Abstract
Expression of the highly conserved replication-dependent histone gene family increases dramatically as a cell enters the S phase of the eukaryotic cell cycle. Requirements for normal histone gene expression in vivo include an element, designated α, located within the protein-encoding sequence of nucleosomal histone genes. Mutation of 5 of 7 nucleotides of the mouse H3.2 α element to yield the sequence found in an H3.3 replication-independent variant abolishes the DNA-protein interaction in vitro and reduces expression fourfold in vivo. A yeast one-hybrid screen of a HeLa cell cDNA library identified the protein responsible for recognition of the histone H3.2 α sequence as the transcription factor Yin Yang 1 (YY1). YY1 is a ubiquitous and highly conserved transcription factor reported to be involved in both activation and repression of gene expression. Here we report that the in vitro histone α DNA-protein interaction depends on YY1 and that mutation of the nucleotides required for the in vitro histone α DNA-YY1 interaction alters the cell cycle phase-specific up-regulation of the mouse H3.2 gene in vivo. Because all mutations or deletions of the histone α sequence both abolish interactions in vitro and cause an in vivo decrease in histone gene expression, the recognition of the histone α element by YY1 is implicated in the correct temporal regulation of replication-dependent histone gene expression in vivo.
The histone genes are among the most highly conserved genes in higher eukaryotes, both within each class of histone protein and across species from plants to mammals. Mammalian histone genes consist of 15 to 20 genes for each class of nucleosomal histone protein. These genes are classified as either replication dependent or replication independent on the basis of their patterns of expression within the cell cycle (44). Replication-independent variants are constitutively expressed at a low level throughout the cell cycle and are also known as replacement-variant histones (41, 43). Conversely, the transcription of replication-dependent histone genes is coordinately up-regulated at the onset of the S phase (11). Posttranscriptional regulation (mRNA processing and stability) plays an important role in the regulation of histone gene expression in the cell cycle, resulting in a 30-fold increase in histone mRNA levels in the S phase (11, 12, 17, 39). Extensive examination of histone gene promoters has identified elements necessary for the normal expression of replication-dependent histone genes through interactions of specific transcription factors with promoter sequences (18, 26, 27, 30, 31), but the promoter sequences that have been identified are class specific (i.e., present in all H2b gene promoters) and not shared between the different histone classes (17, 18, 31).
Previously, we identified a region (110 bp; called the coding region-activating sequence [CRAS]) necessary for the normal expression of H2a.2 and H3.2 histone genes (20, 21). Common elements contained within the CRAS are present in all four nucleosomal histone genes. Ours was the first report of elements common to all four classes of nucleosomal histone genes (4). Using S1 nuclease protection assays of total RNA isolated from asynchronously growing populations of stable transfectant CHO cells, we showed that deletion of the H3 or H2a CRAS caused a 20-fold decrease in the steady-state levels of expression of these genes (20, 21). We also showed that normal steady-state levels of H2a mRNA could be restored to a mouse H2a gene with the CRAS deleted when the CRAS from an H3.2 gene was inserted in frame at the site of the H2a CRAS deletion (20). Subsequently, two subsequences, each consisting of a core of 7 nucleotides (nt) and designated α and Ω, were identified as the sites of CRAS interactions with nuclear proteins in vitro (3, 4, 23). Mutation of the 7 nt of the H3.2 α or Ω element independently caused a fourfold decrease in the level of H3.2 mRNA in vivo and abolished the formation of DNA-protein complexes in vitro. When both elements were mutated, the observed decrease in the mRNA level was comparable to that observed upon deletion of the entire 110-bp H3.2 CRAS.
Mutation of these elements to yield the corresponding sequences from a replication-independent H3.3 gene also abolished the formation of DNA-protein complexes in vitro and caused a decrease in expression in stably transfected CHO cells in vivo identical to that caused by mutating all 7 nt of the H3.2 α or Ω element (23). As with the 7-nt mutations, the decrease in the steady-state level of mRNA with the doubly mutated gene in stable transfectants was similar to that observed upon deletion of the entire 110-bp CRAS.
The coding sequence of the H3.3 replacement-variant histone gene is 67% identical to that of the H3.2 gene at the nucleotide level, and third-base changes account for most of the differences (19, 25, 40); however, the sequence of the H3.3 “α element” is less similar than the rest of the protein-encoding sequence. Five of seven nucleotides differ and in fact encode the amino acid changes (codons 89 and 90) diagnostic of all known H3.3 histones (41, 44).
The interaction of nuclear factors with the two histone CRAS elements is regulated by phosphorylation, but in an opposite manner (22). That is, the α factor must be dephosphorylated to interact with the histone α element, and the Ω factor must be phosphorylated to interact with the histone Ω element. Phosphorylation on both serine/threonine and tyrosine residues is capable of inhibiting the α factor interaction because dephosphorylation events of both types activate the binding activity. Only tyrosine phosphorylation is involved in activation of the Ω DNA-binding activity, because a tyrosine-specific phosphatase abolishes the ability of the Ω factor to bind to its target sequence. The α DNA-binding activity is more abundant in nuclear extracts prepared from cells in late G1, and there is evidence that cyclin-dependent kinases can play a role in the regulation of the CRAS α DNA-binding activity (22).
Here we report that the DNA-binding component of the histone α factor is Yin Yang 1 (YY1) (35, 37). The identification was accomplished by means of a yeast one-hybrid system (28). We find that YY1 interacts specifically with the H3.2 α element in both yeast and mouse extracts. Furthermore, we show that mutation of the histone α element alters the G1-S-phase-specific up-regulation of the mouse H3.2 gene in stable transfectant CHO cells. In these experiments, we used synchronous populations of unperturbed CHO cells obtained with an automated mitotic shakeoff device. Because all mutations or deletions of the histone α sequence result in both the loss of protein-DNA interactions in vitro and a decrease in histone gene expression in vivo, these results are strong evidence that YY1 plays an important role in the regulation of replication-dependent histone gene expression in vivo.
MATERIALS AND METHODS
Construction of yeast strains.
The yeast one-hybrid strategy used was that of Li and Herskowitz (28). Six copies of the H3.2 α oligonucleotides were inserted into the promoter regions of the pHISi-1 gene (GenBank accession no. U89928) and the pLacZi gene (GenBank accession no. U89671) (7) (the double-stranded sequence is shown below with BamHI and BglII overhangs designated by lowercase letters): gatcCTCGGCCGTCATGGCGCTGCAGGAGGCa GAGCCGGCAGTACCGCGACGTCCTCCGtctag
Negative control reporter genes were constructed by insertion of six copies of the H3.3 α oligonucleotides into the promoter regions of the same reporter genes (H3.3 oligonucleotide sequences are shown below with BamHI and BglII overhangs designated by lowercase letters): gatccTGCAGCTATTGGTGCTTTGCAGGAGCa gACGTCGATAACCACGAAACGTCCTCGtctag
Successful insertion and proper orientation were verified by diagnostic restriction analysis. To generate the H3.2 yeast reporter strain, we transformed yeast strain YM4271 (Clontech) sequentially by the lithium acetate procedure with the H3.2 α pLacZi gene and then the H3.2 α pHISi-1 gene. The expression of β-galactosidase was determined by means of the colony lift assay and hydrolysis of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma). A concentration of 30 mM 3-aminotriazole (3-AT) (Sigma) was used to reduce background from basal HIS3 expression from the H3.2 reporter gene. The H3.3 reporter strain was constructed in the same manner, and 3-AT was similarly used to reduce background from basal HIS3 expression. The H3.2 strain was used to screen a directional HeLa cell cDNA library which was cloned into pGAD-GH (a gift from Yue Xiong) and which confers survival on Leu− media.
Rescue of plasmids and analysis.
Plasmids were rescued from activated strains with stationary-phase yeast cultures grown in selective media. Standard methods were used for yeast cell culturing and manipulation unless otherwise indicated (14). Spheroplasts were produced by incubation with 7.5 mg of Zymolyase per ml in 0.1 M NaCl–5% glycerol. Spheroplasts were resuspended in 50 mM Tris (pH 7.4)–20 mM EDTA and lysed with 10% sodium dodecyl sulfate (SDS). Proteins were removed with 5 M potassium acetate, and DNA was precipitated with isopropanol. Plasmid DNAs were purified by passage through Wizard Plus columns (Promega) and transformed into chemically competent Escherichia coli DH10B cells by means of a standard heat shock transformation procedure (29). Library plasmids were harvested from E. coli by alkaline lysis (29) and ethanol precipitation and grouped for further analysis on the basis of cDNA insert size after restriction analysis with BamHI and KpnI.
Extract preparation. (i) Mouse myeloma cell nuclear extracts.
Mouse myeloma cells were grown in Spinner cultures to a density of 5 × 105 cells/ml in Dulbecco’s modified Eagle’s medium (Gibco)–10% horse serum (Gibco)–5% CO2 as previously described (3). Nuclear extracts were prepared with HEPES dialysis buffer (HDB), our modification (20) of the method of Shapiro et al. (34). Aliquots were stored at −80°C for later use.
(ii) Yeast whole-cell extracts.
Yeast cultures were grown in selective medium to the late log phase, pelleted, washed, and resuspended in a buffer (25 mM HEPES [pH 7.0], 100 mM KCl, 10% glycerol, 1 mM EDTA) containing a cocktail of protease inhibitors (1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, leupeptin [1 μg/ml], aprotinin [1 μg/ml], bestatin [1 μg/ml], trypsin inhibitor [1 μg/ml], protease inhibitor [1 μg/ml]). The resuspended cells were passed through a French press twice at 23,000 lb/in2 and then centrifuged at high speed (100,000 × g). Supernatants were stored at −80°C.
Column purification.
Partial purification of the α DNA-binding activity was accomplished by application of nuclear extracts to double-stranded calf thymus DNA-cellulose resin (Pharmacia). Proteins were sequentially eluted with HDB containing 0.1, 0.2, 0.3, 0.4, or 1 M KCl (22). Fractions were analyzed for α DNA-binding activity by electrophoretic mobility shift assays (EMSA) with the H3.2 α oligonucleotides. Active fractions were pooled and dialyzed against 0.1 M KCl. The dialyzed pool was applied to a DEAE-Sepharose resin as previously described (22). Fractions were again tested by EMSA, and α DNA-binding fractions were pooled and dialyzed. The pooled fractions were applied to hydroxyapatite columns (5-ml bed volume; Bio-Rad) and eluted with a phosphate gradient (0 to 700 mM PO43−). Fractions were tested for α DNA-binding activity as described below. Equivalent amounts of α shift activity from each of these columns were also tested for the presence of YY1 by Western analysis.
EMSA.
DNA probes (oligonucleotide duplexes or restriction fragments) were 3′ end labeled by means of the filling-in reaction with the Klenow fragment of DNA polymerase I. The resulting DNA probes (1 ng), poly(dI-dC) (0.2-mg/ml final concentration; Pharmacia), and mouse myeloma cell nuclear extract (4 to 6 μg of protein/reaction) were incubated at 4°C in binding buffer (final concentrations: 10 mM Tris [pH 7.5], 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol). The reaction mixture (10 μl) was incubated for 20 min and then analyzed by electrophoresis on a 4% native polyacrylamide gel at 200 V (9, 20). Radioactivity was quantified by autoradiography of the gel after drying. In the experiment in which mutant H3.2 restriction fragments were used as probes, the CRAS fragment was excised from the appropriate wild-type or mutant gene with SalI and StuI.
Oligonucleotides.
The sequences of the H3.2 and H3.3 oligonucleotides are shown above. The YY1-binding site was synthesized on the basis of the published adeno-associated virus promoter P5-60 sequence (36) (the sequence is shown below, with the BamHI and BglI overhangs indicated by lowercase letters): gatccGTTTTGCGACATTTTGCGACACa gCAAAACGCTGTAAAACGCTGTGtctag
Oct 1 22-mer oligonucleotides comprising the consensus DNA-binding site for the Oct family of transcription factors were obtained from Santa Cruz Biotechnology, Inc., and used to show the specificity of YY1 antibodies for the histone α factor in EMSA (see Fig. 4).
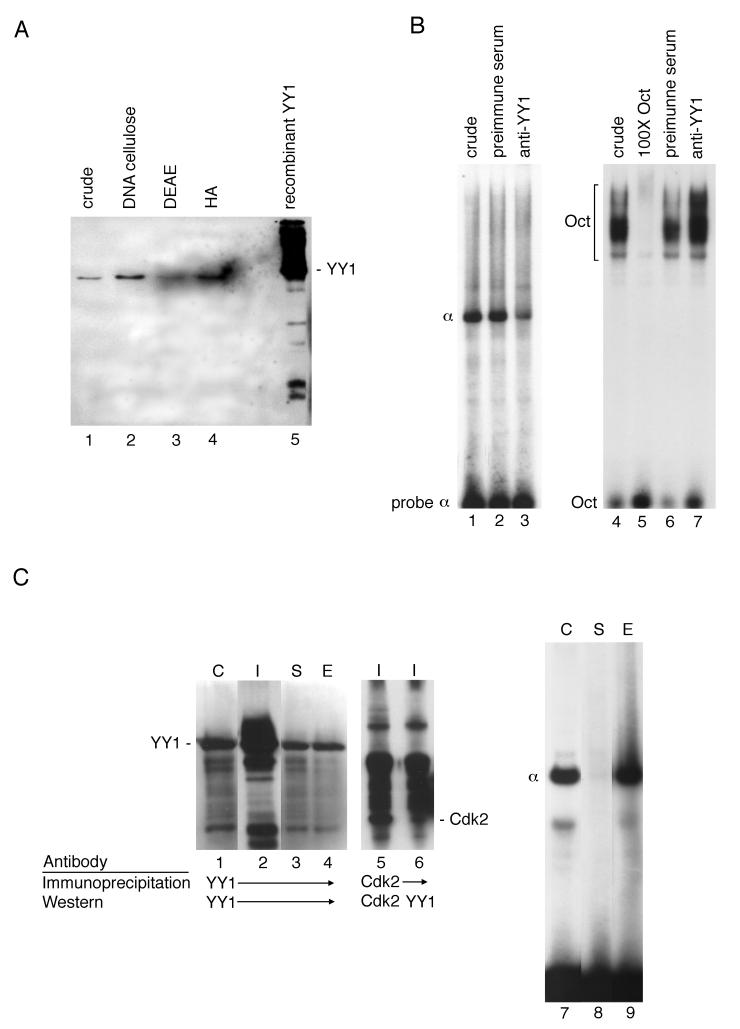
FIG. 4.
(A) YY1 copurifies biochemically with the in vitro α DNA-binding activity. Aliquots of dialyzed pooled fractions containing α DNA-binding activity from the column indicated above each lane were examined for the presence of YY1 by Western analysis. Lane 1, crude nuclear extract; lane 2, double-stranded calf thymus DNA-cellulose; lane 3, DEAE-Sepharose; lane 4, hydroxyapatite (HA); lane 5, YY1 control (human recombinant YY1). Proteins contained in the pooled fractions were separated by SDS-PAGE and electroblotted to a polyvinylidene difluoride membrane for Western analysis with polyclonal anti-human YY1 antibodies. See Materials and Methods for details of column chromatography. (B) YY1 antibodies diminish the histone α shift. Reactions in all lanes contained 4 μg of crude nuclear extract and 2 μg of poly(dI-dC)–poly(dI-dC) and were incubated for 20 min at 4°C. The probe in lanes 1 to 3 was 1 ng of end-labeled H3.2 α oligonucleotides. Lane 1, control α reaction; lane 2, 2 μl of preimmune serum added at the end of the 20-min incubation for an additional 15 min; lane 3, 2 μl of anti-YY1 antibody added after 20 min and incubated for 15 min. The probe in lanes 4 to 7 was 1 ng of end-labeled Oct 1 consensus binding site. Lane 4, control Oct 1 reaction; lane 5, 100-fold molar excess of unlabeled Oct 1 duplex; lane 6, 2 μl of preimmune serum added as in lane 2; lane 7, 2 μl of anti-YY1 antibody added as in lane 3. Reactions were analyzed by EMSA. (C) Immunodepletion of YY1 abolishes α DNA-binding activity in crude nuclear extracts. Details of the immunoprecipitation of YY1 from nuclear extracts are given in Materials and Methods. Lanes 1 to 6 show the results of Western analysis with undepleted crude nuclear extract (C, lane 1, 15 μg of protein), the entire immunoprecipitate fraction (I, lanes 2, 5, and 6, 20 μl of bead mixture), the supernatent fraction (S, lane 3, 15 μg of protein), and the eluate fraction (E, lane 4, 30 μl). Lanes 7 to 9 show the results of EMSA with undepleted crude nuclear extract (C, lane 7, 6 μg of protein), the supernatant fraction (S, lane 8, 6 μg of protein), and protein eluted from the immunoprecipitate (E, lane 9, 1/10 the eluate fraction). Reaction conditions for EMSA duplicated those shown in Fig. 3A, lane 1.
YY1 antibody inhibition and immunodepletion experiments.
In EMSA experiments (see Fig. 4B) with polyclonal anti-human YY1 antibody (affinity-purified rabbit polyclonal antibody; Santa Cruz), 2 μl of YY1 antibody or rabbit polyclonal preimmune serum was added to the binding reaction mixture after incubation for 20 min, and the reaction mixture was incubated for an additional 15 min at 4°C.
Immunoprecipitations were performed with anti-human YY1 antibody (affinity-purified rabbit polyclonal antibody) added to precleared nuclear extracts and incubated at 4°C with rotation for 2 h. Thirty microliters of protein A-agarose beads was added to 200 μl of crude extracts, and incubation was continued for 1 h. The extracts were centrifuged at 3,000 × g for 2 min. The supernatant (210 μl) was removed, and the beads were washed three times with radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) (17) and centrifuged under the same conditions after each wash. Elutions were performed by the addition of 100 μl of elution buffer (ImmunoCruz System; Santa Cruz), incubation on ice for 2 min, centrifugation under the above conditions, and dialysis for 1 h at 4°C against HDB containing 0.5 mM ZnCl2. The eluted fractions were tested for the presence of YY1 by Western analysis and EMSA. Human recombinant YY1 was obtained from Santa Cruz. Mock immunodepletion of nuclear extracts was done with affinity-purified rabbit polyclonal antibody to mouse Cdk2 (Santa Cruz).
Western analysis.
We performed Western analysis of the column fractions by first resolving the appropriate extracts by SDS-polyacrylamide gel electrophoresis (PAGE) (10% gel). After electrophoretic separation, proteins were transferred to a polyvinylidene difluoride membrane (Amersham) by electroblotting for 3 h at 180 mA and 4°C. The blot was blocked with Blotto (4% nonfat dry milk, 40 mM Tris, 20 mM NaCl, 0.1% Tween 20, 0.5 mM sodium azide) for 1 h and incubated with YY1 antibody (1:100) (Santa Cruz) for 2 h at room temperature (16). The blot was washed with TBST (1× Tris-buffered saline with 0.5% Tween 20) five times for 10 min each time. After the washing, secondary antibody (horseradish peroxidase conjugate) (1:2,500) (Amersham) was added for 1 h with rotation at room temperature. The blot was again washed five times with TBST and developed with the Amersham ECL kit by the recommended procedure.
Cell culture and stable transfections.
Chinese hamster ovary cells (CHO cells) were grown in McCoy’s 5A medium supplemented with 10% calf serum as previously described (3, 22, 23). Cells were plated 24 h prior to transfection (5 × 105 cells per 75-cm2 flask), and pools of stable transfectants were selected with the drug G418 beginning 18 h after cotransfection with the mouse wild-type H3.2 gene or the mutant H3.2αXba gene, pSVneo, Polybrene, and dimethyl sulfoxide as previously described (23). The mutant H3.2αXba gene contains seven altered nucleotides, so the histone α sequence is changed from CATGGCG to TTCTAGA; the remainder of the 1,600 nt of the H3.2 5′- and 3′-flanking sequences and the protein-encoding sequence are unchanged from those of the wild-type mouse gene (23). These stable transfectant cell lines were used as the source of mitotic cells in separate shakeoff experiments as described below.
Mitotic selection of cells.
Mitotic cells were collected by the mitotic selection technique described by Terasima and Tolmach (38), Schneiderman et al. (33), and Kaludov et al. (22). We used an automated shakeoff device (patent pending) to obtain mitotic cells. At 24 h before collection, cells were plated (3 × 106 to 5 × 106 cells per 75-cm2 flask) as described above. CHO cells traverse the cell cycle in approximately 12 h under these conditions (17). Mitotic cells were harvested with a completely automated shakeoff device that maintains a constant temperature of 37°C and is programmed to shake the flask platform for 15 s every 10 min. Then, medium containing mitotic cells is automatically withdrawn from the flasks, injected into collection vessels on ice, and replaced with fresh medium. Mitotic cells can be kept on ice for up to 4 h without alteration of the timing of their progression into the S phase (17, 33, 38). In the experiments described here, cells were kept on ice for no longer than 2 h and then were plated at time zero. After plating, mitotic cells were allowed to progress through the cycle under the culture conditions described above for harvesting at the appropriate time. For the 0-h RNA sample, cells were immediately harvested and total RNA was extracted. Cells were plated on glass slides in parallel for each time point and labeled with bromodeoxyuridine (BrdUrd) for 30 min before the end of the time point. Labeled cells were detected with BrdUrd-specific antibody, alkaline phosphatase-conjugated secondary antibody, and FastRed dye (Boehringer Mannheim Biochemicals). We determined the number of labeled cells (cells in the S phase) by counting 400 cells per slide for each time point and calculating the percentage of cells incorporating BrdUrd (cells in the S phase). No zero-hour slide is possible since cells must first attach, so the first time point represented in the BrdUrd experiment is 1 h—cells were allowed to attach to the slide for 30 min and were labeled with BrdUrd for an additional 30 min.
RNA isolation and analysis.
Cells were harvested at the appropriate time as described above, and total RNA was prepared with a BioTecx Laboratories Ultraspec RNA isolation system. The amount of RNA for the gene of interest was quantified by an S1 nuclease protection assay (10). For the assay of H3.2 mRNA levels, the wild-type or mutant H3.2 gene was 3′ end labeled at the SalI site in the mouse H3.2 614 protein-encoding sequence. The exact conditions for the assay and the separation of protected probe fragments are described by Hurt et al. (21). The homologous H3.2 probes map the 3′ half of the mouse H3.2 transcripts (fragments of 285 nt). Because the nucleotide sequences of the mouse and hamster genes are identical in the coding sequence but diverge in the 3′-untranslated region, the wild-type mouse H3.2 probe also maps the stop codon of the endogenous H3.2 transcripts. Because of the 7-nt mutation in the histone α sequence, the mutant H3.2 probe maps the endogenous hamster H3.2 transcripts up to the site of the mutation, producing fragments 100 nt in length. We determined the amount of mRNA encoding the ubiquitous translation factor, elongation factor 1-α (EF-1α), in these timed extracts to show that equivalent amounts of RNA were used (24). A Molecular Dynamics Storm PhosphorImager was used for direct measurement of radioactivity in the probe fragments in dried 6% polyacrylamide gels.
Sequence analysis.
Sequences were obtained from cDNAs by automated DNA sequencing. cDNA sequences were then compared to those in on-line databases. The sequences shown here (see Table 1 and Fig. 2) can be retrieved with the following GenBank accession numbers: human YY1, Z14077 (42) and M77698 (36); mouse YY1, M74590 (15); Xenopus YY1, X77698 (32); and human arginine-rich nuclear protein, M74002 (6).
TABLE 1.
HeLa cell cDNAs activating H3.2 reporter genesa
| cDNA | Size of insert (bp) | Identity (%) | No. of times isolated |
|---|---|---|---|
| 1 | 1,100 | YY1 (100) (human) | 2 |
| 2 | 2,100 | None | 3 |
| 3 | 2,400 | None | 22 |
| 4 | 1,000 | Arginine-rich nuclear protein (99) (human) | 2 |
Plasmids were rescued from yeast colonies showing induced levels of reporter gene expression in the positive yeast strain and individually tested in the negative control yeast strain (H3.3 α element). None of the negative control (H3.3) reporters transformed with H3.2-positive cDNA showed induced levels of expression.
FIG. 2.
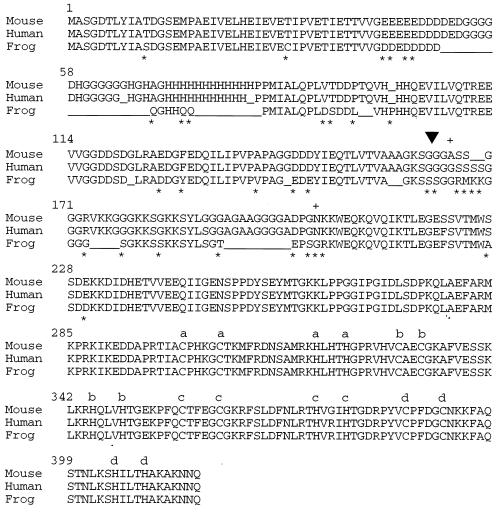
Protein sequence comparison of human, mouse, and frog YY1. Numbers at the beginnings of the lines indicate amino acid numbers. Asterisks indicate amino acid changes between mouse and frog YY1; plus signs indicate amino acid changes between mouse and human YY1. An inverted triangle marks the beginning of human cDNA clone 1 (Fig. 1). The sequence obtained for clone 1 extends from the triangle to the end of the protein. The C and H residues responsible for four potential zinc fingers (a to d) are indicated. GenBank accession numbers for YY1 clones are given in Materials and Methods.
RESULTS
Identification of the nuclear factor that binds to the histone α element.
The yeast one-hybrid strategy was used to identify cDNAs encoding proteins that interact specifically with the histone α element. In this set of experiments, a yeast strain that contained lacZ and HIS3 reporter genes with six copies of the replication-dependent H3.2 α DNA-binding site (see Materials and Methods) cloned into their promoters was constructed. The H3.2 reporter strain was used to screen a HeLa cell cDNA expression library in which plasmids containing directionally cloned cDNAs were fused to DNA encoding the Gal4 transcriptional activation domain. Plasmids encoding fusion proteins capable of interacting with the H3.2 α DNA sequences in reporter gene promoters should produce activated levels of reporter expression in this yeast strain in vivo.
A negative control reporter strain was constructed to test the specificity of plasmids identified by activation of the H3.2 α reporter strain. In this case, six copies of the replication-independent H3.3 α binding site were cloned into the promoter regions of the same two reporter genes, lacZ and HIS3, and these reporter genes were recombined into the yeast genome. Of the 7 nt shown by EMSA, methylation interference, and DNase footprinting (3, 4, 20, 23) to be required for interaction with the histone α factor, 5 nt are different in the comparable H3.3 α sequence. These 5 nt changes completely abolish the in vitro α DNA-protein interaction (see Fig. 5, lane 5), making the H3.3 α sequence an ideal negative control with which to rule out false-positives.
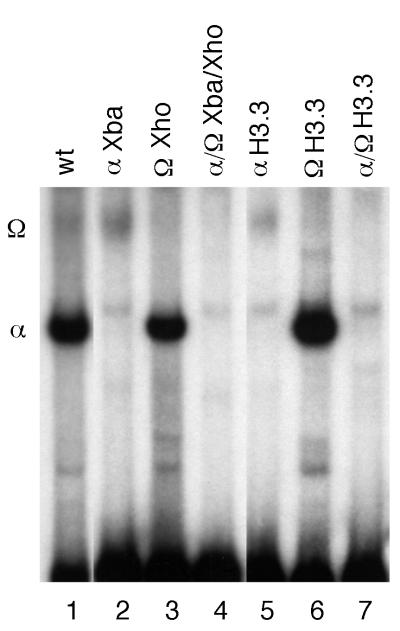
FIG. 5.
Mutation of the histone α element (YY1-binding site) causes the disappearance of the α complex in vitro. Wild-type (wt) or mutant CRAS fragments were excised from the appropriate H3.2 gene with SalI and StuI. The end-labeled fragments were used in EMSA with crude nuclear extract. Reaction conditions exactly duplicated those used in Fig. 3A, lane 1. The mutant fragment used as a probe is indicated above the lane. The nucleotides changed in the H3.2 α sequence in the various mutant genes are shown in Table 2.
The H3.2 reporter strain was transformed with the HeLa cell library, and 106 transformants were plated on media lacking histidine and leucine. The selective media also contained 30 mM 3-AT, a competitive inhibitor of the HIS3 gene product; only yeast cells with high levels of HIS3 expression can survive in the presence of the inhibitor.
Twenty-nine large colonies formed on 3-AT plates, and all of these colonies also showed activated expression of β-galactosidase in the presence of the substrate. Plasmids were rescued from these 29 transformants and then transformed into the H3.3 α negative control yeast strain as a test for nonspecific interactions.
A YY1-encoding plasmid activates the H3.2 α reporter genes.
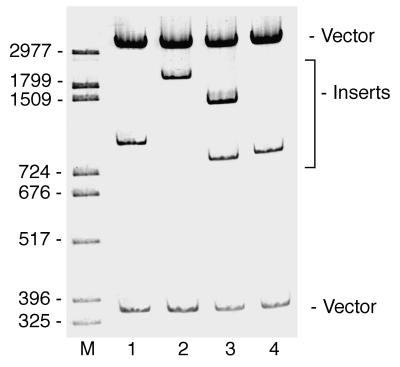
Restriction analysis of the plasmids rescued from the H3.2 reporter strain showed that four different size classes of cDNAs were capable of causing activated levels of reporter gene expression in the positive clones. The restriction digest of the four classes of rescued plasmids is shown in Fig. 1. The cDNA inserts in the four types of plasmids were partially sequenced, and the results are shown in Table 1. One class of these plasmids, shown in lane 1 of Fig. 1, contained cDNA sequences which were 100% identical to those of a human protein, YY1. Two independent clones of the same YY1 cDNA were found among the 29 rescued plasmids. The plasmid shown in lane 4 of Fig. 1 contained an insert that was 99% identical to the human arginine-rich nuclear protein, an mRNA splicing factor (6). The sequences of the other two unique cDNAs (Fig. 1, lanes 2 and 3) were not found in any of the on-line databases.
FIG. 1.
Restriction analysis of rescued plasmids. Plasmids were rescued from positive clones, and DNAs were digested with BamHI and KpnI and compared after separation of fragments by PAGE. The four unique cDNAs identified are shown in lanes 1 to 4. M, markers.
A comparison of the amino acid sequences of human YY1 (36, 42), mouse YY1 (15), and frog YY1 (32) is shown in Fig. 2. The entire DNA sequence that we obtained from the HeLa cell YY1-encoding cDNA was sequenced, including the sequence from codon 162 through the end of the protein. At the amino acid level, the mouse and human YY1 proteins are 99% identical; the carboxyl-terminal 200 amino acids of the frog, mouse, and human YY1 proteins, which include the DNA-binding domain (1), are 98% identical, an unusually high level of identity between frog and human sequences.
The adeno-associated virus P5-60 YY1-binding site competes with the H3.2 α element in EMSA.
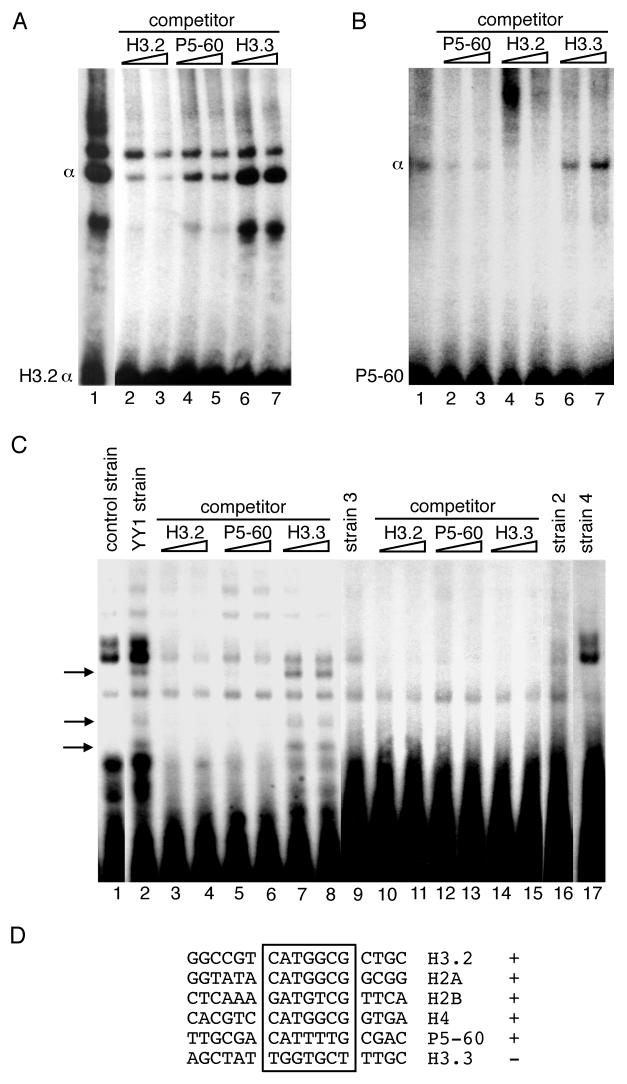
The sequence requirements for α DNA-protein interactions in vitro have been well characterized (3, 4). The histone H3.2 α sequence (CATGGCG) competes for binding of nuclear factors with the α sequences from replication-dependent H2a, H2b, and H4 genes in EMSA (4), but the comparable α sequences from a replication-independent H3.3 gene do not compete with the α sequences from any of the replication-dependent nucleosomal histone genes. In the experiment shown in Fig. 3A, we directly compared the ability of a well-characterized YY1-binding site (the adeno-associated virus P5-60 site) (36) to compete with the H3.2 α element for binding of the histone α factor.
FIG. 3.
Adeno-associated virus P5-60 YY1-binding site competes with the H3.2 α element for binding of the histone α factor. In all reactions, mouse myeloma cell nuclear extract (A and B, 4 μg) or yeast whole-cell extract (C, 15 μg) was incubated with end-labeled H3.2 α duplex oligonucleotides (A and C) or P5-60 duplex oligonucleotides (B) (1 ng) in the presence of the nonspecific competitor poly(dI-dC)–poly(dI-dC) (2 μg) at 4°C for 20 min. (A and B) Unlabeled competitor duplex oligonucleotides were added in a 50- or 100-fold molar excess. Lane 1, control reaction; lanes 2 and 3, 4 and 5, and 6 and 7, 50- and 100-fold excesses of oligonucleotides. (C) Yeast whole-cell extracts were prepared as described in Materials and Methods from the control H3.2 reporter strain and the yeast strains containing cDNAs (Table 1) which activated the H3.2 reporter genes. Competition reactions reproduced those shown in panels A and B. Lanes 9 to 15, results of binding reactions with the H3.2 α probe, to which whole-cell extract from yeast strain 3 was added; lanes 16 and 17, reactions with whole-cell extracts from yeast strains 2 and 4. Reactions were analyzed by EMSA. (D) Comparison of histone α DNA-binding sites with the YY1 P5-60 DNA-binding site. +, interaction of the boxed sequences with the α factor; −, absence of binding activity (3, 4, 36).
Mouse myeloma cell nuclear extract was used as the source of α DNA-binding activity, and end-labeled duplex H3.2 α oligonucleotides were used as probes in the mobility shift experiment shown in Fig. 3A; the control H3.2 α shift is shown in lane 1. The α DNA-binding activity was eliminated by competition from a 50- or 100-fold molar excess of unlabeled H3.2 α oligonucleotides (Fig. 3A, lanes 2 and 3). Unlabeled P5-60 oligonucleotides also competed for the α DNA-binding activity when present in a 50- or 100-fold molar excess (Fig. 3A, lanes 4 and 5), but this DNA was somewhat less effective as a competitor. The α DNA-binding site from the replication-independent H3.3 gene, however, failed to compete for binding of the α sequences (Fig. 3A, lanes 6 and 7), illustrating the specificity of the α interaction with the H3.2 α DNA as well as the P5-60 DNA.
The P5-60 oligonucleotides were then used as probes in an identical competition experiment shown in Fig. 3B; the competitor oligonucleotides were present in a 50- or 100-fold molar excess in lanes 2 to 7. The H3.2 α DNA-binding site competed well with the P5-60 oligonucleotides for binding of the α factor (Fig. 3B, lanes 4 and 5), reproducing the results of the previous experiment (Fig. 3A), but the H3.3 α oligonucleotides failed to compete with the P5-60 oligonucleotides (Fig. 3B, lanes 6 and 7), reproducing the results shown in Fig. 3A, lanes 6 and 7.
Protein extracts were made from the yeast strain containing the YY1-encoding plasmid, as well as from the other three positive strains from the one-hybrid screen (Fig. 1, lanes 2 to 4). These yeast extracts were used in a competition experiment identical to those shown in Fig. 3A and B. Figure 3C shows the results of this experiment. In the reaction shown in Fig. 3C, lane 1, extract from the H3.2 reporter strain (containing no cDNA) was incubated with a radioactively labeled H3.2 α probe. Multiple nonspecific bands resulted. These bands were also observed in reactions containing extracts from the other yeast strains, but incubation of a yeast whole-cell extract from the strain containing the cDNA encoding YY1 with the probe produced three specific bands (Fig. 3C, lane 2). The presence of multiple bands may have been due to proteolysis because of the use of a whole-cell extract. The specific nature of these complexes was shown by competition with unlabeled H3.2 α, P5-60, or H3.3 oligonucleotides present in the binding reactions in a 50- or 100-fold molar excess (Fig. 3C, lanes 3 to 8). The specific bands disappeared in reactions containing H3.2 α or P5-60 oligonucleotides but were unaffected by the addition of the H3.3 oligonucleotides. None of the specific bands observed in lanes loaded with reactions containing YY1 was observed in extracts prepared from the other positive strains (Fig. 3C, lanes 9 to 17). The competition experiment shown in Fig. 3C, lanes 2 to 8, was reproduced with an extract from strain 3 (Table 1). The pattern of nonspecific bands observed in Fig. 3C, lanes 9 to 15, was almost identical to that seen in lanes 2 to 8 (YY1-containing strain), but no specific interactions between proteins in the extract from strain 3 and the H3.2 α probe were detected. This was also true for extracts from strains 2 and 4 (Fig. 3C, lanes 16 and 17). These results indicated that the interaction of proteins encoded by these cDNAs with the H3.2 reporter genes in vivo was nonspecific. The reason for the failure of the negative control reporter yeast strain to distinguish between the YY1-H3.2 α interaction and that of the proteins encoded by cDNAs 2–4 is not clear.
A sequence comparison of nucleosomal histone α elements and the P5-60 YY1-binding site is shown in Fig. 3D. The H3.3 α element is also shown. We previously showed that the H3.2 α oligonucleotides competed specifically with the α DNA complexes formed with the H2a, H2b, and H4 elements but that the H3.3 oligonucleotides did not (4). Here we showed that the human YY1-containing yeast extracts contained a DNA-binding activity with sequence specificity identical to that of the histone α factor found in mouse myeloma cell nuclear extracts (Fig. 3A and B).
YY1 is necessary for in vitro histone α DNA-binding activity. (i) YY1 copurifies with the α DNA-binding activity.
Rabbit polyclonal anti-YY1 (human) antibodies were used to test column fractions containing α DNA-binding activity for the presence of YY1. Crude nuclear extracts were prepared from mouse myeloma cells grown in suspension culture. Nuclear extract was loaded on a double-stranded DNA–cellulose column and eluted with buffer containing KCl in steps of increasing molarity. The fractions containing α DNA-binding activity were pooled, dialyzed, passed over a DEAE-Sepharose column, and eluted with KCl. Fractions containing α DNA-binding activity were pooled, dialyzed, and further purified by passage over a hydroxyapatite column. A Western blot analysis of proteins present in the pooled column fractions containing the in vitro α DNA-binding activity is shown in Fig. 4A. A single polypeptide was identified by reaction with YY1 antibody in the presence of a crude nuclear extract (Fig. 4A, lane 1) as well as in the active fractions from each column (lanes 2 to 4). Equivalent amounts of DNA-binding activity were the source of protein for the Western analysis. Figure 4A shows that YY1 and the α DNA-binding activity were enriched in parallel. The amounts of protein detected in Fig. 4A, lanes 1 to 5, were roughly equivalent. The reactive bands in Fig. 4A, lanes 1 to 4, had apparent molecular weights comparable to that of human recombinant YY1 (lane 5). These results indicated that YY1 was present in all column fractions containing the histone α DNA-binding activity.
(ii) YY1 antibodies diminish the histone α DNA-binding activity.
Next, the effect of YY1 antibodies on the α DNA-binding activity was examined by EMSA (Fig. 4B). Mouse myeloma cell crude nuclear extract was used as the source of α DNA-binding activity, and end-labeled H3.2 α oligonucleotides were used as the probe. The control binding reaction, containing the histone α complex, is shown in Fig. 4B, lane 1. In the reaction shown in Fig. 4B, lane 2, rabbit preimmune serum was added after 20 min of incubation of the probe with the nuclear extract and incubated for an additional 15 min. In the reaction shown in Fig. 4B, lane 3, rabbit polyclonal YY1 antibodies were also added after incubation of the extract with the probe as in lane 2. The presence of YY1 antibodies greatly reduced the formation of the histone α complex (Fig. 4B, lane 3). Similar results were obtained when YY1 antibodies were incubated with the nuclear extract before the probe addition (data not shown). The specificity of the YY1 antibodies for the histone α complex was tested in an experiment with the Oct 1-DNA complex (Fig. 4B, lanes 4 to 7). In this experiment, the Oct 1 consensus binding site was used as the probe in binding reactions with the same nuclear extract as that used in Fig. 4B, lanes 1 to 3. The Oct 1 protein complex formed with the probe is shown in Fig. 4B, lane 4. The addition of a 100-fold molar excess of unlabeled Oct 1 duplex oligonucleotides to the binding reaction totally abolished the formation of this complex (Fig. 4B, lane 5), but the addition of preimmune serum or YY1 antibodies did not have this effect on the complex (lanes 6 and 7); therefore, the effect of YY1 antibodies was specific to the histone α complex.
YY1 antibodies were then used in an immunodepletion experiment (Fig. 4C). Mouse nuclear extract was incubated with affinity-purified rabbit polyclonal anti-YY1 antibodies and then with protein A-agarose beads. The immunoprecipitate was pelleted by centrifugation. A Western blot analysis of the YY1 immunoprecipitation is shown in Fig. 4C, lanes 1 to 4. Most of the YY1 protein was present in the immunoprecipitated fraction (Fig. 4C, lane 2). A very small amount of the YY1 protein was left in the supernatant after immunodepletion (Fig. 4C, lane 3). The eluate from the immunoprecipitation clearly contained YY1 (Fig. 4C, lane 3). Western analysis of a mock immunodepletion with mouse Cdk2 antibodies is shown in Fig. 4C, lanes 5 and 6. The immunoprecipitate contained a reactive peptide of the correct molecular weight (Fig. 4C, lane 5). YY1, however, was not immunoprecipitated with the Cdk2 antibodies (Fig. 4C, lane 6), so YY1 immunoprecipitation was specific (lanes 2 to 4).
The effect of immunodepletion of YY1 from nuclear extracts upon the formation of the α-DNA complex in EMSA is shown in Fig. 4C, lanes 7 to 11. The α-DNA complex detected after incubation of a normal nuclear extract with the H3.2 α DNA-binding site is shown in Fig. 4C, lane 7. The immunodepleted nuclear extract (supernatant, Fig. 4C, lane 8) retained only a small amount of α DNA-binding activity upon incubation with the labeled H3.2 α DNA-binding site, demonstrating the critical role of YY1 in α-DNA complex formation.
YY1 contained in the immunoprecipitated fraction was subsequently eluted from the beads at a high pH after three high-stringency washes (see Materials and Methods) and then added to the binding reaction shown in Fig. 4C, lane 9. In spite of the harsh treatment (detergent washes, high-pH elution), the α-DNA complex was observed, indicating that α DNA-binding activity immunoprecipitated with YY1 and that at least some of this activity could be recovered from the immunoprecipitated fraction.
Mutation of the histone α element reduces H3.2 gene expression in vivo and abolishes α-DNA complex formation in vitro. (i) In vivo effect of mutating the α element upon H3.2 gene expression.
We previously examined the effects of histone coding region deletions and mutations on DNA-protein interactions in vitro and the expression of these genes in vivo in stable transfectants (3, 20, 21, 23). Briefly, gene constructs were introduced by cotransfection into CHO cells with a neomycin resistance marker gene. RNA was isolated from several independent pools of stable transfectant cells for each gene construct examined. Gene expression was examined by a nuclease protection assay with the endogenous (hamster) H3.2 gene serving as an internal control. This hamster gene is identical to the mouse H3.2 gene in the coding sequence but not in the 5′-flanking or 3′-untranslated sequences, allowing quantification of hamster H3.2 RNA and mouse H3.2 RNA with the same probe (21, 23). The probe band resulting from protection by the hamster RNA was approximately 50 nt shorter than that resulting from protection by mouse wild-type H3.2 RNA.
Table 2 shows the results of an in vivo analysis of the steady-state H3.2 mRNAs from 12 different wild-type and mutant H3.2 genes. Table 2 also shows the effect of the particular deletion or mutation upon α-DNA complex formation in EMSA. Gene constructs 2, 4, 5, and 6 contained in-frame deletions. The entire 110 nt of the CRAS was deleted in gene 2. Genes 4 to 6 contained sequential deletions of between 20 and 30 nt across the H3.2 CRAS. In gene 4, 27 nt, including those of the α element, were deleted. Because the deletion in gene 2 deleted both the histone α and histone Ω elements, a 20-fold decrease in expression was observed (20), whereas the gene in which only 27 nt (and only the α element) were deleted showed a 4-fold decrease in expression (3). Only nucleotides within the H3.2 α element were changed in genes 9 and 10, and in both cases, a fourfold decrease in expression relative to that of the wild type was observed (23). Nucleotides in both α and Ω elements were mutated in genes 11 and 12, and the observed decrease in expression approached that observed when the entire CRAS (110 nt) was deleted (gene 2) (23). In genes 10 and 12, nucleotides in the α element (and in the Ω element in the case of gene 12) were changed to the sequence of the comparable region of the replication-independent H3.3 gene.
TABLE 2.
Effects of deletions or mutations of CRAS elements in vivo and in vitro
| Construct | Gene | Relative expression (%) in vivoa | α sequence | α complex observed in vitrob |
|---|---|---|---|---|
| 1 | H3.2 (wild type) | 100 | CATGGCG | + |
| 2 | H3.2Δ58-93 | 4 | Deleted | NA |
| 3 | H3.2CRAS1a | 84 | CATGGCG | + |
| 4 | H3.2CRAS2 | 39 | CATGGCG | + |
| 5 | H3.2CRAS3 | 94 | CATGGCG | + |
| 6 | H3.2CRAS4 | 28 | Deleted | − |
| 7 | H3.2ΩXho (7 nt) | 23 | CATGGCG | + |
| 8 | H3.2ΩH3.3 (3 nt) | 23 | CATGGCG | + |
| 9 | H3.2αXba (7 nt) | 26 | TTCTAGA | − |
| 10 | H3.2αH3.3 (5 nt) | 27 | TTGTGCT | − |
| 11 | H3.2αXbaΩXho (14 nt) | 5 | TTCTAGA | − |
| 12 | H3.2α/ΩH3.3 (8 nt) | <10 | TTGTGCT | − |
Nuclease protection assays were done with total RNA isolated from at least three independent pools of stably transfected CHO cells. Gene 2, in-frame deletion of H3.2 CRAS (110 nt) (20); genes 3 to 6, in-frame site-directed mutations in or deletions of H3.2 CRAS (gene 6 contains a 27-nt in-frame deletion) (3); genes 7 to 12, oligonucleotide-directed mutations (23). The entire sequence of mouse H3.2-614 can be found in GenBank (see Materials and Methods for accession number).
Results obtained by EMSA with the end-labeled CRAS fragment from the gene indicated as a probe and mouse myeloma cell nuclear extract as the source of binding activity. +, observed; −, not observed; NA, not applicable (the entire CRAS was deleted).
(ii) Mutation of the α element causes the disappearance of the α-DNA complex in vitro.
We previously showed that deletion of the H3.2 α sequence (Table 2, gene 6) abolished the formation of the α-DNA complex in EMSA (3). Here we showed that mutation of 7 nt of the α element (Fig. 5, lanes 2 and 4) completely abolished the wild-type α-DNA interaction observed in EMSA when the mutant CRAS fragments (Table 2, genes 9 and 11) were used as probes (130 nt). In addition, mutation of the α element found in the replication-dependent H3.2 gene to yield the comparable sequence of the replication-independent H3.3 gene (5 nt changes; Table 2, genes 10 and 12) duplicated the effect of the 7-nt α element mutation. That is, the α-DNA complex was not observed (Fig. 5, lanes 5 and 7). As was observed in Fig. 5, lanes 3 and 6, however, formation of the α-DNA complex was unaffected by mutations in the CRAS Ω element (Table 2, genes 7 and 8).
In Fig. 3 and 4, we demonstrated that YY1 is necessary for the formation of the α-DNA complex in vitro. We showed in Table 2 and Fig. 5 that the wild-type α sequence (CATGGCG) is required for α-DNA complex formation in vitro and for normal high-level expression in vivo. For example, mutation of 5 nt of the α sequence to yield that of a replication-independent gene (TGGTGCT) abolished α-DNA complex formation in vitro (Fig. 5, lane 5) and caused a fourfold decrease in expression in vivo (Table 2, gene 10).
(iii) Mutation of the histone α element alters up-regulation of the H3.2 histone gene in synchronous populations of unperturbed cycling cells.
As described in the previous sections, we showed that mutation of 7 nt of the histone α element (the H3.2αXba mutation) abolished DNA-protein interactions in vitro (Fig. 5) and reduced the steady-state level of histone mRNA fourfold in asynchronous populations of cells in logarithmic growth (23). Here, we examined the effect of this mutation on G1-S-phase up-regulation of the replication-dependent mouse H3.2 gene. Specifically, synchronous populations of stable transfectant CHO cells containing the wild-type mouse H3.2 gene or the H3.2αXba gene were obtained by use of an automated mitotic shakeoff device as described above. In these experiments, cycling cells progressed unperturbed through the cell cycle.
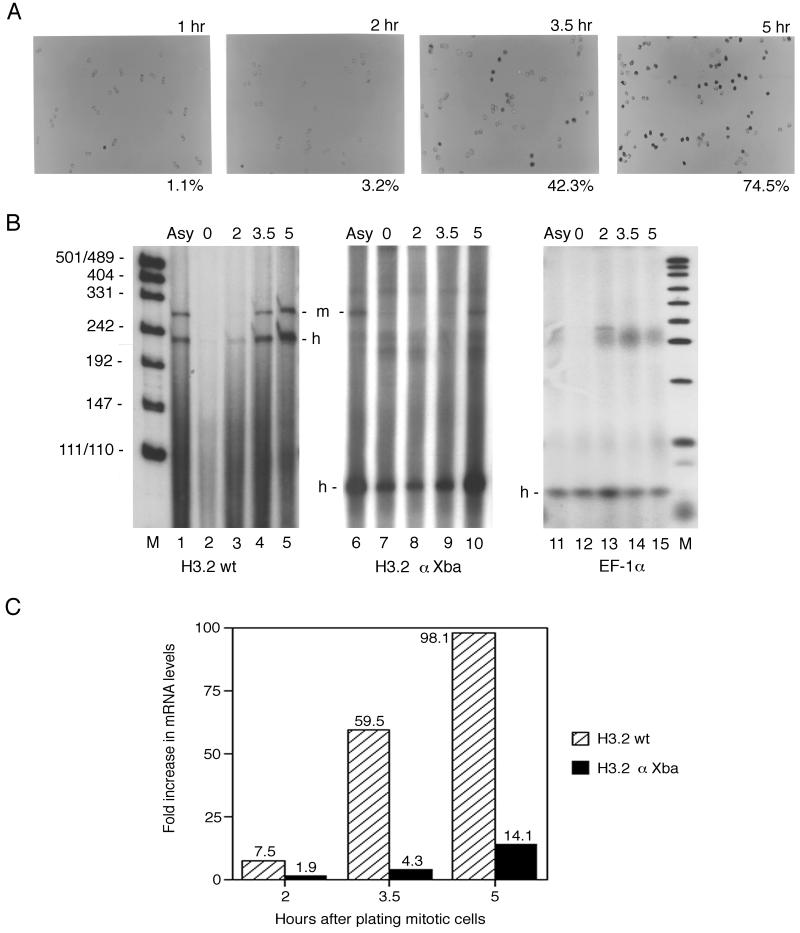
Slides of cell populations used in these experiments are shown progressing from mitosis into the S phase in Fig. 6A. Entry into the S phase was identified by detection of BrdUrd (with BrdUrd-specific antibody, alkaline phosphatase-conjugated secondary antibody, and FastRed dye as described above). Postmitotic pairs of cells were observed at 1 h, at which time very few, if any, S-phase cells were detected. The number of cells in the S phase indicated that the G1-S boundary was between 2 and 3.5 h, as the number of S-phase cells increased sharply at 3.5 h and all cells were in the S phase by 7 h. The timing of S-phase entry in these experiments agreed exactly with the published results of Harris et al. (17), who examined histone gene expression in synchronous CHO cell populations obtained by mitotic shakeoff.
FIG. 6.
Mutation of the YY1-binding site in the H3.2 CRAS α element alters histone gene up-regulation in synchronous populations of cycling CHO cells. (A) Synchronous cell populations obtained by mitotic shakeoff were plated on glass slides, fixed, and stained at the times indicated. Cells were labeled with BrdUrd, and S-phase cells were detected with FastRed dye (see Materials and Methods). The percentages indicate the number of cells in the S phase at the indicated times. (B) S1 nuclease protection assay of H3.2 RNA in synchronous populations of stable transfectant cells plated in parallel with those shown in panel A (lanes 1 to 5, CHO cells stably transfected with the mouse H3.2-614 gene; lanes 6 to 15, CHO cells stably transfected with the H3.2αXba gene). The wild-type (wt) H3.2 gene (lanes 1 to 5) or the H3.2αXba gene (lanes 6 to 10) was 3′ end labeled with the Klenow fragment at the SalI site of the gene and used as a probe, mapping both mouse and endogenous hamster H3.2 transcripts (see Materials and Methods). The EF-1α gene (24) was 3′ end labeled at the NcoI site for use in the assays shown in lanes 11 to 15. The RNA samples used in lanes 6 and 11, 7 and 12, 8 and 13, 9 and 14, and 10 and 15 were identical. M, markers; Asy, asynchronous. (C) Comparison of the up-regulation of histone gene expression from 0 to 5 h for the wild-type and mutant mouse H3.2 genes. Quantitation was done by PhosphorImager analysis of at least three independent experiments.
Cells were plated in parallel with those shown in Fig. 6A for harvest at the same time points. After harvest, total RNA was prepared, and the steady-state level of mRNA was determined by an S1 nuclease protection assay. The results are shown in Fig. 6B; lanes 1 to 5 show the results of a shakeoff experiment with a cell line stably transfected with the mouse wild-type H3.2 gene. This mouse H3.2 DNA probe mapped both the mouse H3.2 transcript and the endogenous hamster H3.2 transcript. Figure 6B, lanes 1 to 5, showed that both the transfected mouse H3.2 gene and the endogenous hamster H3.2 gene were up-regulated sharply in parallel between the plating of mitotic cells and the 5-h time point. These experiments were repeated at least three times with independently transfected cell lines. A shakeoff experiment with a cell line carrying the transfected mutant mouse H3.2 gene is shown in Fig. 6B, lanes 6 to 10. In this experiment, the mutant gene was used as a probe and mapped from the labeled SalI site to the end of the mutant transcript but mapped the endogenous hamster H3.2 transcript to the point of the α mutation, producing probe fragments of about 100 nt. The up-regulation of the endogenous hamster H3.2 gene as the cells reached the G1-S boundary matched that observed in Fig. 6B, lanes 1 to 5. The amount of the specific mouse H3.2 transcript was much lower than that of the wild-type H3.2 transcript, as we previously reported (3, 23). This result appeared to be due to a failure in up-regulation as the cells moved through G1 to the S-phase boundary (Fig. 6B, lanes 7 to 9). The increase in specific transcripts observed between 3.5 and 5 h (Fig. 6B, compare lanes 9 and 10) was due to the change in the histone mRNA half-life (stabilization by the stem-loop binding protein (10, 17, 39). The threefold increase observed here duplicated the results of Harris et al. (17), who directly examined the contribution of the 3′-end sequences to histone gene expression in the cell cycle. Figure 6B, lanes 11 to 15, show the quantitation of the mRNA levels for the ubiquitous translation factor EF-1α in the same RNA samples as those shown in lanes 6 to 10; EF-1α served as a loading control for the experiment.
Figure 6C shows graphically the difference in up-regulation between the mutant gene during the cell cycle and the wild-type H3.2 gene. The levels of wild-type H3.2 mRNA increased eightfold between 0 and 2 h and again between 2 and 3.5 h. The difference between 3.5 and 5 h, when well over half of the cells had entered the S phase, was two- to threefold (because of the stabilization of histone mRNA). For the mutant H3.2 gene, a dramatic change in the levels of mRNA between 0 and 3.5 h was absent. The change in the levels of mRNA between 0 and 2 h and between 2 and 3.5 h was twofold. The threefold change in the levels of mRNA between 3.5 and 5 h for mutant gene expression duplicated that observed for wild-type gene expression. The stem-loop structure of the mutant gene was unaltered, and these results confirmed that the posttranscriptional component of histone gene regulation was normal for both mutant and wild-type H3.2 genes. A major component of cell cycle regulation of histone gene mRNA levels is posttranscriptional (12, 17, 39), the effects of which were seen here, implicating a change in transcription initiation as the explanation for the altered G1-S-phase expression of the mouse H3.2 gene.
DISCUSSION
The processes culminating in commitment to the S phase in metazoan cells include the up-regulation of genes required for duplication of the genome. The histone genes that encode proteins required for packaging of the newly synthesized DNA in the two daughter cells are also up-regulated in this sequence of events. The metazoan histone gene family includes the nucleosomal histone genes H2a, H2b, H3, and H4, as well as the genes encoding linker histones (H1 genes). Histone genes are among the most highly expressed and are among the most highly conserved genes known; within histone classes, the protein sequence is remarkably conserved among all eukaryotes. The cellular resources required for histone synthesis in the duplicating cell parallel those required for making a copy of the genome.
In the mouse, the up-regulation of the histone gene family at the G1-S boundary requires the coordinated activation of the transcription of many genes. Although various histone promoter elements have been implicated in the G1-S-specific up-regulation of vertebrate histone gene expression, none of these is common to more than one or two histone classes. Because genes of all histone classes are coordinately up-regulated, it follows that a common pathway is responsible. Furthermore, a common point of interaction with this pathway must exist for histone genes of all classes.
We have identified a highly conserved transcription factor, YY1, on the basis of its interaction with an element found in the protein-encoding sequence of replication-dependent histone genes of all classes. This transcription factor, implicated as both an activator and a repressor of gene expression in different cellular and viral genetic contexts, is found in all animal cell types. YY1 has not been found in yeast, but cDNAs encoding the factor have been cloned in frogs, mice, humans and, very recently, fruit flies (5). The DNA-binding domain of YY1 is at the carboxy-terminal end of the protein and is comprised of four potential zinc fingers. This region of the protein is 100% identical in frog, mouse, and human sequences, and the fruit fly YY1 DNA-binding domain is 97% identical to that of human YY1 (5), a remarkable degree of identity.
We have proven that the in vitro histone α DNA-binding activity requires YY1 for α DNA-protein complex formation. The addition of YY1 antibodies to the binding reaction greatly diminished the level of the H3.2 α complex. Similarly, the removal of YY1 by immunodepletion of a mouse myeloma cell nuclear extract with YY1 antibodies also abolished the formation of the α complex.
We examined the ability of a well-characterized YY1-binding site, the adeno-associated virus P5-60 element, to compete with the histone α factor for binding of the replication-dependent H3.2 α element. This nonhistone element and unlabeled H3.2 α oligonucleotides both competed effectively, although the H3.2 α DNA was a better competitor than P5-60 for either the H3.2 α or the P5-60 probe. This result may indicate that a viral cofactor that facilitates the YY1 P5-60-binding activity is not present in normal cell extracts or that the histone α element is simply a better binding site for YY1 than the viral element.
In contrast, the replication-independent H3.3 oligonucleotides did not compete with the labeled H3.2 α probe for binding of the α factor, as we previously showed (3, 4, 8), nor did the H3.3 oligonucleotides compete with the labeled P5-60 oligonucleotides. Interestingly, the P5-60 oligonucleotide sequence is 55% different from that of H3.2, whereas the H3.3 oligonucleotides used as competitors are 41% different from those of H3.2 (11 of 27 nt positions differ). The G+C content of the H3.2 oligonucleotides is 70%, whereas the G+C contents of the P5-60 and H3.3 oligonucleotides are lower but very similar (45 and 52%, respectively). The specificity of YY1 interactions with the H3.2 α and adeno-associated virus P5-60 sites confirms the results of the experiments with YY1 antibodies; the highly specific interaction of the histone α DNA-binding activity is due to the factor YY1.
We previously examined the effect of altering the histone α sequence on expression in vivo and DNA-protein interactions in vitro (3, 4, 8, 20, 21, 23). Because all deletions or mutations of the α element abolished the formation of the α-DNA complex in vitro and also caused a significant decrease in gene expression in vivo in stable transfectants, it is very likely that YY1 plays an in vivo role in the regulation of replication-dependent histone gene expression. Specifically, mutation of the replication-dependent H3.2 α element to yield the replication-independent H3.3 sequence caused a fourfold decrease in expression in vivo and a loss of formation of the α-DNA complex in vitro. Further in vivo evidence is that yeast reporter genes incorporating the H3.3 α sequence were not activated by the YY1-GAL4 fusion protein, whereas reporter genes containing the H3.2 α sequence in their promoters showed activated levels of expression in strains containing YY1-GAL4 fusion cDNAs.
In the experiments shown in Fig. 6, we examined in vivo, in unperturbed cycling cells, the role of the histone α element in correct temporal regulation of the replication-dependent mouse H3.2 gene. We showed that the dramatic increase in the amounts of histone mRNA as cells move forward in the cell cycle to the G1-S boundary that is normally observed for replication-dependent histone genes is altered by mutation of the histone α element. A similar result was obtained by Harris et al. (17) using a histone gene construct in which the 5′-flanking sequences were replaced with the promoter of a constitutive U1 snRNA promoter. These experiments can be directly compared to our studies because mitotic shakeoff was used to obtain synchronous cell populations for RNA analyses. They found that the level of transcripts from the U1-histone chimeric gene increased a total of 10-fold between the plating of mitotic cells and entry into the S phase, very similar to the 14-fold increase that we observed for the mutant H3.2αXba gene here. Because the posttranscriptional regulatory events that play an important role in the regulation of histone mRNA in the cell cycle are unaffected by the α mutation, we postulate that the decrease observed in the up-regulation of the mutant histone gene is due to an alteration in events required for transcription initiation, as was clearly the case for the U1-histone chimeric gene described by Harris et al. (17). Because a 100% correlation is observed between sequence requirements for α-DNA complex formation in vitro and effects on gene expression in vivo, a role for YY1 in histone gene expression in vivo is strongly implicated.
This highly conserved transcription factor, YY1, has been the subject of extensive study in recent years. Its abundance and ubiquity make it an excellent candidate for an important global role in gene regulation in the metazoan cell. The 100% correlation between our in vitro studies of the histone α DNA-protein interaction (3, 4, 8, 22; this study) and in vivo studies of the role of the α element in wild-type histone gene expression (3, 23) is strong evidence that YY1 plays just such a role in the coordinated up-regulation of histone genes at the G1-S boundary of the cell cycle.
Because the yeast one-hybrid experimental system is designed to circumvent the possibility that an additional factor(s) is required in the histone α-YY1 interaction, our experiments thus far do not rule out this possibility. Here, we have demonstrated that YY1 is necessary for the α-DNA interaction, but it is our hypothesis that other factors may also participate in the histone α-YY1 interaction. In previous experiments, we demonstrated the dependence of the histone α DNA-binding activity on the state of phosphorylation (22). Phosphorylation on serine/threonine or tyrosine residues results in inhibition of the α DNA-binding activity (22). Kinases present in crude nuclear extracts are capable of inhibitory phosphorylation at 37°C, and the α DNA-binding activity can be recovered by treatment with phosphatases. We have also shown that cyclin D1-dependent complexes are capable of either direct or indirect inhibitory phosphorylation that results in the loss of the histone α DNA-protein complex in vitro (22).
Although there is some evidence that YY1 is a phosphoprotein (35), only one report implicates phosphorylation in the regulation of YY1 interactions with DNA (2). In this case, phosphatase treatment of Jurkat T-cell nuclear extracts abolished YY1 interactions with a viral DNA sequence. This result is the converse of our previous results (22), in which phosphatase treatment restored the histone α DNA-binding activity.
Roles suggested for YY1 in gene expression include activation, repression, and initiation. A role for YY1 in nuclear matrix interactions has also been postulated (13). The results just described, obtained by phosphatase treatment (YY1 DNA-binding activity in nuclear extracts), are examples of the complexity of YY1 interactions in the metazoan cell. One explanation for the variety of reported effects of YY1 on gene expression could be a requirement for interactions with additional factors in a gene- or pathway-specific manner in different genetic contexts. We are currently examining directly the effect on histone gene expression of YY1 overexpression in vivo. The identification of YY1 as the DNA-binding component of the histone α factor will facilitate our ongoing studies of the role of elements in the coding region of replication-dependent histone genes in the coordinated up-regulation of histone gene expression in the proliferating cell.
ACKNOWLEDGMENT
This research was supported by grant R01-GM46768 from the National Institutes of Health to M.M.H.
REFERENCES
- 1.Austen M, Luscher B, Luscher-Firzlaff J M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAF II55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 2.Becker K G, Jedlicka P, Templeton N S, Liotta L, Ozato K. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene. 1994;150:259–266. doi: 10.1016/0378-1119(94)90435-9. [DOI] [PubMed] [Google Scholar]
- 3.Bowman T L, Hurt M M. The coding sequences of mouse H2A and H3 histone genes contain a conserved seven nucleotide element that interacts with nuclear factors and is necessary for normal expression. Nucleic Acids Res. 1995;23:3083–3092. doi: 10.1093/nar/23.16.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman T L, Kaludov N K, Klein M, Hurt M M. An H3 coding region regulatory element is common to all four nucleosomal classes of mouse histone-encoding genes. Gene. 1996;176:1–8. doi: 10.1016/0378-1119(96)00198-9. [DOI] [PubMed] [Google Scholar]
- 5.Brown J L, Mucci D, Whiteley M, Dirksen M-L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell Biol. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary N, McMahon C, Blobel G. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc Natl Acad Sci USA. 1991;88:8189–8193. doi: 10.1073/pnas.88.18.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clontech Laboratories. Yeast protocols handbook. Palo Alto, Calif: Clontech Laboratories; 1996. [Google Scholar]
- 8.Ficzycz A, Kaludov N K, Lele Z, Hurt M M, Ovsenek N. A conserved element in the protein-coding sequence is required for normal expression of replication-dependent histone genes in developing Xenopus embryos. Dev Biol. 1997;182:21–32. doi: 10.1006/dbio.1996.8459. [DOI] [PubMed] [Google Scholar]
- 9.Gilman M Z, Wilson R N, Weinberg R A. Multiple protein-binding sites in the 5′-flanking region regulate c-fos expression. Mol Cell Biol. 1986;6:4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graves R A, Marzluff W F. Rapid reversible alterations in histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984;4:351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves R A, Marzluff W F, Giebelhaus D H, Shultz G A. Quantitative and qualitative changes in histone gene expression during early mouse embryo development. Proc Natl Acad Sci USA. 1985;82:5685–5689. doi: 10.1073/pnas.82.17.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves R A, Pandey N B, Chodchoy N, Marzluff W F. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 13.Guo B, Odgren P, Van Wijnen A, Last T, Nickerson J, Penman S, Lian J, Stein J, Stein G S. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc Natl Acad Sci USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 15.Hariharan N, Kelley D E, Perry R P. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 17.Harris M, Bohni R, Schneiderman M, Ramamurthy L, Schumperli D, Marzluff W F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 19.Hraba-Renevey S, Kress M. Expression of a mouse replacement histone H3.3 gene with a highly conserved 3′ noncoding region during SV40- and polyoma-induced G0 to S-phase transition. Nucleic Acids Res. 1989;17:2449–2461. doi: 10.1093/nar/17.7.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurt M M, Bowman T L, Marzluff W F. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol Cell Biol. 1991;11:2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurt M M, Pandey N B, Marzluff W F. A region in the coding sequence is required for high-level expression of murine histone H3 gene. Proc Natl Acad Sci USA. 1989;86:4450–4454. doi: 10.1073/pnas.86.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaludov N K, Bowman T L, Sikorski E M, Hurt M M. Cell cycle-regulated binding of nuclear proteins to elements within a mouse H3.2 histone gene. Proc Natl Acad Sci USA. 1996;93:4465–4470. doi: 10.1073/pnas.93.9.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaludov N K, Pabon-Pena L, Hurt M M. Identification of a second conserved element within the coding sequence of a mouse H3 histone gene that interacts with nuclear factors and is necessary for normal expression. Nucleic Acids Res. 1996;24:523–531. doi: 10.1093/nar/24.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreig P, Varnum S, Wormington W, Melton D A. The mRNA encoding elongation factor 1-α (EF-1α) is a major transcript at the midblastula transition in Xenopus. Dev Biol. 1989;133:93–100. doi: 10.1016/0012-1606(89)90300-x. [DOI] [PubMed] [Google Scholar]
- 25.Krimer D B, Cheng G, Skoultchi A I. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 1993;21:2873–2879. doi: 10.1093/nar/21.12.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBella F, Heintz N. Histone gene transcription factor binding in the extracts of normal human cells. Mol Cell Biol. 1991;11:5825–5831. doi: 10.1128/mcb.11.12.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaBella F, Sive H L, Roeder R G, Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 28.Li J J, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex, by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 30.Osley M A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 31.Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- 32.Pisaneschi G, Ceccotti S, Falchetti M L, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- 33.Schneiderman M, Dewey W, Leeper D, Nagasawa H. Use of the mitotic selection procedure for cell cycle analysis. Comparison between the X-ray and cycloheximide G2 markers. Exp Cell Res. 1972;74:430–438. doi: 10.1016/0014-4827(72)90398-9. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Lee J S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Seto S E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 37.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terasima T, Tolmach L. Variations in several responses of HeLa cells to X-irradiation during the division cycle. Exp Cell Res. 1963;3:344–351. doi: 10.1016/s0006-3495(63)86801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z F, Whitfield M L, Ingledue III T C, Dominski Z, Marzluff W F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 40.Wellman S E, Casano P J, Pilch D R, Marzluff W F, Sittman D B. Characterization of mouse H3.3-like histone genes. Gene. 1987;59:29–39. doi: 10.1016/0378-1119(87)90263-0. [DOI] [PubMed] [Google Scholar]
- 41.Wells D, Kedes L. Structure of human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylated mRNAs. Proc Natl Acad Sci USA. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitson, R. H., T. Huang, J. Dang, and K. Itakura. Unpublished data.
- 43.Wu R S, Bonner W M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981;27:321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- 44.Zweidler A. Core histone variants of the mouse primary structure and differential expression. In: Stein G S, Stein J L, Marzluff W F, editors. Histone gene expression: structure, organization and regulation. New York, N.Y: Wiley Publishing Co.; 1984. pp. 339–371. [Google Scholar]