Abstract
Nuclear receptor corepressor (CoR)-histone deacetylase (HDAC) complex recruitment is indispensable for the biological activities of the retinoic acid receptor fusion proteins of acute promyelocytic leukemias. We report here that ETO (eight-twenty-one or MTG8), which is fused to the acute myelogenous leukemia 1 (AML1) transcription factor in t(8;21) AML, interacts via its zinc finger region with a conserved domain of the corepressors N-CoR and SMRT and recruits HDAC in vivo. The fusion protein AML1-ETO retains the ability of ETO to form stable complexes with N-CoR/SMRT and HDAC. Deletion of the ETO C terminus abolishes CoR binding and HDAC recruitment and severely impairs the ability of AML1-ETO to inhibit differentiation of hematopoietic precursors. These data indicate that formation of a stable complex with CoR–HDAC is crucial to the activation of the leukemogenic potential of AML1 by ETO and suggest that aberrant recruitment of corepressor complexes is a general mechanism of leukemogenesis.
Chromatin modifications by histone acetylases (HATs) or histone deacetylases (HDACs) represent a fundamental mechanism of transcriptional regulation (reviewed in references 13, 16, 19, 26, 56, and 59). It has been proposed that histone acetylation weakens interactions of histones with DNA and induces alterations in nucleosome structure, enhancing accessibility of targeted promoters to components of the transcription machinery, thus increasing transcription (52, 61). Recently reported results support this model. Several transcriptional coactivators, including GCN5 (5, 6), CBP/p300 (47), PCAF (69), ACTR (8), SRC-1 (55), and TAF(II)250 (43), have intrinsic HAT activity. Moreover, in some cases, HAT activity has been shown to be required for coactivator function (34, 35, 63).
Conversely, decreased histone acetylation due to the action of HDACs is thought to lead to a less accessible chromatin conformation, resulting in repression of transcription (16, 26, 27). Macromolecular complexes containing the yeast HDAC homologue Rpd3 are involved in gene silencing in yeast (30, 53, 54, 62). There are multiple mammalian Rpd3 homologues with HDAC activity (20, 58, 67, 68). Unlike activation complexes, in which coactivators have intrinsic HAT activity, one of the functions of corepressors (CoR), including N-CoR (25), SMRT (9), and mSin3 (2), is to recruit HDAC to large multiprotein repression complexes (1, 18, 22, 37, 44). Recruitment of HDAC-containing complexes is involved in repression by a number of mammalian silencers, including nuclear hormone receptors (1, 22, 44), Mad (18, 37), YY1 (67), CBF (29), and PLZF (14, 17, 21, 24, 39). In addition to N-CoR, SMRT, and Sin3, the corepressor complexes include SAP18 (76), retinoblastoma-associated proteins (76), SUN-CoR (71), and SAP30 (36, 77). Immunoprecipitates of Sin3 contain numerous other polypeptides that are likely to be components of corepressor complexes (36, 77).
Studies of the retinoic acid receptor alpha (RARα) fusion proteins of acute promyelocytic leukemia (APL) provide further evidence of a biological role of the corepressor complex (10, 14, 17, 21, 39). Recruitment of HDACs is indispensable for the capacity of APL fusion proteins (PML-RARα and PLZF-RARα) to block myeloid differentiation and is responsible for the retinoic acid-resistant phenotype of PLZF-RARα APLs (10, 14, 17, 21, 39). In addition, translocations involving p300 and CBP HATs are found in rare cases of acute myeloid leukemia (AML) (12), strengthening the link between tight control of histone acetylation and normal cell growth and differentiation.
Transcription factor AML1 is the most frequent target of chromosomal translocations in AML (23, 46). The AML1 gene encodes various isoforms, sharing a DNA binding domain highly homologous to that of the Drosophila runt transcription factor (3, 28). The AML1B isoform behaves as a transcriptional activator and is able to recruit p300 HAT (32) and activate a set of target genes, including those for macrophage colony-stimulating factor (50), granulocyte-macrophage colony-stimulating factor (11), interleukin-3 (60), neutrophil elastase (45), myeloperoxidase (4, 45), and T-cell receptor subunits (41), that are essential for definitive hematopoiesis of all lineages (49). In the t(8;21) chromosome translocation, AML1 recombines with the ETO (eight-twenty-one or MTG8) zinc finger nuclear protein, a putative transcription factor homologous to the Drosophila gene nervy (23, 42, 46).
The resulting AML1/ETO fusion protein retains the runt homology domain, but the AML1B transactivation domain has been replaced with ETO (23, 38, 46). AML1/ETO antagonizes AML1B function, and the ETO portion of the fusion protein is required for its effect (38, 41, 64, 78). AML/ETO behaves as a transcriptional repressor on AML1 target genes and inhibits differentiation of hematopoietic precursors in vivo (48, 64, 70, 78). We show here that ETO interacts with CoRs and recruits HDAC activity in vivo. The AML1/ETO fusion protein retains the ability of ETO to form stable complexes with CoRs, and CoR binding and HDAC recruitment correlate with the ability of AML1/ETO to inhibit terminal differentiation of hematopoietic precursors.
MATERIALS AND METHODS
Yeast two-hybrid screen.
A yeast two-hybrid screen of a 17-day mouse embryo library (Clontech) with SMRT (amino acids 1 to 483) as bait was performed as previously described (72). We screened 1.4 million independent colonies and obtained 13 identical interacting clones containing the partial ETO cDNA. A partial ETO cDNA was isolated during two-hybrid screening, and additional sequences were obtained by PCR using the cDNA library DNA as the template.
Eukaryotic expression vectors.
Flag-ETO and Flag-ETOΔC (amino acids 1 to 416) constructs were subcloned into the pFlag-CMV2 vector (Eastman Kodak). The AML1/ETO cDNA was kindly provided by S. Nimer. The Gal4-SMRT repression domain (RD; amino acids 1 to 483) and Gal4-N-CoR RD1 (amino acids 1 to 312) were generated by PCR and cloned into the pCMX-Gal4 DBD (DNA binding domain) vector (75). The Gal4-SMRT receptor interaction domain (amino acids 983 to 1485) has been previously described (75). Mutant ETOs were made by PCR or restriction enzyme digestion and cloned into the pCMX-HA vector (71) or a Gal4 DBD expression vector. The green fluorescent protein (GFP)-AML1/ETO and GFP-AML1/ETOΔC fusion proteins were generated by PCR and cloned into a modified PINCO retroviral vector (not containing the cytomegalovirus-GFP cassette) (15). The ETOΔC and AML/ETOΔC constructs were truncated at amino acid 416 of ETO. ETOΔZnF (lacking amino acids 488 to 525) and ETO-C488S and -C508S mutants were constructed by using PCR. All PCR products, mutations, and fusion junctions were confirmed by sequencing.
In vitro interaction assays.
SMRT (amino acids 1 to 483) and N-CoR (amino acids 1 to 312) were generated by PCR and cloned into the pGex4T-1 vector. Glutathione S-transferase (GST) pulldowns were performed as previously described (14, 75). Input lanes show 10% of the total input.
Cell culture and transfection.
293T cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. In coimmunoprecipitation experiments, cells were transfected with Lipofectamine reagent (GIBCO) in accordance with the manufacturer’s instructions. In the luciferase reporter assay, cells were transfected by the calcium phosphate precipitation method. The Gal4 UAS × 5-SV40-luciferase reporter contains five copies of the Gal4 17-mer binding site. Light units were normalized to expression of a cotransfected β-galactosidase expression plasmid. U937 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. The amphotropic packaging Phoenix cell line was transfected by the calcium phosphate-chloroquine method, and the U937 cells were infected as previously described (15).
Coimmunoprecipitation and Western analysis.
Immunoprecipitation assays were performed with 293T cells. Cells were lysed in a buffer (1× phosphate-buffered saline, 10% glycerol, 1% Nonidet P-40, 100 μM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride plus protease inhibitors). After sonication, whole-cell extracts were clarified by centrifugation. Immunoprecipitations were performed at 4°C by using antibodies to Flag (M2; Research Diagnostics), nuclear CoR (N-CoR) (amino acids 150 to 425), or N-CoR (amino acids 1944 to 2453) (71) or to a Myc tag (Oncogene Research), followed by Western blotting as described previously (66). The rabbit polyclonal antibody to HDAC1 used was a generous gift of C. Hassig and S. Schreiber (Harvard University) (20).
HDAC assay.
[3H]acetate-labeled histones were prepared as previously described (7) from 293T cells. Immunoprecipitated complexes on protein G-agarose beads were incubated at 37°C for 1 h with [3H]acetate-labeled 293T histones in 200 μl of HDAC buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol). The reaction was stopped by addition of 50 μl of 1 M HCls 0.16 M acetic acid. Released [3H]acetic acid was extracted with ethyl acetate and quantified by liquid scintillation analysis.
Cell differentiation experiments.
Differentiation experiments with vitamin D3 and transforming growth factor β (TGF-β) were performed as previously described (14). The percentages of GFP-positive cells and antigen-positive cells and the fluorescence intensity were evaluated by FACScan.
RESULTS
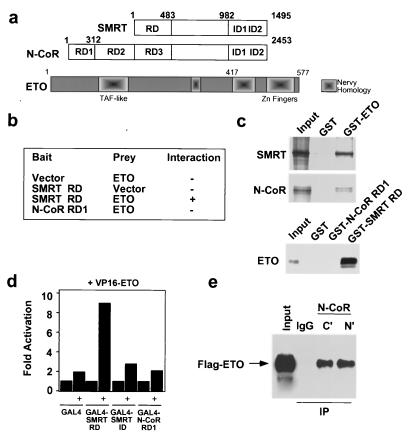
A yeast two-hybrid screen for proteins that interacted with an RD conserved between N-CoR and SMRT (SMRT RD and N-CoR RD3; Fig. 1a) led to the identification of ETO as a CoR-interacting protein (Fig. 1b). Full-length SMRT and N-CoR interacted with ETO in pulldown experiments using a GST-ETO fusion protein and in vitro-translated CoR proteins (Fig. 1c and data not shown). Conversely, GST fusions of the SMRT RD (or N-CoR RD3; data not shown) pulled down in vitro-translated ETO (Fig. 1c). The strength of the interaction between CoR and ETO was at least as great as that which we have observed for nuclear receptor interaction (72). N-CoR RD1 (not present in SMRT, Fig. 1a) did not interact with ETO (Fig. 1b to d), indicating that the ETO-CoR interaction was not a general feature of the CoR RDs. In human 293T cells, a VP16-ETO fusion protein greatly enhanced luciferase activity from a GAL4-based reporter in the presence of the GAL4-SMRT RD, suggesting that the interaction between ETO and SMRT occurred in vivo (Fig. 1d). We also performed coimmunoprecipitation experiments with 293T cells transiently transfected with an epitope-tagged ETO (Flag-ETO) expression vector. Cell lysates were immunoprecipitated with anti-N-CoR antibodies, and the resulting immunocomplexes were analyzed by Western blotting using antibodies directed against the Flag epitope. As shown in Fig. 1e, anti-N-CoR antibodies specifically precipitated ETO, confirming the in vivo interaction between ETO and endogenous N-CoR.
FIG. 1.
In vitro and in vivo interactions of ETO with CoRs. (a) Modular organization of SMRT, N-CoR, and ETO. ID, nuclear receptor interaction domains. TBF-associated factor (TAF)-like and Nervy homology domains of ETO (including Zn fingers) are indicated. (b) Results of a two-hybrid assay using the indicated baits and preys. (c) Pulldown experiments. In vitro-translated SMRT (full-length) and N-CoR (amino acids 1 to 1445) (upper panels) and ETO (lower panel) were precipitated with GST or the indicated GST fusion proteins. (d) Mammalian two-hybrid interaction assay. 293T cells were cotransfected with a VP16ETO expression vector (as indicated) and the appropriate GAL4 fusion protein expression vectors. (e) Coimmunoprecipitations of N-CoR and ETO. Immunoprecipitates with anti-N-CoR antibodies against the N- or C-terminal region and control immunoglobulin G (IgG) were obtained from 293T cells transfected with a Flag-ETO expression vector and blotted with an anti-Flag antibody. The input lane contains 2.5% of the total.
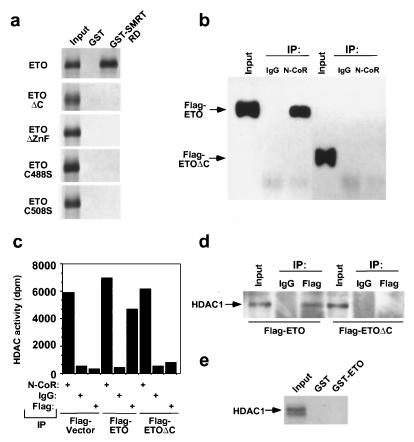
To map the ETO domain(s) required for CoR interaction, we performed yeast two-hybrid (data not shown) and GST pulldown experiments. Truncation of ETO at amino acid 416 (ETOΔC) abrogated the interaction with SMRT (Fig. 2a). Finer mapping of the interaction revealed that deletion of amino acids 488 to 525, containing the zinc fingers of ETO, also prevented CoR interaction. Point mutation of cysteine residues in either zinc finger abolished the interaction (C488S and C508S; Fig. 2a), indicating that both zinc fingers are critical for the interaction between ETO and a CoR. The C-terminal truncation (ETOΔC) that abolished in vitro interaction between ETO and SMRT similarly abolished the ETO–N-CoR interaction in vivo, as demonstrated in coimmunoprecipitation experiments (Fig. 2b).
FIG. 2.
In vivo interaction of ETO with HDAC1 and mapping of the ETO CoR-binding region. (a) Mapping of the CoR interaction domain in ETO. In vitro-translated ETO or mutant ETOs were precipitated with GST or a GST-SMRT RD fusion protein. ETOΔC contains amino acids 1 to 416, ETOΔZnF lacks amino acids 488 to 525, and ETO-C488S and -C508S represent point mutations of the indicated amino acids. All of the ETO proteins were hemogglutinin fusions, except ΔC, which was a Gal4 fusion. (b) In vivo interactions of ETO and endogenous N-CoR. Coimmunoprecipitations of N-CoR and ETO or ETOΔC. Immunoprecipitates with anti-N-CoR antibodies or control immunoglobulin G (IgG) were obtained from 293T cells transfected with a Flag-ETO or Flag-ETOΔC expression vector and blotted with an anti-Flag antibody. The input lane contains 2% of the total. (c) In vivo association of ETO and HDAC. HDAC activity of the indicated immunoprecipitates from 293T cells transfected with the indicated expression vectors. (d) In vivo association of ETO and HDAC. Coimmunoprecipitation of ETO and HDAC1. Immunoprecipitates with an anti-Flag antibody or control immunoglobulin G were obtained from 293T cells transfected with a Flag-ETO or Flag-ETOΔC expression vector and blotted with an anti-HDAC1 antibody. The input lane contains 2% of the total. (e) Lack of direct interaction between HDAC1 and ETO in vitro. In vitro-translated HDAC1 was precipitated with GST or GST-ETO. IP, immunoprecipitation; dpm, disintegrations per minute.
To investigate the possibility that ETO might recruit the HDAC component of the CoR/HDAC complex, anti-Flag immunoprecipitates were analyzed for the presence of HDAC activity and protein (Fig. 2c and d). HDAC activity (Fig. 2c) and HDAC1 protein (Fig. 2d) were undetectable in anti-Flag immunoprecipitates from cells transfected with the empty Flag control vector, although anti-N-CoR antibodies immunoprecipitated levels of HDAC activity comparable to those of Flag-ETO-transfected cells (Fig. 2c and data not shown). HDAC activity was specifically detected in the anti-Flag immunoprecipitates from Flag-ETO-transfected 293T cells (Fig. 2c); likewise, anti-Flag antibodies specifically precipitated significant levels of HDAC1 protein (Fig. 2d). These results confirmed the specific association of ETO with HDAC in vivo. ETO did not interact with HDAC in vitro (Fig. 2e), strongly suggesting that the interaction with HDAC in vivo was indirect and due to the interaction with an endogenous CoR. Moreover, ETOΔC did not recruit HDAC activity or protein in vivo (Fig. 2c and d), further suggesting that the ETO–N-CoR interaction is required for HDAC recruitment in vivo. The ETO point mutants that did not interact with SMRT similarly did not coimmunoprecipitate with N-CoR or HDAC1 in 293T cells (data not shown).
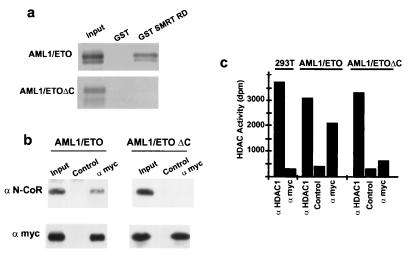
In the AML1/ETO fusion protein, the transcriptional activation domain of AML1 has been replaced with ETO (41, 42). Therefore, we tested whether AML1/ETO retained the ability of ETO to interact with CoRs and HDACs. Both GST-NCoR RD3 and GST-SMRT RD fusion proteins (Fig. 3a and data not shown) interacted with in vitro-translated AML1/ETO. A specific AML1/ETO–N-CoR/HDAC complex was detected in vivo in coimmunoprecipitation experiments performed with 293T cells cotransfected with a Myc-tagged AML1/ETO expression vector. Cell lysates were immunoprecipitated with anti-Myc tag antibodies, and the resulting immunocomplexes were analyzed for the presence of N-CoR protein and HDAC activity. As shown in Fig. 3b and c, N-CoR was specifically detected by Western blotting and anti-Myc tag antibodies precipitated significant levels of HDAC activity from AML1/ETO-transfected cells. In contrast, N-CoR did not interact with AML1/ETOΔC in vitro (Fig. 3a) and it was absent in immunoprecipitates from AML1/ETOΔC-transfected cells (Fig. 3b). Likewise, no detectable HDAC activity was found in the AML1/ETOΔC immunoprecipitates (Fig. 3c).
FIG. 3.
In vitro and in vivo interactions of AML1/ETO and AML1/ETOΔC with CoRs and HDAC. (a) Pulldown experiments. In vitro translated AML1/ETO or AML1/ETOΔC was precipitated with GST or A GST-SMRT RD fusion protein. (b) Coimmunoprecipitation of AML1/ETO or AML1/ETOΔC and N-CoR. Immunoprecipitates were obtained with an anti-Myc antibody or an appropriate control monoclonal antibody from 293T cells transfected with a Myc-tagged AML1/ETO or AML1/ETOΔC expression vector and blotted with anti-N-CoR or anti-Myc antibodies, as indicated. (c) HDAC activity of the indicated immunoprecipitates from 293T cells transfected with the indicated expression vectors. dpm; disintegrations per minute.
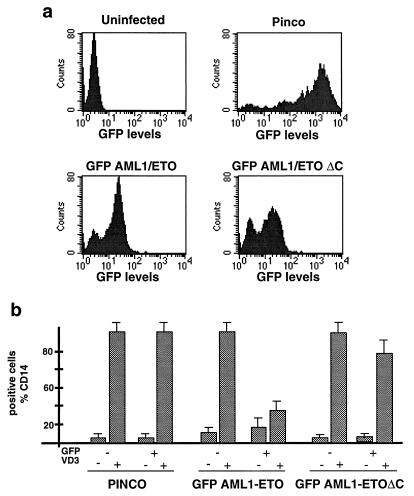
It has been previously demonstrated that the ectopic expression of AML1/ETO into hematopoietic precursor cell lines blocks terminal differentiation (48, 64, 70, 73). To explore the biological relevance of the interaction between AML1/ETO and the N-CoR–HDAC complex, we compared the abilities of AML1/ETO and the AML1/ETOΔC mutant to block terminal differentiation of human promonocytic U937 cells after vitamin D3 and TGF-β treatment. To facilitate the monitoring of ectopic protein expression, the two fusion proteins were fused to the GFP. The parental GFP, GFP-AML1/ETO, and GFP-AML1/ETOΔC cDNAs were cloned under the control of the 5′ long terminal repeat of a derivative of the hybrid Epstein-Barr virus–retroviral PINCO vector (see Materials and Methods) (14). Efficiency of infection, as evaluated by the frequency of GFP-positive cells, varied from 70 to 90% (PINCO control) to 50 to 75% (GFP-AML1/ETO and GFP-AML1/ETOΔC) (Fig. 4a). The intensities of the fluorescence signals were similar in AML1/ETO and AML1/ETOΔC cells, indicating comparable levels of expression that were confirmed by Western analysis (Fig. 4a and data not shown). Evaluation of vitamin D3-induced differentiation in cells infected with either the control, GFP-AML1/ETO, or GFP-AML1/ETOΔC retrovirus was performed by double-fluorescence fluorescence-activated cell sorter (FACS) analysis (see Materials and Methods) of the CD14 differentiation antigen in GFP-positive and -negative cells. In cells infected with the control retrovirus, CD14 expression was low or absent without stimulation but increased progressively during vitamin D3-induced differentiation in both the GFP-positive and -negative cell populations (Fig. 4b). Comparable up-regulation of CD14 expression was also detected in the GFP-negative cells of both the AML1/ETO- and AML1/ETOΔC-infected populations. Differentiation was, instead, inhibited in the GFP-AML1/ETO GFP-positive cells, while it was almost complete in the GFP-AML1/ETOΔC GFP-positive cells (Fig. 4b). The ability of the GFP-AML1/ETO fusion protein to inhibit differentiation was similar to what we have observed for the parental AML1/ETO when it is expressed in U937 or 32D cells (unpublished results). It therefore appears that the integrity of the N-CoR binding region is critical for the capacity of AML1/ETO to block differentiation by vitamin D3 and TGF-β, suggesting that recruitment of the N-CoR–HDAC complex is critical to the biological activity of AML1/ETO.
FIG. 4.
Effects of AML1/ETO and AML/ETOΔC on differentiation. (a) The parental GFP or the GFP-AML1/ETO and GFP-AML1/ETOΔC fusion proteins were expressed in U937 cells by using a derivative of the PINCO Epstein-Barr virus–retroviral vector. At 48 h after infection, cells were evaluated by FACS analysis to determine the frequency of GFP-positive cells and the intensity of fluorescence. (b) Cells were induced to differentiate with vitamin D3 and TGF-β and evaluated for CD14 expression after 4 days by double-fluorescence FACS analysis. CD14 expression data were acquired separately for GFP-positive and -negative cells, as indicated. Results are given as mean values ± the standard deviations from three experiments.
DISCUSSION
The data presented here identify a role for recruitment of the N-CoR/SMRT–HDAC repression complex in the mechanism of transcriptional repression by ETO and, possibly, other ETO family members (31). Most importantly, our results suggest that one crucial mechanism of oncogenic activation of AML1 by the t(8;21) chromosome translocation is its conversion from a transcriptional activator to a repressor. One of the AML1 isoforms (AML1B), in fact, is associated in vivo with the transcriptional coactivator p300 (32). The p300-interacting domain of AML1 is lost in the chromosomal translocation and is replaced with ETO, which retains the N-CoR–HDAC interaction domain. The resulting AML1/ETO fusion protein, therefore, is devoid of the ability of AML1 to recruit one HAT (p300), while it is endowed with that of ETO to recruit the N-CoR/SMRT CoR complex, including HDAC. This would be predicted to alter the chromatin structure of AML1 target genes in a manner that is the opposite of that normally associated with AML1B-dependent activation during hematopoietic differentiation (Fig. 5 shows a model). This hypothesis is supported by recent findings showing that AML1B is a transcriptional activator of some AML1 target promoters, while AML1/ETO behaves as a transcriptional repressor (11, 23, 38, 40, 41, 51). Consistent with our results, the transcriptional repressor function of AML1/ETO was mapped within its C-terminal region, including the two zinc fingers (40).
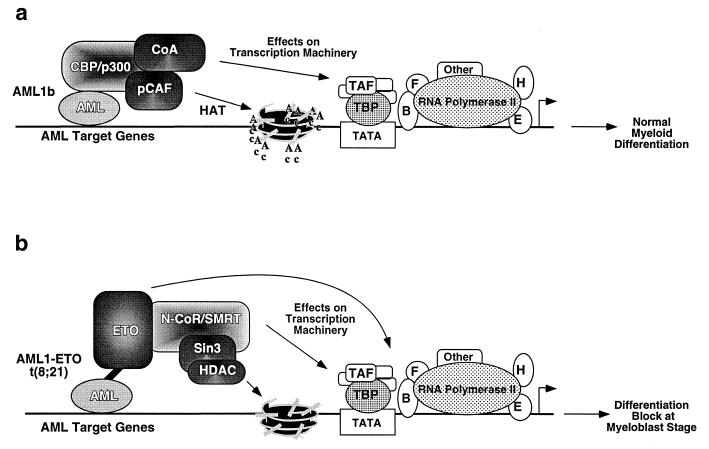
FIG. 5.
Model of the role of interactions of fusion protein AML1/ETO with the N-CoR–SMRT–Sin3–HDAC complex. (a) DNA-bound AML1 interacts with p300 and potentially with pCAF and nuclear receptor coactivators (CoA). This association results in increased histone acetylation, chromatin remodeling, and other effects on the transcriptional machinery resulting in transcriptional activation. (b) AML1/ETO recruits the N-CoR–Sin3–HDAC complex, decreasing histone acetylation and producing repressive chromatin organization and other effects on the transcriptional machinery resulting in transcriptional repression of some genes and, potentially, activation of a subset of other genes. The interaction between AML1/ETO and the N-CoR–Sin3–HDAC complex is mediated by ETO. TAF, TBP-associated factor; TBP, TATA-binding protein. F, TFIIF; B, TFIIB; H, TFIIH; E, TFIIE.
Our data strongly suggest that recruitment of the CoR/Sin3/HDAC complex is important for the function of AML1/ETO. However, it should be pointed out that this CoR complex may mediate transcriptional repression via both HDAC-dependent and HDAC-independent mechanisms. While this paper was under review, Wong and Privalsky reported on SMRT-mediated repression that was unaffected by the HDAC inhibitor trichostatin A (65). Interestingly, we have observed that trichostatin A only very modestly relieves ETO-dependent repression in 293T cells (74). It should also be noted that AML1/ETO has been shown to activate the transcription of some genes, including those for macrophage colony-stimulating factor and BCL-2 (33, 51). This could relate to the recent observation that N-CoR and SMRT can activate a subset of genes that are normally repressed by thyroid hormone (57). Alternatively, this could reflect another function of the AML1/ETO fusion protein. Further understanding of the mechanism(s) mediating deregulation of AML1 target genes by AML1/ETO awaits the analysis of chromatin structure and dynamics of those genes in vivo.
Like AML1/ETO, PML/RARα, the transforming protein of APLs, functions as a repressor in a complex containing N-CoR and HDACs (14, 39). Also, in this case, complex formation is crucial to its biological activities. Thus, CoR recruitment is a common feature of two otherwise unrelated leukemia-specific signaling pathways. AMLs and APLs consist of the accumulation of hematopoietic myeloid precursors arrested at specific stages of differentiation, and their corresponding oncogenic fusion proteins inhibit differentiation in vitro. Therefore, the aberrant recruitment of the CoR-HDAC complex by AML1/ETO or PML/RARα and the consequent alterations in chromatin structure and other effects on transcriptional regulation might be responsible for the differentiation block and contribute to myeloid leukemogenesis (Fig. 5). The cytological, ontological, and pathological differences between AMLs and APLs might reflect qualitative or quantitative differences in the genes targeted by AML1/ETO and PML-RARα. Finally, the involvement of the CoR-HDAC repression complex in two genetically distinct forms of myeloid leukemia underscores the critical importance of this repression pathway in myeloid differentiation.
ACKNOWLEDGMENTS
We thank C. Matteucci for excellent technical help, S. Nimer for the AML1/ETO cDNA, C. Hassig and S. Schreiber for HDAC1 antiserum, and C. Seiser for reagents and helpful discussions. V.G. and J.Z. contributed equally to this work.
This work was supported by NIH grants DK43806 and DK45586 (to M.A.L.), by the DNA Sequencing Core of the Center for Molecular Studies in Digestive and Liver Disease (NIH P30 DK50306) at the University of Pennsylvania, and by grants from AIRC and EC (Biomed program) to S.M. and P.G.P. M.F. is the recipient of a fellowship from INT (Milan).
REFERENCES
- 1.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 3.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britos-Bray M, Friedman A D. Core binding factor cannot synergistically activate the myeloperoxidase proximal enhancer in immature myeloid cells without c-Myb. Mol Cell Biol. 1997;17:5127–5135. doi: 10.1128/mcb.17.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Collins S J. Acute promyelocytic leukemia: relieving repression induces remission. Blood. 1998;91:2631–2633. [PubMed] [Google Scholar]
- 11.Frank R, Zhang H, Uchida H, Meyers S, Hiebert S W, Nimer S D. AML1/ETO blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 12.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Gene. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 13.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 14.Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Grignani F, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 15.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 16.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARα underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 18.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–348. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 19.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1998;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 20.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L Z, Guidex F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat Gen. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 22.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yuang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 23.Hiebert S W, Downing J R, Lenny N, Meyers S. Transcriptional regulation by the t(8;21) fusion protein, AML-1/ETO. Curr Top Microbiol Immunol. 1996;211:253–258. doi: 10.1007/978-3-642-85232-9_25. [DOI] [PubMed] [Google Scholar]
- 24.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 26.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 27.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 29.Kao H-Y, Ordentlich P, Koyano-Nakagawa K, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci USA. 1996;93:14059–14061. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 35.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 37.Laherty C E, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 38.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 39.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 40.Lutterbach B, Sun D, Schuetz J, Hiebert S W. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol Cell Biol. 1998;18:3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML1-B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Naseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 44.Nagy L, Kao H-Y, Chakvarkti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 45.Nuchprayoon I, Meyers S, Scott L M, Suzow J, Hiebert S, Friedman A D. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2 beta/CBR beta proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nucifora G, Rowley J D. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- 47.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 48.Okuda T, Cai Z, Lenny N, Lyu C J, vanDeursen J M, Harada H, Downing J R. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 49.Okuda T, vanDeursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 50.Petrovick M S, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D-E. Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhoades K L, Hetherington C J, Rowley J D, Hiebert S W, Nucifora G, Tenen D G, Zhang D E. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc Natl Acad Sci USA. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 53.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 55.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 56.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 57.Tagami T, Madison L D, Nagaya T, Jameson J L. Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol. 1997;17:2642–2648. doi: 10.1128/mcb.17.5.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 59.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 60.Uchida H, Zhang J, Nimer S D. AML1A and AML1B can transactivate the human IL-3 promoter. J Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- 61.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 62.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong C-W, Privalsky M L. Transcriptional repression by the SMRT-mDin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue J-C, Schwarz E J, Chawla A, Lazar M A. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARγ. Mol Cell Biol. 1996;16:1567–1575. doi: 10.1128/mcb.16.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global repressor RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W-M, Yao Y-L, Sun J-M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 69.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 70.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 71.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazer M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Pro Natl Acad Sci USA. 1997;94:14400–14495. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zamir I, Harding H P, Atkins G B, Hörlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang D E. AML1 gene in human leukemias: dominant negative effects of the chimeric proteins over wild-type AML1. Jpn J Cancer Res. 1997;88:1234–1235. [PubMed] [Google Scholar]
- 74.Zhang, J., and M. A. Lazar. Unpublished data.
- 75.Zhang J, Zamir I, Lazar M A. Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor regulates transcriptional repression. Mol Cell Biol. 1997;17:6887–6897. doi: 10.1128/mcb.17.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Sun Z W, Iratni R, Erdjument B H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y W, Bae S C, Huang G, Fu Y X, Lu J, Ahn M Y, Kanno Y, Kanno T, Ito Y. A novel transcript encoding an N-terminally truncated AML-PEPB2αB protein interferes with transactivation and blocks granulocytic differentiation of 32Dc13 myeloid cells. Mol Cell Biol. 1997;17:4133–4145. doi: 10.1128/mcb.17.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]