ABSTRACT
Histophilus somni is one of the predominant bacterial pathogens responsible for bovine respiratory and systemic diseases in cattle. Despite the identification of numerous H. somni virulence factors, little is known about the regulation of such factors. The post-transcriptional regulatory protein Hfq may play a crucial role in regulation of components that affect bacterial virulence. The contribution of Hfq to H. somni phenotype and virulence was investigated following creation of an hfq deletion mutant of H. somni strain 2336 (designated H. somni 2336Δhfq). A comparative analysis of the mutant to the wild-type strain was carried out by examining protein and carbohydrate phenotype, RNA sequence, intracellular survival in bovine monocytes, serum susceptibility, and virulence studies in mouse and calf models. H. somni 2336Δhfq exhibited a truncated lipooligosaccharide (LOS) structure, with loss of sialylation. The mutant demonstrated increased susceptibility to intracellular and serum-mediated killing compared to the wild-type strain. Transcriptomic analysis displayed significant differential expression of 832 upregulated genes and 809 downregulated genes in H. somni 2336Δhfq compared to H. somni strain 2336, including significant downregulation of lsgB and licA, which contribute to LOS oligosaccharide synthesis and sialylation. A substantial number of differentially expressed genes were associated with polysaccharide synthesis and other proteins that could influence virulence. The H. somni 2336Δhfq mutant strain was attenuated in a mouse septicemia model and somewhat attenuated in a calf intrabronchial challenge model. H. somni was recovered less frequently from nasopharyngeal swabs, endotracheal aspirates, and lung tissues of calves challenged with H. somni 2336Δhfq compared to the wild-type strain, and the percentage of abnormal lung tissue in calves challenged with H. somni 2336Δhfq was lower than in calves challenged with the wild-type strain. In conclusion, our results support that Hfq accounts for the regulation of H. somni virulence factors.
KEYWORDS: Histophilus somni, Hfq, gene regulation, virulence, biofilm, lipooligosaccharide
INTRODUCTION
Histophilus somni is an opportunistic pathogen of cattle and other ruminants, and one of the primary bacterial agents of bovine respiratory disease (BRD) (1). However, unlike most other BRD pathogens, H. somni can disseminate and cause multi-systemic diseases, including septicemia, myocarditis, thrombotic meningoencephalitis, arthritis, and reproductive disease (2). A wide variety of virulence factors have been identified in H. somni that enable this bacterium to escape innate and adaptive host defense mechanisms. Such mechanisms include an endotoxic lipooligosaccharide (LOS), which undergoes compositional and antigenic phase variation (3), biofilm formation (4), a large surface fibrillar immunoglobulin binding protein (IbpA) that also contains cytotoxic Fic motifs (5), histamine formation, platelet activation and adherence to host cells (6), survival inside phagocytic cells (7), and a variety of outer membrane proteins that may contribute to virulence (8). Although these virulence factors have been fairly well characterized, there is no information available regarding how the genes responsible for these virulence factors are regulated. For example, when H. somni forms a biofilm, which is the normal state of growth for this bacterium during BRD and other chronic infections (9), about half of the bacterium’s genes are differentially regulated (10, 11). Therefore, understanding the regulatory mechanisms that control expression of H. somni virulence is essential to develop measures to control BRD and other systemic infections.
Hfq is an RNA chaperone protein that binds small regulatory RNAs (sRNAs), rRNA, tRNA, and other substrates, which can result in translational regulation of mRNA in response to stress and metabolite alterations (12). In Escherichia coli, Hfq-regulation of such stress responses are mediated by sigma factors RpoS, Sigma-E, and Sigma-32 (13). Therefore, binding of Hfq to sRNAs that contribute to the regulation of gene expression at the post-transcriptional level may enhance or inhibit gene expression by blocking the Shine-Dalgarno and/or start codon regions, or through affecting RNA stability (14). Hfq does not have specific target sequences, but binds to sRNAs via unstructured AU-rich regions (15–17). In E. coli, the majority of the sRNAs identified thus far work in association with Hfq (18, 19). The inactivation of Hfq in E. coli leads to impaired growth, elevated carbon source oxidation, negative supercoiling of plasmids in stationary phase, and increased sensitivity to ultraviolet light (20). Hfq is also involved in the virulence and pathogenicity of other bacterial pathogens, including Salmonella, Vibrio cholerae, Neisseria meningitidis, and Pseudomonas aeruginosa (21, 22). The role of Hfq in virulence has also been studied in other members of the family Pasteurellaceae (23–27).
For this work, the role of H. somni Hfq in the expression of phenotypic components, regulation of gene expression, and overall virulence was examined using an Hfq mutant of virulent H. somni strain 2336. As reported with other related bacteria, particularly Haemophilus influenzae, H. somni Hfq appears to play a substantial role in gene regulation, affects the expression of some, but not all, virulence factors, and has some negative effect on bacterial virulence.
RESULTS
Mutagenesis of the hfq gene in H. somni strain 2336
Biofilm formation and other virulence factors are clearly regulated in H. somni, but there is no information regarding such regulatory mechanisms. Hfq is an important translational regulator of mRNA through its binding to sRNAs and other substrates (12). Therefore, we sought to mutagenize hfq to determine if this protein was necessary for full virulence in H. somni. The hfq gene of H. somni strain 2336 (HSM_RS05555) is located at 1235543–1235833 in the genome (NC_010519) and is flanked by gene HSM_RS05550, which encodes for glutathione-disulfide reductase (GSR), and gene HSM_RS05560, encoding for GTPase high frequency of lysogenization X (GTPase HflX) (Fig. S1). The H. somni Hfq open reading frame consists of 291 nucleotides that encode a protein of 96 amino acids. To examine the role of Hfq in H. somni, we constructed an hfq mutant by replacing the hfq gene with a chloramphenicol resistance gene using homologous recombination, as described in Materials and Methods and Fig. S1. The replacement of hfq was confirmed by genomic PCR and DNA sequencing (data not shown). Furthermore, hfq-specific reverse transcriptase-PCR (RT-PCR) and Western blot analysis showed that the hfq mutant, which was designated H. somni 2336Δhfq, expressed no hfq mRNA (Fig. S2A, HS2336Δhfq) or Hfq protein (Fig. S2B, Hs2336Δhfq). Expression of hfq cDNA (Fig. S2A, HS2336ΔhfqC) and the Hfq protein (Fig. S2B, Hs2336ΔhfqComp) was restored in trans in H. somni 2336ΔhfqComp by generating a plasmid-based complementary clone of hfq in the H. somni shuttle vector pNS3K, designated plasmid pNS3K-hfq. However, relatively little Hfq protein was expressed by H. somni 2336ΔhfqComp.
To rule out any potential secondary mutations that may have occurred during the generation of mutant strains, advanced DNA sequencing techniques were used to sequence the entire genomes of H. somni strain 2336 and H. somni 2336Δhfq. Sequencing confirmed that the hfq gene was replaced with a chloramphenicol resistance gene in H. somni 2336Δhfq (Fig. S3). Furthermore, no alterations were noted in the amino acid sequences of the hfq mutant genome when compared to wild-type H. somni strain 2336, except for a single T/G point mutation at position 1,405,304, which was located within an intergenic region and would not impact synthesis of any protein (Fig. S4).
The growth rate of H. somni 2336Δhfq is compromised compared to wild-type H. somni strain 2336
Alteration of growth rate is a common feature following hfq mutagenesis, which can have a substantial impact on virulence (28–30). To compare their growth rates, H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336ΔhfqComp were each grown in Columbia Broth (Becton, Dickinson, Sparks, MD) supplemented with 0.1% Trizma base and 0.01% thiamine monophosphate (Sigma-Aldrich, St. Louis, USA) (CTT) in a 50 mL flask, and growth was measured by absorbance in Klett units for 7 h using a Klett colorimeter. The lag phase growth rate of H. somni 2336Δhfq was reduced in comparison to the wild-type strain for the initial 1–3 h of culture, but log phase growth was only slightly slower (Fig. 1A). To further compare the growth rates of the wild-type and mutant strains over 24 h, each strain was cultured, grown, and monitored in 96-well plates. The optical density at 600 nm (OD600) of H. somni 2336Δhfq was significantly slower than that of H. somni strain 2336 from 10 h through 24 h of culture in 96-well plates (n = 24, P < 0.0001). The exponential growth phase of H. somni 2336Δhfq in 96-well plates occurred between 2 h and 16 h. In small volume wells, the mutant entered stationary phase at about 16 h post-inoculation, whereas H. somni strain 2336 continued in log phase through 24 h (Fig. 1B). The slower growth of H. somni 2336Δhfq was consistently noted, and differences in their colony size on blood agar plates were observed as well (not shown). However, the growth rate was not restored following complementation of the hfq mutant with an intact copy of hfq in a shuttle plasmid (Fig. 1A).
Fig 1.
Growth curves of H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336 ΔhfqComp. (A) H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336ΔhfqComp were grown in side arm flasks and the density measured in Klett units over 7 h; (B) H. somni strain 2336 and H. somni 2336Δhfq were grown in 96-well plates and the density measured at OD600 in a GloMax plate reader spectrophotometer over 24 h. All tests were repeated three times, and standard deviations calculated using GraphPad Prism 7 software.
H. somni strain 2336 and H. somni 2336Δhfq form similar, but not identical, biofilms and biofilm matrix components
Histophilus somni can form a prominent biofilm in vitro and in the lungs and myocardium of infected cattle (9, 31). Biofilms may not make the disease more severe, but they may enhance the resistance of the bacteria to antibiotics and host immunity, thereby prolonging the disease. Biofilm formation by the wild-type and hfq mutant was examined by confocal laser scanning microscopy (CLSM) after fluorescence in situ hybridization (FISH) with a Texas Red labeled-oligonucleotide probe specific for the 16S rRNA gene of H. somni strain 2336. The 5-day-old biofilms of H. somni 2336 (Fig. 2A and B) and H. somni 2336Δhfq (Fig. 2C and D) strains were imaged by CLSM in the orthogonal view (Fig. 2) and topographical view (Fig. 2B and D), respectively. The 3-D structure of the biofilms of H. somni strain 2336 and H. somni 2336Δhfq mutant appeared similar to each other. However, quantification of the biofilm biomass (Fig. 3A), thickness (Fig. 3B), roughness (Fig. 3C), and the ratio of surface to biovolume (Fig. 3D) by COMSTAT analysis with Image J software showed that compared to H. somni strain 2336, H. somni 2336Δhfq had less biomass and average thickness, though not significantly. However, the roughness coefficient and the surface to biovolume ratio of the mutant’s biofilm were significantly greater (P = 0.0006 for the roughness coefficient; P = 0.0474 for the surface to biofilm ratio) than for that of the wild-type strain, indicating the Δhfq mutant’s biofilm was not as dense and had more surface area exposed to the surrounding environment than the biofilm of the wild-type strain. To determine if the differences in biofilm roughness and surface to biofilm ratio could be attributed to differences in the number of cells, viable plate counts from the biofilms of the wild-type and mutant were done at 24 post-biofilm growth. The number of viable cells for H. somni strain 2336 was 1.6 ± 8 × 106 colony forming units (CFU)/mL, and for H. somni 2336Δhfq was 1.5 ± 5 × 106 CFU/mL. As expected, the difference in viable cells in the biofilms was highly variable and not significant (P = 0.86), likely because the samples were taken from different sites in biofilms, and dead cells and live cells predominate in different regions.
Fig 2.
Biofilm architecture of H. somni strain 2336 and H. somni 2336Δhfq by CLSM. The biofilm architecture of H. somni strain 2336 and H. somni 2336Δhfq was visualized using CLSM. The CLSM images shown are representative of three images each for the 5-day-old biofilms formed by H. somni strain 2336 (A and B) and H. somni 2336Δhfq (C and D). The images are displayed under the orthogonal (A and C) and topographical (B and D) views following FISH using an H. somni oligonucleotide 16S rRNA probe labeled with Texas Red.
Fig 3.
COMSTAT analysis of biofilms formed by H. somni strains 2336 and 2336Δhfq. (A) Mean biomass, (B) mean average thickness, (C) roughness coefficient, and (D) surface to biovolume ratio. All tests were repeated in triplicate. The error bars represent the 95% confidence intervals of the mean and standard deviation. Statistical significance was determined by two-tailed t-tests using GraphPad Prism 7; *, P < 0.05; ***, P < 0.001.
As shown in Table 1, proteins and carbohydrates were the two primary components of the H. somni biofilms. Compared to the wild-type biofilm, the amount of protein, carbohydrate, and eDNA in the H. somni 2336Δhfq biofilm matrix was reduced, although not significantly.
TABLE 1.
Total protein, carbohydrate, and eDNA in the biofilm matrix of H. somni strain 2336 and H. somni 2336Δhfq
| Analyte | H. somni strain 2336 | H. somni 2336Δhfq |
|---|---|---|
| Protein | 33.84 ± 2.09 µg/cm2 | 28.10 ± 4.64 µg/cm2 |
| Carbohydrate | 32.93 ± 6.46 µg/cm2 | 22.42 ± 7.27 µg/cm2 |
| eDNA | 0.12 ± 0.03 µg/cm2 | 0.09 ± 0.01 µg/cm2 |
LOS and protein electrophoretic profiles of H. somni strain 2336 and H. somni 2336Δhfq mutant are altered
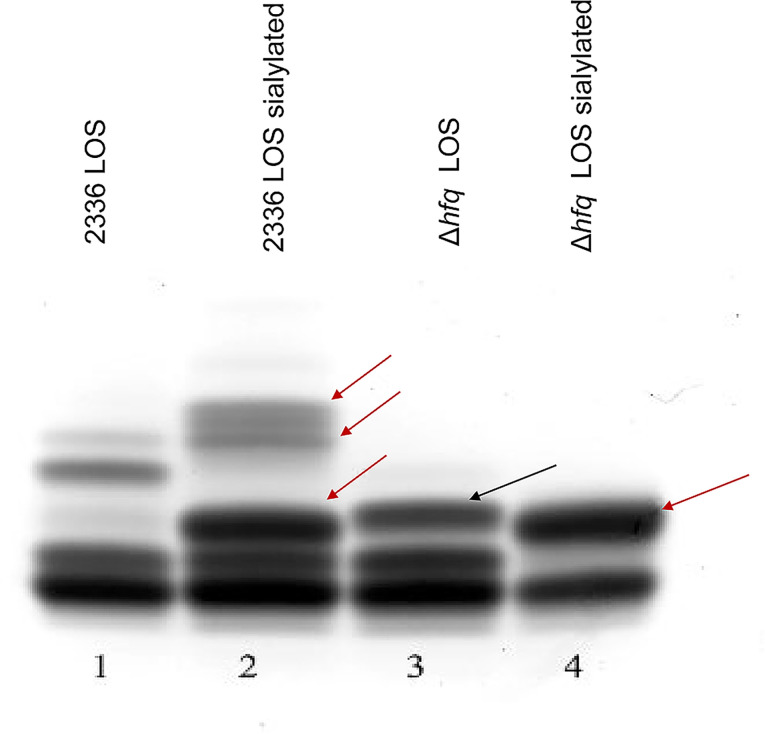
The LOS and IbpA protein are major virulence factors for H. somni. Other outer membrane proteins may also contribute to virulence, particularly to maintain the integrity of the outer membrane, adherence to host cells, and resistance to host defenses (5). There were clear qualitative differences in the electrophoretic profiles of the wild-type and mutant strain’s LOS (Fig. 4). The larger molecular size bands of the LOS were absent in H. somni 2336Δhfq (lane 3), indicating the LOS was truncated. In the presence of N-acetyl-5-neuraminic acid, H. somni can sialylate the terminal galactose residues on its LOS through at least two sialyltransferases (32). LOS sialylation can be identified by an increase in the size of the larger LOS electrophoretic bands (Fig. 4, lane 2, red arrows). If the LOS is truncated and the terminal galactose is absent, this size shift is not seen (33). Although the larger size bands are absent in H. somni 2336Δhfq, there is a size shift in a middle molecular size band when H. somni 2336Δhfq was grown with sialic acid (same sialylated band as in the wild-type strain), indicating that a galactose residue on this LOS residue is sialylated (Fig. 4, lane 4, red arrow). Furthermore, a band slightly larger than a middle sialylated LOS band in H. somni strain 2336 is present in H. somni 2336Δhfq (Fig. 4, lane 3, black arrow), indicating some minor structural change occurred in the oligosaccharide structure. An expression analysis of some genes putatively involved in H. somni virulence indicated that two LOS genes (licA and lsgB) were significantly downregulated in the mutant (Table 2). LicA is a phosphotransferase involved in phosphorylation of choline on the H. somni LOS (34), and lsgB is a sialyltransferase that attaches sialic acid to a terminal galactose, or other sugars required for synthesis of the N-acetyllactosamine residue (32, 35, 36). Upregulation of lob1, which is a galactosyl transferase (37), could account for the enhanced, sialylated LOS band (Fig. 4, lane 4, red arrow). Nuclear magnetic resonance analysis would need to be done to determine the structure of this oligosaccharide chain. Nonetheless, it is clear that there is alteration of oligosaccharide chain synthesis, which may result from alternate regulatory transcription.
Fig 4.
LOS electrophoretic profile of H. somni strain 2336 and H. somni 2336Δhfq. H. somni was grown with or without sialic acid (to sialylate terminal galactose residues) and the LOS extracted by a mini phenol-water protocol. LOS profiles were resolved by electrophoresis through 15% polyacrylamide gels and stained with ammoniacal silver. The red arrows point to sialylated LOS bands, indicating a terminal galactose moiety on the oligosaccharide. The slightly larger highest molecular size band in the LOS of H. somni 2336Δhfq indicates a minor structural change may have occurred. There was not a substantial change in the LOS profile of the complemented mutant compared to H. somni 2336Δhfq (not shown).
TABLE 2.
Genes related to virulence that were significantly upregulated or downregulated in H. somni strain 2336 compared to H. somni 2336Δhfq
| ID | Mutant vs wild-type fold change | Mutant vs wild-type log2-fold change | Mutant vs wild-type adjusted P-value | Gene name | Description | Reference |
|---|---|---|---|---|---|---|
| HSM_RS03835 | 1.548 | 0.630 | 1.6E-35a | tbpB | Transferrin-binding protein | (38) |
| HSM_RS01800 | 1.405 | 0.490 | 2.3E-21a | fur | Ferric iron uptake transcriptional regulator | (35) |
| HSM_RS05070 | 1.475 | 0.560 | 1.8E-22a | lob1 | LOS galactosyl transferase | (37) |
| HSM_RS02730 | 1.499 | 0.584 | 2.9E-05a | licD | LOS putative diphosphonucleoside choline transferase | (34) |
| HSM_RS02715 | 0.546 | −0.874 | 1.4E-03b | licA | LOS phosphotransferase (choline kinase) |
(34) |
| HSM_RS09250 | 0.640 | −0.643 | 6.3E-09b | lsgB | LOS sialyltransferase or other glycosyl transferase | (32, 36, 39) |
Significantly upregulated in H. somni 2336Δhfq.
Significantly downregulated in H. somni 2336Δhfq.
Outside of endotoxin (LOS), only the H. somni IbpA DR2 region containing Fic motifs is toxic for cells (40). IbpA is not readily visible on primary electrophoretic gels, but is readily identified by Western blotting, and is represented by multiple bands greater than 140 kD (41). Western blots of culture supernatants of H. somni strain 2336 and H. somni 2336Δhfq showed that IbpA (multiple protein bands > 140 kD) was present in both the wild-type and mutant strains (Fig. S5).
Multiple qualitative and quantitative differences were seen in the outer membrane protein (OMP) profiles between equivalent numbers of H. somni strain 2336 and H. somni 2336Δhfq (Fig. 5). The predominant differences (twofold loss or greater) were most obvious in the protein bands of about 115 kDa, 68 kDa, 40 kDa, 36 kDa, and 21 kDa in the Δhfq mutant (red arrows). There were no proteins absent in H. somni strain 2336 that were present in H. somni mutant 2336Δhfq, but there were some proteins that were increased at least twofold in the mutant (band numbers 2, 5, 9, 13, 17, and 21).
Fig 5.
Outer membrane protein profiles of H. somni strain 2336 and H. somni 2336Δhfq. Protein-enriched outer membranes were extracted from bacterial total membranes using sodium dodecyl sarcosinate and differential ultracentrifugation. (A) SDS-PAGE gel of protein-enriched outer membranes extracted from H. somni strain 2336 and H. somni 2336Δhfq. Each lane was loaded with 5 µg or 7 µg of protein for improved resolution of specific bands. Red arrows point to protein bands present in H. somni strain 2336, but missing or reduced in quantity in H. somni 2336Δhfq. (B) Relative density analysis of the major outer membrane proteins using ImageJ software. Each number in the left column refers to protein bands labeled by red numbers in the center of Fig. 5A.
H. somni 2336Δhfq is less resistant to the bactericidal action of serum and phagocytic cells
Avirulent strains of H. somni can be differentiated from virulent strains by their differences is susceptibility to killing by serum, particularly immune serum, and survival within professional phagocytic cells. H. somni pathogenic strains are resistant to killing by normal serum (42), but are susceptible to killing by immune serum with fresh complement (37). H. somni 2336∆hfq was significantly more susceptible to killing by 50% immune serum (P = 0.0213) and 70% immune serum (P = 0.0002) (Fig. 6A) than the wild-type strain. Enhanced serum susceptibility is likely due to truncation of the LOS, altered concentrations of outer membrane proteins, or both.
Fig 6.
Diminished serum resistance and phagocytosis of H. somni 2336Δhfq. (A) Serum susceptibility of H. somni strain 2336 and H. somni 2336Δhfq. Log phase cultures of H. somni strain 2336 and H. somni 2336Δhfq were both adjusted to 104 CFU/mL and incubated in various concentrations of bovine serum and 20% precolostral calf serum (as a complement source). Viable plate counts were done at 0 min (before incubation) and after 60-min incubation at 37°C. (B) Phagocytosis and survival of H. somni strain 2336 and H. somni 2336Δhfq in bovine peripheral blood monocytes (BPBMs). H. somni strain 2336 and H. somni 2336Δhfq were incubated with BPBM, extracellular cells were killed with gentamicin, BPBMs were lysed, and viable plate counts were determined after 0-min and 60-min incubation to determine survival within BPBMs. BPBMs incubated with bacteria and cytochalasin D (to prevent phagocytosis) were used as a control to account for any surviving adherent bacteria. Three to five replicates of each assay were completed for statistical analyses using GraphPad Prism 7. *P < 0.05, **P < 0.01, ***P < 0.001.
H. somni pathogenic strain 2336 can survive intracellularly in bovine peripheral blood monocytes (BPBMs), whereas avirulent strain 129Pt is killed within BPBMs (7). Therefore, we sought to determine if deletion of hfq would impact phagocytosis and intracellular survival of H. somni strain 2336 by BPBMs. At 0 h post-infection (1 h after the addition of bacteria), fewer H. somni 2336Δhfq were recovered from BPBMs compared to H. somni strain 2336 (Fig. 6B), indicating that deletion of hfq reduced phagocytosis of H. somni by BPBMs. By 12 h and 24 h post-infection, the CFU of H. somni strain 2336 recovered from the BPBMs were significantly greater (P < 0.001 and 0.01, respectively) than the number of H. somni 2336Δhfq recovered from infected BPBMs. Unlike H. somni strain 2336, there was very little increase in the number of H. somni 2336Δhfq cells from 0 to 12 h post-infection. However, H. somni 2336∆hfq did increase in numbers between 12 h and 24 h, indicating that H. somni 2336Δhfq was significantly compromised in regard to phagocytosis and growth within BPBMs, but not necessarily more sensitive than the wild-type strain to intracellular killing.
RNA sequencing reveals broad differential gene expression between H. somni strain 2336 and H. somni 2336Δhfq
Since Hfq in other bacteria is a global regulator of gene expression, we sought to determine the effects of hfq mutagenesis on gene expression in H. somni. There was significant variation in gene expression between H. somni strain 2336 and H. somni 2336∆hfq, as determined by principal component analysis (PCA). The principal component 1 contributes 96% of the variance, showing that the mutant replicates are distinctly different from the wild-type replicates (Fig. 7A). A total of 832 genes were significantly upregulated, and a total of 809 genes were significantly downregulated in H. somni 2336Δhfq, respectively (Fig. 7B). Many of the genes that were differentially expressed in the mutant compared to the wild-type could be associated with virulence, such as synthesis and sialylation of LOS (see above), and synthesis of biofilm exopolysaccharide (EPS) and sugar transport, which are highlighted in yellow in Fig. 8. The important differentially expressed EPS-associated genes included dctQ (encoding tripartite ATP-independent periplasmic [TRAP] transporter small permease), dctP (encoding TRAP transporter substrate-binding protein), ybhA (encoding pyridoxal phosphatase), rbs2A (encoding sugar ABC transporter ATP-binding protein), araD (encoding L-ribulose-5-phosphate 4-epimerase), rbs1A (encoding sugar ABC transporter ATP-binding protein), glsS (encoding SMP-30/gluconolactonase/LRE family protein), sgbU (L-ribulose-5-phosphate 3-epimerase), xylB (encoding carbohydrate kinase), csrA (encoding carbon storage regulator), rbs1C (encoding ABC transporter permease), and waaA (lipid IV(A) 3-deoxy-D-manno-2-octulosonic acid transferase).
Fig 7.
Differential gene expression of H. somni strain 2336 and H. somni 2336Δhfq. (A) PCA. The PCA plot shows the clustering pattern of gene expression profiles between H. somni strain 2336 and H. somni 2336Δhfq. Each dot represents a sample, and the position of the dots in the plot reflects the overall similarity or dissimilarity of the gene expression patterns. (B) MA plot: The MA plot (log-intensity ratios vs log-intensity averages) displays the mean normalized counts (x-axis) against the log2-fold change (y-axis) for all the genes analyzed. Red dots represent genes that are significantly differentially expressed (adjusted P-value <0.05) in H. somni 2336Δhfq compared to H. somni strain 2336, as determined using DEseq2 package ver. 1.12.3. This plot directly shows the differential expression of genes between H. somni strain 2336 and H. somni 2336Δhfq.
Fig 8.
Gene expression heatmap of genes involved in EPS production by H. somni strain 2336 and H. somni 2336Δhfq. The heatmap displays the hierarchical clustering of differentially expressed genes responsible for expression of the biofilm EPS between H. somni strain 2336 and H. somni 2336Δhfq. Each row represents a gene, and each column represents a sample. The color scale represents the log2-fold change, from yellow to blue indicating the upregulation and downregulation levels of gene expression, respectively.
H. somni 2336Δhfq is attenuated for virulence in animal models
The only natural hosts for H. somni are ruminants, particularly farmed cattle, where disease can occur under stressful conditions (2). Due to the expense, limited numbers of cattle that can be handled at one time, and difficulty in replicating natural challenge, a mouse model has been developed to assess virulence through bacteremia/morbidity.
Mice
All mice challenged intraperitoneally (IP) with 5.5 × 106 CFU of H. somni strain 2336 in 2% mucin had large numbers of bacteria in the blood (mouse 1: 2.01 × 105 CFU/mL; mouse 2: 6 × 103 CFU/mL; mouse 3: 5.01 × 105 CFU/mL) at 5 h post-challenge (Table 3). The latter mouse was severely moribund and was euthanized shortly after the collection time. The other two mice also showed clinical signs of lethargy, lack of movement unless prompted, and ears pulled back. At 29 h post-challenge, only 200 CFU/mL (two colonies from 10 µL of blood) of H. somni strain 2336 could be recovered from mouse 1 and 0 CFU/mL from mouse 2, and neither mouse showed any of the morbidity that was present the day before. In contrast, four of six mice challenged with a higher dose (1.6 × 107 CFU/mL) of H. somni 2336Δhfq had 0 CFU/mL in the blood at 5 h post-challenge, and one had 100 CFU/mL (one colony from 10 µL of blood). Culture results were contaminated for one mouse and were therefore not counted. None of the six mice challenged with H. somni 2336∆hfq showed any clinical signs. At 29 h post-challenge, there were 0 CFU/mL of bacteria recovered from any of these six mice (Table 3). There were no clinical signs and no bacteria recovered from control mice inoculated with mucin alone.
TABLE 3.
Bacteremia in mice following intraperitoneal challenge with H. somnid
| Animals | H. somni 2336 (CFU/mL) 6 h post-challenge | H. somni 2336 (CFU/mL) 29 h post-challenge | H. somni 2336Δhfq (CFU/mL) 6 h post-challenge | H. somni 2336Δhfq (CFU/mL) 29 h post-challenge |
|---|---|---|---|---|
| 1 | 2 × 105 /mL | 200 /mL | 0 | 0 |
| 2 | 6 × 103 /mL | 100 /mL | 100 /mL | 0 |
| 3 | 5 × 105/mLa | a | c | 0 |
| 4 | n/ab | 0 | 0 | |
| 5 | n/a | 0 | 0 | |
| 6 | n/a | 0 | 0 |
This mouse was very moribund at 6 h post-challenge and was euthanized after blood collection. No sample was available at 29 h post-challenge.
n/a, not applicable. There were only three mice in the challenge control group to minimize numbers.
Culture was contaminated and H. somni could not be detected.
A control group of three mice inoculated with mucin alone showed no clinical symptoms and no bacteria were recovered from their blood at any time point.
Cattle
Respiratory rates, rectal temperatures, and clinical scores were obtained before and after challenge for calves in all three groups (Fig. 9, and Fig. S6 and S7, respectively). At most time points after challenge, which occurred on the morning of day 0, the average respiratory rates (Fig. 9) and temperatures (Fig. S6) were highest for calves challenged with H. somni strain 2336, the next highest for calves challenged with H. somni 2336Δhfq, and lowest for the mock calves challenged with only sterile saline + 5% fetal calf serum. The clinical scores were variable among all groups (Fig. S7). However, there was no statistically significant difference in temperatures, respiratory rates, or clinical scores between the three groups (P > 0.05). There was a significant effect of time following challenge on temperatures, respiratory rates, and clinical scores (P-values = 0.0018, 0.001, and 0.0066, respectively), and the interaction between the time after challenge and the treatment group significantly affected the respiratory rate (0.0336).
Fig 9.
Average (±SD) respiratory rates of calves before and after challenge with H. somni strain 2336, H. somni 2336Δhfq, or sterile saline only (mock). Calves were challenged with 1.4–2 × 109 CFU/mL on day 0 by intrabronchial instillation of 10 mL of the specific inoculum. Respiratory rates were not statistically significantly different between groups (P > 0.05).
Microbiologic and postmortem results of calves
Cattle are one of the few natural hosts for H. somni, and calves are particularly susceptible to disease by this opportunistic pathogen. Results of culture to identify H. somni from nasopharyngeal swabs (NPS), endotracheal aspirates (ETA), and lung tissue collected from calves are shown in Table 4. The NPS from all calves were negative for H. somni on study days −8 and 0, before challenge. All three calves challenged with H. somni strain 2336 were culture-positive for H. somni by NPS on all days post-challenge, while calves challenged with H. somni 2336∆hfq were negative on NPS culture on some days. Similarly, all calves challenged with H. somni strain 2336 were positive for H. somni on lung culture at necropsy, while only two of four calves challenged with H. somni 2336∆hfq were positive on lung culture; these results suggested that H. somni 2336∆hfq could not persist in the respiratory tract as effectively as the wild-type strain. However, the difference in the proportion of calves culture-positive for H. somni that were challenged with H. somni 2336∆hfq was not statistically significantly different from calves challenged with H. somni strain 2336 for any sample or time point tested. Histophilus somni was not isolated from any sample from either of the two mock-challenged calves. Pasteurella multocida was isolated from occasional samples from calves in all groups, which was presumed to be of endogenous (commensal) origin. Trueperella pyogenes, which is also an opportunistic commensal bacterium, was identified from the NPS of one calf challenged with H. somni 2336∆hfq on day 5, and from lung tissue of one calf challenged with H. somni strain 2336.
TABLE 4.
Proportion of calves positive by bacterial culture for H. somni on NPS, ETA, or lung tissue collected on various days post-challengea
| Group | Day −8 NPS | Day 0 NPS | Day 1 NPS | Day 1 ETA | Day 2 NPS | Day 3 NPS | Day 4 NPS | Day 4 ETA | Day 5 NPS | Day 5 lung |
|---|---|---|---|---|---|---|---|---|---|---|
| Mock (n = 2) |
0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 H | 0/2 | 0/2 | 0/2 | 0/2 |
| Mutant (n = 4) |
0/4 | 0/4 | 4/4 | 0/4 | 2/4 | 3/4 | 4/4 | 1/4 | 4/4 | 2/4 |
| Wild-type (n = 3) |
0/3S | 0/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 |
Calves were challenged with sterile saline + 5% fetal calf serum alone (mock), H. somni strain 2336 (wild-type), or H. somni 2236Δhfq (mutant).The difference in the proportion of calves positive for H. somni for calves in the wild-type group vs the mutant group was not significantly different (P > 0.05).
The results of gross postmortem assessment of the lungs of calves at necropsy on day 5 are shown in Fig. 10 and 11. The total percentage of abnormal lung for the two calves in the mock challenge group was 2% and 5%. The lungs of calf 1 challenged with H. somni 2336∆hfq were essentially normal, which could indicate that the challenge inoculum was inadvertently administered into the gastrointestinal tract rather than into the lung of that calf. The total percentage of abnormal lung from the remaining three calves challenged with H. somni 2336Δhfq was 12%, 18%, and 31%, and the total percentage of abnormal lung for the three calves challenged with H. somni strain 2336 was 17%, 20%, and 23% (Fig. 10); the percent overall abnormal lung in the calves challenged with H. somni strain 2336 was not statistically significantly different from the percent overall abnormal lung in the calves challenged with H. somni 2336∆hfq (P > 0.05). Microscopic evaluation of the grossly abnormal lung in the two mock-challenged calves revealed bronchioles that were filled with neutrophils that extended into adjacent alveoli, and areas of alveolar collapse (Fig. 11A and D). Lung tissue collected from calf 1 challenged with H. somni 2336Δhfq was normal on microscopic evaluation. Mock-challenged calves had rare areas of alveolar collapse and bronchioles that were filled with neutrophils, but alveoli were largely normal (Fig. 11C and F). One calf challenged with H. somni 2336∆hfq and all calves challenged with H. somni strain 2336 had more severe gross and microscopic pathology that included marked consolidation, predominantly of the diaphragmatic lobe, which corresponded histologically to locally extensive areas of necrosis surrounded by degenerate neutrophils, fibrin, and edema (Fig. 11B and E). The location within the diaphragmatic lobe of this severe inflammation and necrosis was likely associated with bronchial administration of the challenge material. Although H. somni infection can also cause lesions in the myocardium and/or joints, no abnormalities were seen in these organs in any calf challenged in this study.
Fig 10.
Percent abnormal lung in calves challenged with H. somni strain 2336, H. somni 2336Δhfq, or mock-challenged calves. Calves were challenged by intrabronchial instillation with a log phase culture of H. somni strain 2336 (wild-type) or H. somni 2336Δhfq (mutant), or sterile saline + 5% fetal calf serum (mock). The percent abnormal lung by lobe (A) and for all lung lobes (B) is shown. The percent abnormal lung in the wild-type and mutant groups was not significantly different (P > 0.05).
Fig 11.
Gross and microscopic lung lesions in calves challenged with H. somni strain 2336, H. somni 2336Δhfq, or mock-challenged calves. Calves were challenged by intrabronchial instillation with a log phase culture of H. somni strain 2336 (wild-type), H. somni 2336Δhfq (mutant), or sterile saline + 5% fetal calf serum (mock). Representative gross lung pathology from calves inoculated with mutant (A) or wild-type (B) compared to mock (C) is shown. Grossly, the mutant and wild-type lung had multifocal to locally extensive dark red depressed areas, which corresponded histologically to abundant neutrophils within bronchioles and alveoli with alveolar collapse (D). In all three calves challenged with the wild-type, and one calf challenged with the mutant, there was marked consolidation of the diaphragmatic lobe which corresponded histologically to large areas of necrosis with abundant fibrin and edema (E). Mock-infected calves had rare areas of alveolar collapse with largely normal alveoli (F). Hematoxylin and eosin stain, scale bar = 100 µm.
DISCUSSION
A wide variety of virulence factors have been described for H. somni, including decoration of the LOS with phosphorylcholine and sialic acid, LOS phase variation, biofilm formation, expression of IbpA containing a cytotoxic Fic motif, histamine production, intracellular survival, cellular adherence, and others (3–8). We have established that half of the bacterial genome is differentially regulated when the bacteria form a biofilm compared to planktonic growth (10, 11). However, virtually nothing is known about how such virulence factors, or metabolic processes, are regulated in H. somni. sRNAs have been shown to be important regulators of bacterial gene expression, and Hfq is a chaperone protein for sRNAs that can interact directly with mRNA to enhance or suppress gene expression, likely driven by environmental factors (12).
Mutagenesis of hfq in the related bacterium P. multocida resulted in less hyaluronic acid capsule production, reduced fitness in vivo, and altered transcription of 128 genes (26). Mutagenesis of hfq in different serotypes of the swine pathogen Actinobacillus pleuropneumoniae did not affect growth in most strains, but did affect adherence, stress response, and virulence in larvae of the greater wax moth (23). In addition, an A. pleuropneumoniae Hfq mutant was reported to be deficient in biofilm formation, poly-β-1,6-N-acetylglucosamine (major component of the biofilm matrix) production, and was less tolerant to oxidative stress (27). Mutation of hfq in Haemophilus ducreyi resulted in altered expression of 16% of the bacterial genes. Genes encoding for several virulence factors were downregulated in the H. ducreyi mutant, which was attenuated for virulence in humans. The authors concluded that Hfq contributes to gene regulation in the absence of RpoS, particularly in stationary phase growth, and is required for virulence in humans (24). In Haemophilus influenzae, mutagenesis of hfq had no effect on growth rate or survival under stress conditions in vitro. Although no apparent differences in virulence were noted, there was reduced competitive fitness in a chinchilla model of otitis media. In an infant rat model of bacteremia, the H. influenzae Hfq mutant persisted for less time in the blood with fewer numbers of bacteria in comparison to the wild-type strain (25). All of these studies concluded that Hfq can influence virulence, and is an important contributor to the expression of some genes. However, which phenotypic properties are affected by Hfq that contribute to overall bacterial fitness varies between species and possibly strains.
An hfq-deficient mutant of H. somni strain 2336 was created by replacing hfq with a chloramphenicol resistance gene. The aerobic growth rate of the mutant was compromised during the initial log phase of growth and by entering stationary phase earlier, thus limiting its maximum density. These observations may indicate that deletion of hfq may compromise H. somni growth in vivo as well, resulting in decreased fitness in the host. Our observations align with previous studies on Brucella melitensis, Legionella pneumophila, and Salmonella typhimurium, in which hfq mutants of these species exhibited reduced growth rates and longer lag phases in certain in vitro media (28–30).
The effect of Hfq mutagenesis on factors that affect H. somni fitness, stress responses, and virulence was examined. Initially, the resistance of H. somni 2336Δhfq to killing by bovine serum and to phagocytosis and intracellular survival within bovine monocytes was examined. H. somni 2336Δhfq was significantly more susceptible to killing by 50% to 70% immune serum (P = 0.0213 to 0.0002) and to phagocytosis by monocytes the initial 12 h post-infection (P < 0.001 and 0.01), compared to wild-type H. somni strain 2336. Moreover, the mutant was significantly less capable of intracellular growth within the monocytes, indicating a potential decrease in virulence resulting from the deletion of hfq. Examination of H. somni surface components demonstrated that there were substantial modifications to the OMP profile of H. somni 2336Δhfq, as well as truncation and lack of sialylation of some LOS oligosaccharides. Previous studies have established that sialylation of H. somni LOS is associated with enhanced resistance to killing by serum and phagocytic cells, and strains that cannot sialylate their LOS are avirulent in animal models (32, 43). Furthermore, truncation of the H. somni LOS alone is associated with enhanced serum susceptibility (44).
Gene expression analysis showed that at least two genes (lsgB and licA) directly involved in LOS synthesis and sialylation were significantly downregulated in H. somni 2336Δhfq. In H. influenzae, lsgB is required for synthesis of the terminal galactose-GlcNAc moiety (N-acetyllactosamine), which is also present in H. somni (45). However, lsgB is also an established sialyltransferase (36, 39). The inability to synthesize and sialylate the terminal lactosamine of the H. somni LOS would truncate the LOS. Furthermore, fur has been shown to regulate a wide variety of genes, and not just those involved in iron utilization (35). The fur gene was significantly upregulated in H. somni 2336Δhfq implying that Hfq downregulated fur in H. somni. These results support further investigation into the role of fur in H. somni gene regulation, virulence, and interaction with Hfq. The amount of biofilm formed by the wild-type and mutant appeared similar, though COMSTAT analysis demonstrated that the biofilm of the Δhfq mutant had a rougher and larger surface area compared to H. somni strain 2336. Although not quite significant, there was less polysaccharide in the biofilm matrix, which is predominately EPS, of H. somni 2336Δhfq. These overall results indicated that deletion of hfq in H. somni strain 2336 could affect its virulence, as was reported for Legionella pneumophila (29) and Salmonella typhimurium (30).
Due to the variety of phenotypic changes noted in H. somni 2336Δhfq, the hfq mutation was complemented and the genome of the mutant sequenced. Complementation of hfq in other bacterial species have had their growth rates restored to levels close to that of the wild-type strains, such as B. melitensis (28) and P. multocida (46). Notably, in both cases, hfq was found to be overproduced compared to the wild-type, likely due to the high efficiency of the promoter and the multicopy nature of the plasmid. In our study, we were unable to utilize a high copy number expression plasmid for H. somni, and therefore constructed a previously characterized plasmid (47) carrying the hfq gene with its own promoter and terminator. However, the hfq protein appeared to be expressed at a low level and was unable to significantly reverse the phenotypic changes. Similar results were obtained in a previous study to complement a luxS gene deletion in H. somni (10). The inability to adequately reverse phenotypic changes in H. somni may be attributed to several factors, including the low efficiency of hfq expression and the low copy number of the plasmid. Furthermore, competition of RNA transcription promoters between the plasmid and the genome in H. somni could be contributing factors. These issues may collectively hinder the successful restoration of phenotypic changes in the hfq mutant. However, the replacement of hfq with a chloramphenicol resistance gene in H. somni 2336Δhfq was confirmed, and whole genome sequencing of the mutant verified that there were no additional amino acid mutations in the genome, except for a T/G mutation in an intergenic region, which would not affect protein synthesis. Therefore, the phenotypic changes expressed by H. somni 2336Δhfq were confirmed to be due to inactivation of hfq.
Hfq has gained attention for its important role in post-transcriptional regulation of bacterial genes (48, 49). Hfq can modulate translation initiation frequency and mRNA stability, allowing bacteria to adapt to changing environments. In this study, a comparative RNA-seq analysis between H. somni strain 2336 and H. somni 2336Δhfq was carried out to investigate the impact of hfq on mRNA transcription in H. somni. Eight hundred thirty-two genes were upregulated, and 809 genes were downregulated in the mutant compared to the parent strain. Many of the differentially expressed genes in the H. somni 2336Δhfq mutant were associated with virulence, extracellular polysaccharide production, and sugar transport. Of note was that rbs1A, rbs2A, dctQ, dctP, araD, ybhA, glsS, and sgbU genes, which are involved in polysaccharide biosynthesis and are presumed crucial for EPS and biofilm formation, were significantly upregulated in H. somni 2336Δhfq compared to H. somni strain 2336. The production of the galactomannan EPS polymer by H. somni is known to promote biofilm growth, and EPS may play a role in virulence during systemic infections. Therefore, the upregulation of these genes is likely favorable for biofilm formation, which is consistent with the findings of our previous study that showed significantly higher expression levels of these genes in biofilm cultures than in planktonic cultures (4, 50). Conversely, H. somni 2336Δhfq downregulated csrA and manB, which are responsible for the synthesis of the D-mannan polymer, and other genes involved in EPS and biofilm formation, such as xylB, rbs1C, and rbs2C. Notably, these genes were expressed at higher levels in biofilm cultures than in planktonic cultures (50), which suggests that their downregulation may hinder biofilm formation. The variable effect of upregulation and downregulation on the many genes associated with biofilm formation may explain why there was only a minimal overall difference in biofilm production between the wild-type and mutant. The impact of these genes on the virulence of H. somni remains unclear and requires further investigation.
Direct evidence that H. somni 2336Δhfq was less virulent than wild-type H. somni strain 2336 was demonstrated in a mouse model of bacteremia and mortality (10, 44, 51). However, mice are not the natural host for H. somni, and may not be a reliable model for evaluating H. somni virulence. Therefore, a calf challenge study was carried out to compare the virulence of H. somni 2336Δhfq to virulent H. somni strain 2336 in the bacterium’s natural host. Endotracheal aspirates and lung tissue collected at necropsy from calves challenged with the wild-type strain were culture-positive more frequently than aspirates from calves challenged with H. somni 2336Δhfq. The proportion of positive samples was not statistically significantly different between the challenge groups, likely due to the small number of calves challenged. Furthermore, H. somni 2336Δhfq challenge resulted in fewer calves having severe lung lesions (microscopic necrosis with abundant fibrin and edema); this lesion was observed in only one of four calves challenged with H. somni 2336Δhfq, versus all three calves challenged with H. somni strain 2336. These results suggest that H. somni 2336Δhfq was less virulent in calves, though the small number of calves challenged likely precluded identification of statistically significant differences. Additionally, it is important to note that for this challenge, the bacteria were inoculated directly into the lungs. The predominant inflammatory component of H. somni is the lipid A component of the LOS (52). Therefore, it is not surprising that both the wild-type and mutant induced substantial inflammation following challenge. Whether the mutant was less capable of bypassing the innate immune defenses of the upper respiratory tract is difficult to assess because some type of immunosuppression is required for even the wild-type strain to invade the lower respiratory tract (53). However, fewer bacteria were recovered from ETA of calves challenged with H. somni 2336Δhfq than with H. somni strain 2336, suggesting the mutant may be less competent at colonization. Further evaluation of the virulence of H. somni 2336Δhfq in larger numbers of cattle, perhaps with different routes of challenge, is warranted.
MATERIALS AND METHODS
Bacterial strains and culture conditions
H. somni strain 2336 was grown on Columbia agar containing 5% sheep blood (CBA) and incubated at 37°C overnight in 5% CO2. H. somni was grown to mid-logarithmic phase in CTT at 37°C with shaking at 180 rpm. H. somni 2336Δhfq was grown in the same medium supplemented with 2 µg/mL of chloramphenicol, and the complemented H. somni mutant with 10 µg/mL of kanamycin. Escherichia coli Top10 cells were cultured in Luria-Bertani broth (Becton, Dickinson and Company, Franklin Lakes, NJ), or with chloramphenicol or kanamycin depending on the antibiotic resistance marker in the plasmid they carried. All bacterial cells were sedimented by centrifugation at 5,000 × g for 15 min, and resuspended in the appropriate buffer at exactly the same density for all assays.
Mutagenesis and complementation of H. somni hfq
One single-stranded region of DNA (ΔhfqCm gBlock), containing a 738 nt region of the 5´ end upstream of hfq (part of GSR gene), a chloramphenicol resistance cassette (CmR), and a 798 nt region of the 3' end downstream of hfq (part of GTPase Hflx gene), was synthesized by Integrated DNA Technologies Inc. (Coralville, IA). A PCR product of H. somni 2336ΔhfqCm gBlock DNA was generated with the primer pair HfqHomU263F (5´ AGGTGTAGAAACTAACGAACGTGG 3´)/HfqHomL2037R (5´ GAGTTGCAAATAATTGATCTGC CACA 3´) using Q5 DNA polymerase (New England Biolabs, Ipswich, MA) (Fig. S1). The PCR cycling conditions were 98°C for 30 s, 98°C for 10 s, 67°C for 30 s, 72°C for 1 min 15 s, 30 cycles, and 72°C for 5 min. After methylation with HpaII, the H. somni 2336ΔhfqCm PCR product (about 17 µg) was electroporated into H. somni strain 2336 competent cells (44). One hundred microliters of transformed H. somni strain 2336 was seeded onto CBA plates containing 2 µg/mL chloramphenicol and incubated overnight at 37°C with 5% CO2. Chloramphenicol-resistant colonies were selected to confirm replacement of hfq with the CmR gene, as described below.
For complementation of the hfq deletion, a 2,062 bp fragment of the H. somni strain 2336 hfq genomic region, encompassing an EcoRI fragment containing the hfq gene and its potential promotor and terminator regions, was amplified by PCR with HfqHomU263F/HfqHomL2037R primers. The cycling conditions were 98°C for 30 s, 98°C for 10 s, 60°C for 30 s, 72°C for 2 min for 35 cycles, and 72°C for 5 min. H. somni shuttle vector pNS3K (47), containing a kanamycin resistance gene, was digested with EcoRI and dephosphorylated with rSAP sequentially. The 513 bp EcoRI-digested hfq PCR product was then ligated into pNS3K-EcoRI using T4 ligase (ThermoFisher Scientific, Waltham, MA), and transformed into Top10 competent cells using standard protocols (54). Plasmid DNA from kanamycin-resistant colonies of the transformed Top10 cells was digested with EcoRI and analyzed by agarose gel electrophoresis for the 513 bp DNA fragment, and the presence of the hfq gene in the plasmid was confirmed by DNA sequencing. The resulting plasmid, pNS3K-2336hfq, was used to transform H. somni 2336Δhfq, and kanamycin-resistant colonies were screened for expression of hfq by RT-PCR and for Hfq by gel electrophoresis (Fig. S2A and S2B, respectively). One positive clone was selected to obtain H. somni 2336ΔhfqComp.
Genome sequencing and bioinformatic analysis of H. somni 2336Δhfq
Genomic DNA from H. somni strain 2336 and H. somni 2336Δhfq were extracted from broth cultures using a MasterPure DNA purification kit (EpiCentre, Madison, WI, USA) according to the manufacturer’s instructions. The genomic DNA concentration and purity was determined using NanoDrop OneC (Thermo Scientific) and was then used for library preparation and Illumina DNA sequencing at SEQCENTER (Pittsburgh, PA, USA). In brief, the Illumina DNA Prep kit was used with primers that contain unique dual index (UDI) sequences from Integrated DNA Technologies (IDT) that were 10 bp long, which were then sequenced on an Illumina NextSeq 2000, yielding 2 × 151 bp reads. The bcl-convert was used for demultiplexing, quality control, and adapter trimming (v.3.9.3). The genomic sequencing analysis of H. somni strain 2336 and H. somni 2336Δhfq was carried out by importing the Illumina reads and the reference genomic sequence of H. somni strain 2336 into Geneious Prime. The trimmed Illumina reads were mapped to the reference genome using the Geneious assembler, and consensus sequences were generated for each sample using Geneious.
Determination of growth rates for H. somni strain 2336 and H. somni 2336Δhfq
To determine the planktonic bacterial growth rate, bacterial colonies of H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336ΔhfqComp were picked from CBA (from frozen, −80°C stocks) and suspended in 50 mL of CTT in a 125 mL flask to the same density (~25 Klett units). The suspensions were shaken at 180 rpm at 37°C, and the densities measured using a Klett Colorimeter at 1-h intervals for 7 h. Bacterial cultures of H. somni strain 2336 and H. somni 2336Δhfq grown on CBA were also inoculated into 2 mL of CTT to an OD600 of 0.1. Two hundred microliters of each culture was then added to 8 wells of a 96-well microtiter plate. The OD600 of each well was determined automatically each hour using the GloMax Discover Microplate Reader (Promega Corporation, Madison, WI) during overnight incubation. GloMax Discover Microplate Reader growth rates were supported by viable plate count. Each assay was repeated three times. The multiple t-test was used to compare the growth rate of H. somni strain 2336 and H. somni 2336Δhfq at each time point during bacterial culture.
Biofilm and matrix component analyses
The biofilms of H. somni strain 2336 and H. somni 2336Δhfq were formed on coverslips or in microtiter wells. Briefly, a loop of bacterial culture from an overnight CBA plate was suspended in 5 mL of CTT broth and incubated at 37°C at 180 rpm until each culture reached mid-log phase (109 CFU/mL), which was determined using a spectrophotometer and confirmed by viable plate count. The bacterial suspension was then diluted 1:100 with CTT broth. One milliliter of the diluted culture was placed into wells of a 24-well plate containing a coverslip, or 200 µL was placed into wells of a 96-well polystyrene microtiter plate (Costar; ThermoFisher Scientific), followed by stationary incubation at 37°C in 5% CO2 for 5 days; half of the bacterial medium was replaced with fresh medium after 2–3 days. After 1.5-day incubation, a sample was obtained from each biofilm for viable plate count in triplicate. Biofilm formation was quantified in the 96-well plate by staining with 1% crystal violet for 15 min at room temperature. After washing three times with sterile distilled water, the crystal violet bound to the biofilm was solubilized with 30% acetic acid and the absorbance at 600 nm was measured with the GloMax Discover System. Each experiment was performed in triplicate.
Biofilms were statically grown for 5 days on round Chemglass Life Sciences coverslips (# 50-121-5159, ThermoFisher Scientific) in CTT. Half of the culture medium was replaced with fresh medium on the third day. FISH was performed as described 55) with modifications to detect H. somni in biofilms. Briefly, mature biofilms were rinsed, dried, and fixed with 4% (wt/vol) paraformaldehyde/phosphate buffered saline, pH 7.2 (PBS), rinsed, and incubated with 200 ng of specific oligonucleotide 16S rRNA probe labeled with TEX615 (5´- /5TEX615/GTT CCC ACC CTA ACA TGC TGG -3´) (Integrated DNA Technologies Inc., Coralville, IA) per coverslip in 20 µL hybridization buffer (900 mM NaCl, 20 mM Tris-pH 7.5, 0.01% SDS, and 25% formamide), followed by incubation with washing buffer (150 mM NaCl, 5 mM EDTA, 20 mM Tris-pH 7.5, and 0.01% SDS), and rinsed with cold distilled water. Glass coverslips were embedded biofilm-side down with 8 µL of Invitrogen ProLong Glass Antifade Mountant (#P36982, ThermoFisher Scientific) onto glass slides, sealed with Biotium coverslip sealant (#NC0154994, ThermoFisher Scientific), and stored in the dark at 4°C. Three coverslips of H. somni strain 2336 biofilms and three coverslips of H. somni 2336Δhfq biofilms were examined by CLSM using a ZEISS LSM 900 Airyscan 2 laser-scanning microscope, mounted on an inverted Axiocam Color Camera (Carl Zeiss, Oberkochen, Germany) at 20× magnification using a laser line of 561 nm for excitation and the emission filter for TEXRe fluorophore (575 nm–700 nm). Z-stack images were analyzed using the image-processing software COMSTAT (56). Images were acquired from the center of the coverslip in 1.1 µm sections throughout the biofilm depth, with the number of sections varying depending on the thickness of the biofilm. For analysis of biofilm matrix components, 5-day-old biofilms of H. somni strain 2336 and H. somni 2366Δhfq were suspended in 1 mL of PBS, and vortexed rapidly for 20 min to disperse the biofilm matrix prior to assay. The protein and carbohydrate content of the biofilms were determined using the bicinchoninic acid (BCA) (ThermoFisher Scientific) and anthrone (57) assays, respectively. Following gel electrophoresis, the approximate concentration of extracellular DNA was determined by reference to the band intensity of DNA molecules of known concentration in a DNA ladder on the gel using ImageJ software (https://imagej.net/ij/index.html).
Extraction and electrophoretic analysis of LOS and OMPs
LOS was extracted from H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336ΔhfqComp using a hot phenol-water microextraction method, as previously described (58). All H. somni strains were in mid-log phase and were adjusted to the same OD600 density prior to LOS extraction. Protein-enriched outer membranes were prepared by sodium sarcosinate extraction and differential ultracentrifugation (59). Briefly, the bacterial cells were lysed by sonication, intact bacteria were removed by low-speed centrifugation, and total membranes were pelleted by ultracentrifugation at 120,000 × g for 60 min. The pellet was resuspended in 0.01 M HEPES, pH 7.4, with 2 mM MgCl2 and 2% sodium lauryl sarcosinate to solubilize the inner membranes. After 30 min, the insoluble protein-enriched outer membranes were pelleted by centrifugation at 120,000 × g for 60 min. Extraction of the pellet was repeated, and the protein-enriched outer membranes were resuspended in water. The electrophoretic profile of H. somni LOS was carried out by SDS-PAGE, followed by silver staining, as described (58). OMP profiles were resolved by SDS-PAGE using NuPAGE 4%–12% Bis-Tris mini protein polyacrylamide gels (Invitrogen, Waltham, MA), at 180 V for 45 min, and stained by PageBlue protein staining solution (ThermoFisher Scientific).
Immunoprecipitation and Western blotting
Hfq, if present, was immunoprecipitated from lysates of H. somni strain 2336, H. somni 2336Δhfq, and H. somni 2336ΔhfqComp that were adjusted to the same OD600 density using anti-Hfq antibody covalently coupled to N-hydroxysuccinimide-activated magnetic beads (ThermoFisher Scientific, Waltham, MA) as described (60). Briefly, 300 µL of magnetic beads were coupled to 132 µg of anti-Hfq antibody at 4°C overnight. After washing, the reaction was stopped with quenching buffer (3 M ethanolamine, pH 9.0). Twenty-five microliters of antibody-coupled beads was incubated with lysates containing 1 mg total protein from H. somni strain 2336, H. somni 2336Δhfq, or H. somni 2336ΔhfqComp for 2 h at room temperature. After washing, bound proteins were eluted from the beads with 3.5 M MgCl2 and immediately neutralized with 1M Tris-HCl buffer, pH 8. The eluted proteins were resolved by SDS-PAGE using NuPAGE 4%–12% Bis-Tris gel (ThermoFisher Scientific, Waltham, MA). For Western blotting, the protein bands were transferred to nitrocellulose blotting membrane and blocked with 5% skim milk in phosphate buffered saline containing 0.05% Tween 20. The membranes were treated with affinity-purified IgG raised against Hfq followed by incubation with affinity-purified peroxidase-labeled goat anti-rat IgG secondary antibody (SeraCare, Milford, MA). The immunoreactive bands were detected by chemiluminescence (ThermoFisher Scientific, Waltham, MA).
Bactericidal assay
Resistance to the bactericidal activity of H. somni convalescent bovine antiserum for H. somni 2336Δhfq in comparison to the wild-type strain was determined as previously described (61). Briefly, log phase bacteria (109 CFU/mL, determined spectrophotometrically and confirmed by viable plate count for wild-type and mutant) were incubated with convalescent antiserum (30%, 40%, 50%, 60%, and 70%) and 20% precolostral calf serum (an antibody-free source of complement). Viable plate counts were determined at 0 min and after 60-min incubation at 37°C, and the percent survival was determined.
Intracellular survival of H. somni strain 2336 and H. somni 2336Δhfq in BPBMs
Peripheral blood was collected from the jugular vein of adult Holstein cows into a blood bag containing citrate phosphate dextrose adenine solution as an anticoagulant. The whole blood was diluted 1:1 with PBS and the red blood cells pelleted by centrifugation at 2,000 × g for 15 min. The buffy coat containing mononuclear cells was carefully transferred into fresh tubes, resuspended in Hanks’ balanced salt solution (Life Technologies, Carlsbad, CA), and then laid over Ficoll-Paque Premium (MilliporeSigma, Rockville, MD). The BPBMs were collected after centrifugation at 1,200 × g for 30 min at 15°C. The viability of the isolated BPBMs was determined by trypan blue staining. The cells were seeded into six-well plates in RPMI 1640 medium supplemented with 10% fetal bovine serum. Floating cells were removed after 2 h of incubation at 37°C, leaving the attached cells, which were primarily BPBMs. The BPBMs were cultured overnight in six-well tissue culture plates before being inoculated with bacteria.
Bacterial uptake and intracellular survival were determined in BPBMs as previously described (7). Briefly, H. somni strain 2336 and H. somni 2336Δhfq in mid-log phase (109 CFU/mL) were co-incubated with BPBMs for 1 h at a multiplicity of infection of 100:1 (bacteria to monocytes) at 37°C. The extracellular bacteria were killed by incubating the BPBMs with 50 µg/mL gentamicin for 30 min, followed by washing the cells three times with PBS and incubating for an additional 24 h at 37°C. At 0 h (1 h after the addition of bacteria and 30 min after the addition of gentamicin), 12 h, and 24 h time points, the BPBMs were lysed with distilled water, neutralized with 2× PBS, and the lysate cultured on CTT to determine the number of viable intracellular bacteria. BPBMs incubated with cytochalasin D, which inhibits phagocytosis, were used as a control to detect surviving extracellular bacteria. A total of nine assay replicates were performed at each time point.
RNA sequencing and analysis
Total RNA was extracted from H. somni strain 2336 and the hfq deletion mutant using RNeasy mini Kit (Qiagen, Germantown, MD) after growing the bacteria in CTT to mid-exponential phase and adjusting to the same density (109 CFU/mL). The isolated RNA was stored at −80°C until use. All RNA sequencing and transcriptome alignment procedures were carried out at the Center for Genomics and Bioinformatics, Indiana University, Bloomington, IN. The library construction was performed using TruSeq stranded mRNA library preparation kit (Illumina Baltimore, Baltimore, MD) and the sequencing was performed using Illumina NextSeq 75 cycles High Output kit (Illumina Baltimore, Baltimore, MD). Reads were adapter trimmed and quality filtered using Trimmomatic 0.38 with the cutoff threshold for average base quality score set at 20 over a window of three bases. Reads shorter than 20 bases post-trimming were excluded. Cleaned reads were mapped to the H. somni strain 2336 reference genome sequence retrieved from NCBI (NC_010519.1) using bowtie2 version 2.4.2. Counts of reads mapping to each of the annotated genes and intergenic regions divided into 100 bp bins were generated using the FeatureCounts tool of the Subread package.
To evaluate the divergence between H. somni strain 2336 and H. somni 2336Δhfq, PCA was carried out utilizing normalized differences, and subsequently visualized using the DEseq2 package. Differential gene expression between H. somni strain 2336 and H. somni 2336Δhfq was further examined through the generation of an MA plot, employing logarithmic ratio (M) and mean average (A) scales via the DEseq2 tool. In addition, a heatmap illustrating expression changes in genes associated with EPS production in H. somni strain 2336 and H. somni 2336Δhfq was generated using the DEseq2 package ver. 1.12.3. The mRNA sequences of the H. somni wild-type and hfq deletion mutant have been deposited into the NCBI database under accession no. PRJNA882837 .
Mouse challenge and determination of bacteremia
Three Swiss Webster mice (one male, two females) were inoculated IP with 5.5 × 106 CFU of H. somni wild-type strain 2336 in 0.5 mL of PBS containing 2% hog gastric mucin (HGM) (44). Six Swiss Webster mice (three males, three females) were inoculated IP with 1.6 × 107 CFU of H. somni 2336Δhfq, also in 0.5 mL of PBS containing 2% HGM. Three control mice (one male, two females) were inoculated IP with 0.5 mL of PBS containing 2% HGM only. All mice were 8 weeks old. All bacterial suspensions were estimated spectrophotometrically and the exact number confirmed by viable plate count. Ten microliters of blood was obtained from the tail vein of each mouse at 0 h, 5 h, and 29 h after challenge, and was diluted into PBS, inoculated onto CBA plates, and incubated overnight at 37°C to determine bacteremia levels per milliliter of blood in each mouse.
Calf challenge and clinical assessments
Conventionally reared male Holstein steer calves ranging from 106 to 148 days of age were blocked by age and randomly assigned to be challenged with H. somni strain 2336 (n = 3, average age 132 days, range 117–145) or H. somni 2336Δhfq (n = 4, average age also 132 days, range 106–148 days). Two calves that were slightly younger (88 and 127 days old) were assigned to receive mock challenge with sterile saline + 5% fetal calf serum alone to assess any effects due to the challenge procedure or medium only. Prior to enrollment, calves were confirmed to be free of persistent bovine viral diarrhea virus infection by antigen capture enzyme-linked immunosorbent assay (ELISA) of a skin biopsy. Serum samples collected from calves prior to challenge were negative for IgG to H. somni whole cells, as measured by ELISA (62).
At 22 days and at 8 days before challenge, immediately prior to challenge, and every 12 h after challenge until necropsy, calves were examined by one of two veterinarians to identify signs of respiratory disease. Signs evaluated included rectal temperature, heart rate, respiratory rate, attitude, appetite, and the presence or absence of nasal discharge, ocular discharge, enlarged lymph nodes, abnormal respiration pattern, and abnormal respiratory sounds, as determined by thoracic auscultation. A previously described scoring system (63) was used to calculate a clinical score based on all assessed signs at each time point for each calf.
H. somni strain 2336 and H. somni 2336Δhfq were grown on CBA plates at 37°C, in 5% CO2, for 24 h. For each calf to be challenged, all colonies were swabbed from a blood agar plate and transferred to 10 mL of pre-warmed CTT in a 50 mL tube. The tube was capped tightly and incubated for 18 h–24 h on a plate shaker at 37°C at 200 rpm. The 10 mL of broth culture was then transferred to 90 mL of pre-warmed CTT in a tightly capped bottle, which was then incubated for 3 h on a plate shaker at 37°C. Beginning at 3 h of incubation, the optical density (OD) of each culture was determined hourly at 540 nm. When the OD was between 0.219 and 0.258 (consistent with a concentration of 1.4—2.0 × 109 CFU/mL, as determined by viable plate count), the cultures were maintained on ice. The cells were pelleted at 10,000 × g for 10 min at 4°C, and the bacterial pellet was resuspended in 10 mL of sterile 0.9% NaCl containing 5% fetal calf serum. The total dose of H. somni strain 2336 administered to each calf was 9.5 × 109 CFU, and the total dose of H. somni 2336Δhfq administered to each calf was 3.0 × 109 CFU.
Calves were challenged with H. somni strain 2336 or H. somni 2336Δhfq by intrabronchial instillation. Calves were restrained in a cattle chute, 5 mL of 2% lidocaine was instilled into the nostril, then a bronchoalveolar lavage catheter (Large Animal Broncho-Alveolar Lavage Catheter, BAL240, MILA International Inc., Florence, KY) was advanced through the nostril, into the trachea, and advanced until the catheter wedged in a bronchus. The balloon on the catheter was inflated and the catheter was held into position while 10 mL of the bacterial inoculum was instilled, followed quickly by 60 mL of sterile 0.9% NaCl, and then 120 mL of air. The balloon on the catheter was deflated and the tube was quickly removed from the calf’s airway.
Prior to challenge and every 12 h after challenge until necropsy, a guarded NPS (E9-5200, Continental Plastic, Delavan, WI) was collected and submitted to the Mississippi State University Veterinary Diagnostic Laboratory (MSU VDC) for aerobic culture to identify endogenous H. somni, P. multocida, Mannheimia haemolytica, or other relevant bacterial species. At 24 h and 96 h after challenge, an ETA was also collected using a cold, sterilized 1-m endoscope passed through the nostril of the calf and into the trachea. A single-use sterile aspiration catheter was threaded through the biopsy port of the endoscope, 30 mL of sterile 0.9% NaCl was instilled into the trachea, then the fluid was immediately withdrawn, usually yielding 2 mL to 4 mL of tracheal exudate in the saline. The tracheal material in a sterile tube was immediately transferred to the MSU VDC for aerobic culture.
Postmortem evaluation of lung pathology
At 120 h after challenge, clinical signs were assessed and a nasopharyngeal swab was collected. The calves were euthanized with a combination of phenobarbital sodium (390 mg/mL) and phenytoin sodium (50 mg/mL) (Beuthanasia-D, Merck Animal Health, Millsboro, DE) at 87 mg/kg phenobarbital by rapid intravenous infusion. Lungs were scored for gross lesions, as previously described (63), by a board-certified veterinary pathologist (A.O.) who was unaware of the treatment each calf had received at the time of lesion scoring. Briefly, each lung lobe was assessed and the percent abnormal lung was estimated. A total gross pathology score was calculated based on the proportion of abnormal lung in each lobe, and the percentage of the total lung volume represented by each lobe. A section of lung with gross changes consistent with H. somni infection was submitted to MSU VDC for aerobic culture, and an adjacent section was fixed in formalin, then processed for microscopic evaluation.
Statistical analyses
For data from bacterial and mouse experiments, the unpaired t-test was used to calculate two-tailed P-values using Prism 7 software (GraphPad, Inc., La Jolla, CA). P-values of <0.05 were considered significant.
For data from calf experiments, the temperature, respiratory rate, and clinical score data were assessed for normality using the D'Agostino-Pearson omnibus normality test; clinical score data were square root transformed to achieve a normal distribution before analysis, and values for each of the three treatment groups were compared with two-way analysis of variance (ANOVA) with repeated measures. The proportions of calves in groups challenged with H. somni strain 2336 or H. somni 2336Δhfq that were positive for H. somni on nasopharyngeal swabs over time were compared by two-way ANOVA with repeated measures. The proportions of calves from which H. somni could be isolated from endotracheal aspirates on day 1 or day 4 post-challenge, and from lung tissue collected at necropsy on day 5 post-challenge, were compared between the H. somni strain 2336 and the H. somni 2336Δhfq groups by Fisher’s exact test. The percent total abnormal lung found at necropsy of each calf was compared between the wild-type and the H. somni 2336Δhfq groups by Mann-Whitney test. For all comparisons, significance was set at P ≤ 0.05. Analyses were conducted in Prism 10 [version 10.1.1 (270)].
ACKNOWLEDGMENTS
This work was supported by USDA-NIFA grant number 2017-67015-29560 to T.J.I., and by Long Island University College of Veterinary Medicine. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank James Q. Robinson at College of Veterinary Medicine, Long Island University, for obtaining bovine blood, and Ram Podicheti at the Center for Genomics and Bioinformatics, Indiana University, Bloomington, IN, for bioinformatics analysis of the RNA-Seq results.
Contributor Information
Thomas J. Inzana, Email: Thomas.Inzana@liu.edu.
Guy H. Palmer, Washington State University, Pullman, Washington, USA
ETHICS APPROVAL
Research involving mice was approved by the Long Island University Institutional Animal Care and Use Committee (protocol #2021–001). The research involving calves was approved by the Mississippi State University Institutional Animal Care and Use Committee (protocol #20-016).
DATA AVAILABILITY
The raw data of H. somni strain 2336 and H. somni 2336Δhfq were deposited into the National Center for Biotechnology Information (NCBI) (BioProject accession PRJNA954873).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00038-24.
Legends Fig. S1 to S7.
Construction of hfq mutation.
RT-PCR and Western blot of Hfq.
Replacement of hfq with chloramphenicol gene.
Sole mutation in Hfq mutant outside of Cm gene replacement.
Western blot of IbpA in hfq mutant.
Rectal temperatures of challenged calves.
Clinical scores of challenged calves.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Griffin D, Chengappa MM, Kuszak J, McVey DS. 2010. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 26:381–394. doi: 10.1016/j.cvfa.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 2. O’Toole D, Sondgeroth KS. 2016. Histophilosis as a natural disease. Curr Top Microbiol Immunol 396:15–48. doi: 10.1007/82_2015_5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inzana TJ. 2016. The many facets of lipooligosaccharide as a virulence factor for Histophilus somni. Curr Top Microbiol Immunol 396:131–148. doi: 10.1007/82_2015_5020 [DOI] [PubMed] [Google Scholar]
- 4. Petruzzi B, Inzana TJ. 2016. Exopolysaccharide production and biofilm formation by Histophilus somni. Curr Top Microbiol Immunol 396:149–160. doi: 10.1007/82_2015_5013 [DOI] [PubMed] [Google Scholar]
- 5. Corbeil LB. 2016. Histophilus somni surface proteins. Curr Top Microbiol Immunol 396:89–107. doi: 10.1007/82_2015_5011 [DOI] [PubMed] [Google Scholar]
- 6. Behling-Kelly E, Rivera-Rivas J, Czuprynski CJ. 2016. Interactions of Histophilus somni with host cells. Curr Top Microbiol Immunol 396:71–87. doi: 10.1007/82_2015_5010 [DOI] [PubMed] [Google Scholar]
- 7. Pan Y, Tagawa Y, Champion A, Sandal I, Inzana TJ. 2018. Histophilus somni survives in bovine macrophages by interfering with phagosome-lysosome fusion but requires IbpA for optimal serum resistance. Infect Immun 86:e00365-18. doi: 10.1128/IAI.00365-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corbeil LB. 2016. Host immune response to Histophilus somni, p 109–129. In Inzana TJ (ed), Curr top Microbiol Immunol, 2016/01/06 Ed. Vol. 396. Springer, Switzerland. [DOI] [PubMed] [Google Scholar]
- 9. Sandal I, Shao JQ, Annadata S, Apicella MA, Boye M, Jensen TK, Saunders GK, Inzana TJ. 2009. Histophilus somni biofilm formation in cardiopulmonary tissue of the bovine host following respiratory challenge. Microbes Infect 11:254–263. doi: 10.1016/j.micinf.2008.11.011 [DOI] [PubMed] [Google Scholar]
- 10. Pan Y, Siddaramappa S, Sandal I, Dickerman A, Bandara AB, Inzana TJ. 2021. The role of luxS in Histophilus somni virulence and biofilm formation. Infect Immun 89:e00567-20. doi: 10.1128/IAI.00567-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan Y, Subhadra B, Sandal I, Dickerman A, Inzana TJ. 2021. The role of uspE in virulence and biofilm formation by Histophilus somni. Vet Microbiol 263:109267. doi: 10.1016/j.vetmic.2021.109267 [DOI] [PubMed] [Google Scholar]
- 12. Dos Santos RF, Arraiano CM, Andrade JM. 2019. New molecular interactions broaden the functions of the RNA chaperone Hfq. Curr Genet 65:1313–1319. doi: 10.1007/s00294-019-00990-y [DOI] [PubMed] [Google Scholar]
- 13. Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. 2007. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol 189:1963–1973. doi: 10.1128/JB.01243-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cech GM, Szalewska-Pałasz A, Kubiak K, Malabirade A, Grange W, Arluison V, Węgrzyn G. 2016. The Escherichia coli Hfq protein: an unattended DNA-transactions regulator. Front Mol Biosci 3:36. doi: 10.3389/fmolb.2016.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. 2003. Identification of the Hfq-binding site on Dsra RNA: Hfq binds without altering DsrA secondary structure. RNA 9:33–43. doi: 10.1261/rna.2570803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev 14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell 9:11–22. doi: 10.1016/s1097-2765(01)00437-3 [DOI] [PubMed] [Google Scholar]
- 18. Papenfort K, Vogel J. 2010. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127. doi: 10.1016/j.chom.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 19. Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x [DOI] [PubMed] [Google Scholar]
- 20. Tsui HC, Leung HC, Winkler ME. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x [DOI] [PubMed] [Google Scholar]
- 21. Fantappiè L, Metruccio MME, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. 2009. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun 77:1842–1853. doi: 10.1128/IAI.01216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4:e1000163. doi: 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crispim JS, da Silva TF, Sanches NM, da Silva GC, Pereira MF, Rossi CC, Li Y, Terra VS, Vohra P, Wren BW, Langford PR, Bossé JT, Bazzolli DMS. 2020. Serovar-dependent differences in Hfq-regulated phenotypes in Actinobacillus pleuropneumoniae. Pathog Dis 78:ftaa066. doi: 10.1093/femspd/ftaa066 [DOI] [PubMed] [Google Scholar]
- 24. Gangaiah D, Labandeira-Rey M, Zhang X, Fortney KR, Ellinger S, Zwickl B, Baker B, Liu Y, Janowicz DM, Katz BP, Brautigam CA, Munson RS, Hansen EJ, Spinola SM. 2014. Haemophilus ducreyi Hfq contributes to virulence gene regulation as cells enter stationary phase. mBio 5:e01081-13. doi: 10.1128/mBio.01081-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hempel RJ, Morton DJ, Seale TW, Whitby PW, Stull TL. 2013. The role of the RNA chaperone Hfq in Haemophilus influenzae pathogenesis. BMC Microbiol 13:134. doi: 10.1186/1471-2180-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mégroz M, Kleifeld O, Wright A, Powell D, Harrison P, Adler B, Harper M, Boyce JD. 2016. The RNA-binding chaperone Hfq is an important global regulator of gene expression in Pasteurella multocida and plays a crucial role in production of a number of virulence factors, including hyaluronic acid capsule. Infect Immun 84:1361–1370. doi: 10.1128/IAI.00122-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subashchandrabose S, Leveque RM, Kirkwood RN, Kiupel M, Mulks MH. 2013. The RNA chaperone Hfq promotes fitness of Actinobacillus pleuropneumoniae during porcine pleuropneumonia. Infect Immun 81:2952–2961. doi: 10.1128/IAI.00392-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui M, Wang T, Xu J, Ke Y, Du X, Yuan X, Wang Z, Gong C, Zhuang Y, Lei S, Su X, Wang X, Huang L, Zhong Z, Peng G, Yuan J, Chen Z, Wang Y, Kwaik YA. 2013. Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PLoS ONE 8:e71933. doi: 10.1371/journal.pone.0071933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McNealy TL, Forsbach-Birk V, Shi C, Marre R. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol 187:1527–1532. doi: 10.1128/JB.187.4.1527-1532.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sandal I, Hong W, Swords WE, Inzana TJ. 2007. Characterization and comparison of biofilm development by pathogenic and commensal isolates of Histophilus somni. J Bacteriol 189:8179–8185. doi: 10.1128/JB.00479-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howard MD, Willis L, Wakarchuk W, St Michael F, Cox A, Horne WT, Hontecillas R, Bassaganya-Riera J, Lorenz E, Inzana TJ. 2011. Genetics and molecular specificity of sialylation of Histophilus somni lipooligosaccharide (LOS) and the effect of LOS sialylation on Toll-like receptor-4 signaling. Vet Microbiol 153:163–172. doi: 10.1016/j.vetmic.2011.02.054 [DOI] [PubMed] [Google Scholar]
- 33. Inzana TJ, Glindemann G, Cox AD, Wakarchuk W, Howard MD. 2002. Incorporation of N-acetylneuraminic acid into Haemophilus somnus lipooligosaccharide (LOS): enhancement of resistance to serum and reduction of LOS antibody binding. Infect Immun 70:4870–4879. doi: 10.1128/IAI.70.9.4870-4879.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elswaifi SF, St Michael F, Sreenivas A, Cox A, Carman GM, Inzana TJ. 2009. Molecular characterization of phosphorylcholine expression on the lipooligosaccharide of Histophilus somni. Microb Pathog 47:223–230. doi: 10.1016/j.micpath.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrison A, Santana EA, Szelestey BR, Newsom DE, White P, Mason KM. 2013. Ferric uptake regulator and its role in the pathogenesis of nontypeable Haemophilus influenzae. Infect Immun 81:1221–1233. doi: 10.1128/IAI.01227-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones PA, Samuels NM, Phillips NJ, Munson RS, Bozue JA, Arseneau JA, Nichols WA, Zaleski A, Gibson BW, Apicella MA. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J Biol Chem 277:14598–14611. doi: 10.1074/jbc.M110986200 [DOI] [PubMed] [Google Scholar]
- 37. McQuiston JH, McQuiston JR, Cox AD, Wu Y, Boyle SM, Inzana TJ. 2000. Characterization of a DNA region containing 5'-(CAAT)(n)-3' DNA sequences involved in lipooligosaccharide biosynthesis in Haemophilus somnus. Microb Pathog 28:301–312. doi: 10.1006/mpat.1999.0351 [DOI] [PubMed] [Google Scholar]
- 38. Pogoutse AK, Moraes TF. 2020. Transferrin binding protein B and transferrin binding protein A2 expand the transferrin recognition range of Histohilus somni. J. Bacteriol 202:e00177–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips NJ, Miller TJ, Engstrom JJ, Melaugh W, McLaughlin R, Apicella MA, Gibso BW. 2000. Characterization of chimeric lipopolysaccharides from Escherichia coli strain Jm109 transformed with lipooligosaccharide synthesis genes (Lsg) from haemophilus influenzae. J Biol Chem 275:4747–4758. [DOI] [PubMed] [Google Scholar]
- 40. Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE. 2009. The fic domain: regulation of cell signaling by adenylylation. Mol Cell 34:93–103. doi: 10.1016/j.molcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yarnall M, Widders PR, Corbeil LB. 1988. Isolation and characterization of Fc receptors from Haemophilus somnus. Scand J Immunol 28:129–137. doi: 10.1111/j.1365-3083.1988.tb02424.x [DOI] [PubMed] [Google Scholar]
- 42. Corbeil LB, Blau K, Prieur DJ, Ward ACS. 1985. Serum susceptibility of Haemophilus somnus from bovine clinical cases and carriers. J Clin Microbiol 22:192–198. doi: 10.1128/jcm.22.2.192-198.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Inzana TJ, Balyan R, Howard MD. 2012. Decoration of Histophilus somni lipooligosaccharide with N-acetyl-5-neuraminic acid enhances bacterial binding of complement factor H and resistance to killing by serum and polymorphonuclear leukocytes. Vet Microbiol 161:113–121. doi: 10.1016/j.vetmic.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 44. Wu Y, McQuiston JH, Cox A, Pack TD, Inzana TJ. 2000. Molecular cloning and mutagenesis of a DNA locus involved in lipooligosaccharide biosynthesis in Haemophilus somnus. Infect Immun 68:310–319. doi: 10.1128/IAI.68.1.310-319.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. St Michael F, Li J, Howard MD, Duncan AJ, Inzana TJ, Cox AD. 2005. Structural analysis of the oligosaccharide of Histophilus somni (Haemophilus somnus) strain 2336 and identification of several lipooligosaccharide biosynthesis gene homologues. Carbohydr Res 340:665–672. doi: 10.1016/j.carres.2004.12.029 [DOI] [PubMed] [Google Scholar]
- 46. Mégroz M, Kleifeld O, Wright A, Powell D, Harrison P, Adler B, Harper M, Boyce JD. 2016. The RNA-binding chaperone Hfq is an important global regulator of gene expression in Pasteurella multocida and plays a crucial role in production of a number of virulence factors, including hyaluronic acid capsule. Infect Immun 84:1361–1370. doi: 10.1128/IAI.00122-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siddaramappa S, Duncan AJ, Brettin T, Inzana TJ. 2006. Comparative analyses of two cryptic plasmids from Haemophilus somnus (Histophilus somni). Plasmid 55:227–234. doi: 10.1016/j.plasmid.2005.11.004 [DOI] [PubMed] [Google Scholar]
- 48. Park S, Prévost K, Heideman EM, Carrier M-C, Azam MS, Reyer MA, Liu W, Massé E, Fei J. 2021. Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. eLife 10:e64207. doi: 10.7554/eLife.64207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Teng Y, Geng J, Long J, Yang H, Duan G, Chen S. 2023. Involvement of RNA chaperone Hfq in the regulation of antibiotic resistance and virulence in Shigella sonnei. Res Microbiol 174:104047. doi: 10.1016/j.resmic.2023.104047 [DOI] [PubMed] [Google Scholar]
- 50. Sandal I, Inzana TJ, Molinaro A, De Castro C, Shao JQ, Apicella MA, Cox AD, St Michael F, Berg G. 2011. Identification, structure, and characterization of an exopolysaccharide produced by Histophilus somni during biofilm formation. BMC Microbiol 11:186. doi: 10.1186/1471-2180-11-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geertsema RS, Kimball RA, Corbeil LB. 2007. Bovine plasma proteins increase virulence of Haemophilus somnus in mice. Microb Pathog 42:22–28. doi: 10.1016/j.micpath.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 52. Inzana TJ, Todd J. 1992. Immune response of cattle to an Haemophilus somnus lipid A-protein conjugate vaccine and efficacy in a mouse model. Am J Vet Res 53:175–179. [PubMed] [Google Scholar]
- 53. Elswaifi SF, Scarratt WK, Inzana TJ. 2012. The role of lipooligosaccharide phosphorylcholine in colonization and pathogenesis of Histophilus somni in cattle. Vet Res 43:49. doi: 10.1186/1297-9716-43-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Green MR, Sambrook J. 2012. Molecular cloning: A laboratory manual. 4th ed. Vol. 1, 2, and 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 55. Thurnheer T, Gmur R, Guggenheim B. 2004. Multiplex FISH analysis of a six-species bacterial Biofilm. J. Microbiol Methods 56:37–47. [DOI] [PubMed] [Google Scholar]
- 56. Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology (Reading) 146 ( Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395 [DOI] [PubMed] [Google Scholar]
- 57. ROE JH. 1954. The determination of dextran in blood and urine with anthrone reagent. J Biol Chem 208:889–896. [PubMed] [Google Scholar]
- 58. Inzana TJ. 1983. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis 148:492–499. doi: 10.1093/infdis/148.3.492 [DOI] [PubMed] [Google Scholar]
- 59. Barenkamp SJ, Munson RS, Granoff DM. 1981. Comparison of outer-membrane protein subtypes and biotypes of isolates of Haemophilus influenzae type B. J Infect Dis 144:480. doi: 10.1093/infdis/144.5.480 [DOI] [PubMed] [Google Scholar]
- 60. Subhadra B, Cao D, Jensen R, Caswell C, Inzana TJ. 2023. Identification and initial characterization of Hfq-associated sRNAs in Histophilus somni strain 2336. PLoS One 18:e0286158. doi: 10.1371/journal.pone.0286158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Inzana TJ, Anderson P. 1985. Serum factor-dependent resistance of Haemophilus influenzae type B to antibody to lipopolysaccharide. J Infect Dis 151:869–877. doi: 10.1093/infdis/151.5.869 [DOI] [PubMed] [Google Scholar]
- 62. Howard MD, Cox AD, Weiser JN, Schurig GG, Inzana TJ. 2000. Antigenic diversity of Haemophilus somnus lipooligosaccharide: phase-variable accessibility of the phosphorylcholine epitope. J Clin Microbiol 38:4412–4419. doi: 10.1128/JCM.38.12.4412-4419.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang X, Outlaw C, Olivier AK, Woolums A, Epperson W, Wan XF. 2019. Pathogenesis of co-infections of influenza D virus and Mannheimia haemolytica in cattle. Vet Microbiol 231:246–253. doi: 10.1016/j.vetmic.2019.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends Fig. S1 to S7.
Construction of hfq mutation.
RT-PCR and Western blot of Hfq.
Replacement of hfq with chloramphenicol gene.
Sole mutation in Hfq mutant outside of Cm gene replacement.
Western blot of IbpA in hfq mutant.
Rectal temperatures of challenged calves.
Clinical scores of challenged calves.
Data Availability Statement
The raw data of H. somni strain 2336 and H. somni 2336Δhfq were deposited into the National Center for Biotechnology Information (NCBI) (BioProject accession PRJNA954873).