Abstract
In humans, gut microbiota can determine the health status. The regulatory mechanisms of the gut microbiota in asthma must be elucidated. Although antibiotics (ABXs) can clear infections, they markedly alter the composition and abundance of gut microbiota. The present study used ABX-treated mice to examine the time-dependent effects of ABX administration on the gut microbiota and intestinal mucosal barrier. The mouse asthma model was established using ovalbumin (OVA) and gavaged with an ABX cocktail for different durations (1 or 2 weeks) and stacked sequences. The pathology of the model, model 2, OVA-ABX, OVA-ABX 2, ABX-OVA and ABX-OVA was severe when compared with the control group as evidenced by the following results: i) significantly increased pulmonary and colonic inflammatory cell infiltration; ii) enhanced pause values and iii) OVA-induced immunoglobulin E (IgE) and TGF-β expression levels, and significantly downregulated Tight Junction Protein 1 (TJP1), claudin 1 and Occludin expression levels. Furthermore, the intestinal bacterial load in the OVA-ABX and OVA-ABX 2 groups was significantly lower than that in the ABX-OVA and ABX-OVA 2 groups, respectively. The predominant taxa were as follows: phyla, Firmicutes and Proteobacteria, genera, Escherichia-Shigella, Lactobacillus and Lachnospira. The abundances of Lachnospira and Escherichia-Shigella were correlated with the expression of OVA-induced IgE and TJPs. These findings indicated that ABX administration, which modifies microbiome diversity and bacterial abundance, can disrupt colonic integrity, downregulate TJ proteins, damage the intestinal barrier, enhance enterocyte permeability, and promote the release of inflammatory factors, adversely affecting asthma alleviation and long-term repair.
Keywords: asthma, intestinal flora, inflammation, tight junction proteins
Introduction
Allergic asthma, a complex chronic allergy-related inflammatory disease, is characterized by airway hyperresponsiveness, inflammation and remodeling. The clinical manifestations of allergic asthma include cough, chest tightness and croup (1). Currently, allergic asthma is one of the most prevalent diseases, affecting more than 300 million individuals worldwide. The incidence of allergic asthma is projected to reach 400 million by 2025(2). Asthma is frequently characterized by the upregulation of systemic allergen-specific immunoglobulin E (IgE) levels and pulmonary eosinophil infiltration. Various hypotheses have been proposed to explain the pathophysiology of asthma. According to one hypothesis, asthma onset is associated with the disruption of the intestinal milieu. The ecological dysregulation of colonic bacteria (such as thick-walled bacteria and mycobacteria) promotes their transfer from the intestine to the lungs via the lymph and blood, promoting the development of respiratory diseases, such as lung cancer, asthma, tuberculosis and cystic fibrosis (3). Usage of antibiotics (ABX), which markedly modifies the abundance and composition of the bacterial population, is among the major factors that influence gut microbiota composition (4). In particular, ABX usage alters the microbiome and decreases microbial diversity. Previous studies have demonstrated that ABX usage exerts adverse effects on health (5,6). ABX usage during early pregnancy determines the gut microbial composition of the mother and fetus and increases the risk of atopic rhinitis and asthma in the fetus (7).
An aberrant intestinal milieu increases the susceptibility of the intestine to foreign bacterial colonization and consequently disrupts gut microbiota homeostasis, promotes intestinal immune dysfunction, and modifies host immunity. Intestinal mucosal barrier function is essential for maintaining a balanced response between the host and its microbiome (8). The intestinal mucosal barrier comprises chemical, biological, immune and mechanical barriers, which are essential for its function (9). Occludin (OCLN) and claudin 1 (CLDN1) are transmembrane proteins predominantly expressed in the intercellular space. The band closure protein Tight Junction Protein 1 (TJP1), which is essential for the aggregation of CLDN1, is a crucial TJP. Additionally, TJP1 is an adaptor protein that promotes the binding between OCLN and CLDN1 at the cell junctions, as well as the binding of transmembrane proteins with the actin cytoskeleton (10,11).
Although the correlation between ABXs, gut microbiota and asthma has been identified, the underlying mechanisms have not been completely elucidated (12). Based on a previous study (13), the present study examined the long-term effects of ABXs on the gut microbiota and the pathogenesis of allergic inflammation using the ovalbumin (OVA)-induced asthma mouse model.
Materials and methods
Experimental materials and reagents
Materials and reagents used in the present study were as follows: OVA (cat. no. A5503; Sigma-Aldrich; Merck KGaA), aluminum hydroxide (cat. no. 77161; Thermo Fisher Scientific, Inc.), methacholine (MCh; cat. no. A2251; Sigma-Aldrich; Merck KGaA), enzyme-linked immunosorbent assay (ELISA) kits for TGF-β (cat. no. MB-6138A) and OVA-sIgE (cat. no. MB-6148A) (both from Jiangsu Enzyme Biological Co., Ltd.), total protein extraction kit (RIPA Lysis Buffer:Phenylmethanesulfonyl fluoride:Protease inhibitor cocktail for general use, 100X=100:1:1; cat. nos. P0013B, ST506 and P1005; Beyotime Institute of Biotechnology), bicinchoninic acid (BCA) kit (cat. no. P0010; Beyotime Institute of Biotechnology), rabbit polyclonal antibodies for TJP1 (cat. no. AF5145; Affinity Biosciences), rabbit polyclonal antibodies for CLDN1 (cat. no. AF0127; Affinity Biosciences), rabbit polyclonal antibodies for OCLN (cat. no. DF7504; Affinity Biosciences), rabbit anti-actin antibodies (cat. no. 10966R; BIOSS), and horseradish peroxidase (HRP)-conjugated Affinipure goat anti-Rabbit IgG (H + L) (cat. no. SA00001-2; Proteintech Group, Inc.). 2X Taq plus master kit (cat. no. P211-02; Nanjing Novozan Biotechnology Co., Ltd.), TruSeq DNA Library Preparation Kit v2 (cat. no. FC-121-2001; Illumina Inc.), primer synthesis [Sangong Bioengineering (Shanghai) Co., Ltd.] and ChamQ SYBR Color qPCR Master Mix (2X) (cat. no. Q421-02; Vazyme Biotech Co, Ltd.).
Experimental animals
A total of 40 female BALB/c mice (age: 6-8 weeks; body weight: 20 g) of specific pathogen-free grade were purchased from Chengdu Dashuo Co. [Production license: SCXK (chuan) 2020-030]. The mice were reared under the following conditions: Temperature, 22-25˚C; humidity, ~60%; 12-h light/dark cycle; access to food and water, ad libitum. Mice treated with ABXs for 1 week were assigned to the control, model, OVA-ABX, and ABX-OVA groups, while those treated with ABXs for 2 weeks were assigned to the control 2, model 2, OVA-ABX 2 and ABX-OVA 2 groups. The number of mice in each group was 6. All animal experiments were approved by the Animal Ethics Committee of Yunnan University of Traditional Chinese Medicine (approval no. R-06202085; Kunming, China). After the detection of enhanced pause (Penh), the mice were anesthetized using an instrument via isoflurane inhalation under the following conditions: Exposure rate, 300-500 ml/min; isoflurane concentration, 4-5%; duration, 3-4 min. The orbital plexus blood sample (1 ml) was collected from the anesthetized mice. After blood sample collection, mice were euthanized via decapitation. The heartbeat in the chest cavity, the nerve reaction, and the body temperature were examined to confirm the death of mice. The remaining tissues were extracted and immediately transferred to the animal room of Yunnan University of Traditional Chinese Medicine for centralized treatment.
Drug preparation
Based on previous studies (14,15), mice were administered with the following ABX cocktail in physiological saline: Amoxicillin (100 mg/kg bodyweight) (to eliminate gram-negative bacteria); neomycin sulfate (100 mg/kg bodyweight) (to eliminate gram-positive and gram-negative bacteria); metronidazole (100 mg/kg bodyweight) (to eliminate gram-positive anaerobic and gram-negative anaerobic bacteria); amphotericin (1 mg/kg bodyweight) (to eliminate fungi); vancomycin (50 mg/kg bodyweight) (to eliminate gram-positive bacteria) (Beijing Solarbio Science & Technology Co., Ltd.).
Establishment of animal models
The asthma mouse model was established using OVA and aluminum hydroxide suspension. The model group was intraperitoneally administered with 50 µg OVA + 50 µl aluminum hydroxide (volume: 0.2 ml) during the first and second weeks of the experiment. Asthma was stimulated through nebulization with 2% OVA (6 ml/12 animals) for 30 min every other day beginning from the third week (for the model 2 group, the time of stimulation was at weeks 3 and 4). The control group was nebulized with saline.
Mice in the OVA-ABX groups were gavaged with an ABX cocktail at the beginning of the third week of the experiment (the third to the fourth week for the OVA-ABX 2 group) twice a day (0.2 ml/dose) and nebulized every second day.
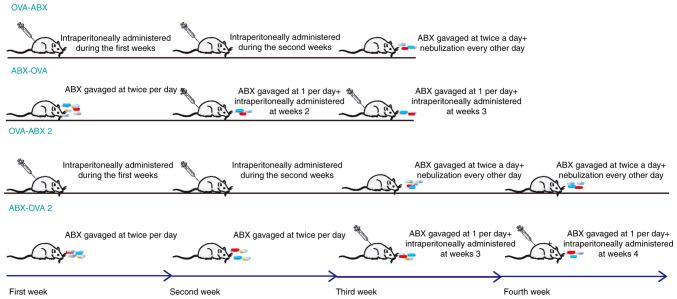
Meanwhile, mice in the ABX-OVA groups were gavaged with the ABX cocktail at week 1, twice per day (weeks 1-2 for the ABX-OVA 2 group), intraperitoneally administered with OVA at weeks 2-3 (weeks 3-4 for the ABX-OVA 2 group), and nebulized on alternate days between day 21 and 29. The precise modeling timing and methodology is illustrated in Fig. 1.
Figure 1.
Establishment of the asthma model and administration of antibiotics in different groups. OVA, ovalbumin; ABX, antibiotics.
Whole Body Plethysmography (WBP) of Penh levels in the respiratory system of mice
The room temperature and tranquility were maintained throughout the experiment. The experiment was performed in a WBP (cat. no. 27389; EMMS; http://www.electromedsys.com/wbp.html). A nebulizer was used to administer different concentrations of MCh (0, 6.25, 12.5, 25, 50 and 100 mg/ml) for 2 min. The change in the respiratory movement after inhalation of the nebulized gas caused corresponding changes in the pressure and flow rate outputs in the box. The pressure sensors (flow rate sensors) connected to the body descriptor box transmitted the collected signals to the signal processing system. The signal amplifiers converted these signals to values of various respiratory dynamic indices. The Penh values were recorded for 3 min. The duration between the administration of two concentrations was 4 min.
Hematoxylin and eosin (H&E) staining of pathological samples
The right lower lung lobe and colon tissue 1 cm from the rectum were rapidly excised after blood sampling. The samples were placed in EP tubes, fixed with 4% paraformaldehyde for 24 h at room temperature, dehydrated, embedded, sectioned (~5-µm thick), stained with H&E, and sealed. The pathological morphological changes in the lung and intestinal tissues were evaluated under a light microscope (Nikon Eclipse 2000 equipped with Nikon DS-Fi2; Nikon Corporation).
ELISA
The levels of OVA-specific IgE in the mouse serum and TGF-β in the lung tissue homogenates were examined using ELISA. At the end of the experiment, the blood sample (~1 ml per mouse) was obtained from mice at the time of euthanasia. The blood sample was allowed to naturally clot at room temperature for 1 h and centrifuged at 4˚C and 2,000 g x for 15 min. The supernatant was collected and stored at -80˚C. The lung tissue samples (50 mg) were homogenized with 450 ml phosphate-buffered saline. The homogenate was centrifuged for 15 min at 626 x g. The supernatant was collected and stored at -80˚C until analysis.
The samples were spiked, incubated and washed, following the ELISA kit instructions. The optical density at 450 nm of the reaction mixture was measured using an enzyme marker. The levels of TGF-β expression in the lung homogenates and OVA-specific IgE in the serum were measured based on enzyme standardization and a standard curve using a microplate reader (SYNERGY H1; BioTek Instruments, Inc.).
Western blotting
The protein expression levels of TJP1, CLDN1 and OCLN in the colon were analyzed using western blotting. Briefly, total proteins were extracted using the total tissue protein extraction kit, following the manufacturer's instructions, and quantified using the BCA kit. The extracted proteins (20 µg/lane) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 10% gel. The resolved proteins were electro-transferred onto a polyvinyl difluoride membrane (0.45 µm) at 300 mA for 45 min. The membrane was then incubated with anti- TJP1, anti-CLDN1, anti-OCLN and anti-β-actin (1:1,000 for all antibodies) antibodies at 4˚C for 12 h. After blocking with 5% skimmed milk for 1 h at room temperature, the membrane was washed several times with Tris-buffered saline containing 0.1% Tween 20. After washing, the membrane was incubated with secondary antibodies (1:6,000) for 1 h at room temperature. Immunoreactive signals were developed and visualized using a digital imaging and analysis system Panasonic WV-CP230 (Panasonic Corp). The expression levels of target proteins were normalized to those of β-actin using ImageJ V1.8.0 (National Institutes of Health).
Quantification of mouse gut microbiota using 16S rDNA-specific primers
Genomic DNA was extracted from the fecal samples using the fast DNA® spin kit (cat. no. 116570200; MP Biomedicals), following the manufacturer's instructions. The quality of the isolated DNA was evaluated using agarose gel electrophoresis with a 1% gel (SYBR Green). The DNA concentration and purity were assessed using NanoDrop 2000 (Thermo Fisher Scientific, Inc.). DNA concentration ≥10 was ng/µl; while DNA purity was assessed on the A260/A280 nm ratio being 1.8-2.0. Amplification was performed using ABI GeneAmp®, 9700 with TransStart® FastPfu DNA Polymerase (cat. no. AP221-02; TransGen Biotech Co., Ltd.), following the manufacturer's instructions. The amplicons from the same sample were pooled and subjected to agarose gel electrophoresis with a 2% gel. High fidelity PCR was utilized to amplify bacterial 16S rDNA hypervariable region 3 (V3) and 4 (V4) with the primers 338F, 5'ACTCCTACGGGAGGCAGCAG-3' and 806R, 5'-GGACTACHVGGGTWTCTAAT-3'. Next, the amplicons were recovered from the gel using the AxyPrepDNA gel recovery kit (cat. no. AP-GX-500; Axygen; Corning, Inc.), eluted with Tris-HCl, and confirmed using agarose electrophoresis with a 2% gel. Based on the preliminary quantitative results of electrophoresis, the amplicons were quantified using Qubit 4.0 (Thermo Fisher Scientific, Inc.). Determination of loading concentration was conducted using Quantus™ Fluorometer (Promega Corporation); the range of the loading concentration of the final library was 54.13 nmol and sequencing libraries were generated with TruSeqTM DNA Sample Prep Kit (Illumina Inc.). Purified amplicons were pooled in equimolar and paired-end sequenced (2x300) on an PE300 platform (Illumina, Inc.) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. The average sequence length was 420 bp.
The relative abundance of bacterial taxa was used to rank the taxa. Operational taxonomic units were analyzed using Uparse (16) (version 7.0.1090; http://drive5.com/uparse/). The species composition of the community was determined for each sample and compared with the Silva database (Release138 http://www.arb-silva.de) with confidence threshold of 70%. Principal coordinate analysis (PCoA) was performed using R (version 3.3.1) to generate PCoA statistics and plots. Community composition at the phylum and genus levels was analyzed using R (version 3.3.1) tools to obtain the statistical data and plots. Comparative analysis of colonies between the groups was performed using the stats package in R (version 3.3.1) and the Scipy package in Python. Environmental factors were compared using the heatmap package in R (version 3.3.1) to generate the correlation heatmaps. Linear discriminant analysis effect size (LEfSe) analysis was performed using the LEfSe software (http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=lefse_upload).
Absolute fluorescence quantification of mouse colonic contents
DNA extraction and primer sequences were consistent with the aforementioned. The OD260 values of the constructed plasmids were determined by UV spectrophotometer (NanoDrop2000; Thermo Fisher Scientific, Inc.), and converted to copy number (copies/µl) by the following formula: Plasmid Name: 16S rDNA, Concentration (ng/µl): 103.61, Number of copies/µl: 2.99x1010; Plasmid vector name: pMD18-T; Plasmid vector size: 2692 bp; Plasmid starting copy number conversion formula (copies/µl)=concentration (ng/µl) x10-9 x6.02x1023/(molecular weight x660). It should be noted that molecular weight refers to the size of the vector plus the fragment size of the target gene.
Standard curve samples were prepared as follows: 10-fold gradient dilutions of each constructed plasmid (90 µl dilution + 10 µl plasmid) was used to prepare a standard curve comprising 4-6 points. Based on pre-experiments, 10-2 to 10-6 dilutions of 16S standard were selected for the preparation of the standard curve.
Regarding the fluorescence quantitative PCR assay, the amplification was performed in a fluorescent quantitative PCR instrument Model ABI7300 (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the instruction manual of ChamQ SYBR Color qPCR Master Mix 2X (cat. no. Q421-02; Vazyme Biotech Co, Ltd.). The copy values were calculated based on the number of cycles after obtaining the following data: 16S gene fluorescence PCR amplification curve and melting curve, amplification curve and melting curve of 16S gene quantitative standard, slash diagram of 16S gene quantification standards and hybridization efficiency of 16S standard [Slope: -3.4601; Y-Inter: 42.231; R2: 0.9996; Efficiency (%): 94.54].
Statistical analysis
Data are presented as mean ± standard deviation. The experiment was repeated three times. The mean between groups was compared using one-way analysis of variance (ANOVA), followed by Tukey's post hoc test. The relative levels of OVA-IgE and microbial community species were analyzed using the Multivariate Association with Linear Models (MaAslin). The correlation between OVA-IgE and the relative abundance of microbial community species (data) was analyzed using MaAslin (R version 3.3.1; MaAsLin2 package; R Core Team), which constitutes a multivariate linear model. The correlation coefficients were calculated and analyzed using one-way ANOVA. Heatmap analysis was performed to calculate Spearman rank correlation coefficients between TJPs and microorganisms, as well as to obtain the R and P values of the correlations. The heatmap was plotted with R values distinguished by different color shades. *P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS 26.0 software (IBM Corp.). Graphing was performed using GraphPad Prism 8.0.2 software (Dotmatics).
Results
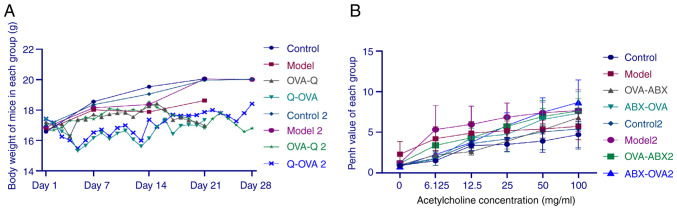
Effect of ABXs on the bodyweight of the OVA-induced asthma mouse model. The bodyweight of mice was not significantly different between the control, model and model 2 groups. However, the bodyweight of mice in the OVA-ABX, ABX-OVA, OVA-ABX 2, and ABX-OVA 2 groups significantly decreased after ABX administration (Fig. 2A). These findings indicated that ABXs decrease the bodyweight of the OVA-induced asthma mouse model.
Figure 2.
Antibiotic administration decreases bodyweight and increases the enhanced pause (Penh) value in the asthma mouse model. (A) Changes in the bodyweight of mice in different groups. (B) Changes in the Penh values of mice in different groups. OVA, ovalbumin; ABX, antibiotics.
Effect of ABX administration on the Penh values in the OVA-induced asthma mouse model
Compared with those in the control group, the Penh values were higher in the model, OVA-ABX, ABX-OVA, model 2, OVA-ABX 2 and ABX-OVA 2 groups. Furthermore, MCh dose-dependently increased the Penh value (Fig. 2B). Thus, ABXs increased the Penh values in the OVA-induced asthma mouse model.
Histopathological pulmonary changes in different groups
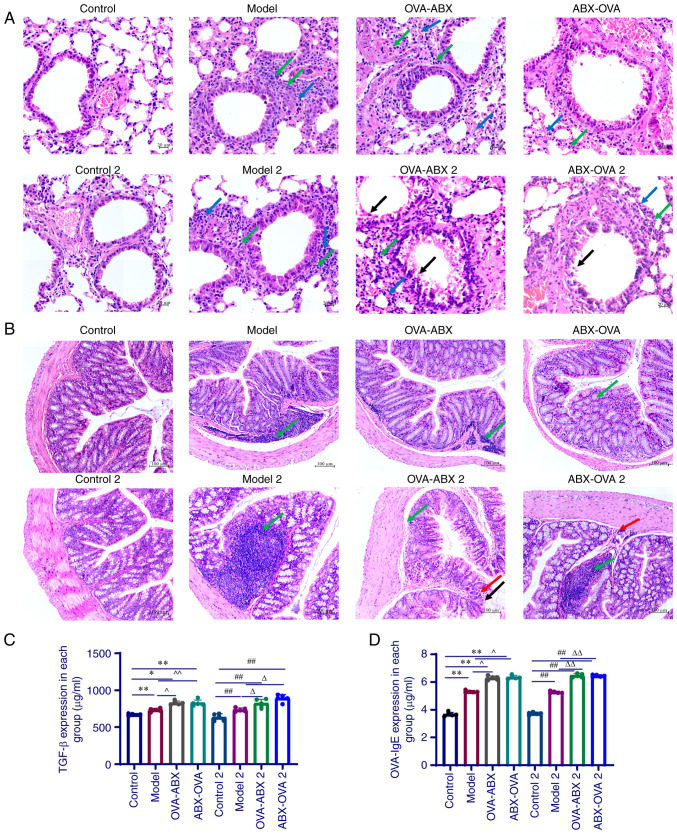
Mice in the control and control 2 groups did not exhibit marked inflammatory cell infiltration in the lung tissue. Additionally, the tracheal lumen was unobstructed with no marked thickening of the tracheal basement membrane. Furthermore, congestion and edema in the tracheal mucosa epithelium and epithelial damage were not observed, and the alveolar structure was intact. By contrast, the other groups exhibited an increased incidence of inflammatory cell infiltration, bronchial mucosa congestion and edema, smooth muscle and basement membrane thickening, alveolar collapse, widening of intervals, partial alveolar expansion, detached epithelial cells in the alveolar lumen and mucus in the tracheal lumen (Fig. 3A). These observations suggested the successful establishment of the asthma model.
Figure 3.
Antibiotic administration exacerbates inflammation and upregulates IgE expression in the asthma mouse model. (A) H&E staining of the lung sections of mice in different groups. Blue arrow, alveolar wall thickening; green arrow, neutrophils; black arrow, bronchial epithelial cells that are shed; (B) H&E staining of the colon sections of mice in different groups. Green arrow, lymphocytic infiltration; red arrow, granulocyte infiltration; black arrow, mucosal epithelial cells of intestinal tissue that are shed. (C) The levels of TGF-β in the lung tissue homogenate of mice in different groups. (D) The expression of OVA-induced IgE in the serum of mice in different groups (n=5). *P<0.05 and **P<0.01, control group vs. model group; ##P<0.01 control 2 group vs. control group; ^P<0.05 and ^^P<0.01, model group vs. OVA-ABX and ABX-OVA group; ∆P<0.05 and ∆∆P<0.01, model 2 group vs. OVA-ABX 2 and ABX-OVA 2 group. IgE, immunoglobulin E; H&E, Hematoxylin and eosin OVA, ovalbumin; ABX, antibiotics.
As shown in Fig. 3B, mice in the control and control 2 groups exhibited an intact colon mucosal tissue structure with no inflammatory infiltration in the mucosal, submucosal and muscle layers. In the other groups, the colonic tissue within the lamina propria exhibited atrophy of large intestinal glands that were loosely arranged, which was accompanied by inflammatory cell infiltration. The levels of inflammatory cell infiltration were upregulated in the model 2 and OVA-ABX 2 groups, while the lamina propria was severely damaged in the ABX-OVA 2 group (Fig. 3B). These observations indicated that inflammatory lesions are induced in the colon of the OVA-induced asthma mouse model and that ABXs may differentially affect the development of asthma.
Pulmonary TGF-β and serum OVA-induced IgE levels in different groups
The lung tissue homogenate levels of TGF-β in all experimental groups were significantly upregulated when compared with those in the control and control 2 groups (P<0.05 and P<0.01) (Fig. 3C). The expression of TGF-β in the OVA-ABX, OVA-ABX 2, ABX-OVA and ABX-OVA 2 groups was significantly upregulated when compared with that in the model and model 2 groups (P<0.05 and P<0.01).
The serum levels of OVA-induced IgE in the model, OVA-ABX and ABX-OVA groups were higher than those in the control group (P<0.01). Similarly, the serum levels of OVA-induced IgE in the model 2, OVA-ABX 2 and ABX-OVA 2 groups were upregulated when compared with those in the control 2 group (P<0.01). Compared with those in the model and model 2 groups, the increased IgE levels were significantly elevated in the OVA-ABX, OVA-ABX 2, ABX-OVA and ABX-OVA 2 groups (P<0.05 and P<0.01). Thus, ABX administration upregulated OVA-induced IgE expression (Fig. 3D). These results demonstrated that long-term treatment of ABX can upregulate the expression of inflammatory markers and the production of immunoglobulins in the OVA-induced asthma mouse model.
Effect of ABXs on the expression of TJPs in the asthma mouse model
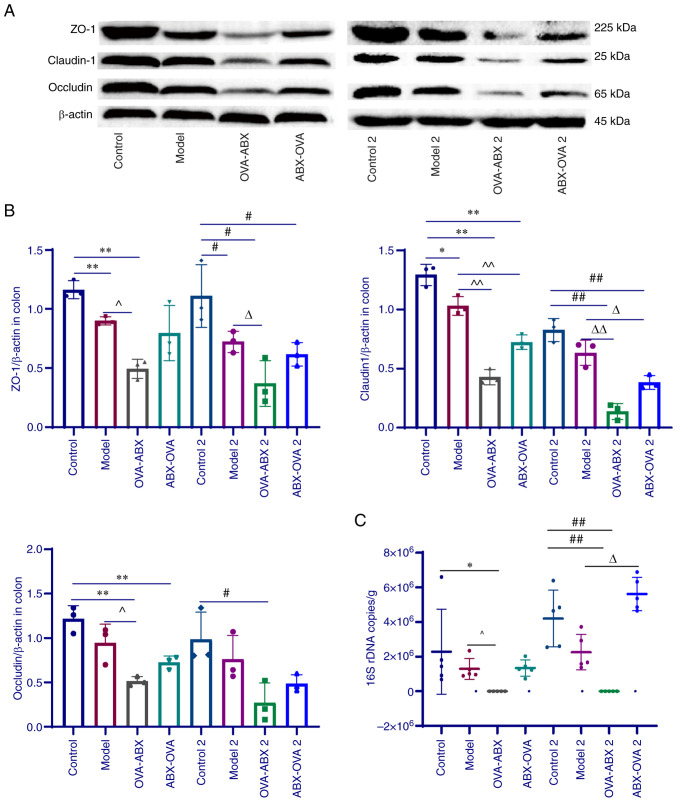
The colonic expression levels of TJP1, CLDN1 and OCLN in the OVA-ABX, OVA-ABX 2, ABX-OVA and ABX-OVA 2 groups were downregulated when compared with those in the control and control 2 groups (P<0.05 and P<0.01) (Fig. 4A and B). Compared with those in the model and model 2 groups, the expression levels of TJPs were significantly downregulated in the OVA-ABX and OVA-ABX 2 groups but the downregulation was less significant in the ABX-OVA and ABX-OVA 2 groups. Furthermore, the downregulation of TJPs in the group treated with ABX for 2 weeks was lower than that in the group treated with ABX for 1 week. These findings suggested that the expression of TJPs is downregulated in the OVA-induced asthma model and that ABX administration further downregulates their expression.
Figure 4.
Antibiotic administration downregulates the expression of tight junction proteins and suppresses gut microbial colonization. (A) The protein expression levels of TJP1, CLDN1 and OCLN in different groups. (B) The ratio of TJP1, CLDN1, or OCLN grayscale values to β-actin grayscale values in different groups (n=3). (C) Bacterial load in the colonic contents of different groups (n=5). *P<0.05 and **P<0.01, compared with the model group; ^P<0.05 and ^^P<0.01, compared with the control 2 group; #P<0.05 and ##P<0.01, compared with the model 2 group; IP<0.05 and IIP<0.01. CLDN1, claudin-1; OCLN, Occludin; OVA, ovalbumin; ABX, antibiotics.
Effect of ABX administration on the colonic bacterial load
Absolute fluorescence quantification of mouse colon contents revealed that the fecal bacterial load in the OVA-ABX and OVA-ABX 2 groups was significantly lower than that in the other groups (Fig. 4C). Furthermore, the bacterial load recovered one week and two weeks after ABX discontinuation with recovery being directly proportional to time. Thus, the bacterial load of mice was found to be markedly decreased after ABX administration and can progressively be restored after the discontinuation of ABX treatment.
Analysis of the diversity, structure and correlation of gut microbiota in different groups. Effect of ABXs on the intestinal microbial diversity in the asthma mouse model
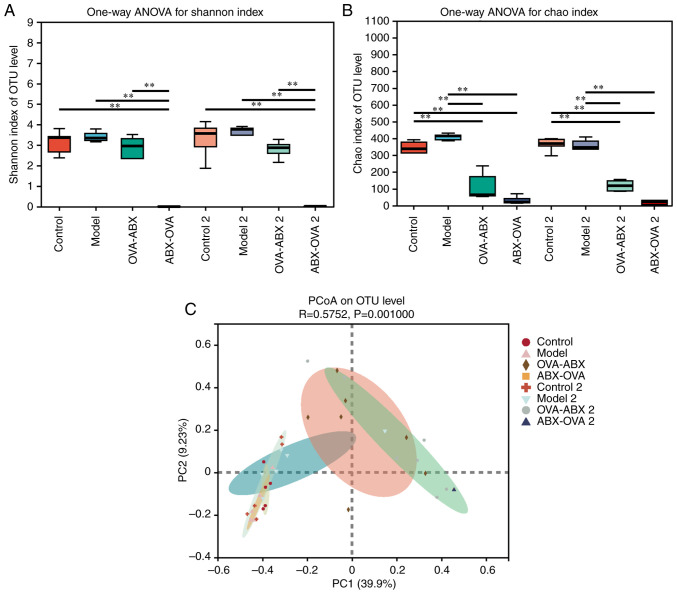
Shannon index in the OVA-ABX and ABX-OVA groups was lower than that in the control group. Furthermore, the Shannon index in the ABX-OVA group was significantly lower than that in the control, model and OVA-ABX groups (P<0.01). ABX treatment after 2 weeks exerted similar effects on the Shannon index (P<0.01) (Fig. 5A). Chao index in the OVA-ABX and ABX-OVA groups was significantly lower than that in the control and model groups. ABX treatment after 2 weeks exerted similar effects on the Chao index (Fig. 5B). These findings suggested that ABX administration alters the variety and richness of gut microbiota in the OVA-induced mouse model, especially when the asthma model is established after ABX administration.
Figure 5.
ABX administration significantly decreases gut microbiota diversity in the asthma mouse model. (A) Shannon index of mouse gut microbiota. (B) Chao index of mouse gut microbiota. Alpha diversity refers to the diversity within a specific region or ecosystem. Typical measurements include Shannon and Chao indices. The Shannon index primarily reflects the species diversity in a sample. The Shannon index value is directly proportional to the diversity of species. (C) Principal coordinate analysis. The X-axis and Y-axis represent the two selected principal axes, while the percentage indicates the degree of difference in sample composition between the principal axes. The scales of the X-axis and Y-axis are relative distances with no practical significance. The different colors or shapes represent samples of different groupings. The closeness of the two sample points indicates that the species composition is similar). **P<0.01. OVA, ovalbumin; ABX, antibiotics; OTU, operational taxomic unit.
The similarities and differences in community composition in different groups were analyzed using beta diversity analysis and comparison group analysis of intestinal diversity in various mice (Fig. 5C and Table I). The community composition was similar in the control and model groups but dissimilar in the OVA-ABX, ABX-OVA, OVA-ABX 2 and ABX-OVA 2 groups. Analysis of similarity revealed that the R value was >0, suggesting that the difference between the groups was more significant than that within each group (P<0.05). Furthermore, the differences were significant between the ABX-treated, control and model groups.
Table I.
Anosim analysis results.
| Method | ANOSIM |
|---|---|
| Statistic | 0.6129 |
| P-value | 0.001 |
| Permutation number | 999 |
The distance between samples was calculated using the Unifrace distance method, and Anoism analysis was used primarily to detect whether the between-group differences were greater than the within-group differences and to determine whether the grouping was successful based on the R and P-values, where R>0 indicated that the between-group differences were greater than the within-group differences, and P<0.05 was considered to indicate statistically significant differences.
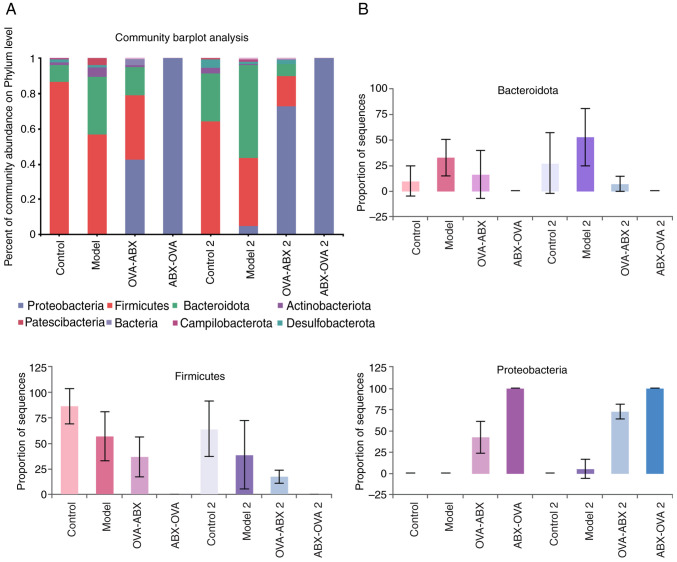
Effect of ABX administration on the abundance of gut microbiota at the phylum level. The predominant phyla were Proteobacteria, Firmicutes and Bacteroidota. Compared with those in the control and control 2 groups, the abundance of Firmicutes decreased and the abundance of Proteobacteria increased in the model, model 2, ABX-OVA, ABX-OVA 2 and OVA-ABX 2 groups. Additionally, the relative abundance of Bacteroidota in the model group was higher than that in the control group but was downregulated in the ABX-OVA and OVA-ABX groups (Fig. 6A and B). Thus, ABX administration was revealed to alter the abundance of gut microbiota in the OVA-induced asthma mouse model.
Figure 6.
Antibiotics significantly alter the gut microbiota structure in the asthma mouse model. (A) The abundance of mouse gut microbiota at the phylum level in different groups. (B) The top three phyla with abundance values in different groups. OVA, ovalbumin; ABX, antibiotics.
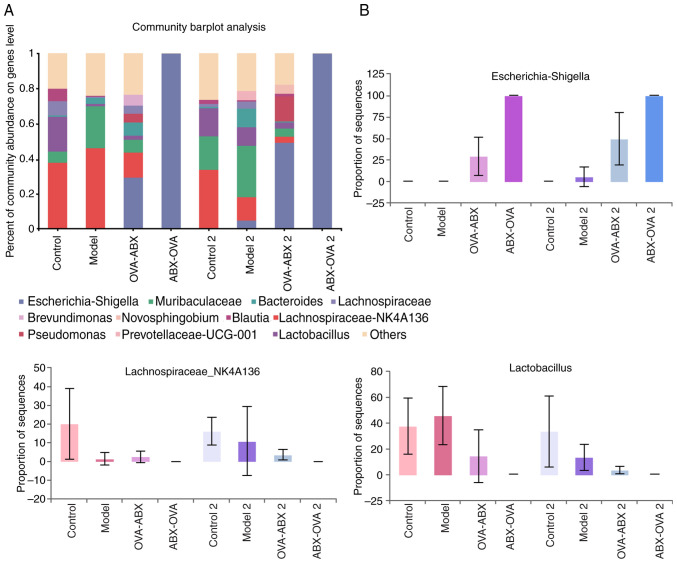
Effect of ABX administration on the abundance of gut microbiota at the genus level. Escherichia-Shigella, Lactobacillus and Lachnospira were the predominant genera in the mouse intestinal contents. Compared with those in the control group, the relative abundances of Escherichia-Shigella were higher and the relative abundances of Lactobacillus were lower in the model 2, OVA-ABX and ABX-OVA groups (Fig. 7A and B). Thus, broad-spectrum ABXs markedly affected the diversity and composition of healthy mouse intestinal microbiota.
Figure 7.
Structural changes in gut microbiota at the genus level in different groups. (A) The abundance of mouse gut microbiota at the genus level in different groups. (B) The top three genera with abundance values in different groups. OVA, ovalbumin; ABX, antibiotics.
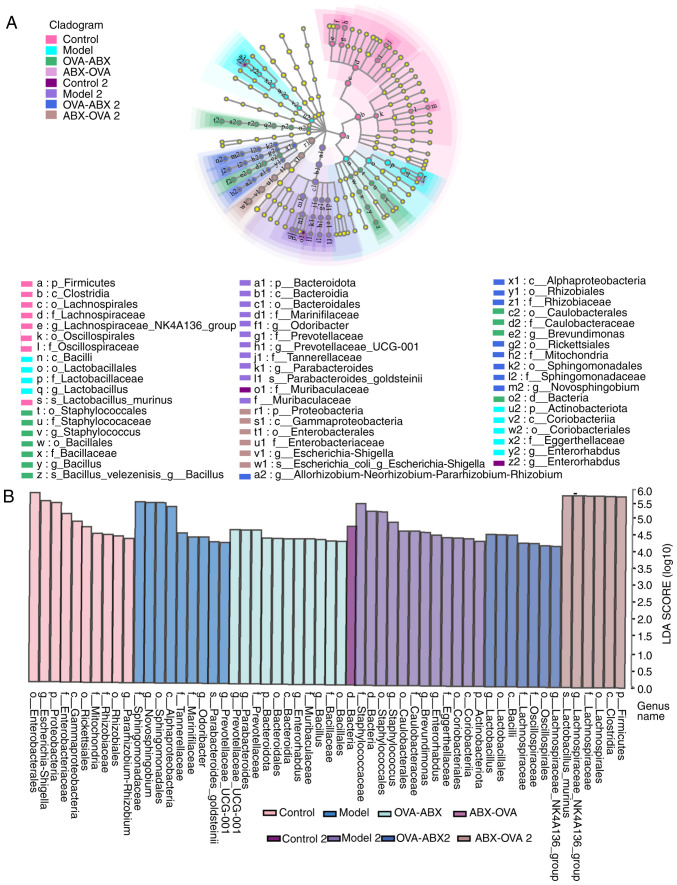
Major differential genera of gut microbiota in mice. Based on the initial findings, the distinct species affected by ABX treatment were identified. Thus, LEfSe analysis was performed to identify differential species (Fig. 8A and B). The distinctive bacteria in different groups were as follows: Control group, Lachnospira; model group, Lactobacillus and Enterorhabdus; OVA-ABX group, Brevundimonas, Staphylococcus and Bacillus; control 2 group, Enterorhabdus; model 2 group, Parabacteroides, Prevotellaceae and Odoribacter; OVA-ABX 2 group, Novosphingobium and Allorhizobium; ABX-OVA 2 group, Escherichia-Shigella. These findings suggested that asthma and ABX usage can alter the composition of the gut microbiota and impair the equilibrium of the microbiome.
Figure 8.
Antibiotic administration alters the abundance of gut microbiota in the asthma mouse model. (A) Evaluation of differential species in the mouse gut microbiota using LDA and LEfSe analysis. The non-parametric factorial Kruskal-Wallis sum-rank test was used to determine significant differences in abundance. (B) LEfSe analysis examines the degree of contribution of the abundance of each component (species) to different effects. In this experiment, a log LDA score threshold of 4.0 was utilized to determine significant taxonomic differences between groups. LDA was performed to analyze species information associated with a significantly increased abundance of species within each group relative to other species. LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; OVA, ovalbumin; ABX, antibiotics.
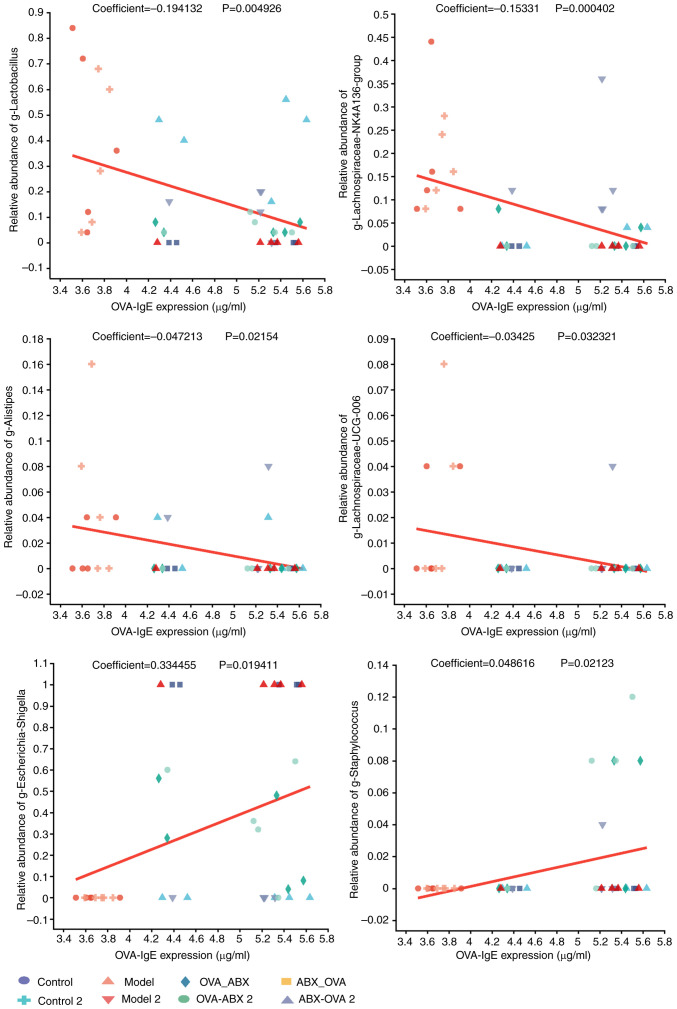
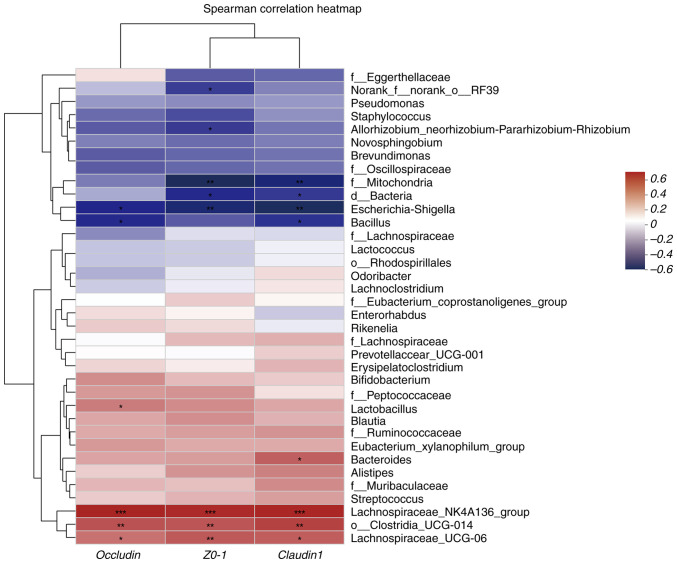
Analysis of the correlation between distinct mouse species and environmental factors. Correlation heatmap plots were used to evaluate the correlation between the proportion of genera and the expression of TJPs. Additionally, MaAslin was used to determine the linear correlation between environmental factors and the relative abundance of the differential microbial species. IgE expression was negatively correlated with Lactobacillus, Lachnospira NK4A136 group, Alistipes and Lachnospira UCG-006 abundances and positively correlated with Escherichia-Shigella and Staphylococcus abundances in the model and ABX-treated groups relative to the control group (P<0.05 and P<0.01) (Fig. 9). TJP expression was positively correlated with Lachnospira NK4A136 group and Lachnospira UCG-006 abundances and negatively correlated with Escherichia-Shigella abundances (P<0.05 and P<0.01) (Fig. 10). These findings indicate the correlation between gut microbiota composition and the production of TJPs and IgE in the model and ABX-treated groups.
Figure 9.
OVA-induced IgE expression is correlated with enterobacterial abundance. Linear correlation between distinct genera and environmental variables. The X-axis represents environmental variables, while the Y-axis indicates the relative abundance of a species. Coefficient value indicates the magnitude of the correlation coefficient between environmental factor variables and species abundance. A value greater than 0 indicates that environmental factors are positively correlated wit h the relative abundance of a species, while a value equal to 0 indicates that there is no correlation. The P-value was used to measure the reliability of the test. P<0.05 indicates that the environmental variables are significantly correlated with the species' abundance. OVA, ovalbumin; IgE, immunoglobulin E.
Figure 10.
Tight junction protein expression was negatively correlated with Escherichia-Shigella abundance and positively correlated with Lachnospira abundance. Heatmap of the correlation between gut microbiota and environmental factors in mice. *P<0.05, **P<0.01 and ***P<0.001
Discussion
Allergic asthma is a chronic inflammatory disease of the airways involving several inflammatory cells, including eosinophils, mast cells and macrophages. Recent studies have demonstrated that the gut microbiota plays a vital role in immune development, maturation and regulation (17,18). Genetic factors, delivery methods, living environment and ABX abuse can disrupt the gut microbiota, leading to the development of allergic rhinitis and asthma. Gut microbes are involved in maintaining local (19) and systemic immune balance (12,20). Dysbiosis promotes the colonization and proliferation of potentially pathogenic bacteria, induces the production of inflammatory factors, and adversely affects the immune barrier and homeostasis (21), contributing to the development of immune diseases, such as asthma (22,23). According to the theory of the lung-gut axis, pulmonary diseases are associated with gut microorganisms (12). Additionally, a microbe-rich environment positively affects the maturation of the gut microbiota of infants during the first year of life, modulating the ability of gut microbiota to produce the short-chain fatty acid butyric acid and consequently exert protective effects on childhood asthma (24). Microorganisms affect the immune response of intestinal and respiratory epithelial cells. Patients with chronic obstructive pulmonary disease have been reported to be associated with an increased prevalence of inflammatory bowel disease (25). Various factors affect the gut microbiota, and controlling these variables in human studies evaluating the correlation between gut microbiota and asthma is challenging. Thus, the use of experimental models enables the monitoring of all variables and provides reliable evidence.
To establish the asthma model with ABX usage, mice were administered with ABXs for different durations and stacked sequences. Fluorescence measurement of bacterial load in the intestinal contents of mice demonstrated that the administration of ABXs efficiently altered gut microbiota. However, the bacterial load was restored when the number of ABX was decreased. Next, the composition and abundance of gut microbiota in different groups were examined using 16S rDNA sequencing. ABX administration decreased the diversity and abundance of gut microbiota in mice, especially in the ABX-OVA and ABX-OVA 2 groups. Furthermore, Proteobacteria was the dominant phylum, while Escherichia-Shigella and Lachnospira were the dominant genera. Escherichia-Shigella and Lachnospira were not detected in the control group but their abundance gradually increased in the model, OVA-ABX and ABX-OVA groups. Proteobacteria is the predominant bacterial phylum, comprises gram-negative bacteria with lipopolysaccharides in the outer membrane, exerts pro-inflammatory effects, and dysregulates immunological reactions (26). Additionally, Proteobacteria is prevalent in respiratory illnesses (27), including chronic obstructive pulmonary disease, asthma (28) and bronchiectasis (29). The abundance of Proteobacteria was reported to be upregulated in the gut microbiota of patients with asthma and rhinitis (30). Wu et al (31) demonstrated similar findings in the OVA-sensitized asthma mouse model and reported a raised abundance of Proteobacteria. Elevated levels of Proteobacteria were accompanied by inflammatory cell infiltration in lung and intestinal tissues and Th2 cell subsets and the levels of Th2-related cytokines interleukin-4 (IL-4, IL-5 and IL-13) raised in asthmatic mice. Proteobacteria is likely to exacerbate asthma by stimulating a th2-type inflammatory response. In the present study, the abundance of Proteobacteria was upregulated in the model group and further upregulated upon ABX administration. Additionally, ABX administration exacerbated airway inflammation in mice. Dysbiosis and decreased Lachnospira abundance during the first 100 days of life were reported in infants at risk of developing asthma. The transplantation of Lachnospira in germ-free mice alleviated airway inflammation in adulthood (32). Lachnospira reduce serum levels of IL-17A, and IL-6 in lung homogenates and OVA-specific IgG2a, and significantly reduce the amount of Th17 in the immune response to achieve a lower inflammatory response (33). In the present study, the abundance of Lachnospira was downregulated in the model group and further downregulated upon treatment with ABX. Additionally, the abundance of Lachnospira was positively correlated with the expression of TJPs.
ABXs disrupt the gut microbiota composition by increasing the number of pathogenic bacteria or decreasing the number of beneficial bacteria, contributing to the development of atopic rhinitis, asthma, and other allergic illnesses.
A consensus on the correlation between ABXs, asthma and the intestinal barrier has not been achieved. In the present study, ABX administration affected the intestinal mucosa and lamina propria of mice. Additionally, intestinal tissue disruption was severe with prolonged ABX treatment. ABXs downregulated the expression of TJPs (TJP1, CLDN1 and OCLN), especially in the OVA-ABX and OVA-ABX 2 groups. This suggested that ABXs impair tight junctions and the protective function of the intestinal barrier in vivo.
Previous studies have demonstrated that clindamycin affects the function of the intestinal barrier by modulating the diversity of the animal intestinal microbiota or through other mechanisms (34-36). TJPs are crucial for maintaining the integrity of the intestinal mucosal barrier. CLDN1 and OCLN proteins play a vital role in maintaining barrier integrity and permeability by sealing and tightening the tight junctions. CLDN1-deficient animals do not survive owing to a compromised mucosal barrier (37). OCLN protein interacts with TJP1 protein to modulate intracellular and extracellular signaling and intestinal mucosal permeability (36). Inflammatory mediators, intestinal microbes and cytokines dysregulate the expression of CLDN1, OCLN and TJP1. Lachnospira is positively correlated with the expression of TJPs and negatively correlated with the proportion of Escherichia-Shigella. Electroacupuncture restored TJP expression and the balance of thick-walled bacteria, such as Lachnospira and Ehrlichia in rats with drowsy colitis (35). By contrast, Escherichia-Shigella, which are adnexal invasive bacteria, exacerbated the development of colitis in the ulcerative colitis group, which was accompanied by the downregulation of TJPs (38). Impaired intestinal barrier function and intestinal mucosal immunity exacerbate allergic disorders, such as asthma.
Intestinal mucosal barrier dysfunction promotes the entry of pathogenic bacteria, endotoxins and metabolic wastes in the gut wall into the bloodstream, resulting in systemic inflammatory responses, severe infections and immunological dysfunction. ABX administration exacerbated the inflammatory cell infiltration into the lung and colonic tissues in the asthma mouse model. Additionally, ABX administration upregulated the expression of OVA-induced IgE and TGF-β in the asthma mouse model. Previous studies have reported that dysbiosis contributes to the breakdown of the intestinal mucosal barrier and the enhanced production of inflammatory markers, aggravating the related illness. One ulcerative colitis study reported that P2RY13 disrupts the intestinal mucosal barrier, increases the production of I-6 and other components, and aggravates the inflammatory response (39). The enhanced production of inflammatory factors, including IL6, TNF and TGF-β disrupts the immunological balance of Th1/Th2 and Th17/Treg cells (40). The disruption of the gut microbiota structure promotes inflammation and increases the number of pathogenic microorganisms in the host (41). Additionally, the prolonged use of ABXs time-dependently exacerbates the inflammatory response during asthma attacks.
Most recent studies on asthma have focused on symptom relief during acute asthma exacerbation. Meanwhile, the present study focused on asthma triggers and mid-stage and late-stage repair. ABXs are often used to treat infections during asthma exacerbation. The present study demonstrated that the administration of ABXs can disrupt the homeostasis of gut microbiota, adversely affecting the recovery of asthma. Additionally, the abundance of intestinal bacteria was significantly downregulated, while that of opportunistic pathogens, such as Proteobacteria and Lachnospira was upregulated in the ABX-OVA, ABX-OVA 1, OVA-ABX and OVA-ABX 2 groups. Moreover, the inflammatory response and asthma symptoms were aggravated upon ABX treatment. Prolonged ABX treatment disrupted the balance of gut microbiota and the expression of TJP1, CLDN1 and OCLN as evidenced by the manifestations of the ABX-OVA 2 and OVA-ABX 2 groups. These changes may induce inflammatory responses and are not conducive to asthma alleviation during the recovery period. Dysbiosis increases the risk and severity of acute asthma attacks. The OVA-IgE and TGF-β expression levels and the pathological scores in the ABX-OVA, ABX-OVA 2, OVA-ABX and OVA-ABX 2 groups were higher than those in the model and model 2 groups. These findings indicated that the maintenance of gut microbiota balance can prevent acute asthma attacks.
Acknowledgements
The authors would like to thank Yunnan Provincial Key Lab of Molecular Biology for Sinomedicine for platform support and use of their research site for the experiments.
Funding Statement
Funding: The present study was supported by the National Nature Science Foundations of China (grant nos. 81660765 and 81860778), the Yunnan Applied Basic Research Projects (grant no. 2018FF001-017), and the Science and Technology Plan of Yunnan Science and Technology Department (grant no. 202101AZ070001-171).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information repository (https://www.ncbi.nlm.nih.gov/sra/PRJNA946646).
Authors' contributions
CLX, JC and CW designed the experiments and completed the study. CLX, GBL and TZ established the asthma model. CLX, CW and GBL were responsible for RT-qPCR and western blotting. CLX, RLZ and GBL wrote the manuscript. CLX, CW, GBL, TZ and RLZ made substantial contributions to the acquisition of data and confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All animal experiments were approved (approval no. R-06202085) by the Animal Ethics Committee of Yunnan University of Traditional Chinese Medicine (Kunming, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shipp CL, Gergen PJ, Gern JE, Matsui EC, Guilbert TW. Asthma management in children. J Allergy Clin Immunol Prac. 2023;11:9–18. doi: 10.1016/j.jaip.2022.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Voskamp AL, Kormelink TG, van Wijk RG, Hiemstra PS, Taube C, de Jong EC, Smits HH. Modulating local airway immune responses to treat allergic asthma: Lessons from experimental models and human studies. Semin Immunopathol. 2020;42:95–110. doi: 10.1007/s00281-020-00782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trivedi R, Barve K. Gut microbiome a promising target for management of respiratory diseases. Biochem J. 2020;477:2679–2696. doi: 10.1042/BCJ20200426. [DOI] [PubMed] [Google Scholar]
- 4.Ye X, Wang A, Lin W, Xu Y, Dong X, Zhou Y, Tian K, Xu X. The role of intestinal flora in anti-tumor antibiotic therapy. Front Biosci (Landmark Ed) 2022;27(281) doi: 10.31083/j.fbl2710281. [DOI] [PubMed] [Google Scholar]
- 5.Francino MP. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front Microbiol. 2015;6(1543) doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cryan JF, O'Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, Codagnone MG, Cussotto S, Fulling C, Golubeva AV, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 7.McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur J Immunol. 2018;48:39–49. doi: 10.1002/eji.201646721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017;12(e0173004) doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan H, Wang A, Wang Y, Sun Y, Han J, Chen W, Wang S, Wu Y, Lu Y. Innate lymphoid cells: Regulators of gut barrier function and immune homeostasis. J Immunol Res. 2019;2019(2525984) doi: 10.1155/2019/2525984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner JR. Molecular basis of epithelial barrier regulation: From basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo WT, Odenwald MA, Turner JR, Zuo L. Tight junction proteins occludin and ZO-1 as regulators of epithelial proliferation and survival. Ann N Y Acad Sci. 2022;1514:21–33. doi: 10.1111/nyas.14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hufnagl K, Pali-Schöll I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42:75–93. doi: 10.1007/s00281-019-00775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia W, Xu C, Zhao T, Fan Q, Qiao B, Wu Y, Yuan J, Chen J. Integrated network pharmacology and gut microbiota analysis to explore the mechanism of sijunzi decoction involved in alleviating airway inflammation in a mouse model of Asthma. Evid Based Complement Alternat Med. 2023;2023(1130893) doi: 10.1155/2023/1130893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo W, Zhou X, Li X, Zhu Q, Peng J, Zhu B, Zheng X, Lu Y, Yang D, Wang B, Wang J. Depletion of gut microbiota impairs gut barrier function and antiviral immune defense in the liver. Front Immunol. 2021;12(636803) doi: 10.3389/fimmu.2021.636803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6(e17996) doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 17.Brodin P. Immune-microbe interactions early in life: A determinant of health and disease long term. Science. 2022;376:945–950. doi: 10.1126/science.abk2189. [DOI] [PubMed] [Google Scholar]
- 18.Leonardi I, Gao IH, Lin WY, Allen M, Li XV, Fiers WD, De Celie MB, Putzel GG, Yantiss RK, Johncilla M, et al. Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell. 2022;185:831–846.e814. doi: 10.1016/j.cell.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han P, Gu JQ, Li LS, Wang XY, Wang HT, Wang Y, Chang C, Sun JL. The association between intestinal bacteria and allergic diseases-cause or consequence? Front Cell Infect Microbiol. 2021;11(650893) doi: 10.3389/fcimb.2021.650893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodin P. Immune-microbe interactions early in life: A determinant of health and disease long term. Science. 2022;376:945–950. doi: 10.1126/science.abk2189. [DOI] [PubMed] [Google Scholar]
- 21.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52:241–255. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambrecht BN, Hammad H, Fahy JV. The cytokines of asthma. Immunity. 2019;50:975–991. doi: 10.1016/j.immuni.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Zou XL, Wu JJ, Ye HX, Feng DY, Meng P, Yang HL, Wu WB, Li HT, He Z, Zhang TT. Associations between gut microbiota and asthma endotypes: A cross-sectional study in south china based on patients with newly diagnosed asthma. J Asthma Allergy. 2021;14:981–992. doi: 10.2147/JAA.S320088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, Schmausser-Hechfellner E, Roduit C, Frei R, Lauener R, et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. 2020;26:1766–1775. doi: 10.1038/s41591-020-1095-x. [DOI] [PubMed] [Google Scholar]
- 25.Raftery AL, Tsantikos E, Harris NL, Hibbs ML. Links between inflammatory bowel disease and chronic obstructive pulmonary disease. Front Immunol. 2020;11(2144) doi: 10.3389/fimmu.2020.02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nieuwdorp M, Gilijamse PW, Pai N, Kaplan LM. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146:1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Satokari R. High intake of sugar and the balance between pro- and anti-inflammatory gut bacteria. Nutrients. 2020;12(1348) doi: 10.3390/nu12051348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dicker AJ, Huang JTJ, Lonergan M, Keir HR, Fong CJ, Tan B, Cassidy AJ, Finch S, Mullerova H, Miller BE, et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2021;147:158–167. doi: 10.1016/j.jaci.2020.02.040. [DOI] [PubMed] [Google Scholar]
- 29.Guo MY, Chen HK, Ying HZ, Qiu FS, Wu JQ. The role of respiratory flora in the pathogenesis of chronic respiratory diseases. Biomed Res Int. 2021;2021(6431862) doi: 10.1155/2021/6431862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan J, Song J, Lv Q, Zhang H, Xiang Q, Dai H, Zheng H, Lin X, Zhang W. Alterations in the gut microbiome of young children with airway allergic disease revealed by next-generation sequencing. J Asthma Allergy. 2023;16:961–972. doi: 10.2147/JAA.S422537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Chen Y, Li Q, Ye X, Guo X, Sun L, Zou J, Shen Y, Mao Y, Li C, Yang Y. Tetrahydrocurcumin alleviates allergic airway inflammation in asthmatic mice by modulating the gut microbiota. Food Funct. 2021;12:6830–6840. doi: 10.1039/d1fo00194a. [DOI] [PubMed] [Google Scholar]
- 32.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307ra152) doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 33.Arrieta MC, Sadarangani M, Brown EM, Russell SL, Nimmo M, Dean J, Turvey SE, Chan ES, Finlay BB. A humanized microbiota mouse model of ovalbumin-induced lung inflammation. Gut Microbes. 2016;7:342–352. doi: 10.1080/19490976.2016.1182293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, McDowell A, Seo H, Kim S, Min TK, Jee YK, Choi Y, Park HS, Pyun BY, Kim YK. Diagnostic models for atopic dermatitis based on serum microbial extracellular vesicle metagenomic analysis: A pilot study. Allergy Asthma Immunol Res. 2020;12:792–805. doi: 10.4168/aair.2020.12.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tulstrup MV, Christensen EG, Carvalho V, Linninge C, Ahrné S, Højberg O, Licht TR, Bahl MI. Antibiotic treatment affects intestinal permeability and gut microbial composition in wistar rats dependent on antibiotic class. PLoS One. 2015;10(e0144854) doi: 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoedt EC, Hueston CM, Cash N, Bongers RS, Keane JM, van Limpt K, Amor KB, Knol J, MacSharry J, van Sinderen D. A synbiotic mixture of selected oligosaccharides and bifidobacteria assists murine gut microbiota restoration following antibiotic challenge. Microbiome. 2023;11(168) doi: 10.1186/s40168-023-01595-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouaud F, Vasileva E, Spadaro D, Tsukita S, Citi S. R40.76 binds to the α domain of ZO-1: Role of ZO-1 (α+) in epithelial differentiation and mechano-sensing. Tissue Barriers. 2019;7(e1653748) doi: 10.1080/21688370.2019.1653748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan L, Qi Y, Qu S, Chen X, Li A, Hendi M, Xu C, Wang L, Hou T, Si J, Chen S. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes. 2021;13:1–17. doi: 10.1080/19490976.2020.1826746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Wei S, Chen M, Li J, Wei Y, Zhang J, Dong W. P2RY13 Exacerbates intestinal inflammation by damaging the intestinal mucosal barrier via activating IL-6/STAT3 pathway. Int J Biol Sci. 2022;18:5056–5069. doi: 10.7150/ijbs.74304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad R, Sorrell MF, Batra SK, Dhawan P, Singh AB. Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol. 2017;10:307–317. doi: 10.1038/mi.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baltazar-Díaz TA, González-Hernández LA, Aldana-Ledesma JM, Peña-Rodríguez M, Vega-Magaña AN, Zepeda-Morale ASM, López-Ro RI, Toro-Arreola SD, Martínez-López E, Salazar-Montes AM, Bueno-Topete MR. Escherichia/Shigella, SCFAs, and metabolic pathways-the triad that orchestrates intestinal dysbiosis in patients with decompensated alcoholic cirrhosis from Western Mexico. Microorganisms. 2022;10(1231) doi: 10.3390/microorganisms10061231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information repository (https://www.ncbi.nlm.nih.gov/sra/PRJNA946646).