Abstract
t(8;21) is one of the most frequent translocations associated with acute myeloid leukemia. It produces a chimeric protein, acute myeloid leukemia-1 (AML-1)–eight-twenty-one (ETO), that contains the amino-terminal DNA binding domain of the AML-1 transcriptional regulator fused to nearly all of ETO. Here we demonstrate that ETO interacts with the nuclear receptor corepressor N-CoR, the mSin3 corepressors, and histone deacetylases. Endogenous ETO also cosediments on sucrose gradients with mSin3A, N-CoR, and histone deacetylases, suggesting that it is a component of one or more corepressor complexes. Deletion mutagenesis indicates that ETO interacts with mSin3A independently of its association with N-CoR. Single amino acid mutations that impair the ability of ETO to interact with the central portion of N-CoR affect the ability of the t(8;21) fusion protein to repress transcription. Finally, AML-1/ETO associates with histone deacetylase activity and a histone deacetylase inhibitor impairs the ability of the fusion protein to repress transcription. Thus, t(8;21) fuses a component of a corepressor complex to AML-1 to repress transcription.
The gene for acute myeloid leukemia-1 (AML-1) is one of the most frequently translocated genes in human cancer. It is targeted by t(8;21) and t(3;21) in AML and by t(12;21) in acute lymphocytic leukemia (39). AML-1 is also indirectly targeted by inv(16), which disrupts core binding factor beta, an AML-1-interacting protein. AML-1 binds the enhancer core motif (TGT/cGGT) and regulates a variety of viral and cellular genes in concert with other factors (31). t(8;21) is one of the most frequent translocations found in AML, comprising 10 to 15% of cases with discernible translocations (39). The t(8;21) fusion protein AML-1/ETO acts as a repressor of transcription in transient-transfection assays (10, 31, 32, 43). When expressed during development, the t(8;21) fusion protein yielded the same phenotype as AML-1 deficiency (37, 45).
Although eight-twenty-one (ETO; also known as MTG8 [myeloid tumor gene 8] [8, 34]) was identified at the breakpoint of t(8;21), little is known about the normal function of the protein. ETO is the human homologue of the Drosophila Nervy protein (9), and it shares four homologous domains with the Nervy protein. These include a region with extensive homology to a Drosophila coactivator, transcription-activating factor 110 (TAF110), a predicted hydrophobic heptad repeat (HHR), a small domain with no other homology, termed the Nervy domain (27), and the MYND (myeloid–Nervy–DEAF-1 [12]) domain. The MYND domain is present in numerous human, murine, Caenorhabditis elegans, and Drosophila proteins and contains two putative zinc finger (ZF) motifs (9, 12, 31). ETO is expressed in hematopoietic cells and in the brain, but another closely related family member is ubiquitously expressed (19). A third closely related factor, MTG16, is fused to AML-1 by t(16;21) (20).
AML-1 is a site-specific DNA binding protein that can both activate and repress transcription (2, 28, 36). The t(8;21) fusion protein AML-1/ETO contains the N-terminal 177 amino acids of AML-1, including the DNA binding domain, fused to nearly all of ETO (7, 8, 34). The fusion protein inhibits AML-1-dependent transactivation (10, 32). AML-1/ETO also repressed both basal transcription and Ets-1-dependent activation of the multidrug resistance 1 promoter (27). Similarly, AML-1/ETO inhibited both AML-1 and C/EBPα-dependent transactivation of the neutrophil protein 3 (NP-3) promoter (44). AML-1/ETO-mediated repression is dependent on both the DNA binding domain of AML-1 and ETO sequences (24). AML-1/ETO acts at substoichiometric levels and thus does not compete with AML-1 for DNA binding sites within promoters, nor does it act to “squelch” transcription (24). Thus, we hypothesized that ETO recruits a corepressor or normally functions as a corepressor to inhibit transcription (30, 31, 33).
Several corepressor proteins have been recently described that associate with histone deacetylases (HDACs) to repress transcription (3, 5, 13, 17, 38, 40, 42). The nuclear hormone corepressor N-CoR was identified through interactions with the thyroid hormone receptor and associates with mSin3 proteins and HDACs (16, 17). N-CoR and the related protein SMRT are released from nuclear hormone receptors upon ligand binding, allowing transcriptional activation (5, 16, 22, 35).
Here we demonstrate that ETO associates with corepressors. ETO physically interacts with N-CoR, mSin3A, and HDACs and resides in cells as a component of large protein complexes. Deletion of the domains of ETO that are homologous to Nervy indicate that mSin3A can bind ETO independently of N-CoR. AML-1/ETO also associates with these corepressors and with HDAC activity. Single amino acid substitutions in the MYND domain that affect the ability of ETO to interact with the central portion of N-CoR impair AML-1/ETO-mediated repression of the NP-3 promoter. Finally, an HDAC inhibitor impairs AML-1/ETO-mediated repression. Thus, t(8;21) fuses the DNA binding domain of AML-1 to a putative corepressor, ETO.
MATERIALS AND METHODS
Yeast two-hybrid assays.
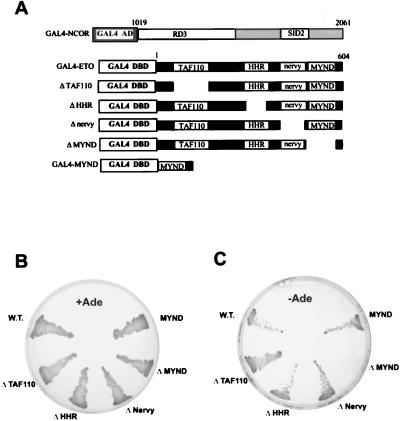
ETO residues 1 to 604 were subcloned in frame with GAL4 residues 1 to 147 in the pASII vector. Human N-CoR residues 1019 to 2061 linked to the transcriptional activation domain of GAL4 was isolated previously (18), and this plasmid was cotransformed with pASII-ETO into yeast strains PJ69-4A and Y190. Protein interactions in strains PJ69-4A were measured by growth on adenine-deficient media (Fig. 1B and C) and by growth on histidine-deficient media containing 2 mM 3-aminotriazole. Interactions in strain Y190 were measured by growth on histidine deficient media in the presence of 50 mM aminotriazole. Deletion and point mutations in ETO have been described previously (27).
FIG. 1.
ETO interacts with N-CoR. (A) Schematic diagram of the GAL4–N-CoR, GAL4-ETO, and GAL4-ETO deletion mutant fusion proteins used in the yeast two-hybrid interaction system. Although a fragment of the human N-CoR cDNA was used in these assays, the numbering is in relation to the murine sequence. (B) Growth on complete media of yeast strain PJ69-4A expressing the indicated ETO proteins together with the GAL4 activation domain (AD)–N-CoR fusion protein. (C) Growth of the yeast shown in panel B on adenine (Ade)-deficient media. W.T.; wild type; DBD, DNA binding domain.
Glutathione agarose precipitation assays.
N-CoR was transcribed and translated in vitro in the presence of [35S]methionine and cysteine (PROMIX; Amersham) by using the T7 coupled reticulocyte lysate system (Promega) in accordance with the manufacturer’s instructions. For precipitation assays, equal amounts of the glutathione S-transferase (GST) fusion proteins were incubated with 25 μl of the N-CoR in vitro transcription-and-translation reaction mixture in 200 μl of phosphate-buffered saline (PBS) containing 0.5% Triton X-100. After 1 h at 4°C, the beads were washed three times with PBS–0.5% Triton X-100 and resuspended in 2× sodium dodecyl sulfate (SDS) sample buffer, and proteins were separated by SDS–8% polyacrylamide gel electrophoresis (PAGE) prior to autoradiography.
Cell culture.
C33A and Cos-7 cells were maintained in Dulbecco modified Eagle medium (BioWhittaker Inc., Walkersville, Md.) containing 10% fetal calf serum, 50-U/ml penicillin, 50-μg/ml streptomycin, and 2 mM l-glutamine (all from BioWhittaker). Human erythroleukemia (HEL) cells were cultured in RPMI 1640 medium (BioWhittaker) containing 10% fetal calf serum, antibiotics, and l-glutamine. NIH 3T3 cells were cultured in Dulbecco modified Eagle medium containing 10% calf serum.
Coimmunoprecipitations, sucrose gradient sedimentation, and immunoblotting.
Cos-7 cells (3 × 106/100-mm-diameter dish) were cotransfected by using Lipofectamine (Bethesda Research Laboratories) with 3.5 μg of plasmid CMX-NCOR (Flag epitope tagged) (16) and 1.5 μg of plasmid CMV5-ETO (32), or cells were transfected with 4 μg of each plasmid individually. For endogenous proteins, 5 × 106 HEL cells were extracted with PBS supplemented with 1 mM EDTA, 1.5-mg/ml iodoacetamide, 0.2 mM phenylmethylsulfonyl fluoride, 0.1-trypsin IU/ml aprotinin, and 0.5% Triton X-100 unless otherwise noted. Lysates were sonicated and incubated with 100 μl of formalin-fixed Staphylococcus aureus (Pansorb; CalBiochem) for 30 min to remove nonspecific protein binding. After centrifugation for 5 min at 4°C, the supernatants were collected and immunoprecipitated for 1 h with an affinity-purified primary antibody (K-20 anti-mSin3A or C-19 anti-HDAC-2, Santa Cruz Biotechnology; anti-HDAC-1 has already been described [23]). A 15-μl volume of a 50% slurry of protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden) was then added, the mixture was incubated for 30 min to collect the immune complexes, and the immune complexes were then washed three times at 4°C with lysis buffer. A 100-μg sample of protein (quantitated with the Bio-Rad DC protein assay) or the immune complexes were boiled in Laemmli buffer for 2 min, fractionated by SDS-PAGE, and transferred to nitrocellulose. Blots were blocked for 1 h with 5% milk and incubated with the indicated primary antibody overnight at 4°C. Proteins were visualized by ECL (Pierce).
Sucrose gradient sedimentation analysis was performed by lysing HEL cells in PBS containing 0.5% Triton X-100 and 1 mM EDTA. Cell lysates were separated by centrifugation at 28,000 rpm for 15 h in an SW50 rotor. Sedimentation standards (Bio-Rad broad-range markers supplemented with ferritin and thyroglobulin) were analyzed on parallel gradients. Fractions (250 μl) were collected, and 40 μl of each fraction was analyzed by SDS–8% PAGE. Immunoblot analysis was sequentially performed with ETO, mSin3A, and HDAC-1 antibodies. The upper portion of the blot was independently probed with N-CoR antibodies.
HDAC assays.
Cells were lysed in PBS containing 0.5% Triton X-100 and protease inhibitors, and immunoprecipitations were performed as described above. Immune complexes were resuspended in 20 mM Tris [pH 8]–150 mM NaCl–10% glycerol and assayed for HDAC activity by using [3H]acetate-labeled chicken erythrocytes as previously described (16).
Transcription assays.
C33A cells were transfected with 5 μg of an NP-3-137-Luciferase plasmid, 1 μg of a pCMV5-AML-1B plasmid, 0.5 μg of an MSV-C/EBPα plasmid, 1 μg of a Rous sarcoma virus long terminal repeat (LTR)-chloramphenicol acetyltransferase plasmid, and 10 μg of the indicated pCMV5-AML-1/ETO plasmid (44) (Fig. 1 and reference 28 contain details of the mutations). Luciferase activity was measured as previously described (44) and normalized to chloramphenicol acetyltransferase activity, which was quantitated by using a Molecular Dynamics PhosphorImager. NIH 3T3 cells were transfected by using the Superfect reagent (Qiagen) with 1 μg of WWP-Luciferase (p21Waf1/Cip1 [6]), 2 μg of pCMV5-AML-1/ETO and 1 μg of the Rous sarcoma virus LTR-renilla luciferase plasmid. Firefly and renilla luciferase activities were measured by using the Duel Luciferase Assay System (Promega).
RESULTS
ETO interacts with N-CoR.
Our transcriptional analysis of the t(8;21) fusion protein indicated that AML-1/ETO repressed transcription by recruiting a corepressor. Therefore, we adopted a candidate gene approach to test whether known corepressors could physically interact with ETO in a yeast two-hybrid assay. ETO was fused to the GAL4 DNA binding domain and tested for interaction with a chimeric protein containing the central portion of N-CoR fused to the Gal4 transcriptional activation domain (the latter was obtained in a yeast two-hybrid screen for proteins interacting with the repressor BCL-6 [18]) (Fig. 1A). In this assay, interaction of the two proteins generates a functional activator capable of inducing the Ade2 and His3 reporter genes. As measured by growth on adenine-deficient media (Fig. 1B and 1C) and growth on histidine-deficient media containing 3-aminotriazole (data not shown), the nuclear hormone corepressor N-CoR interacted with ETO.
We next determined which of the conserved domains in ETO are required for N-CoR interaction (Fig. 1B and C). Deletion of the MYND motif, but not the other conserved domains, eliminated the ability of ETO to interact with N-CoR. Moreover, the MYND domain alone fused to the GAL4 DNA binding domain associated with N-CoR (Fig. 1B and C).
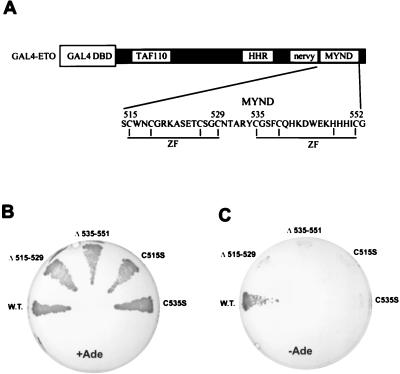
The MYND domain contains two predicted zinc finger motifs (Fig. 2A). When either of these putative motifs was disrupted by small deletions, the ETO–N-CoR interaction was lost (Fig. 2B and C). Furthermore, serine substitutions of either of two conserved cysteine residues within the putative ZF motifs also abrogated the ETO–N-CoR interaction (Fig. 2B and C), indicating that these ZF motifs act together to form a protein interaction domain.
FIG. 2.
N-CoR interacts with the ZFs of the MYND domain. (A) Schematic diagram of the GAL4-ETO fusion protein. The amino acid sequence of the MYND domain of ETO is shown. Conserved cysteine and histidine residues that form the predicted ZF motifs are indicated by vertical lines. (B) Growth on complete media of PJ69-4A yeast expressing the indicated ETO proteins together with the GAL4 activation domain–N-CoR fusion protein. (C) Growth of the yeast shown in panel B on adenine (Ade)-deficient media. W.T., wild type; DBD, DNA binding domain.
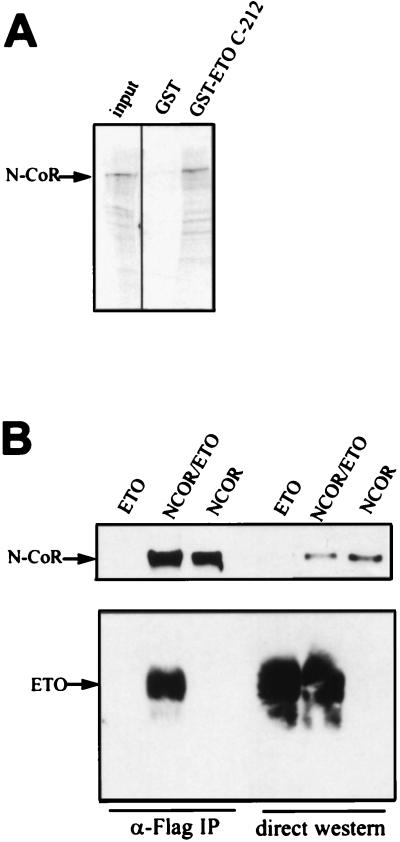
To determine whether the MYND domain was sufficient for interaction with full-length N-CoR, we tested whether a GST fusion protein containing the C-terminal one-third of ETO, including the MYND domain, could associate with N-CoR synthesized in vitro. As shown in Fig. 3A, the C-terminal 212 amino acids of ETO fused to GST bound significantly more N-CoR that did GST alone.
FIG. 3.
ETO interacts with N-CoR in vitro and in mammalian cells. (A) ETO interacts with N-CoR in vitro. GST and GST-ETO containing the C-terminal 283 residues of ETO were linked to glutathione agarose beads and used to purify in vitro-transcribed and -translated N-CoR. (B) ETO interacts with N-CoR in mammalian cells. Cos-7 cells were cotransfected with ETO and Flag-tagged N-CoR plasmids. Cell extracts were prepared in PBS containing 0.5% Triton X-100, 0.5% Na deoxycholate, and 0.2% SDS and immunoprecipitated with Flag antibodies. ETO and N-CoR were detected by Western immunoblot analysis using antibodies directed against the C terminus of ETO or the Flag epitope, respectively. Twenty-fold more extract was used for immunoprecipitation (IP) than was analyzed by direct immunoblotting.
To demonstrate that ETO and N-CoR interact in mammalian cells, we coexpressed ETO and Flag epitope-tagged N-CoR in Cos-7 cells and measured their association by using immunoprecipitation assays. Cell extracts were immunoprecipitated by using anti-Flag antibodies, and N-CoR and ETO were detected by immunoblot analysis using antibodies directed against the Flag epitope and the C terminus of ETO (Fig. 3B). ETO was coimmunoprecipitated with anti-Flag antibodies only when Flag–N-CoR was coexpressed, indicating a specific physical interaction.
Endogenous ETO coimmunoprecipitates with mSin3A.
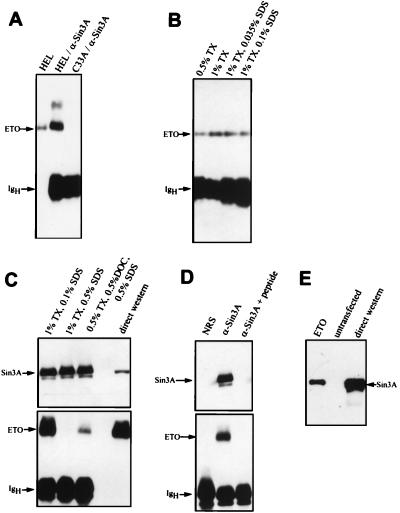
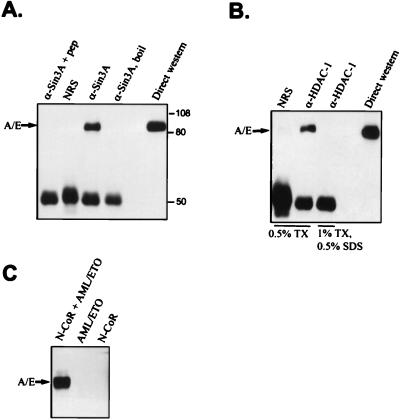
N-CoR interacts with mSin3 proteins and HDACs to repress transcription (1, 14, 16, 23, 35, 47). Therefore, we investigated whether endogenous ETO also associates with these proteins. HEL cells express easily detectable levels of ETO protein (27); therefore, we immunoprecipitated HEL cell extracts with mSin3A antibodies and determined whether ETO was a component of mSin3A complexes by using immunoblot analysis (Fig. 4A). A significant proportion of the ETO in HEL cells coimmunoprecipitated with mSin3A antibodies (Fig. 4A). Lesser amounts of protein coimmunoprecipitated with mSin3B antibodies (data not shown). This could be because of lower levels of mSin3B in these cells or because the antibodies to mSin3B were directed against PAH2 and this domain may not be accessible in the complex. The ETO-mSin3A complex remained intact when the cells were lysed in PBS containing both 1% Triton X-100 and 0.1% SDS but was dissociated when cell lysates were prepared with 1% Triton X-100 and 0.5% SDS (Fig. 4B and C). As an additional control, we added the mSin3A antigenic peptide prior to immunoprecipitation, and this peptide eliminated the coprecipitation of ETO with mSin3A (Fig. 4D).
FIG. 4.
Endogenous ETO associates with mSin3A. (A) HEL cell extracts were analyzed directly, or 25-fold more extract was immunoprecipitated with mSin3A antibodies and then subjected to immunoblot analysis by using antibodies to ETO. C33A cells express undetectable levels of ETO protein and were used as an antibody control. (B and C) HEL cell extracts were prepared, and mSin3A immunoprecipitations were performed by using increasing amounts of the detergents Triton X-100 (TX), sodium deoxycholate (DOC), and SDS as indicated. A portion of the blot panel C was probed with anti-ETO antibodies, and the high-molecular-weight portion of the blot was probed with anti-mSin3A IgG. (D) A 30-μg sample of antigenic peptide was used to block the mSin3A antibody prior to immunoprecipitation of HEL cell extracts. The upper portion of the blot was probed with anti-mSin3A IgG, and the lower portion was probed with anti-ETO IgG. (E) mSin3A coimmunoprecipitated with ETO. Flag-ETO was expressed in Cos-7 cells and immunoprecipitated with anti-Flag IgG. Copurified mSin3A was detected by immunoblot analysis. Twenty-fold more extract was used for immunoprecipitation than was analyzed by direct immunoblotting. NRS, normal rabbit serum.
Our anti-ETO serum is directed to the C terminus of ETO, which contains the MYND domain. We were unable to coprecipitate mSin3A by using this serum, likely because the epitope is obscured when ETO is in protein complexes. Therefore, we created a Flag-tagged ETO cDNA and investigated whether this protein could coimmunoprecipitate endogenous mSin3A. As shown in Fig. 4E, mSin3A was precipitated with anti-Flag antibodies only when Flag-ETO was expressed, indicating a specific association between mSin3A and ETO.
Endogenous ETO interacts with endogenous HDAC-2.
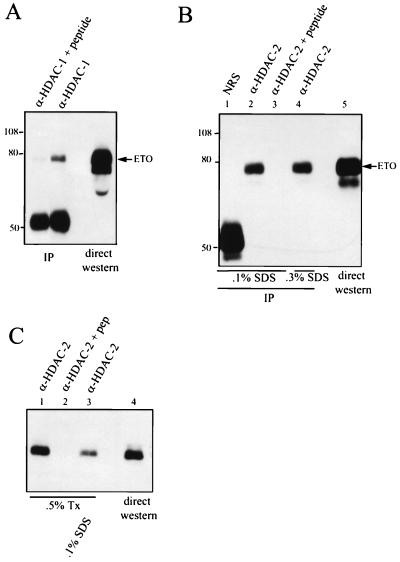
Both N-CoR and mSin3A act as corepressors by linking site-specific DNA binding proteins to HDACs (1, 14, 16, 23, 35, 47). Therefore, we investigated whether ETO associates with HDAC-1 or HDAC-2. ETO was transiently expressed in Cos-7 cells, and cell lysates were immunoprecipitated with anti-HDAC-1 antibodies prior to immunoblot analysis with anti-ETO immunoglobulin G (IgG). ETO copurified with HDAC-1, and addition of the HDAC-1 antigenic peptide eliminated the association of ETO with HDAC-1 (Fig. 5A). The association was not observed under conditions of higher stringency (1% Triton X-100–0.1% SDS; data not shown). Thus, either the ETO–HDAC-1 interaction is relatively weak or the association is indirect and is mediated by mSin3A or N-CoR. Similarly, transiently expressed ETO was detected in HDAC-2 immune complexes (Fig. 5B). However, in contrast to that with HDAC-1, the association of ETO with HDAC-2 was stable under more stringent conditions.
FIG. 5.
ETO interacts with HDACs. (A) ETO interacts with endogenous HDAC-1. Cos-7 cells were transfected with pCMV5-ETO, and cell extracts were prepared in PBS containing 0.5% Triton X-100 and immunoprecipitated with anti-HDAC-1 IgG in the presence or absence of the antigenic peptide. ETO was detected by immunoblot analysis using antibodies directed against the C terminus of ETO. IP, immunoprecipitation. (B) ETO interacts with endogenous HDAC-2. Cos-7 cells were transfected with pCMV5-ETO, and cell extracts were prepared in PBS containing 0.5% Triton X-100, 0.5% Na deoxycholate, and either 0.1% (lanes 1 to 3) or 0.3% (lane 4) SDS and subjected to immunoprecipitation (IP) with 4 μg of anti-HDAC-2 IgG or anti-HDAC-2 IgG that had been blocked with 20 μg of antigenic peptide. Proteins were separated by SDS–8% PAGE prior to autoradiography. NRS, normal rabbit serum. Note that the anti-HDAC-2 is a goat polyclonal antibody and is not detected by the anti-rabbit secondary antibody. (C) Endogenous ETO interacts with endogenous HDAC-2. HEL cell lysates were prepared in PBS containing 0.5% Triton X-100 (Tx) with or without 0.1% SDS and immunoprecipitated with anti-HDAC-2 IgG. The presence of ETO in immune complexes was detected by immunoblot analysis using anti-ETO IgG. Twenty-fold more extract was used for immunoprecipitation than was analyzed by direct immunoblotting. The values on the left of panels A and B are molecular sizes in kilodaltons.
Given the association of overexpressed ETO with endogenous HDAC-1 and HDAC-2, we investigated whether endogenous ETO interacts with either of these enzymes. HEL cell lysates were immunoprecipitated with antibodies to the HDACs, and the immune complexes were analyzed by immunoblotting for the presence of ETO. From HEL cells, ETO coimmunoprecipitated with HDAC-2 (Fig. 5C) but not HDAC-1 (negative data not shown). Because HDAC-1 can associate with transiently expressed ETO (Fig. 5A), this may suggest that the levels of HDAC-1 are too low in HEL cells for detection in this assay, that the antisera have different affinities, or that in HEL cells ETO preferentially associates with HDAC-2.
ETO interacts with mSin3A in the absence of N-CoR binding.
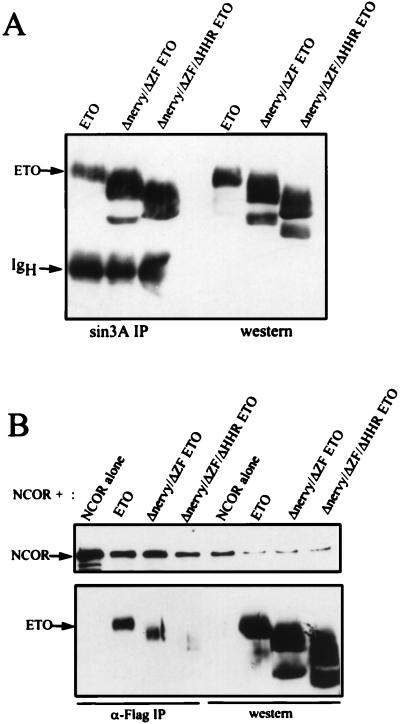
The MYND of ETO interacts with the central portion of N-CoR (Fig. 1), but other regions of ETO could also associate with N-CoR. When mutant ETO proteins lacking each of the domains conserved with Nervy were tested for N-CoR interaction in vivo, these proteins, including a deletion of the MYND domain, still bound to N-CoR (negative data not shown; Fig. 1 contains a schematic diagram of these mutant proteins). Therefore, we expressed an ETO protein lacking both the Nervy and MYND (ZF) domains and a mutant protein lacking the HHR, Nervy, and MYND (ZF) domains and tested these mutant proteins for interaction with mSin3A (Fig. 6A) and N-CoR (Fig. 6B). By comparison to wild-type ETO, somewhat less of the Nervy/MYND mutant bound to N-CoR (Fig. 6B). The further deletion of the HHR motif greatly reduced the affinity of ETO for N-CoR. Given that the MYND domain interacts with the central portion of N-CoR in yeast two-hybrid assays, these results suggest a second interaction domain between ETO and N-CoR, outside of residues 1019 to 2061 of N-CoR. By contrast, the mSin3A association was not impaired by the deletion of the individual conserved domains (data not shown) or by the combined deletion of the HHR, Nervy, and MYND domains (Fig. 6A). Thus, mSin3A can bind ETO independently of N-CoR. Moreover, this result indicates that N-CoR does not bind ETO through the mSin3A-ETO interaction.
FIG. 6.
ETO contacts mSin3A independently of N-CoR. The indicated ETO mutants were expressed in Cos-7 cells and tested for the ability to coimmunoprecipitate with mSin3A (A) or N-CoR (B) as described in the legends to Fig. 3 and 4. Cell lysates were immunoprecipitated by using anti-mSin3A IgG or anti-Flag antibodies (for N-CoR), and the ETO mutants were detected by immunoblot analysis by using anti-ETO IgG. Twenty-fold more extract was used for immunoprecipitation (IP) than was analyzed by direct immunoblotting.
ETO cosediments with mSin3A and N-CoR.
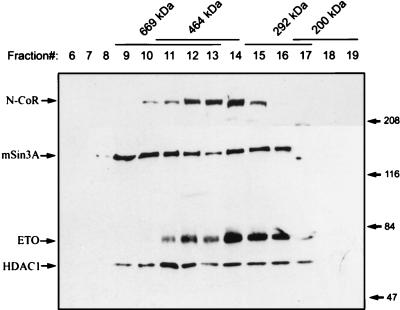
N-CoR and mSin3A have been suggested to be components of a large complex containing HDACs and other associated proteins (1, 14, 16, 23, 35, 47). Therefore, we tested whether endogenous ETO is a component of a high-molecular-weight complex. HEL cell lysates were prepared in PBS containing 0.5% Triton X-100 and fractionated over a 10 to 30% sucrose gradient. By comparison to standards separated on parallel gradients, ETO, N-CoR, mSin3A, and HDAC-1 cosedimented with an apparent molecular mass of 300 to 600 kDa (Fig. 7). As well, mSin3A and HDAC-1 were found in fractions corresponding to an apparent molecular mass of greater than 600 kDa. The apparent size of the ETO-containing complex is much larger than the expected size of free ETO, ETO dimers, or ETO–N-CoR and ETO-mSin3A heterodimers. Coupled with the physical association of ETO with mSin3A and N-CoR, we propose that endogenous ETO is a component of one or more corepressor complexes.
FIG. 7.
Endogenous ETO cosediments with mSin3A and N-CoR. HEL cell lysates (2 mg of total protein) were fractionated on a 10 to 30% sucrose gradient. A 40-μl sample of each fraction was separated on an 8% polyacrylamide gel and then subjected to sequential immunoblot analysis using ETO, mSin3A, and HDAC-1 antibodies. The upper portion of the blot was independently probed with N-CoR antibodies. The standards are as follows: 669 kDa, thyroglobulin; 464 kDa, a tetramer of β-galactosidase; 292 kDa, a trimer of phosphorylase b; 200 kDa, myosin. The values on the right are molecular sizes in kilodaltons.
AML-1/ETO interacts with corepressors.
Cell lines containing t(8;21) are difficult to culture, likely due to the ability of the fusion protein to inhibit the cell cycle-promoting activities of AML-1 (41). To determine whether the t(8;21) fusion protein associates with corepressors, we transiently expressed the AML-1/ETO fusion protein in Cos-7 cells, immunoprecipitated mSin3A or HDAC-1, and detected coimmunoprecipitating AML-1/ETO by immunoblot analysis. The fusion protein copurified with both mSin3A (Fig. 8A) and HDAC-1 (Fig. 8B). Similar results were obtained with antibodies directed to HDAC-2 (data not shown). Finally, AML-1/ETO was coimmunoprecipitated with Flag antibodies when AML-1/ETO was cotransfected with Flag-tagged N-CoR (Fig. 8C). Thus, as expected, AML-1/ETO associates with the same corepressors as does wild-type ETO.
FIG. 8.
The t(8;21) fusion protein interacts with mSin3A, HDAC-1, and N-CoR. (A) AML-1/ETO coimmunoprecipitates with anti-mSin3A IgG. AML-1/ETO was transfected into Cos-7 cells, and cell extracts were immunoprecipitated with mSin3A antibodies in PBS containing 0.5% Triton X-100. As a marker, 50 μg of lysate was analyzed by immunoblotting for AML-1/ETO expression (direct Western blot). The values on the right are molecular sizes in kilodaltons. (B) AML-1/ETO associates with HDAC-1. Cos-7 cell lysates containing transiently expressed AML-1/ETO were prepared in a buffer containing PBS and the indicated detergents. These lysates were immunoprecipitated with HDAC-1 antibodies prior to immunoblot analysis with antibodies directed to ETO. (C) AML-1/ETO interacts with N-CoR. Flag-N-CoR was coexpressed with AML-1/ETO, and cell lysates were immunoprecipitated with anti-Flag IgG. Copurifying AML-1/ETO was detected by immunoblot analysis by using anti-ETO IgG. A/E, AML-1/ETO; NRS, normal rabbit serum; boil, sample heated to 100°C for 2 min prior to immunoprecipitation; TX, Triton X-100; pep, antigenic peptide. Twentyfold more extract was used for immunoprecipitation than was analyzed by direct immunoblotting.
ETO and AML-1/ETO associate with HDAC activity.
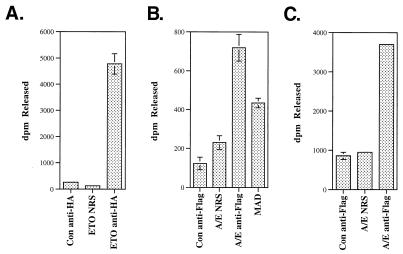
To determine whether ETO and AML-1/ETO associate with HDAC activity, we transiently expressed hemagglutinin (HA)-ETO in 293 cells or Flag–AML-1/ETO in Cos-7 cells, immunoprecipitated it with anti-HA or anti-Flag antibodies, and measured the associated HDAC activity. The HA antibodies immunoprecipitated HDAC activity only when ETO was expressed (Fig. 9A). Likewise, the anti-Flag antibodies immunoprecipitated HDAC activity only when Flag–AML-1/ETO was expressed (Fig. 9B). This level of activity was greater than that observed for Max dimerization (MAD) (Fig. 9B), which served as a positive control in this assay. Similar results were obtained for AML-1/ETO in 293 cells (Fig. 9C). Thus, ETO and AML-1/ETO interact with corepressor complexes containing active HDAC.
FIG. 9.
ETO and AML-ETO are associated with HDAC activity. (A) L293 cells were transfected with an HA-tagged ETO expression plasmid, and cell lysates were immunoprecipitated with anti-HA antibodies or with normal rabbit serum (NRS). (B) Cos-7 or L293 (C) cells were transfected with Flag-tagged AML-1/ETO (A/E) or MAD expression plasmids and immunoprecipitated with anti-Flag or anti-MAD antibodies or with normal rabbit serum. Untransfected Cos-7 or L293 cells were also immunoprecipitated with anti-Flag or anti-HA antibodies. HDAC activity was assayed in the immune complexes. Con, control; dpm, disintegrations per minute.
AML-1/ETO domains that are required for repression of AML-1B- and C/EBPα-dependent transactivation.
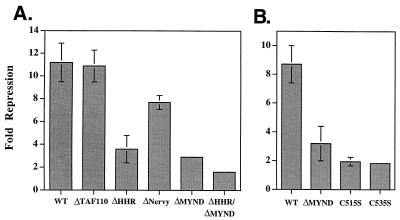
The C-terminal 283 amino acids of ETO are required for transcriptional interference with AML-1B (the largest transcriptionally active isoform of AML-1 [32]) and for inhibition of C/EBPα and Ets-1 transactivation (24, 27, 44). To test the role of N-CoR interactions in repression, we transferred the ETO deletions depicted in Fig. 1 into AML-1/ETO (27) and tested these mutant proteins for the ability to repress the transactivation of the differentiation-specific NP-3 promoter. NP-3 transcription was activated over 50-fold by a combination of AML-1B and C/EBPα. The ability of wild-type AML-1/ETO and deletion mutant AML-1/ETO to inhibit this activation was assessed by using luciferase activity as a reporter. As demonstrated previously (44), AML-1/ETO efficiently blocked AML-1B-C/EBPα synergistic activation (Fig. 10A). Deletion of the TAF110 homology domain and the Nervy domain had little or no effect, but deletion of either the MYND domain or the HHR motif significantly impaired AML-1/ETO-mediated repression. Deletion of both the HHR motif and the MYND domain completely inactivated AML-1/ETO. Finally, the single amino acid changes that are predicted to disrupt the putative ZF motifs in the MYND domain and which eliminate the ETO–N-CoR interaction in yeast two-hybrid assays (Fig. 2) significantly impaired transcriptional repression (Fig. 10B). Given that deletion of the MYND domain did not ablate the association of ETO with N-CoR or mSin3A in mammalian cells, it appears that specific contacts between the MYND domain and N-CoR are required for repression.
FIG. 10.
AML-1/ETO-mediated repression cosegregates with the ability to interact with the central domain of N-CoR. The rat NP-3 promoter was activated approximately 50- to 60-fold by a combination of AML-1B and C/EBPα and repression by AML-1/ETO, and the indicated AML-1/ETO mutants were assessed. (A) Mapping of the domains required for transcriptional repression. The mutant ETOs used in Fig. 1 were transferred into AML-1/ETO, and the ability of these mutant ETOs to repress AML-1B-C/EBPα-dependent activation was measured and is expressed as fold repression. (B) Both predicted ZF motifs are required for AML-1/ETO function. Small deletions and single amino acid changes in the MYND domain of ETO (depicted as GAL4-ETO mutations in Fig. 2A) were transferred into AML-1/ETO and tested for repression of NP-3-activated transcription. The bars indicate average results of duplicate (A) or triplicate (B) experiments. WT, wild type.
An HDAC inhibitor affects AML-1/ETO-mediated repression of the p21Waf1/Cip1 promoter.
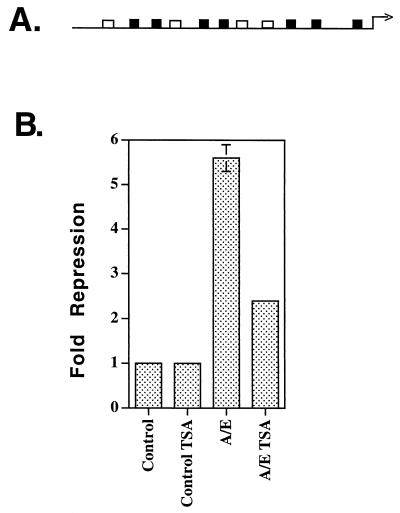
Because transcriptionally impaired forms of AML-1/ETO still bind mSin3A and N-CoR, we sought to determine the role of HDACs in AML-1/ETO-mediated repression by using trichostatin A (TSA), an HDAC inhibitor. In NIH 3T3 cells, the NP-3 promoter was affected by TSA and could not be used for this analysis. However, we have previously demonstrated that the p21Waf1/Cip1 promoter (Fig. 11A) is a transcriptional target of AML-1 (28) and that the p21Waf1/Cip1 promoter was not activated by 300 nM TSA in NIH 3T3 cells (28). Therefore, we determined the effect of TSA on AML-1/ETO-mediated repression of the p21Waf1/Cip1 promoter. Cells were transiently transfected, and TSA was added to the culture media immediately after transfection. AML-1/ETO inhibited expression from the p21Waf1/Cip1 promoter by 5.6-fold, and the addition of TSA reduced repression by nearly 60% (Fig. 11B). Taken together with the physical association of the t(8;21) fusion protein with mSin3A, N-CoR, HDAC-1, and HDAC-2 (Fig. 8) and the association of AML-1/ETO with HDAC activity (Fig. 9), this result confirms the role of HDACs in AML-1/ETO-mediated repression. Moreover, we have recently demonstrated that TSA inactivates AML-1/ETO in a biological assay (41).
FIG. 11.
An HDAC inhibitor impairs AML-1/ETO-mediated repression of the p21Waf1/Cip1 promoter. (A) Schematic diagram of the p21Waf1/Cip1 promoter. Open boxes represent perfect matches for the AML-1 consensus binding site, and dark boxes represent 5-of-6-bp matches for the consensus binding site. (B) TSA blocks AML-1/ETO-mediated repression. Repression assays were performed by using 2 μg of AML-1/ETO in the absence or presence of 300 nM TSA. The control levels were arbitrarily set to 1. A Rous sarcoma virus LTR-renilla luciferase plasmid was used as an internal control. The bars represent average results of duplicate experiments. A/E, AML-1/ETO.
DISCUSSION
Analysis of the transcriptional regulatory activity of the t(8;21) fusion protein led us to propose that AML-1/ETO interacts with a corepressor (24, 32). We have demonstrated that in mammalian cells, ETO associates with N-CoR, mSin3A, and HDACs. We have demonstrated that N-CoR interacts with ETO in yeast two-hybrid assays, in vitro, and in mammalian cells. ETO deletion mutants that have lost the ability to interact with N-CoR still bind mSin3A, indicating that N-CoR does not associate with ETO through mSin3A. Thus, the cumulative evidence suggests that the N-CoR interaction with ETO is direct. As well, the interaction of endogenous ETO with endogenous mSin3A under stringent conditions and the association of mSin3A with AML-1/ETO mutants that fail to bind N-CoR suggest that this interaction is also direct. However, these data do not preclude the possibility that an unidentified protein (that is conserved from yeast to humans) mediates the association of ETO and these corepressors. Based on the association of endogenous ETO with endogenous mSin3A at high stoichiometry and the observation that ETO is found only in high-molecular-weight complexes by sucrose gradient sedimentation analysis, we propose that ETO is a component of one or more complexes containing mSin3A, N-CoR, and HDACs in vivo.
The four domains of ETO that are conserved in its Drosophila homologue Nervy appear to be protein interaction motifs (e.g., the HHR [27] and MYND [Fig. 1] regions). We have been unable to demonstrate that wild-type ETO binds DNA cellulose or that it binds DNA specifically (28). Therefore, we propose that ETO functions in a corepressor complex as an adapter protein, perhaps linking N-CoR, mSin3, and other proteins. These interactions could occur within the complex, for instance, to stabilize N-CoR/mSin3A complexes, or ETO may link the corepressors to site-specific DNA binding proteins to regulate transcription. In the latter case, ETO would be analogous to the retinoblastoma protein, which represses transcription by linking an HDAC complex to DNA binding proteins (4, 26, 29). t(8;21) takes advantage of this activity to create an AML-1 repressor by fusing the DNA binding domain of AML-1 to ETO.
The MYND domain of ETO interacts with the central portion of N-CoR, including repression domain 3, in yeast two-hybrid assays (Fig. 1 and 2). However, when the MYND domain was deleted, the mutant ETO retained the ability to interact with both N-CoR and mSin3A in mammalian cells. This result suggests the presence of a second N-CoR binding domain on ETO. Moreover, because an ETO protein lacking the HHR, Nervy, and MYND domains retained the ability to interact with mSin3A, we conclude that mSin3A can bind ETO in the absence of an ETO–N-CoR interaction. Because deletion of the TAF110 domain also did not affect mSin3A interactions (data not shown), mSin3A may contact ETO through more than one domain or the interaction site may be outside of the conserved domains. However, deletion of the MYND and HHR domains did impair the ability of the fusion protein to repress transcription. Therefore, the interaction of the fusion protein with mSin3A and/or N-CoR is not sufficient for repression, and specific interactions, such as the MYND domain contacting the central portion of N-CoR, may be required for full activity.
Although AML-1/ETO represses the transcription of most of the promoters tested, in two cases, transactivation has been observed. AML-1/ETO synergized with wild-type AML-1 to activate the M-CSF1 receptor promoter (46). Because the cooperativity was mediated by a single AML-1 binding site, it was proposed that the fusion protein was acting indirectly, perhaps by titrating a corepressor, to active transcription (46). Our current results are consistent with this interpretation. In the second report, AML-1/ETO was demonstrated to activate transcription of the BCL-2 promoter through an AML-1 binding site that resides within a negative regulatory region of the promoter (21). While AML-1/ETO appears to strongly bind mSin3A, N-CoR, and HDACs, we cannot rule out the possibility that the fusion protein also can act to activate transcription through an undefined mechanism.
The observation that AML-1/ETO functions by interacting with an HDAC-containing complex(es) may also have therapeutic implications. In acute promyelocytic leukemia, t(11;17) and t(15;17) target the gene for retinoic acid receptor alpha. Both of these translocation fusion proteins interact with the N-CoR and SMRT corepressors and use HDACs to inhibit transcription. Leukemic blasts or cell lines containing t(15;17) differentiate in response to all-trans-retinoic acid, but cells expressing the t(11;17) fusion protein differentiated only when all-trans-retinoic acid was supplemented with the HDAC inhibitors (11, 15, 25). Recently, we have observed that high-level expression of AML-1/ETO disrupts normal cell cycle control in hematopoietic cells. TSA completely ablated AML-1/ETO function in this biological system (41). The association of t(8;21) with N-CoR, mSin3 corepressors, and HDACs, coupled with the ability of TSA to transcriptionally impair AML-1/ETO (Fig. 11), indicates that HDAC inhibitors may have more general application for chemotherapeutic intervention in acute myeloid leukemia.
ACKNOWLEDGMENTS
We thank Dana King and Yue Hou for technical assistance and the members of the Hiebert laboratory for critical evaluation of the manuscript. We thank Shari Meyers (LSUMC) for the GAL4-ETO pASII plasmid.
This work was supported by the Vanderbilt Cancer Center; NIH/NCI grants RO1-AG13726, ROI-CA64140, and RO1-CA77274; American Cancer Society grant JFRA-591 (to S.W.H.); F32-CA77167 (to J.J.W.); a Center grant from the National Cancer Institute (CA68485); Medical Research Council of Canada grant MT-9186 (to J.D.); and grant MCB9631067 from the National Science Foundation (to E.S.). B.L. is a fellow of the Leukemia Society of America (5669-99).
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression [see comments] Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer D, Lawrence Q, Eisenman R. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 4.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 5.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors [see comments] Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 8.Erickson P F, Robinson M, Owens G, Drabkin H A. The ETO portion of acute myeloid leukemia t(8;21) fusion transcript encodes a highly evolutionarily conserved, putative transcription factor. Cancer Res. 1994;54:1782–1786. [PubMed] [Google Scholar]
- 9.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 11.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Grignani F, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 12.Gross C T, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 13.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 14.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 15.He L Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Distinct interactions of PML-RARalpha and PLZF-RARalpha with corepressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 17.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor [see comments] Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 18.Huynh, K., and V. Bardwell. The BCL-6 POZ domain and other POZ domains interact with the co-repressor N-CoR and SMRT. Oncogene, in press. [DOI] [PubMed]
- 19.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid leukemia is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- 21.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci USA. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor [see comments] Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 23.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 24.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 25.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 26.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 27.Lutterbach B, Sun D, Schuetz J, Hiebert S W. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol Cell Biol. 1998;18:3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutterbach, B., J. Westendorf, B. Linggi, E. Seto, and S. Hiebert. AML-1, a target of multiple chromosomal translocations in acute leukemia, interacts with mSin3 and represses transcription from the p21Waf1/Cip1 promoter. Submitted for publication.
- 29.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase [see comments] Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 30.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers S, Hiebert S W. Direct and indirect alteration of transcriptional regulation in cancer. Crit Rev Eukaryot Gene Expr. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 32.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers S, Lenny N, Sun W, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 34.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEPB2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuda T, Cai Z, Yang S, Lenny N, Lyu C, van Deursen J, Harada H, Downing J. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 38.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 39.Rubnitz J E, Crist W M. Molecular genetics of childhood cancer: implications for pathogenesis, diagnosis, and treatment. Pediatrics. 1997;100:101–108. doi: 10.1542/peds.100.1.101. [DOI] [PubMed] [Google Scholar]
- 40.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 41.Strom, D., J. Westendorf, B. Lutterbach, J. Nip, J. Downing, H. Harada, N. Lenny, and S. Hiebert. Enforced expression of the AML-1 oncogene shortens the G1 phase of the cell cycle. Submitted for publication. [DOI] [PubMed]
- 42.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p [see comments] Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 43.Uchida H, Zhang J, Nimer S D. AML1A and AML1B can transactivate the human IL-3 promoter. J Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- 44.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1/ETO, associates with C/EBPα, inhibits C/EBPα-dependent transcription, and blocks granuloctyic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]