, Proctor M, St. Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 7.Carpick B W, Graziano V, Schneider D, Maitra R K, Lee X, Williams B R G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 8.Chong K L, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colthurst D R, Campbell D G, Proud C G. Structure and regulation of eukaryotic initiation factor eIF-2. Sequence of the site in the alpha subunit phosphorylated by the haem-controlled repressor and by the double-stranded RNA-activated inhibitor. Eur J Biochem. 1987;166:357–363. doi: 10.1111/j.1432-1033.1987.tb13523.x. [DOI] [PubMed] [Google Scholar]
- 10.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig A W, Cosentino G P, Donze O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 12.Davies M V, Elroy-Stein O, Jagus R, Moss B, Kaufman R J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der S D, Yang Y L, Weissmann C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois M F, Hovanessian A G. Modified subcellular localization of interferon-induced p68 kinase during encephalomyocarditis virus infection. Virology. 1990;179:591–598. doi: 10.1016/0042-6822(90)90126-c. [DOI] [PubMed] [Google Scholar]
- 15.Feng G S, Chong K, Kumar A, Williams B R G. Identification of double-stranded RNA-binding domains in the interferon-induced double-stranded RNA-activated p68 kinase. Proc Natl Acad Sci USA. 1992;89:5447–5451. doi: 10.1073/pnas.89.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galabru J, Hovanessian A G. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 17.George C X, Thomis D C, McCormack S J, Svahn C M, Samuel C E. Characterization of the heparin-mediated activation of PKR, the interferon-inducible RNA-dependent protein kinase. Virology. 1996;221:180–188. doi: 10.1006/viro.1996.0364. [DOI] [PubMed] [Google Scholar]
- 18.Green S R, Mathews M B. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 19.Hershey J W B. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 20.Hovanessian A G. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 21.Hovanessian A G, Galabru J. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur J Biochem. 1987;167:467–473. doi: 10.1111/j.1432-1033.1987.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 22.Imani F, Jacobs B L. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc Natl Acad Sci USA. 1988;85:7887–7891. doi: 10.1073/pnas.85.21.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judaware R, Petryshyn R. Partial characterization of a cellular factor that regulates the double-stranded RNA-dependent eIF2a kinase in 3T3-F442A fibroblasts. Mol Cell Biol. 1991;11:3259–3267. doi: 10.1128/mcb.11.6.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 25.Katze M G. The war against the interferon-induced dsRNA-activated protein kinase: can viruses win? J Interferon Res. 1992;12:241–248. doi: 10.1089/jir.1992.12.241. [DOI] [PubMed] [Google Scholar]
- 26.Katze M G, Wambach M, Wong M L, Garfinkel M, Meurs E, Chong K, Williams B R, Hovanessian A G, Barber G N. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharrat A, Macias M J, Gibson T J, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitajewski J, Schneider R J, Safer B, Munemitsu S M, Samuel C E, Thimmappaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 29.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 33.Lee T G, Tomita J, Hovanessian A G, Katze M G. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci USA. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manche L, Green S R, Schmedt C, Mathews M B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maran A, Maitra R K, Kumar A, Dong B, Xiao W, Li G, Williams B R, Torrence P F, Silverman R H. Blockage of NF-kappa B signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 37.Marcus P I, Sekellick M J. Interferon induction by viruses. XVI. 2-Aminopurine blocks selectively and reversibly an early stage in interferon induction. J Virol. 1988;69:1637–1645. doi: 10.1099/0022-1317-69-7-1637. [DOI] [PubMed] [Google Scholar]
- 38.McCormack S J, Ortega L G, Doohan J P, Samuel C E. Mechanism of interferon action motif I of the interferon-induced, RNA-dependent protein kinase (PKR) is sufficient to mediate RNA-binding activity. Virology. 1994;198:92–99. doi: 10.1006/viro.1994.1011. [DOI] [PubMed] [Google Scholar]
- 39.McCormack S J, Thomis D C, Samuel C E. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology. 1992;188:47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- 40.McMillan N A, Carpick B W, Hollis B, Toone W M, Zamanian D M, Williams B R G. Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J Biol Chem. 1995;270:2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- 41.Meurs E, Chong K, Galabru J, Thomas N S, Kerr I M, Williams B R, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 42.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meurs E F, Watanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5804–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murre C, McCaw S P, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 45.Olsen H S, Cochrane A W, Dillon P J, Nalin C M, Rosen C A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 46.O’Malley R P, Mariano T M, Siekierka J, Mathews M B. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 47.Ortega L G, McCotter M D, Henry G L, McCormack S J, Thomis D C, Samuel C E. Mechanism of interferon action. Biochemical and genetic evidence for the intermolecular association of the RNA-dependent protein kinase PKR from human cells. Virology. 1996;215:31–39. doi: 10.1006/viro.1996.0004. [DOI] [PubMed] [Google Scholar]
- 48.Patel R C, Sen G C. Characterization of the interactions between double-stranded RNA and the double-stranded RNA binding domain of the interferon induced protein kinase. Cell Mol Biol Res. 1994;40:671–682. [PubMed] [Google Scholar]
- 49.Patel R C, Sen G C. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J Biol Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 50.Patel R C, Stanton P, McMillan N M J, Williams B R G, Sen G C. The interferon-inducible double-stranded RNA-activated protein kinase, PKR, self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel R C, Stanton P, Sen G C. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 52.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 53.Petryshyn R, Chen J-J, London I M. Growth-related expression of a double-stranded RNA-dependent protein kinase in 3T3 cells. J Biol Chem. 1984;259:14736–14742. [PubMed] [Google Scholar]
- 54.Romano P R, Green S R, Barber G N, Mathews M B, Hinnebusch A G. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF2a kinase DAI in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadowski I, Ptashne M. A vector for expressing GAL4 (1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuel C E. The eIF-2α protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 57.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonenberg N. Measures and countermeasures in the modulation of initiation factor activities by viruses. New Biol. 1990;2:402–409. [PubMed] [Google Scholar]
- 59.St. Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Tan S-L, Gale M J, Jr, Katze M G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiwari R K, Kusari J, Kumar R, Sen G C. Gene induction by interferons and double-stranded RNA: selective inhibition by 2-aminopurine. Mol Cell Biol. 1988;8:4289–4294. doi: 10.1128/mcb.8.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitaker-Dowling P A, Youngner J S. Characterization of a specific kinase inhibitory factor produced by vaccinia virus which inhibits the interferon-induced protein kinase activity. Virology. 1984;137:171–181. doi: 10.1016/0042-6822(84)90020-5. [DOI] [PubMed] [Google Scholar]
- 62.Wong A H, Tam N W, Yang Y L, Cuddihy A R, Li S, Kirchhoff S, Hauser H, Decker T, Koromilas A E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu S, Kaufman R J. Double-stranded (ds) RNA binding and not dimerization correlates with the activation of the dsRNA-dependent protein kinase (PKR) J Biol Chem. 1996;271:1756–1763. doi: 10.1074/jbc.271.3.1756. [DOI] [PubMed] [Google Scholar]
- 64.Zapp M L, Hope T J, Parslow T G, Green M R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zinn K, Keller A, Whittemore L-A, Maniatis T. 2-Aminopurine selectively inhibits the induction of β-interferon, c-fos, and c-myc gene expression. Science. 1988;240:210–213. doi: 10.1126/science.3281258. [DOI] [PubMed] [Google Scholar]