Abstract
Secretory proteins in eukaryotic cells are transported to the cell surface via the endoplasmic reticulum (ER) and the Golgi apparatus by membrane-bounded vesicles. We screened a collection of temperature-sensitive mutants of Saccharomyces cerevisiae for defects in ER-to-Golgi transport. Two of the genes identified in this screen were PRP2, which encodes a known pre-mRNA splicing factor, and RSE1, a novel gene that we show to be important for pre-mRNA splicing. Both prp2-13 and rse1-1 mutants accumulate the ER forms of invertase and the vacuolar protease CPY at restrictive temperature. The secretion defect in each mutant can be suppressed by increasing the amount of SAR1, which encodes a small GTPase essential for COPII vesicle formation from the ER, or by deleting the intron from the SAR1 gene. These data indicate that a failure to splice SAR1 pre-mRNA is the specific cause of the secretion defects in prp2-13 and rse1-1. Moreover, these data imply that Sar1p is a limiting component of the ER-to-Golgi transport machinery and suggest a way that secretory pathway function might be coordinated with the amount of gene expression in a cell.
The isolation of conditional mutants has greatly facilitated study of the secretory pathway in the yeast Saccharomyces cerevisiae. Operationally, the defining characteristic of secretory pathway mutations is that they allow protein synthesis to continue but block the export of newly synthesized secretory proteins at some stage during their transit from the ER to the plasma membrane. The first secretion mutants were isolated from mutagenized cells that had been enriched for high density, a property of cells that continue to make protein and RNA but cannot increase their surface areas. Temperature-sensitive sec mutants were then identified by detection of their inability to deliver invertase, acid phosphatase, and sulfate permease to the cell surface. This screen yielded a set of mutants that were defective at different steps along the secretory pathway and remarkably specific for functions involved in vesicle trafficking (29, 30). Secretion mutants have also been found by a selection for mutants that fail to incorporate high levels of mannose into N-linked carbohydrate chains in the Golgi apparatus by a [3H]mannose suicide technique (25) and by a colony immunoblot screen for mutants that fail to carry out Golgi processing of pro-α-factor (42). Many of the genes isolated in these screens were represented by only a single allele, indicating that additional SEC genes may have been overlooked.
We have used a brute-force approach to find new mutants with ER-to-Golgi transport defects by screening a large collection of random temperature-sensitive mutants for the conditional accumulation of the ER forms of both invertase and the vacuolar protease CPY. Surprisingly, among the new secretion mutants isolated in this screen were mutants primarily defective in pre-mRNA processing. These have mutations in PRP2, a known pre-mRNA splicing factor, or in a novel gene, RSE1 (named for RNA splicing and ER-to-Golgi transport), which we show here to have a role in pre-mRNA processing. We show that the secretion defects of prp2-13 and rse1-1 mutants appear to result from the decreased activity of SAR1, an intron-containing gene that encodes a small GTPase required for vesicle formation from the ER (24). The secretion defects in these mutants can be suppressed by either increasing the amount of SAR1 or deleting the intron from the chromosomal SAR1 locus. Thus, pre-mRNA splicing mutants exert their effects on the secretory pathway by perturbing Sar1p synthesis.
MATERIALS AND METHODS
Strains, media, and plasmids.
S. cerevisiae strains used in this study are listed in Table 1. Preparation of rich medium (YEP) and minimal medium (SD), standard genetic manipulations, and yeast transformations were performed as described (1).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| CKY294 | MATa ura3-52 leu2-3,112 | 6 |
| CKY39 | MATα sec12-4 ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY566 | MATa SAR1-Δi ura3-52 leu2-3,112 | This study |
| CKY567 | MATα rse1-1 ura3-52 leu2-3,112 | This study |
| CKY568 | MATa rse1-1 ura3-52 leu2-3,112 | This study |
| CKY569 | MATα rse1-1 SAR1-Δi ura3-52 leu2-3,112 | This study |
| CKY570 | MATα prp2-13 ura3-52 | This study |
| CKY571 | MATa prp2-13 ura3-52 leu2-3,112 | This study |
| CKY572 | MATa prp2-13 SAR1-Δi ura3-52 leu2-3,112 | This study |
| 368 | MATa prp2-1 ade1 ade2 ura1 his7 tyr1 gal1 | YGSCa (a) |
| 125 | MATa prp3-1 ade1 ade2 ura1 his7 tyr1 gal1 | YGSC (a) |
| 108 | MATa prp5-1 ade1 ade2 ura1 his7 tyr1 gal1 | YGSC (a) |
| 382 | MATa prp11-1 ade1 ade2 ura1 his7 tyr1 gal1 | YGSC (a) |
| YAK22 | MATa ura3 leu2 trp1 ade2 ΔCUP1 ΔU1:HIS3 [TRP CEN U1-C8U] | A. Kistler (UCSF) |
YGSC, Yeast Genetic Stock Center.
pRH262 carries SAR1 (the A364A allele; see Discussion) in pRS316. pAF52 is a pCT3 library plasmid containing PRP2. pAF56 contains PRP2 within an EcoRI fragment in pRS306. For the linkage analysis of PRP2, pAF56 was linearized with SnaBI and transformed into wild type (CKY294). When integrants were crossed to CKY570, the temperature sensitivity and uracil auxotrophy were completely linked in 16 tetrads each from crosses with two different transformants. A 1.8-kb ClaI fragment adjacent to RSE1 from the original rescuing YCp50 library clone, pEC2, was inserted into pRS306 to generate pEC12. To test linkage of the cloned gene to the RSE1 locus, pEC12 was linearized with SnaBI and transformed into CKY567. When integrants were crossed to wild type, the temperature sensitivity was linked to uracil prototrophy in 15 of the 16 tetrads dissected (8 each from two different crosses).
To construct an allele of SAR1 without an intron (SAR1-Δi), a cDNA copy of the SAR1 gene was digested with BamHI and ClaI and ligated into BamHI- and ClaI-digested pRH262, creating pEC23. The insert in pEC23 was sequenced to verify the presence of a wild-type SAR1 gene with the intron precisely deleted. pEC23 was digested with HindIII and KpnI, and the fragment containing the entire intronless SAR1 gene was ligated into a pRS306 derivative with the polylinker from the EcoRI site to the NotI site deleted, forming pEC24. pEC24 was cut with BamHI and integrated into the chromosome of CKY294, and the chromosomal SAR1 allele in CKY294 was replaced with SAR1-Δi using two-step gene replacement (1). Clones were screened for a chromosomal copy of SAR1-Δi by comparing the sizes of PCR fragments from the SAR1 gene (intronless SAR1 is 139 bases shorter than genomic SAR1) and by testing for the absence of the BglII site internal to the SAR1 intron. Once the wild-type strain containing intronless SAR1 (CKY566) was generated, it was crossed to both the rse1-1 (CKY567) and prp2-13 (CKY570) strains to generate the sister spores rse1-1 (CKY568) and rse1-1 SAR1-Δi (CKY569) and the sister spores prp2-13 (CKY571) and prp2-13 SAR1-Δi (CKY572).
Protein extracts and immunoblotting.
Cultures were grown to exponential phase at 24°C in YEP plus 2% glucose (YPD) or in SC medium lacking the appropriate auxotrophic supplements plus 2% glucose. Cells (4 × 107) were collected by centrifugation, suspended at 2 × 107 cells/ml in YEP plus 0.1% glucose to induce invertase, and shifted to restrictive temperature for 2.5 h. Protein extracts were prepared by boiling in sample buffer (80 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 1.5% dithiothreitol, 10% glycerol, 0.1% bromophenol blue), lysing by agitation with glass beads, and diluting to a total volume of 0.1 ml with sample buffer. For Sar1p Western blotting, 4 × 107 cells grown to exponential phase at 24°C were either harvested (at t = 0) or collected by centrifugation, suspended in YPD at concentrations of 5 × 106 to 8 × 106 cells/ml, and incubated at restrictive temperature. After 2 or 4 h at restrictive temperature, 4 × 107 cells were harvested to make protein extracts as described above.
Protein extracts (15 μl) were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose filters. For Western blot detection, the following antibodies were used: rabbit anti-invertase at 1:1,000 dilution, rabbit anti-CPY antibody at 1:7,000 dilution, rabbit anti-Sar1p antibody (gift of A. Nakano) at 1:500 dilution, and horseradish peroxidase-coupled donkey anti-rabbit immunoglobulin G (Amersham) at 1:10,000 dilution. Western blots were developed using chemiluminescence (ECL kit; Amersham).
Northern blotting.
Cultures (25 ml) were grown at 24°C in YPD to exponential phase and were either used immediately (at t = 0) or diluted with 25 ml of pre-warmed YPD at 48°C and incubated in a 36°C bath for 3 h. Cells were harvested by centrifugation in prechilled tubes, and total RNA was extracted by agitation with glass beads (31, 35).
For Northern blots, RNA (∼20 μg) was electrophoresed on 2% agarose-formaldehyde gels (34) and transferred to Hybond N nylon membrane with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). 32P-labeled gene-specific DNA probes were synthesized by random priming (Multiprime DNA labeling; Amersham) of a ClaI-EcoRI fragment from SAR1. Hybridization was done using the method of Church and Gilbert (3).
β-Galactosidase assays.
Cells were grown to exponential phase in the appropriate SC media plus 2% raffinose overnight at 24°C. Cells were shifted to a 37°C water bath for 15 min before galactose was added to 2%. Cells were further incubated at 37°C for 1.5 h before 0.8-ml aliquots were removed and β-galactosidase activity was assayed in cells permeabilized with SDS and chloroform (1). Background readings obtained with untransformed cells were subtracted, and β-galactosidase activity was expressed in Miller units (21): 1,000 × OD420/min of reaction per 1 ml of culture at OD600 of 1. In at least two assays, three independent transformants were tested in parallel, and the results were averaged for each data point.
Invertase assays.
Cultures grown to exponential phase in YPD at 24°C were collected by centrifugation, suspended in YEP plus 0.1% glucose at 2 × 107 cells/ml, and aerated at restrictive temperature for 2 h to induce invertase. Cells were washed with a mixture of 50 mM Tris-HCl (pH 7.5) and 10 mM NaN3, washed with distilled water, incubated in 0.3 ml of 100 mM Tris-SO4 (pH 9.4)–50 mM β-mercaptoethanol for 10 min, washed with spheroplasting buffer (1.2 M sorbitol, 10 mM Tris-HCl [pH 7.5]), and converted to spheroplasts by incubation for 30 min at 30°C in 60 μl of spheroplasting buffer containing 60 U of recombinant lyticase. Efficient spheroplasting was judged to be present when there was >85% lysis upon dilution with 1% Triton X-100. Centrifugation at 500 × g for 5 min yielded a supernatant fraction containing extracellular invertase and a spheroplast pellet. The spheroplast pellet was washed once more with spheroplasting buffer. Then, both the supernatant fraction and the spheroplast pellet were diluted to yield a final volume of 1 ml in a mixture of 10 mM Tris-HCl (pH 7.5) and 1% Triton X-100. Invertase activity was assayed in 5 μl of each sample (7).
RESULTS
Splicing mutants are defective in secretion.
To identify new secretion mutants, we screened a collection of 1,200 temperature-sensitive mutants (10) by Western blotting for the intracellular accumulation of ER forms of invertase and CPY. After backcrossing until the secretion defect and temperature sensitivity cosegregated and complementation testing against our collection of known sec mutants, temperature-sensitive mutations in several new genes required for ER-to-Golgi transport were identified. Two of the new mutants are described in this report.
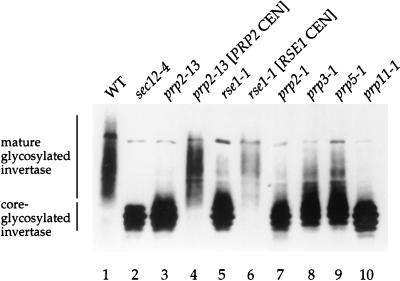
Strain CKY570 failed to grow at temperatures above 30°C. In this strain, invertase accumulated in the core-glycosylated form (Fig. 1, lane 3), and CPY accumulated in the ER form at temperatures above 30°C (data not shown). To identify the affected gene, CKY570 was transformed with a genomic library in the vector pCT3 (38) and screened for rescue of temperature sensitivity. From among 40,000 transformants, 8 rescuing clones representing two different library plasmids were isolated. Both plasmids contained the same 2.2-kb genomic segment which rescued the temperature sensitivity and secretion defect of CKY570 (Fig. 1, lane 4). PRP2 was shown to be the complementing gene in this genomic segment by the following two criteria. In a complementation test in which CKY570 was mated to a known prp2-1 mutant (9), the resulting diploid strain was temperature sensitive. In a linkage test, we found complete linkage between the temperature sensitive mutation and a URA3 marker integrated at the PRP2 locus. Since 12 other prp2 alleles have been reported (9, 39), we named this allele prp2-13.
FIG. 1.
Splicing mutants accumulate the ER form of invertase at restrictive temperature. Wild-type strain (CKY294) (WT) and sec12-4 (CKY39), prp2-13 (CKY570), prp2-13 [PRP2 CEN], rse1-1 (CKY567), rse1-1 [RSE1 CEN], prp2-1, prp3-1, prp5-1, and prp11-1 mutants were shifted to 37°C for 2.5 h in YEP plus 0.1% glucose to induce invertase. Invertase was detected in protein extracts by SDS-PAGE and immunoblotting with anti-invertase antibody. Mature glycosylated invertase migrates heterogeneously at ∼140 kDa. Core-glycosylated invertase, the ER form, migrates at ∼90 kDa.
PRP2 encodes an RNA-dependent ATPase that interacts directly with pre-mRNA before the first cleavage-ligation reaction of splicing (14, 19, 37). Given the known function of Prp2p as a nuclear splicing factor, it seemed unlikely that PRP2 plays a direct role in ER-to-Golgi transport, and we asked whether a general connection between pre-mRNA splicing and protein secretion might exist. To test whether other pre-mRNA processing mutants have secretion defects, we examined invertase maturation in several other prp mutants (9). From the Yeast Genetic Stock Center, we obtained 4 of the 10 original prp mutants (previously called rna mutants), prp2-1, prp3-1, prp5-1, and prp11-1, which are defective in different steps of spliceosome formation (33). All four mutants accumulated the ER form of invertase at 37°C, indicating that each had a defect in ER-to-Golgi transport (Fig. 1, lanes 7 to 10). Thus, defects in protein secretion may be a general feature of pre-mRNA processing mutants.
RSE1 is involved in pre-mRNA splicing.
A second gene identified in our screen was a novel gene we named RSE1 (for RNA splicing and ER-to-Golgi transport). rse1-1 mutants grew poorly at temperatures above 30°C and accumulated core-glycosylated invertase (Fig. 1, lane 5) and p1 CPY (data not shown) at 37°C, indicating an ER-to-Golgi secretion defect. To identify the affected gene, 15,000 transformants of the YCp50 library (32) were screened, and three complementing clones were isolated. Two of the clones were identical, and the sequence of the third clone overlapped those of the first two clones by about 7 kb. By subcloning, YML049c, a 4.1-kb open reading frame (ORF), was identified as the complementing gene which rescued the temperature sensitivity and secretion defect of the rse1-1 strain (Fig. 1, lane 6). We confirmed that YML049c is RSE1 by demonstrating tight linkage between rse1-1 and a URA3 marker integrated adjacent to the YML049c locus.
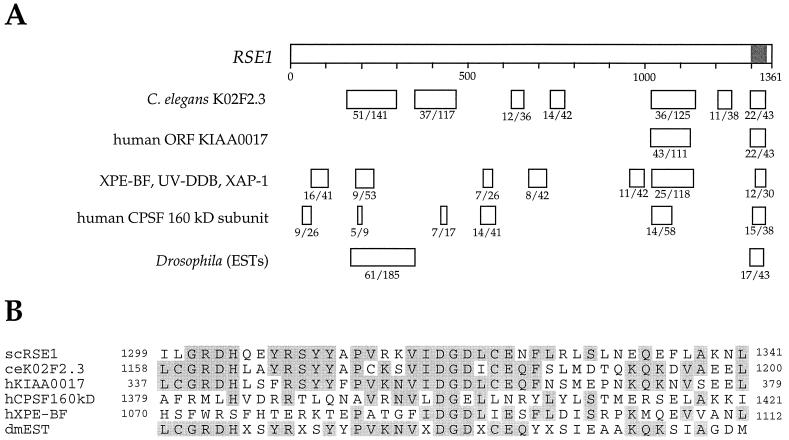
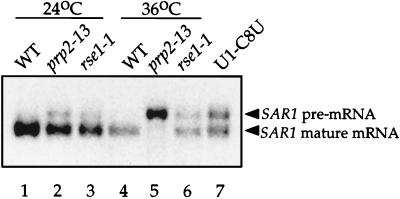
As shown in Fig. 2A, RSE1 has regions of homology to a Caenorhabditis elegans ORF (GenBank accession number 2804455), a human ORF, the genes for putative human and monkey DNA repair proteins (XPE-BF, UV-DDB, and XAP-1), and the genes encoding the 160-kDa subunits of human and bovine cleavage and polyadenylation specificity factor (CPSF), which is involved in 3′ processing of pre-mRNA (11, 12, 16, 23, 28, 36). Rse1p has no known protein motifs, but the most highly conserved region of Rse1p, 43 amino acids located near the C terminus, may represent a novel motif. An alignment of this region with the corresponding regions from the other sequences is shown in Fig. 2B. The sequence relationships shown in Fig. 2 suggest a function for Rse1p in DNA repair or pre-mRNA processing. The rse1-1 mutant appeared to have a normal capacity for DNA repair since it exhibited sensitivity to UV light identical to that of an isogenic wild-type strain (data not shown). To test whether rse1-1 was deficient in mRNA processing, we examined the processing of SAR1 mRNA in an rse1-1 mutant at 24 and 36°C. The SAR1 gene, which contains a single short intron, encodes a small GTP-binding protein required for COPII vesicle formation from the ER (24). As shown in Fig. 3, lane 6, the rse1-1 mutant accumulated intron-bearing SAR1 pre-mRNA after 3 h at 36°C. The defect in SAR1 pre-mRNA splicing in the rse1-1 mutant is similar in strength to the defect seen in a U1-C8U snRNA mutant strain, which was included as a control. The prp2-13 mutant also has a defect in SAR1 pre-mRNA splicing which is stronger than the defect seen in either of the other two mutants.
FIG. 2.
RSE1 contains conserved domains related to mRNA processing and DNA repair proteins. (A) Under each box indicating a region of homology to the RSE1 gene, the number of identical amino acids and the total number of amino acids in the boxed region are given. The amino acids in the shaded area of the RSE1 gene are shown in the alignment below. The C. elegans and mammalian homologies were identified by using BLAST. The Drosophila sequences (GenBank accession nos. AA44069, AA393017, AA263279, and AA142215) were identified in a BLAST search against the NCBI database of expressed sequences tags. (B) At the C terminus of the sequence for the RSE1 gene is a highly conserved domain that may represent a novel motif. The alignment of the amino acids at positions 1299 to 1341 for the RSE1 gene with corresponding sequences for S. cerevisiae (sc), C. elegans (ce), human (h), and Drosophila melanogaster (dm) is shown. Residues that are identical in three or more of the sequences are highlighted. The sequence labelled “dmEST” is a predicted translation from GenBank accession no. AA142215.
FIG. 3.
Intron-bearing SAR1 pre-mRNA accumulates in rse1-1 and prp2-13 mutants. Wild-type strain (CKY294) (WT) and prp2-13 (CKY570) and rse1-1 (CKY567) strains were grown overnight at 24°C (lanes 1 to 3) or shifted to 36°C for 3 h (lanes 4 to 6). Lane 7, a U1-C8U mutant strain (YAK22) grown at 30°C provides a positive control for a SAR1 pre-mRNA splicing defect. Total RNA was prepared, and ∼20 μg was loaded per lane on a 2% agarose-formaldehyde gel. RNA was transferred to a nylon membrane with 10× SSC, and the blot was probed with a 32P-labelled ClaI-EcoRI fragment of the SAR1 gene.
To test whether rse1-1 had more general effects on splicing, we examined its effects on two other introns in a splicing reporter assay. The splicing reporters contain lacZ interrupted by either the ribosomal protein 51a (RP51a) intron or Acc, an artificial intron with an intrinsically low splicing efficiency derived from the RP51a intron (17, 18). Since active β-galactosidase is expressed only when the intron has been spliced correctly, the efficiency of splicing for a given strain can be evaluated by comparing β-galactosidase activity from an intronless construct to the activities from constructs with introns. As shown in Table 2, the rse1-1 mutant at restrictive temperature was about three times less efficient at splicing the RP51a intron, and about six times less efficient at splicing the Acc intron, than an isogenic wild-type strain. In agreement with the results of the Northern analysis of SAR1 splicing, the splicing reporter assay showed that the rse1-1 mutant had a substantial splicing defect, although it was not as strong as the defect in the prp2-13 mutant at restrictive temperature. Thus, RSE1 appears to have a general role in pre-mRNA splicing.
TABLE 2.
Splicing defects in rse1-1 and prp2-13 strains in a reporter assay
| Host strain | % β-galactosidase activitya of:
|
||
|---|---|---|---|
| Construct with no intron | RP51 intron | Acc intron | |
| Wild type | 100 | 37.3 ± 2.9 | 10.6 ± 0.6 |
| rse1-1 | 100 | 13.1 ± 2.8 | 1.7 ± 0.2 |
| prp2-13 | 100 | 0 ± 0 | 0 ± 0 |
β-Galactosidase activity is expressed as the percentage of activity from the intronless β-galactosidase construct under the same conditions. The means and standard deviations for at least two assays of each of three different transformants are given. The absolute values of β-galactosidase activity for the intronless β-galactosidase transformants in a typical experiment were as follows: wild-type, 640 units; rse1-1, 830 units; and prp2-13, 1,000 units. One β-galactosidase unit is defined as 1,000 × OD420/min of reaction for 1 ml of culture at OD600 of 1.
Pre-mRNA splicing and 3′ end formation have been shown to be coupled processes in mammalian cells (8, 20, 26, 27, 40), and the homology of RSE1 to the genes encoding the 160-kDa subunits of human and bovine CPSF suggested that the primary defect in rse1-1 mutants could be in 3′ end formation. CUP1 mRNA has previously been shown to be a particularly sensitive reporter for 3′ processing defects, since unprocessed CUP1 transcripts accumulate quickly and migrate much more slowly in gels than processed transcripts (4). On a Northern blot, the mobility of CUP1 mRNA was not affected by rse1-1 (data not shown). Given this result, it seems unlikely that RSE1 is directly involved in 3′ end processing. However, we cannot exclude the possibility that 3′ end formation of genes other than CUP1 is affected in rse1-1 mutants.
SAR1 is limiting for secretion in prp2-13 and rse1-1.
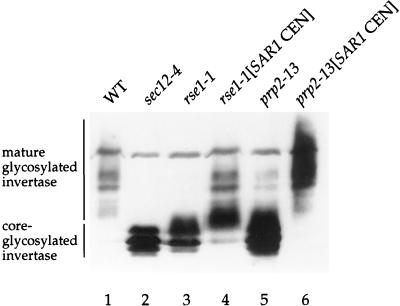
When we screened plasmids for rescue of prp2-13 and rse1-1, we found that the SAR1 gene on a centromere plasmid could partially rescue the temperature sensitivity of both mutants. The extra copies of SAR1 increased the threshold temperature for growth of both strains from 30°C to 33°C (data not shown). To evaluate the effect of SAR1 overexpression on the secretion defect in splicing mutants, we assayed invertase maturation in prp2-13 and rse1-1 strains containing SAR1 on a centromere-containing plasmid (pRH262). As shown in Fig. 4 (lanes 4 and 6) and Table 3, the increased amount of the SAR1 gene restores invertase secretion in both mutants, allowing the mature glycosylated form to be produced and secreted at 30°C for the prp2-13 strain and at 37°C for the rse1-1 strain. The transport of CPY to the vacuole in both strains was also restored by inclusion of an extra copy of SAR1 (data not shown).
FIG. 4.
Increased SAR1 dosage suppresses the invertase secretion defect in rse1-1 and prp2-13 strains. Wild-type (CKY294) (WT), sec12-4 (CKY39), and rse1-1 (CKY567) strains were incubated at 37°C for 2.5 h in YEP plus 0.1% glucose to induce invertase; the prp2-13 (CKY570) strain was shifted to 30°C for 2.5 h in YEP plus 0.1% glucose. Invertase was detected in protein extracts by SDS-PAGE and immunoblotting with anti-invertase antibody.
TABLE 3.
Restoration of invertase secretion in rse1-1 and prp2-13 strains by overexpression of SAR1a
| Strain | % Invertase secreted | Total invertase activity (units/OD600)b |
|---|---|---|
| Wild-type | 87 | 0.26 ± 0.03 |
| sec12-4 | 5 | 1.31 ± 0.07 |
| rse1-1 | 50 | 0.50 ± 0.06 |
| rse1-1 [SAR1 CEN] | 80 | 0.33 ± 0.02 |
| prp2-13 | 42 | 1.40 ± 0.15 |
| prp2-13 [SAR1 CEN] | 83 | 0.66 ± 0.04 |
To induce invertase, wild-type (CKY294), sec12-4 (CKY39), and rse1-1 (CKY567) strains were shifted to 37°C for 2 h in YEP plus 0.1% glucose and the prp2-13 (CKY570) strain was shifted to 30°C for 2 h in YEP plus 0.1% glucose.
The means and standard deviations from four assays (two each from two independent cultures) are given. One unit of invertase activity releases 1 μmol of glucose from sucrose at 30°C.
The finding that extra copies of SAR1 could partially suppress the temperature sensitivity of splicing mutants suggests that Sar1p is the limiting component for secretion when mRNA processing is blocked. To test this idea, we removed the intron from the SAR1 gene to bypass the dependence of Sar1p synthesis on pre-mRNA splicing. SAR1 in our wild-type strain (CKY294) was replaced with the intronless SAR1 allele (SAR1-Δi) by two-step gene replacement, forming CKY566. prp2-13 SAR1-Δi (CKY572) and rse1-1 SAR1-Δi (CKY569) double mutants were then generated by crossing either CKY570 or CKY567 to CKY566.
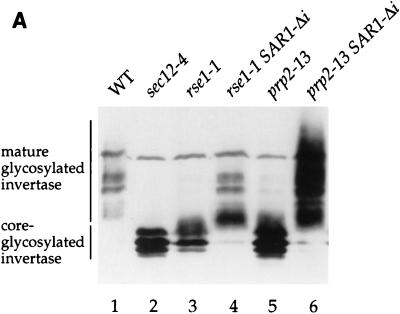
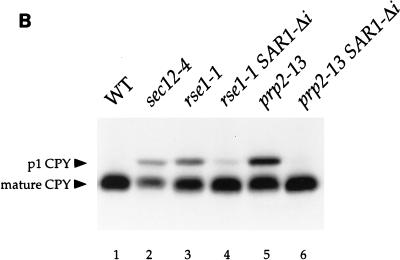
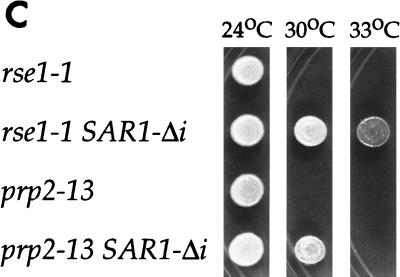
As expected, the intronless allele of SAR1 suppressed the ER-to-Golgi transport defects of both mutants. Mature glycosylated invertase was secreted in prp2-13 SAR1-Δi at 30°C and in rse1-1 SAR1-Δi at 37°C (Fig. 5A, lanes 4 and 6, and Table 4). CPY was processed at temperatures that would be restrictive for the single mutants (Fig. 5B, lanes 4 and 6). Also, the substitution of SAR1-Δi for intron-containing SAR1 partially rescued the temperature sensitivities of the prp2-13 and rse1-1 strains; the threshold temperature for growth of the prp2-13 strain was increased from 30 to 33°C, and the threshold temperature of the rse1-1 strain was increased from 30 to 37°C (Fig. 5C). Thus, it appears that the secretion defects in prp2-13 and rse1-1 mutants resulted directly from the failure of these strains to process pre-mRNA from SAR1. Although delivery of invertase to the cell surface was restored in the rse1-1 SAR1-Δi mutant (Table 4), the invertase produced by this strain (Fig. 5A, lane 4), as well as by the rse1-1 [SAR1 CEN] strain (Fig. 4, lane 4), appeared to have less outer-chain glycosylation than invertase expressed in wild-type cells. This residual glycosylation defect suggested that the rse1-1 mutation alters the function of the Golgi in a way that is not corrected by uncoupling SAR1 expression from pre-mRNA splicing.
FIG. 5.
An intronless SAR1 allele suppresses the secretion defects of rse1-1 and prp2-13 strains. (A and B) rse1-1 (CKY568), rse1-1 SAR1-Δi (CKY569), prp2-13 (CKY571), and prp2-13 SAR1-Δi (CKY572) strains were grown to exponential phase at 24°C and were shifted to 37°C (or 30°C for the prp2-13 strains) for 2.5 h in YEP plus 0.1% glucose to induce invertase. Invertase and CPY were detected in protein extracts by SDS-PAGE and immunoblotting with either anti-invertase or anti-CPY antibody. p1 CPY (ER form) and mature CPY (vacuolar form) are indicated. (C) Suspensions of cells at 107 cells/ml were spotted onto YPD plates and photographed after incubation at 24, 30, or 33°C for 3 days. None of the strains were viable at 37°C. WT, wild type.
TABLE 4.
Restoration of invertase secretion in rse1-1 and prp2-13 strains by intronless SAR1 (SAR1-Δi)a
| Strain | % Invertase secreted | Total invertase activity (units/OD600)b |
|---|---|---|
| Wild type | 87 | 0.26 ± 0.03 |
| sec12-4 | 5 | 1.31 ± 0.07 |
| rse1-1 | 48 | 0.42 ± 0.03 |
| rse1-1 SAR1-Δi | 80 | 0.42 ± 0.05 |
| prp2-13 | 29 | 1.28 ± 0.07 |
| prp2-13 SAR1-Δi | 90 | 0.81 ± 0.10 |
To induce invertase, wild-type (CKY294), sec12-4 (CKY39), and rse1-1 (CKY568 and 569) strains were shifted to 37°C for 2 h in YEP plus 0.1% glucose and prp2-13 (CKY571 and 572) strains were shifted to 30°C for 2 h in YEP plus 0.1% glucose.
The means and standard deviations from four assays (two each from two independent cultures) are given. One unit of invertase activity releases 1 μmol of glucose from sucrose at 30°C.
SAR1 protein levels decline in splicing mutants.
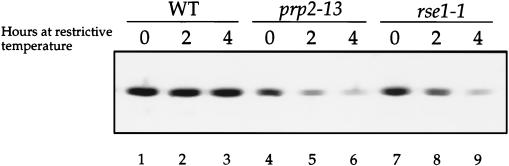
To evaluate the effects of splicing mutations on levels of SAR1 protein, we measured the amount of Sar1p in splicing mutants that had been grown at the restrictive temperature for 0, 2, or 4 h. In prp2-13 and rse1-1 mutants grown at permissive temperature, Sar1p was present at lower steady-state levels than in an isogenic wild-type strain (Fig. 6, lanes 1, 4, and 7). When growth was continued at restrictive temperature, Sar1p levels in the prp2-13 and rse1-1 mutants decreased even further. After 4 h at restrictive temperature, the Sar1p levels in the mutants declined to about one-quarter of the levels in the wild-type strain as determined by quantitative Western blotting and densitometry (Fig. 6, lanes 3, 6, and 9). Four repetitions of this experiment gave similar results. The finding that Sar1p levels decline within 2 h after Sar1p synthesis is blocked by a splicing defect implies that Sar1p is a moderately unstable protein. More direct measurements of the half-life of Sar1p were attempted in pulse-chase experiments, but Sar1p turnover could not be reliably detected by this protocol. Nevertheless, the low levels of Sar1p in prp2-13 and rse1-1 mutants at restrictive temperature indicate how a decrease in Sar1p synthesis could quickly result in an ER-to-Golgi transport defect.
FIG. 6.
prp2-13 and rse1-1 have reduced amounts of Sar1p at restrictive temperature. Wild-type (CKY294) (WT), prp2-13 (CKY570), and rse1-1 (CKY567) strains were grown to exponential phase at 24°C. Cells were either harvested (at t = 0) or shifted to 37°C (30°C for prp2-13) for 2 or 4 h. Protein extracts were prepared and proteins were resolved by SDS-PAGE on a 12% gel. Protein extract from cells (0.3 OD600 equivalent) was loaded in each lane. Sar1p was detected by immunoblot with anti-Sar1p antibody.
DISCUSSION
In this report, we show that (i) RSE1 is a novel gene involved in pre-mRNA splicing, (ii) mutants with defects in pre-mRNA processing, such as rse1-1 and prp2-13 mutants, have ER-to-Golgi secretion defects, and (iii) these splicing mutants exert their effects on secretion specifically because they fail to process SAR1 pre-mRNA.
The observed link between splicing defects and secretion defects can be explained by the fact that Sar1p appears to be moderately unstable in rse1-1 and prp2-13 mutants; consequently, a block of Sar1p synthesis in these mutants allows Sar1p levels to fall. The secretory pathway responds rapidly to a reduction of Sar1p synthesis (the block of secretion caused by a failure to splice the SAR1 intron occurs in about 2 h, well before translation ceases due to a failure to splice the introns in ribosomal subunit genes). Thus, Sar1p may normally be present at a level close to the threshold level required for continued ER-to-Golgi transport.
Previously, it was shown that overexpression of SAR1 could suppress the cold-sensitivity of a U1 snRNP mutation, indicating that Sar1p was limiting for growth in the U1 mutant at restrictive temperature (13). This finding suggested that the SAR1 intron may be very inefficiently processed. However, the possibility that SAR1 processing may be particularly sensitive to leaky splicing defects cannot explain the rapid loss of SAR1 function that occurs in splicing mutants, since a block in secretion occurs for both tight splicing mutants, such as the prp2-13 mutant, and leaky mutants, such as the rse1-1 mutant. Apparently it is either the stability or the activity of the SAR1 gene product itself that is limiting for secretory pathway function.
In addition to SAR1, nine other genes that are involved in secretion also contain introns (SEC17, BET1, BOS1, SEC27, SEC14, ERD2, SFT1, APS3, and SNC1). Inefficient splicing of pre-mRNA from one or more of these nine genes may also affect secretion in splicing mutants, since incomplete glycosylation of invertase is observed in an rse1-1 SAR1-Δi mutant and in an rse1-1 mutant that overexpresses SAR1 (Fig. 4, lane 4, and Fig. 5A, lane 4). However, our data indicate that SAR1 is the key secretion gene affected by splicing defects. In this report, we show that secretion can be largely restored in splicing mutants by deleting the intron from SAR1 or by increasing the amount of SAR1. These results indicate that, of the intron-containing secretion genes, SAR1 is the one whose function is most rapidly compromised after a splicing defect has been imposed. Moreover, the connection between splicing defects and secretion defects mediated by Sar1p is consistent with the putative regulatory role for Sar1p in vesicle budding from the ER, the first step in vesicle trafficking.
Our finding that secretion slows within 2 h after a block in SAR1 pre-mRNA processing raises the possibility that inhibition of other steps in Sar1p synthesis might also lead to a defect in ER-to-Golgi transport. The existence of a more general connection between SAR1 expression and the function of the secretory pathway suggests a mechanism by which the membrane flux within the secretory pathway could be coordinated with the rate at which new cellular proteins are synthesized. Accordingly, we have tried to examine the effect of a general decrease in the rate of translation on ER-to-Golgi transport. However, we were unable to detect the accumulation of ER precursors of secretory proteins after slowing translation by growing cells in sublethal concentrations of cycloheximide or by starving cells for amino acids. A technical problem with this type of experiment is that reducing the rate of translation also inhibits synthesis of the marker proteins used to monitor the rate of transit through the secretory pathway. The observed effect of splicing defects on secretion may depend on their unique property of causing an immediate block in Sar1p synthesis while allowing the synthesis of CPY and invertase to continue unabated for at least several hours. Ultimately, it may be necessary to find a way to assay transport through the secretory pathway that does not rely on the de novo synthesis of marker proteins in order to address a possible connection between the rate of ER-to-Golgi transport and the rate of protein synthesis.
Our findings show how splicing mutations can affect the function of the secretory pathway. A deeper connection between secretion and protein synthesis is implied by the results obtained by Mizuta and Warner, who showed that blocks at various steps in the secretory pathway cause a dramatic reduction in the transcription of rRNA and ribosomal protein genes (22). Decreased transcription of ribosomal protein genes also resulted from treatment with the transport inhibitor brefeldin A. The regulatory interactions that make ribosome synthesis sensitive to the activity of the secretory pathway are not yet understood.
Given that many genes are known to be required for pre-mRNA processing in S. cerevisiae, one may ask why mutations in these genes were not isolated in previous screens for secretion mutants. The most likely explanation stems from the fact that our screen was carried out in a different genetic background than that used in previous screens for secretion mutants. The parent strain used in our screen was A364A, which carries a different allele at the SAR1 locus than the S288C background used for most studies of the secretory pathway (2). The endogenous A364A allele, which our lab previously described as sar1-5, differs from the S288C allele by three nucleotide substitutions: a T instead of a G at position 533, changing Met42 to Ile; an A instead of a G at position 318, in the intron; and an A instead of a G at position 836, preserving an Ala143 codon (6). Based on work in our laboratory (2, 6), the allele of SAR1 in A364A appears to have less activity than the corresponding allele in S288C. Thus, our screen for new SEC genes was likely more sensitive than previous screens to mutations that compromise the expression of SAR1. We first became aware of the importance of genetic background in our secretion assays when we found that the secretion defect of prp mutants did not segregate cleanly in crosses between A364A and S288C strains. Once we had traced the source of this genetic heterogeneity to the SAR1 locus, we were careful to keep the SAR1 allele constant in all subsequent genetic manipulations. In the work presented here, all of the prp2-13 and rse1-1 strains were constructed so that they carry the endogenous allele found in A364A (the sar1-5 allele).
RSE1 shows homology to several genes, and regions near the N and C termini of RSE1 show homology to the predicted translation of Drosophila expressed sequence tag (EST) sequences. The known mammalian proteins related to RSE1 are all thought to bind to nucleic acids. One of these proteins, the 160-kDa subunit of human CPSF, is the subunit which binds the AAUAAA polyadenylation signal in pre-mRNA, but this subunit contains no clear match to a known nucleic-acid-binding domain (23). UV-DDB, XAP-1, and XPE-BF, which appear to be the same protein, have the most extensive similarity to RSE1, but their functions are not well defined. UV-DDB, which was isolated from a monkey cDNA library, has high affinity for UV-damaged DNA, although its sequence lacks any known DNA binding motifs (36). XPE-BF, the human counterpart of UV-DDB, is deficient in a subset of patients in xeroderma pigmentosum complementation group E and also binds to damaged DNA (11). The same protein, called XAP-1, was identified in a two-hybrid screen for proteins that interact with hepatitis B virus X protein (16). And recently, XAP-1 has been found to be closely related to BRF-2, a transactivator of the apolipoprotein B gene (15). Taken together, the mammalian homologies to RSE1 suggest that Rse1p also interacts with nucleic acids, perhaps through a novel nucleic-acid-binding domain, which could lie in the highly conserved region shown in Fig. 2B. Recently, RSE1 was identified as an ORF that interacts with PRP9 in a two-hybrid assay (5). Since Prp9p is required for U2 snRNP addition during spliceosome assembly (41), Rse1p might also act at an early step in spliceosome assembly with Prp9p. The homologies and two-hybrid interaction are consistent with a role for Rse1p in pre-mRNA processing as described in this report, but the specific role of Rse1p in pre-mRNA processing remains to be elucidated.
ACKNOWLEDGMENTS
We are grateful to the members of the Kaiser lab for their help, advice, and encouragement, especially to N. Rowley for technical assistance. We thank M. Winey for providing us with the collection of temperature-sensitive yeast mutants, A. Nakano for the anti-Sar1p antibody, F. Stutz from the Rosbash lab for the splicing reporter plasmids, and A. Kistler from the Guthrie lab for the U1-C8U mutant strain and for helpful discussions.
This work was supported by grants from the National Institute of General Medical Sciences and the Searle Scholars Program (to C. A. Kaiser), a Howard Hughes Medical Institute predoctoral fellowship (to E. J. Chen), and a National Institutes of Health predoctoral fellowship (to A. R. Frand). C. A. Kaiser is a Lucille P. Markey Scholar, and this work was funded in part by the Lucille P. Markey Charitable Trust.
REFERENCES
- 1.Adams A, Gottschling D, Kaiser C. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 2.Chitouras, E., and C. A. Kaiser. Unpublished data.
- 3.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′ end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 5.Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 6.Gimeno R E, Espenshade P, Kaiser C A. SED4 encodes a yeast endoplasmic reticulum protein that binds Sec16p and participates in vesicle formation. J Cell Biol. 1995;131:325–338. doi: 10.1083/jcb.131.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein A, Lampen J O. β-d-Fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- 8.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj I W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell L H, McLaughlin C S, Warner J R. Identification of ten genes that control ribosome formation in yeast. Mol Gen Genet. 1970;109:42–56. doi: 10.1007/BF00334045. [DOI] [PubMed] [Google Scholar]
- 10.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang B J, Chu G. Purification and characterization of a human protein that binds to damaged DNA. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 12.Jenny A, Keller W. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res. 1995;23:2629–2635. doi: 10.1093/nar/23.14.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao H-Y, Siliciano P G. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S-H, Smith J, Claude A, Lin R-J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamoorthy R R, Lee T H, Butel J S, Das H K. Apolipoprotein B gene regulatory factor-2 (BRF-2) is structurally and immunologically highly related to hepatitis B virus X associated protein (XAP-1) Biochemistry. 1997;36:960–969. doi: 10.1021/bi961407c. [DOI] [PubMed] [Google Scholar]
- 16.Lee T H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 18.Liao X C, Tang J, Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 19.Lin R-J, Lustig A J, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- 20.Lutz C S, Murthy K G K, Schek N, O’Connor J P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160 kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Mizuta K, Warner J R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy K G, Manley J L. The 160 kD subunit of human cleavage and polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 24.Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman A P, Ferro-Novick S. Characterization of new mutants in the early part of the yeast secretory pathway isolated by a [3H]mannose suicide selection. J Cell Biol. 1987;105:1587–1594. doi: 10.1083/jcb.105.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niwa M, Berget S M. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 27.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 28.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001–KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 29.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 31.Penn M D, Thireos G, Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984;4:520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 33.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takao M, Abramic M, Moos M, Jr, Otrin V R, Wooton J C, McLenigan M, Levine A S, Protic M. A 127 kD component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime mold protein. Nucleic Acids Res. 1993;21:4111–4118. doi: 10.1093/nar/21.17.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teigelkamp S, McGarvey M, Plumpton M, Beggs J D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, C. Unpublished data.
- 39.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 40.Wassarman K M, Steitz J A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 41.Wiest D K, O’Day C L, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- 42.Wuestehube L J, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]