Abstract
Objective
Spinocerebellar ataxia type 2 (SCA2) is one of the most common autosomal dominant ataxias in the world. Several reports revealed that CAG repeats in some polyQ-containing genes may affect the age at onset (AAO) of patients with SCA2, however, little studies were conducted among Chinese patients with SCA2. Thus, the aim of this study is to evaluate the effect of CAG repeats on the AAO of patients with SCA2 in China.
Methods
A total of 119 patients with SCA2 were enrolled and were divided into 2 groups according to their major phenotype: 17 patients from 9 families with Parkinson’s syndrome were grouped as the Parkinson’s disease-SCA2 (PD-SAC2); 91 patients from 66 SCA2 families and 11 sporadic SCA2 patients were grouped as the ataxia-SCA2 (A-SCA2). Blood samples were obtained from the subjects, and the CAG repeat length in ATXN2 and other (CAG)n-containing genes was screened using fluorescent PCR. The Spearman’s rank correlation between the CAG repeat length in (CAG)n-containing genes and AAO was analyzed. Regression analysis was performed to investigate whether the CAG repeat length could explain the variant of AAO. A t-test was used to compare the difference of CAG repeat length in (CAG)n-containing genes between the PD-SAC2 and A-SCA2 groups.
Results
The CAG repeat length in the longer allele of ATXN2 was negatively correlated with AAO of SCA2 (R=-0.251, P<0.05), and the CAG repeat length could explain 41.7% of the variation of AAO. AAO negatively correlated with the CAG repeat length in the shorter allele of ATXN7 (R=-0.251, P=0.006) or in the longer allele of TBP gene (R=-0.197, P=0.034). A tendency of delay in the AAO was also observed in patients with SCA2 carrying the CAG repeat within the ATXN3, CACNA1A, ATXN7, TBP, and RAI1. In addition, we found that the CAG repeat length in ATXN7 and ATXN2 between the A-SCA2 and the PD-SCA2 groups was significantly different (both P<0.05).
Conclusion
The CAG repeat in ATXN2 is a major genetic factor for the AAO of patients with SCA2 in China. The CAG repeat length in ATXN3, CACNA1A, ATXN7, TBP, and RAI1 genes might be a potential factor associated with the AAO of SCA2. The CAG repeat in ATXN7 might be a potential factor affecting the Parkinson's syndrome in SCA2.
Keywords: spinocerebellar ataxia type 2, Parkinson’s syndrome, (CAG)n-containing genes, CAG repeat length
Abstract
目的
脊髓小脑共济失调2型(spinocerebellar ataxia type 2,SCA2)是世界上最常见的常染色体显性遗传的共济失调之一。多篇报道显示某些含polyQ基因的CAG重复序列可能影响SCA2患者的发病年龄(age at onset,AAO),但在中国SCA2患者中进行研究的较少。因此,本研究旨在探讨CAG重复序列的长度对中国SCA2患者AAO的影响。
方法
纳入119例SCA2患者,根据其主要表型分为2组:17例来自9个帕金森综合征家庭的SCA2患者作为帕金森病-SCA2(Parkinson's disease-SCA2,PD-SAC2)组,91例来自66个SCA2家庭和11例散发的SCA2患者作为共济失调-SAC2(ataxia-SCA2,A-SCA2)组。使用荧光PCR筛查ATXN2和其他含(CAG)n基因中CAG重复序列的长度。采用Spearman's等级相关的方法分析含(CAG)n基因中CAG重复序列的长度与AAO的相关性,采用回归分析评估CAG重复序列的长度对AAO变异的贡献,采用t检验比较PD-SAC2组与A-SCA2组间含(CAG)n基因中CAG重复序列的长度。
结果
ATXN2基因中含较长CAG重复序列的等位基因的CAG重复序列的长度与SCA2的AAO呈负相关 (R=-0.251,P<0.05),可解释41.7%的AAO变异。AAO与ATXN7基因中含较短CAG重复序列的等位基因(R=-0.251,P=0.006)及TBP基因中含较长CAG重复序列的等位基因(R=-0.197,P=0.034)的CAG重复序列的长度均呈负相关。在携带含CAG重复序列的ATXN3、CACNA1A、ATXN7、TBP和RAI1基因的SCA2患者中也检测到AAO延迟的趋势。此外,ATXN7基因和ATXN2基因的CAG重复序列的长度在A-SCA2组和PD-SCA2组之间的差异有统计学意义(均P<0.05)。
结论
ATXN2中的CAG重复序列是影响中国SCA2患者AAO的主要遗传因素。ATXN3、CACNA1A、ATXN7、TBP和RAI1基因的CAG重复序列的长度可能是与SCA2的AAO相关的因素。ATXN7基因中的CAG重复序列的长度可能是SCA2患者表现为帕金森综合征的影响因素之一。
Keywords: 脊髓小脑共济失调2型, 帕金森综合征, 含(CAG)n基因, CAG重复序列的长度
http://xbyxb.csu.edu.cn/xbwk/fileup/PDF/202108793.pdf
Autosomal dominant cerebellar ataxia type 2, also called spinocerebellar ataxia type 2 (SCA2), is one of the most common autosomal dominant ataxias in the world, characterized by progressive cerebellar symptoms of imbalance, gait and limb ataxia, dysarthria accompanied with multiple peripheral neuropathy and extrapyramidal symptoms in some patients. Clinically, SCA2 is highly heterogeneous with a few cases presented with Parkinson's syndrome simultaneously with a good L-dopa response.
The CAG repeat length in SCA2 is inversely related to age at onset (AAO) and directly related to disease severity[1-2]. However, only 48% to 76% of onset variance has been attributed to CAG repeat length, indicating that other factors such as genetic modifiers or environmental might also affect AAO[1-2].
SCA2 is one of the most common subtypes of SCAs in China, accounting for approximately 7.23% of SCAs[3]. Several studies[2, 4-5] have revealed that CAG repeats in some poly Q-containing genes may affect the AAO of SCA2, such as the ATXN3, CACAN1A, ATXN7, and RAI1 genes. However, studies on Chinese patients with SCA2 regarding candidate modifying factors involved in the variability in AAO are rare.
Therefore, to elucidate the genetic factors of the SCA2 subtype in China, we summarized 10 (CAG)n-containing genes (ATXN1, ATXN3, CACNA1A, ATXN7, TBP, ATN1, IT15, HDL2, RAI1, and KCNN3) that are related to neurodegenerative diseases and enrolled 119 SCA2 patients, the largest cohort in China so far, to explore the influence of CAG repeat length on the variability of AAO.
1. Subjects and methods
1.1. Subjects
A total of 119 patients with a molecular diagnosis of SCA2 were recruited from Xiangya Hospital, Central South University, and divided into 2 groups according to their major phenotype: 17 patients from 9 families with Parkinson's syndrome were grouped as Parkinson's disease- SCA2 (PD-SAC2); 91 patients from 66 SCA2 families and 11 sporadic SCA2 patients were grouped as ataxia-SCA2 (A-SCA2). The AAO was defined by the age at first appearance of the phenotype of ataxia or Parkinson's syndrome fulfilled the Movement Disorder Society (MDS) clinical diagnostic criteria for PD.
1.2. Specimen collection and genetic analyses
Blood samples were obtained from the subjects after obtaining informed consent. Written informed consent was obtained from all subjects, as approved by the Ethics Committee and the Expert Committee of Xiangya Hospital, Central South University (equivalent to an Institutional Review Board).
Two alleles of 10 genes that contain (CAG)n in their coding sequences were selected/suspected as potential modifiers of AAO, including 1) ATXN1, ATXN3, CACNA1A, ATXN7, TBP, and ATN1, 6 SCA pathogenic genes; 2) IT15 and HDL2,2 genes associated with other neurological diseases; 3) RAI1 and KCNN3, 2 genes associated with the AAO of SCA2 or other subtypes of SCAs. The length of CAG repeat in 2 alleles of the ATXN2 gene was also screened in all SCA2 patients. Here, we defined the allele contained CAG repeat expansion in ATXN2 as an expanded allele, the smaller one as a normal allele; the allele contained larger repeat number without expansion in other 10 genes as the longer allele, and the smaller one as the shorter allele. The longer and shorter alleles were analyzed separately.
The target sequences covering the (CAG)n were amplified by fluorescent PCR, with the primers and conditions listed in the supplementary materials (Supplementary Table 1 and 2, https://doi.org/10.11817/j.issn.1672-7347.2021.210230T1). The PCR products were analyzed using an ABI-Prism 3 730 Genetic Analyzer, and the data were examined using GeneMapper software.
Table 1.
Correlation analysis between the AAO and the CAG repeat length of (CAG)n-containing genes
| Genes | CAG repeat length of shorter allele | CAG repeat length of longer or expanded allele | ||
|---|---|---|---|---|
| R | P | R | P | |
| ATXN1 | 0.067 | 0.490 | 0.112 | 0.266 |
| ATXN2 | -0.046 | 0.623 | -0.598 | <0.001 |
| ATXN3 | -0.060 | 0.524 | 0.011 | 0.906 |
| CACNA1A | -0.057 | 0.544 | 0.026 | 0.787 |
| ATXN7 | -0.251 | 0.006 | -0.151 | 0.104 |
| TBP | 0.006 | 0.952 | -0.197 | 0.034 |
| ATN1 | -0.029 | 0.762 | -0.012 | 0.897 |
| IT15 | -0.039 | 0.676 | -0.002 | 0.979 |
| HDL2 | -0.045 | 0.628 | -0.162 | 0.082 |
| RAI1 | -0.121 | 0.195 | -0.071 | 0.451 |
| KCNN3 | -0.041 | 0.664 | -0.031 | 0.741 |
Table 2.
Coefficients of regression equation by using ATXN2E2 and ATXN2E3, ATXN3, CACNA1A, or RAI1 as independent variables and lgAAO as dependent variables
| Models | Unstandardized Coefficients | Beta* | t | P | |
|---|---|---|---|---|---|
| B | SE | ||||
| using ATXN2E2 and ATXN2E3 as independent variables | |||||
| (Constant) | 2.931 | 0.197 | 14.854 | <0.001 | |
| ATXN2E3 | 0.016 | <0.001 | 3.967 | 5.475 | <0.001 |
| ATXN2E2 | -0.002 | <0.001 | -4.463 | -6.160 | <0.001 |
| Multiple linear regression with ATXN3 longer allele | |||||
| (Constant) | 3.023 | 0.210 | 14.368 | <0.001 | |
| ATXN2E2 | -0.002 | <0.001 | -4.630 | -6.298 | <0.001 |
| ATXN2E3 | 0.017 | <0.001 | 4.132 | 5.622 | <0.001 |
| ATXN3 | -0.002 | 0.002 | -0.089 | -1.242 | 0.217 |
| Multiple linear regression with CACNA1A longer allele | |||||
| (Constant) | 3.048 | 0.206 | 14.798 | <0.001 | |
| ATXN2E2 | -0.002 | <0.001 | -4.504 | -6.273 | <0.001 |
| ATXN2E3 | 0.017 | <0.001 | 3.998 | 5.569 | <0.001 |
| CACNA1A | -0.007 | 0.004 | -0.127 | -1.799 | 0.075 |
| Multiple linear regression with RAI1 longer allele | |||||
| (Constant) | 3.261 | 0.260 | 12.537 | <0.001 | |
| ATXN2E2 | -0.002 | <0.001 | -4.525 | -6.311 | <0.001 |
| ATXN2E3 | 0.017 | <0.001 | 4.034 | 5.625 | <0.001 |
| RAI1 | -0.027 | 0.014 | -0.135 | -1.921 | 0.057 |
*Standardized coefficients. B: Coefficient; SE: Standard error.
1.3. Statistical analyses
Statistical analysis was performed using the data analysis software SPSS 22.0. The Spearman's rank correlation of the CAG repeat length in (CAG)n-containing genes and AAO was analyzed. Regression analysis was performed to investigate whether the CAG repeat length could explain the variant of AAO. The Mann-Whitney U test was used to compare the CAG repeat length of the 2 alleles of the ATXN1, ATXN3, CACNA1A, ATXN7, TBP, ATN1, IT15, HDL2, RAI1, and KCNN3 genes and the normal allele of ATXN2 between the A-SCA2 and PD-SCA2 groups. The 2 independent samples t-test was used to test the difference in the ATXN2 expanded allele. All continuous data are presented as mean±standard deviation (SD). P<0.05 was considered statistically significant.
2. Results
2.1. AAO and repeat length distribution of patients
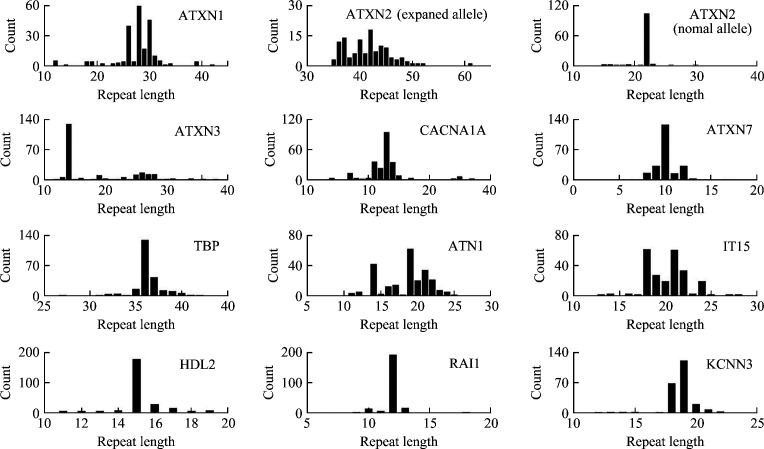
The AAO of the SCA2 patients was (35.01±12.05) years. The CAG repeat length was 41.50±4.31 in the expanded allele of the ATXN2 gene, and 21.77±1.62 in the normal allele. The CAG repeat length of ATXN2 and the other 10 (CAG)n-containing genes are showed in supplementary materials (Supplementary Table 3, https://doi.org/10.11817/j.issn.1672-7347.2021.210230T2) and Figure 1.
Figure 1. Frequency distribution of the CAG repeat length in (CAG)n-containing genes.
2.2. Effect of ATXN2 expanded allele on AAO
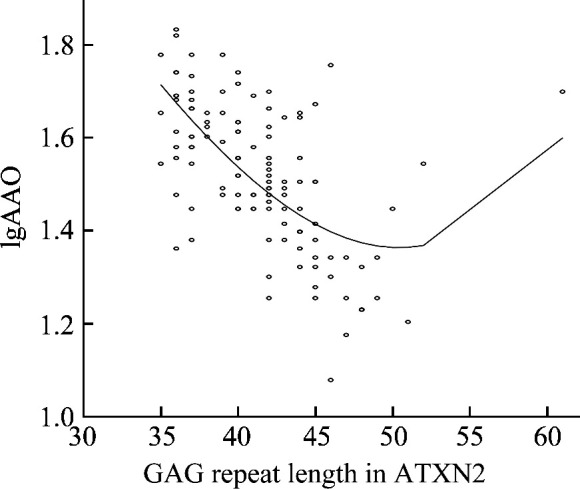
Using the logarithmically transformed AAO denoted as lgAAO and the expanded repeat length (ATXN2E) as an independent variable, Spearman’s rank correlation analysis was conducted to analyze the correlation between ATXN2 and lgAAO (R=-0.598, P<0.001; Table 1). The linear equations, quadratic equations, and cubic equations showed that the cubic equation had the best fit with an R 2 value of 0.408 (P< 0.001).
To further investigate the relationship between ATXN2 and AAO, ATXN2 was subjected to a square transformation (denoted as ATXN2E2) and to a cubic transformation (denoted as ATXN2E3). Multiple linear regression analysis was performed using ATXN2, ATXN2E2, and ATXN2E3 as independent variables and lgAAO as the dependent variable. As a result, we constructed a bivariate linear equation involving ATXN2E2 and ATXN2E3, and then the adjusted R 2 increased from 0.408 to 0.417, which suggests that the CAG repeat length could explain 41.7% of the AAO variation (Table 2 and Figure 2). Meanwhile, when excluded PD-SCA2 group patients, the adjusted R 2 decreased to 0.390.
Figure 2. Curve of the CAG repeat length in expanded allele of ATXN2 gene and lgAAO.
2.3. Effect of ATXN7 and TBP on AAO
Interestingly, there was also a correlation between AAO and the shorter allele of ATXN7 (R=-0.251, P=0.006) and the longer allele of TBP gene (R=-0.197, P=0.034) by using Spearman’s rank correlation analysis (Table 1). Unexpectedly, the following regression analysis fails to explain the variation of AAO of SCA2. And there were no correlations between these 2 genes and AAO when the PD-SCA2 patients were excluded (ATXN7 gene shorter allele, R=-1.44, P=0.148; TBP gene longer allele, R=-0.153, P =0.124).
2.4. Effects of other (CAG)n-containing genes on the AAO of SCA2 patients
The CAG repeat length, including the longer and shorter alleles of ATXN1, ATXN3, CACNA1A, ATN1, IT15, HDL2, RAI1, KCNN3, and the normal allele of ATXN2 was used as the third independent variable for multiple linear regression with ATXN2E2 and ATXN2E3, respectively. We found that R 2 increased when the CAG repeat length of a longer allele of ATXN3 or CACNA1A gene or the shorter allele of the RAI1 gene was used as the third independent variable. Unfortunately, there was no statistically significant P-value detected in the ATXN3 (R 2=0.435, P=0.217), CACNA1A (R 2=0.433, P=0.075), and RAI1 (R 2=0.435, P=0.119) genes (Table 2). Additionally, we did not find any other increase in R 2 within other (CAG)n-containing genes.
2.5. Differences in CAG repeat length between the PD-SCA2 and A-SCA2 groups
Except the difference in the CAG repeat length of ATXN2 expanded allele (P<0.001, 95% CI 3.967 to 7.896) and both alleles of ATXN7 (longer allele, P=0.042; shorter allele, P=0.003) within these 2 groups, we did not find other diversity between them (data not shown).
3. Discussion
SCA2 is a neurodegenerative disease with a high degree of hereditary and clinical heterogeneity. According to the reports, many factors may affect the AAO of SCA2. For the first time, we investigated the modifying factors of AAO within a group of SCA2 patients in China. The results showed that the CAG repeat length of an expanded allele in the ATXN2 gene could explain approximately 41.7% of the AAO variance, suggesting that the expanded CAG repeat length in the ATXN2 gene was the main genetic factor affecting AAO. Filla et al[6] investigated the CAG repeat length in 85 patients from 30 SCA2 families and found that the expanded CAG repeat length in the ATXN2 gene could explain 76% of the AAO variation, which is the largest proportion reported so far. In contrast, Hayes et al[5] found that the value is only 48% in the research including 46 patients in 10 families and 47 unrelated patients. Our result showed a lower rate of AAO variation, which only explains 41.7%. One reason may be different genetic background, the other reason may be sporadic patients recruited or not[6-7, 2]. Considering that uninterrupted CAG repeats could impactfully affect the AAO of Huntington's disease (HD)[8], the potential role of CAG repeat interruptions of ATXN2 gene in AAO needs to be confirmed carefully in our further research.
Of interest, we found that the CAG repeat length in the shorter allele of the ATXN7 gene was negatively correlated with the AAO of SCA2. Even more striking, the CAG repeat length in the 2 alleles of the ATXN7 gene was different between the PD-SCA2 and A-SCA2 groups. The facts about the relationship between ATXN7 and the AAO of SCA2 were already reported by Tezenas et al[4] who found that the SCA2 patients with a CAG repeat expansion more than 12 in the ATXN7 gene have earlier disease onset than those with CAG repeat length less than 12. Notably, as far as we know, this is the first time we discovered that the ATXN7 gene could influence the parkinsonian features of SCA2. In an SCA7 patient from another study[9], the researcher observed neuronal loss in the substantia nigra, which may offer a possible mechanism to explain our result.
Moreover, we found that the CAG repeat length in the longer allele of the TBP was also negatively correlated with the AAO of SCA2, although regression analysis failed to find an effective ratio to explain the variant. The previous study[10] that the number of neurons containing neuronal intranuclear inclusions (NIIs) immunolabeled with anti-TBP protein antibody twice that with the anti-ataxin-2 in SCA2 suggested the important role of TBP protein in SCA2. In addition, some SCA2 patients always present with ataxia and extrapyramidal symptoms[11]. The above findings suggest a potential common pathologic pathway between the TBP and ATXN2 genes. However, little information exists regarding the relationship between these 2 genes and the role of the TBP protein in the pathogenesis of SCA2, and it needs to be elucidated in the future.
Furthermore, within the other 10 (CAG)n-containing genes, there is some association between the CAG repeat length and the AAO in ATXN3, CACNA1A, and RAI1, although a statistically significant P-value could not be reached. The CAG repeat length in the ATXN3 gene is associated with the AAO of SCA2[4], and the CAG repeat length in the CACNA1A gene could explain 5.81% of the AAO variation in 64 unrelated patients[2]. Hayes et al[5] reported that approximately 4.1% of the AAO variation was contributed by the CAG repeat length of the RAI1 gene in 46 SCA2 patients. Our results showed the similar tendency, indicating that the CAG repeats within the ATXN3, CACNA1A, and RAI1 might influence the AAO of SCA2, but more studies with large scale samples are needed to confirm the results.
In conclusion, our results validated that the CAG repeat in the ATXN2 expanded allele, which accounts for 41.7% of variation of AAO, is the major genetic factor of the AAO of SCA2 patients in China. We provided an evidence that the CAG repeat length in the ATXN3, CACNA1A, ATXN7, TBP, and RAI1 genes might be a potential factor associated with the AAO of SCA2. The CAG repeat length of the ATXN7 gene might be a potential factor affecting the parkinsonian syndrome in SCA2.
Appendix.
Supplementary Table 1 Primers used for PCR
| Genes | Subtypes | Primers | |
|---|---|---|---|
| ATXN1 | SCA1 | F | 5'-FAM-AACTGGAAATGTGGACGTA-3' |
| R | 5'-CAACATGGGCAGTCTGAG-3' | ||
| ATXN2 | SCA2 | F | 5'-FAM-GGGCCCCTCACCATGTCG-3' |
| R | 5'-GAGGACGAGGAGACCGAGGAC-3' | ||
| ATXN3 | SCA3 | F | 5'-FAM-CCAGTGACTACTTTGATTCG-3' |
| R | 5'-CTTACCTAGATCACTCCCAA-3' | ||
| CACNA1A | SCA6 | F | 5'-FAM-CACGTGTCCTATTCCCCTGTGATCC-3' |
| R | 5'-TGGGTACCTCCGAGGGCCGCTGGTG-3' | ||
| ATXN7 | SCA7 | F | 5'-FAM-TGTTACATTGTAGGAGCGGAA-3' |
| R | 5'-CACGACTGTCCCAGCATCACTT-3' | ||
| TBP | SCA17 | F | 5'-FAM-ATGCCTTATGGCACTGGACTG-3' |
| R | 5'-CTGCTGGGACGTTGACTGCTG-3' | ||
| ATN1 | DRPLA | F | 5'-FAM-TGACCAACAGCAATGCCCATCCAG-3' |
| R | 5'-TCAGAGACCCAGGGAGGGAGACAT-3' | ||
| RAI1 | GD | F | 5'-FAM-GGGGCAGCGGGTCCAGAATCTTC -3' |
| R | 5'-CTGGCCTTGCTGCCCGTAGTGCT-3' | ||
| IT15 | HD | F | 5'-FAM-CGACCCTGGAAAAGCTGATG-3' |
| R | 5'-GGCTGAGGAAGCTGAGGAG-3' | ||
| HDL2 | \ | F | 5' -FAM-GGTTCCCTGCACAGAAACCATC-3' |
| R | 5'-AGATGCCACCGCATTCGG-3' | ||
| KCNN3 | \ | F | 5'-FAM-GCAGCCCTGGGACCCTCGCT-3' |
| R | 5'-ACATGTAGCTGTGGAACTTGGAGAGT-3' | ||
Supplementary Table 2 PCR condition for amplified (CAG)n in each (CAG)n contained genes
| Genes | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 |
|---|---|---|---|---|---|
| ATXN1 | 95 ℃, 5 min | 95 ℃, 30 s; 67 ℃, 30 s; 72 ℃, 30 s; 25 cycles | 72 ℃, 10 min | 4 ℃, hold | |
| ATXN2 | 95 ℃, 5 min | 95 ℃, 45 s; 66 ℃(-0.5 ℃/cycle), 30 s; 72 ℃, 1 min; 10 cycles | 95 ℃, 45 s; 62 ℃, 30 s; 72 ℃, 1 min; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| ATXN3 | 95 ℃, 5 min | 95 ℃, 30 s; 62 ℃(-0.5 ℃/cycle), 30 s; 72 ℃, 30 s; 10 cycles | 95 ℃, 30 s; 52 ℃, 30 s; 72 ℃, 30 s; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| CACNA1A | 95 ℃, 5 min | 95 ℃, 45 s; 62 ℃(-1 ℃/cycle), 50 s; 72 ℃, 1 min; 10 cycles | 95 ℃, 45 s; 52 ℃, 50 s; 72 ℃, 1 min; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| ATXN7 | 95 ℃, 5 min | 95 ℃, 45 s; 70 ℃(-1 ℃/cycle), 50 s; 72 ℃, 1 min; 15 cycles | 95 ℃, 45 s; 55 ℃, 50 s; 72 ℃, 1 min; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| TBP | 95 ℃, 5 min | 95 ℃, 30 s; 65 ℃, 30 s; 72 ℃, 30 s; 40 cycles | 72 ℃, 10 min | 4 ℃, hold | |
| ATN1 | 95 ℃, 5 min | 95 ℃, 1 min; 68 ℃, 30 s; 72 ℃, 1 min; 40 cycles | 72 ℃, 10 min | 4 ℃, hold | |
| BAI1 | 95 ℃, 5 min | 95 ℃, 30 s; 68 ℃, 30 s; 72 ℃, 30 s; 40 cycles | 72 ℃, 10 min | 4 ℃, hold | |
| IT15 | 95 ℃, 5 min | 95 ℃, 45 s; 66 ℃(-0.5 ℃/cycle), 30 s; 72 ℃, 1 min; 10 cycles | 95 ℃, 45 s; 62 ℃, 30 s; 72 ℃, 1 min; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| HDL2 | 95 ℃, 5 min | 95 ℃, 45 s; 70 ℃(-1 ℃/cycle), 50 s; 72 ℃, 1 min; 15 cycles | 95 ℃, 45 s; 55 ℃, 50 s; 72 ℃, 1 min; 35 cycles | 72 ℃, 10 min | 4 ℃, hold |
| KCNN3 | 95 ℃, 5 min | 95 ℃, 30 s; 70 ℃, 30 s; 72 ℃, 30 s; 40 cycles | 72 ℃, 10 min | 4 ℃, hold |
Supplementary Table 3 CAG repeat length within the (CAG)n-containing genes
| Genes | CAG repeat length of shorter allele | CAG repeat length of longer or expanded allele | ||
|---|---|---|---|---|
| Mean±SD | Range | Mean±SD | Range | |
| ATXN1 | 26.40±4.38 | 12-39 | 28.83±3.37 | 12-39 |
| ATXN2 | 21.77±1.62 | 15-30 | 41.50±4.31 | 35-61 |
| ATXN3 | 15.40±3.80 | 12-30 | 21.96±6.65 | 14-38 |
| CACNA1A | 11.91±3.68 | 4-27 | 13.74±2.71 | 7-27 |
| ATXN7 | 9.93±1.01 | 8-13 | 10.33±1.35 | 8-17 |
| TBP | 35.84±1.49 | 27-40 | 36.86±1.49 | 32-42 |
| ATN1 | 17.45±2.92 | 11-22 | 19.42±3.02 | 11-25 |
| IT15 | 19.39±1.79 | 27-40 | 21.10±2.16 | 14-28 |
| HDL2 | 15.01±0.78 | 11-18 | 15.53±1.03 | 14-19 |
| RAI1 | 11.72±0.79 | 9-13 | 12.37±0.86 | 11-18 |
| KCNN3 | 18.44±1.35 | 12-21 | 19.02±0.92 | 15-22 |
SD: Standard deviation.
Funding Statement
This work was supported by the National Key Research and Development Program (2018YFC1312003), the National Natural Science Foundation (81671120, 81300981), the Natural Science Foundation for Distinguished Young Scholars of Hunan Province(2020JJ2057), the Degree & Postgraduate Education Reform Project of Central South University (2020JGB136), and the Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital) (2020LCJJ13), China.
Conflict of Interest
The authors declare that they have no conflicts of interest to disclose.
Note
http://xbyxb.csu.edu.cn/xbwk/fileup/PDF/202108793.pdf
References
- 1. Cali F, Chiavetta V, Ragalmuto A, et al. Comparative multiplex dosage analysis in spinocerebellar ataxia type 2 patients[J]. Genet Mol Res, 2013, 12(2): 1176-1181. [DOI] [PubMed] [Google Scholar]
- 2. Pulst SM, Santos N, Wang D, et al. Spinocerebellar ataxia type 2: polyQ repeat variation in the CACNA1A calcium channel modifies age of onset[J]. Brain, 2005, 128(Pt10): 2297-2303. [DOI] [PubMed] [Google Scholar]
- 3. Chen Z, Wang P, Wang C, et al. Updated frequency analysis of spinocerebellar ataxia in China[J]. Brain, 2018, 141(4): e22. [DOI] [PubMed] [Google Scholar]
- 4. Tezenas du Montcel S, Durr A, Bauer P, et al. Modulation of the age at onset in spinocerebellar ataxia by CAG tracts in various genes[J]. Brain, 2014, 137(Pt9): 2444-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayes S, Turecki G, Brisebois K, et al. CAG repeat length in RAI1 is associated with age at onset variability in spinocerebellar ataxia type 2 (SCA2)[J]. Hum Mol Genet, 2000, 9(12): 1753-1758. [DOI] [PubMed] [Google Scholar]
- 6. Filla A, De Michele G, Santoro L, et al. Spinocerebellar ataxia type 2 in southern Italy: a clinical and molecular study of 30 families[J]. J Neurol, 1999, 246(6): 467-471. [DOI] [PubMed] [Google Scholar]
- 7. Laffita-Mesa JM, Velazquez-Perez LC, Santos Falcon N, et al. Unexpanded and intermediate CAG polymorphisms at the SCA2 locus (ATXN2) in the Cuban population: evidence about the origin of expanded SCA2 alleles[J]. Eur J Hum Genet, 2012, 20(1): 41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright GEB, Collins JA, Kay C, et al. Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease[J]. Am J Hum Genet, 2019, 104(6): 1116-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rüb U, Brunt ER, Seidel K, et al. Spinocerebellar ataxia type 7 (SCA7): widespread brain damage in an adult-onset patient with progressive visual impairments in comparison with an adult-onset patient without visual impairments[J]. Neuropathol Appl Neurobiol, 2008, 34(2): 155-168. [DOI] [PubMed] [Google Scholar]
- 10. Uchihara T, Fujigasaki H, Koyano S, et al. Non-expanded polyglutamine proteins in intranuclear inclusions of hereditary ataxias—triple-labeling immunofluorescence study[J]. Acta Neuropathol, 2001, 102(2): 149-152. [DOI] [PubMed] [Google Scholar]
- 11. Choubtum L, Witoonpanich P, Kulkantrakorn K, et al. Trinucleotide repeat expansion of TATA-binding protein gene associated with Parkinson's disease: A Thai multicenter study[J]. Parkinsonism Relat Disord, 2016, 28: 146-149. [DOI] [PubMed] [Google Scholar]