Abstract
Objective
Our previous study has verified that high level of SET and MYND domain-containing protein 2 (SMYD2) plays an important role in acquiring aggressive ability for liver cancer cells in hepatocellular carcinoma. MiR-200b as a tumor suppressor gene involves in a variety of cancers. This study aims to investigate the correlation between miR-200b and SMYD2 in hepatocellular carcinoma and the underlying mechanism.
Methods
Firstly, the levels of SMYD2 and miR-200b in hepatocellular carcinoma tissues and matched adjacent non-tumor liver tissues were tested with real-time reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting. Secondly, we evaluated the interaction between miR-200b and SMYD2 using dual-luciferase reporter assay. Thirdly, we elucidated the effect of miR-200b on SMYD2 and its downstream targets p53/CyclinE1. Finally, we silenced SMYD2 in hepatocellular carcinoma cell lines to investigate its effect on tumor proliferation and cell cycle progression, and further confirmed the correlation among SMYD2 and p53/CyclinE1.
Results
Compared with the matched adjacent non-tumor liver tissues, miR-200b was obviously decreased, and SMYD2 was significantly increased in hepatocellular carcinoma (both P<0.05). Spearman’s rank correlation revealed that miR-200b expression was negatively correlated with SMYD2 (P<0.01). Computer algorithm and dual-luciferase reporter assay revealed that miR-200b directly targeted and suppressed SMYD2 in HEK 293T cells. The down-regulated miR-200b expression promoted hepatoma cell proliferation (P<0.05) and increased SMYD2 expression(P<0.01), while the up-regulated expression of miR-200b had an opposite effect. The knockdown of SMYD2 suppressed the proliferation of MHCC-97L cells (P<0.01), down-regulated CyclinE1, and up-regulated p53 expression (both P<0.05).
Conclusion
MiR-200b is involved in hepatocellular carcinoma progression via targeting SMYD2 and regulating SMYD2/p53/CyclinE1 signaling pathway and may be used as a potential target for hepatocellular carcinoma treatment.
Keywords: hepatocellular carcinoma, miR-200b, SET and MYND domain-containing protein 2, p53, CyclinE1
Abstract
目的
本课题组前期研究证实组蛋白甲基转移酶2(SET and MYND domain-containing protein 2,SMYD2)高水平表达在肝癌细胞进展过程中发挥重要作用。MiR-200b作为肿瘤抑制基因参与多种肿瘤的发生和发展。本研究旨在探究miR-200b与SMYD2在肝癌中的作用机制。
方法
采用实时反转录聚合酶链反应(real-time reverse transcription-polymerase chain reaction,RT-PCR)及蛋白质印迹法分别检测癌组织及癌旁组织中SMYD2和miR-200b的表达水平。通过双荧光素酶报告验证SMYD2与miR-200b的靶向关系。通过沉默及过表达miR-200b研究其对SMYD2及下游p53/CyclinE1通路的影响,并通过沉默SMYD2研究其对肝癌细胞株的增殖及细胞周期蛋白(Cyclin)E1的影响,进一步证实SMYD2与p53/CyclinE1的关系。
结果
与癌旁组织相比,肝癌组织中miR-200b表达显著减少,而SMYD2表达显著增加(均P<0.05),SMYD2与miR-200b呈负相关(P<0.01)。双荧光素酶报告结果显示:在工具细胞HEK 293T细胞中,miR-200b直接靶向结合SMYD2并抑制其表达。在肝癌细胞株MHCC-97L中下调miR-200b可增加SMYD2的表达(P<0.01)并促进肝癌细胞的增殖(P<0.05),上调miR-200b则得出相反的结果。在MHCC-97L细胞株中沉默SMYD2可抑制细胞的增殖(P<0.01),并导致p53水平的升高和CyclinE1表达的下降(均P<0.05)。
结论
MiR-200b通过靶向结合SMYD2及调控p53/CyclinE1通路参与肝癌的进展,有望成为治疗肝癌细胞的新靶点。
Keywords: 肝细胞癌, miR-200b, 组蛋白甲基转移酶2, p53, 细胞周期蛋白E1
Hepatocellular carcinoma (HCC) is a primary malignant tumor which is very prevalent worldwide[1]. In China, HCC is the most common malignant tumor of the digestive system caused by endemic hepatitis B virus infection[2-3], and its morbidity and mortality are just less than lung cancer[4]. The diagnosis of HCC relies on histopathological diagnosis, and the prognosis of surgical resection and liver transplantation is poor with a 5-year survival rate of 10.1 to 16.6%[5]. Therefore, new biomarkers and targets are urgently needed for the diagnosis and treatment of HCC.
MicroRNAs (miRNAs) belong to small non-coding RNAs that have been investigated to play vital roles in various diseases mainly through negatively regulating their cognate target genes at a post-transcriptional level[6-7]. In tumors, miRNAs participate in regulating cell proliferation, tumor invasion, and migration of cancer cells. Increasing evidence indicates miRNAs may play a dual role in inducing tumor and suppressing cancer in different diseases. Several recent studies[8-10] have reported that miRNAs serve as potential biomarkers and targets for the diagnosis and therapy of HCC, but the specific function of miRNAs and their targets in HCC need further research.
SET domain and MYND domain-containing protein 2 (SMYD2), is a lysine methyltransferase and a member of SMYD family[11]. Initially, SMYD2 is thought to be involved in myogenic differentiation[12]. However, several recent studies[13-16] have reported that SMYD2 also promotes the development of various tumors, which may be related to the inhibition effects of SMYD2 on the tumor suppressor proteins PTEN[17], p53[18], cyclin D[19] and Rb[20]. Proteomic and genomics studies[21] have shown that SMYD2 not only dimethylates histone H3K4, but also methylates a variety of non-histone proteins associated with cancer. For example, SMYD2 could methylate lysine residue 370 (p53K370mel) of tumor suppressor protein p53 and inhibit p53 activity[18]. SMYD2 could methylate lysine313 (PTEN-K313me1) of phosphate and tension homology deleted on chromsome ten (PTEN) in vitro and vivo[17]. Recent studies[22] have shown that SMYD2 is also associated with the metastatic and invasive ability of cells. SMYD2 is a possible oncogene and closely related to cancer progression. The above studies indicate that SMYD2 could be a new target for cancer therapy.
We chose bioinformatics websites such as miRanda and TargetScan to predict the upstream miRNAs of SMYD2. Predication results have suggested that miR-200b has 2 binding sites with the 3'-UTR region of SMYD2 mRNA, indicating that miR-200b may have a regulatory effect on SMYD2. MiR-200b belongs to the miR-200 family, which mainly includes 5 members. They are divided into 2 groups, miR-200b/200c/429 (miR-200b subfamily) and miR-200a/141 (miR-200a subfamily) based on their function determined by seed sequence homology. MiR-200a/200b/429 are located on the first autosomes and miR-200c/141 are located on the twelfth autosomes[23]. MiR-200 family plays important roles in the progression and metabolism of several tumors. Several studies[24-28] have indicated that miR-200 family members are down-regulated in various cancers, such as ovarian, breast, bladder, pancreatic, and liver cancers. Over-expressed miR-200b can inhibit the migration, proliferation and tumor formation of HCC both in vivo and in vitro[29-30].
However, the specific role of SMYD2 and its regulatory mechanisms in HCC remain poorly understood. Our previous study[19] find that the high level of SMYD2 is associated with the acquisition of an aggressive phenotype and poor prognosis in HCC, but the molecular mechanisms of SMYD2 in promoting the development of HCC have not been investigated in depth.
This study aims to investigate the correlation between miR-200b and SMYD2 and their regulation mechanism in HCC.
1. Materials and methods
1.1. Samples and liver tissue specimens
HCC and tumor-adjacent normal tissues were collected from 25 paired patients undergoing hepatectomy at Third Xiangya Hospital of Central South University (Changsha, China). All samples were in accordance with the following principles: 1) Pathologically confirmed primary HCC; 2) no other malignant diseases before diagnosis and no second concurrent tumor; 3) without serious complications before obtaining biopsy specimens; 4) with complete medical history; 5) involved patients signed informed consent. The samples were grouped by peripheral and tumor. This study was approved by the Ethics Committee of Third Xiangya Hospital of Central South University (NO. 2016S198).
1.2. Dual-luciferase reporter assay
Human embryonic kidney cells (HEK-293T) provided by RiboBio (Guangzhou, China) were tool cells for Luciferase report analysis. The 3'-UTR regions of the SMYD2 mRNA containing predicted miR200b target sites or mutant binding site were designed by RiboBio (Guangzhou, China). The 3'-UTR region of the SMYD2 gene was amplificated and inserted into the pMiR-RB-REPORT™ Luciferase plasmid to provide pMIR-SMYD2 and pMIR-SMYD2-Mut vectors. HEK 293T cells were seeded in 96-well culture plates and were divided into 4 groups: 1) SMYD2-WT+NC; 2) SMYD2-WT+has-miR-200b-3q; 3) SMYD2-Mut+NC; 4) SMYD2-Mut+has-miR-200b-3q. The cells were transfected with pMIR-SMYD2 or pMIR-SMYD2-Mut together with miR-200b mimic (50 nm) or control using Lipofectamine 2 000 (Invitrogen, USA). Cells were collected and fluorescence activity were tested with a Dual Luciferase Reporter Assay System (Promega) in accordance with the manufacturer’s protocol after 48 h transfection. Data were normalized by pMiR-RB-REPORTβ-gal control plasmid.
1.3. Cell culture
MHCC-97H and MHCC-97L cells were purchased from Jennio Biotech (Guangzhou, China). SMCC-7721 and HepG2 cells were provided by GenePharma (Suzhou, China). MHCC-97H, MHCC-97L and SMCC-7721 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco, Invitrogen). HepG2 cells were maintained in DMEM medium with high glucose and 10% fetal bovine serum (Gibco, Invitrogen). All cells were incubated at 37 ℃ in a humidified 5% CO2 incubator. For transient transfection assay, the cells were divided into a control group, 3 miR-200b mimic groups (10, 20, 40 nmol/L), 3 miR-200b inhibitor groups (50, 100 200 nmol/L), a nonspecific miR-mimics (NS-miR) group, and a mock group. For cell proliferation and subsequent assays, the cells were divided into 5 groups: A mock group, an miR-200b mimic group, an NS-miR group, an miR-200b inhibitor group, and an nonspecific miR-inhibitor (NS-miR inhibitor) group.
1.4. Transient transfection
MiR-200b mimic, inhibitor, NS-miR, NS-miR inhibitor, mock and control were synthesized and obtained from RiboBio (Guangzhou, China). The sequences of siRNA oligonucleotide duplexes and si-Negative control (si-NC) were designed by GenePharma (Suzhou, China), described in Table 1. Lipofectamine 2000TM (Life Technologies) and riboFECTTM CP (RiboBio, Guangzhou, China) were used to transfect cells with plasmid and siRNA or miRNA mimic/inhibitor according to the manufacturer’s instructions respectively.
Table 1.
siRNA sequences
| Name | Sequences (5'-3') | |
|---|---|---|
| Sense | Antisense | |
| si-Negative control | UUCCCGAACGUGUCACGU | ACGUGACACGUUCGGGAA |
| si-SMYD2 | GGUUAAGAGAUUCUUAUUU | AAAUAAGAAUCUCUUAACC |
1.5. Cell proliferation assay
MHCC-97L cells were seeded in 96-well plates and were incubated in 50 μm EdU labeling medium according to the manual (RiboBio, Guangzhou, China) for 12 h at 37 ℃ under 5% CO2. Afterwards, cells were immobilized with paraformaldehyde and stained with anti-EdU working solution. Then we observed and photographed under a fluorescence microscope (Operetta, PerkinElmer, USA). Image J software was used for counting the percentage of EdU-positive cells.
1.6. Real-time reverse transcription-polymerase chain reaction
TRIzol reagent (Omega, USA) was used to extract total RNA from liver tissues and cells following the manufacturer’s recommendations. ReverTra Ace Qpcr RT Master Mix (Toyobo, Osaka, Japan) was used to reverse transcribed into cDNA. MiRNAs were isolated from liver tissues and cells using miRNeasy Mini Kit (Toyobo, Osaka, Japan). cDNA from miRNA was synthesized using Rever Tra Ace QPCR RT kit (Toyobo, Osaka, Japan). Real-time reverse transcription-polymerase chain reaction (RT-PCR) was conducted with SYBR Green Real-Time PCR Master Mix (Toyobo, Osaka, Japan) in a total volume of 20 μL on the ABI 7500 (Applied Biosystems, Thermo Fisher Scientific, USA) as following procedures: Pre-denaturing at 95 ℃ for 1 min, denaturing for 40 cycles at 95 ℃ for 15 s, and annealing at 60 ℃ for 20 s. The relative levels of gene expression were calculated with the 2-ΔΔCt method and were normalized to β-actin and U6. The primers used for RT-PCR were described in Table 2.
Table 2.
Sequences of primers used real-time PCR
| Genes | Sequences (5'-3') | |
|---|---|---|
| Forward | Reverse | |
| SMYD2 | ATCTCCTGTACCCAACGGAAG | CACCTTGGCCTTATCCTTGTCC |
| β-actin | TGGCACCCAGCACAATGAA | CTAAGTCATAGTCCGCCTAGAAGCA |
| miR-200b | ATCGTACGTGGGTAATACTGCCTGGTA | GCAGGGTCCGAGGTATTC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
1.7. Western blotting
RIPA lysis buffer containing 1% PMSF was used to extract protein from cells, shanghai, China after centrifugation. BCA protein assay kit (Beyotime, Shanghai, China) was employed to measure protein concentrations of the samples. Antibodies including SMYD2 (Abcam, USA), CyclinE1 (Proteintech, USA), p53 (Cell Signaling, USA), and GAPDH (Abcam, USA) were used in this study. 10% SDS polyacrylamide gel electrophoresis was used to separate the total protein (20 µg) and a polyvinylidene fluoride (PVDF) membrane was used to transfer the resolved proteins. The membrane was then blocked for 3 h at room temperature with 5% skim milk in TBST. The membrane was incubated with the following antibodies overnight at 4 ℃: anti-SMYD2, anti-CyclinE1, anti-p53 (1꞉1 000), and anti-GAPDH (1꞉10 000) after washing with TBST. We detected the target protein at room temperature through incubating with the secondary antibodies (Jackson, USA) (goat anti-rabbit and goat anti-mouse IgG horseradish peroxidase-conjugated antibody 1꞉4 000). The immunoreactive proteins were detected with luminol-based chemiluminescence reagent (Invitrogen, USA) and brand densities were analyzed by Image Lab software.
1.8. Statistical analysis
Data were analyzed using SPSS 18.0 software. Significant associations between miR-200b and SMYD2 expression were assessed using a Spearman’s rank correlation. The miR-200b expressions in different groups were analyzed through one-way ANOVA. All data were expressed as mean±standard deviation ( ±s) or median (IQR), and the t-test or rank sum test analysis was used to determine the significance of differences between 2 groups. P<0.05 was considered statistically significant.
2. Results
2.1. High transcription levels of SMYD2 in HCC specimens
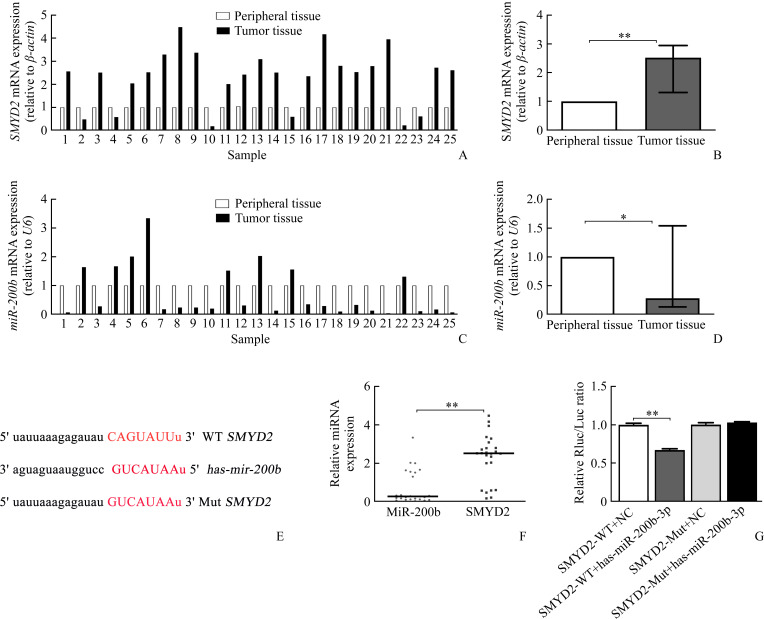
SMYD2 transcription levels in 25 paired HCC and matched adjacent normal tissues were analyzed by performing RT-PCR. We observed that SMYD2 mRNA level was highly expressed in 76% (19/25) of the HCC patients (Figure 1A). A significant difference of SMYD2 mRNA level was observed in tumor tissues compared with matched adjacent normal tissues (P<0.01,Figure 1B). In contrast, the low expression of miRNA-200b was observed in 68% (17/25) of patients (Figure 1C). We found miRNA-200b expression was significantly down-regulated in HCC tissues compared with the adjacent normal tissues (P<0.05, Figure 1D). Furthermore, according to the data from bioinformatics website (Figure 1E), we predicted that SMYD2 may serve as a target gene of miR-200b. The results of Spearman’s rank correlation showed that miR-200b expression was negatively correlated with SMYD2 (P<0.01, Figure 1F). Dual luciferase reporter assay was used to determine whether miRNA-200b targets SMYD2. MiRNA-200b significantly inhibited SMYD2 luciferase activity in HEK 293T cells compared with the control group (P<0.01, Figure 1G). However, 3'-UTR mutation of SMYD2 blocked this effect.
Figure 1. Low expression of miRNA-200b correlated with the high transcription levels of SMYD2 in the HCC specimens.
A-B: SMYD2 mRNA was examined with RT-PCR in 25 paired HCC and matched adjacent normal tissues. C-D: MiR-200b level was examined with RT-PCR in 25 paired HCC and matched adjacent normal tissues. E-F: Correlations between the miR-200b and SMYD2 expressions. G: Putative miR-200b binding sequence in the 3'-UTR of SMYD2. HEK-293T cells were transfected with SMYD2-WT+NC, SMYD2-WY+has-miR-200b-3p, SMYD2-Mut+NC, or SMYD2-Mut+has-miR-200b-3p. Relative luciferase activity was measured using fluorescence illuminometer. Data were presented as the mean±SD (n=4). *P<0.05, **P<0.01. SMYD2: SET and MYND domain-containing protein 2.
2.2. MiR-200b inhibits HCC proliferation invitro
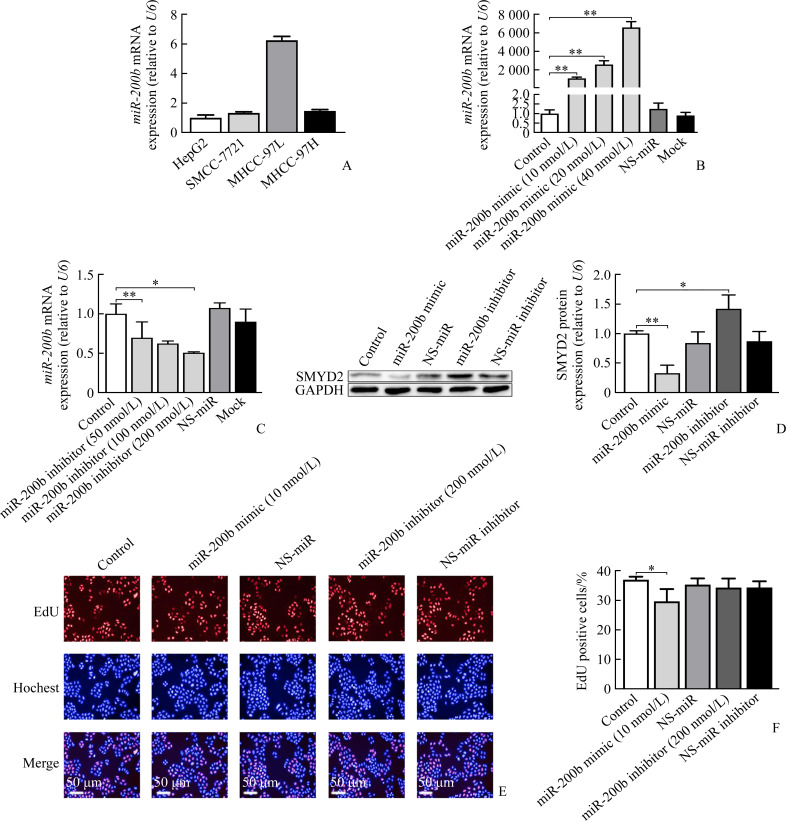
We first observed basal levels of miR-200b in different HCC cell lines, including HepG2, SMMC-7721, MHCC-97H, and MHCC-97L cells. The results showed that MHCC-97L cells had the highest expression of miR-200b at baseline relative to other 3 cell lines and therefore MHCC-97L cells was chosen for next step (Figure 2A). We next investigated the effects of mimic or inhibitor of miR-200b in MHCC-97L cells and found that treatment of MHCC-97L cells with a miR-200b mimic resulted in the increased expression of endogenous miR-200b in a dose-dependent manner (all P<0.01, Figure 2B), while miR-200b inhibitor suppressed the mRNA levels of miR-200b (P<0.01, Figure 2C). Both NS-miR and NS-miR inhibitor had no such effects.
Figure 2. MiR-200b inhibits the hepatocellular carcinoma proliferation in MHCC-97L cells.
A: MHCC-97L cells have the highest expression of miRNA-200b at baseline relative to other 3 cell lines. B-C: Expression levels of miR-200b were measured with RT-PCR in MHCC-97L cell line transfected with miR-200b mimic (10, 20, 40 nmol/L), miR-200b inhibitor (50, 100, 200 nmol/L), nonspecific miR-mimics (NS-miR), nonspecific miR-inhibitor (NS-miR inhibitor), transfection reagent (mock), and control group for 24 h (n=3). D: Expression levels of SMYD2 protein were measured with Western blotting in MHCC-97L cell line transfected with the miR-200b mimic (10 nmol/L), miR-200b inhibitor (200 nmol/L), NS-miR, NS-miR inhibitor, and control groups for 36 h (n=3). E-F: Cell proliferations of the miR-200b mimic (10 nmol/L), miR-200b inhibitor (200 nmol/L), NS-miR, NS-miR inhibitor, and control groups for 24 h (n=3). *P<0.05, **P<0.01. SMYD2: SET and MYND domain-containing protein 2.
We also evaluated effects of miRNA-200b mimic or inhibitor on SMYD2 protein expressions in MHCC-97L cells. Overexpression of miRNA-200b resulted in the reduced SMYD2 expression, while the miRNA-200b inhibitor increased SMYD2 expression (P<0.01, Figure 2D). Moreover, the proliferation of MHCC-97L cells was further confirmed by 5-ethynyl-2'-deoxyuridine (EdU) incorporation and quantification. The results demonstrated that the viability and proliferation of MHCC-97L cells were significantly decreased after miR-200b mimic treatment compared with the control group (P<0.05, Figure 2E and 2F). No significant difference was found after administration of miR-200b inhibitor cornpared with the control group (P>0.05).
2.3. MiR-200b inhibits proliferation by p53 up-regulation and cell cycle arrest
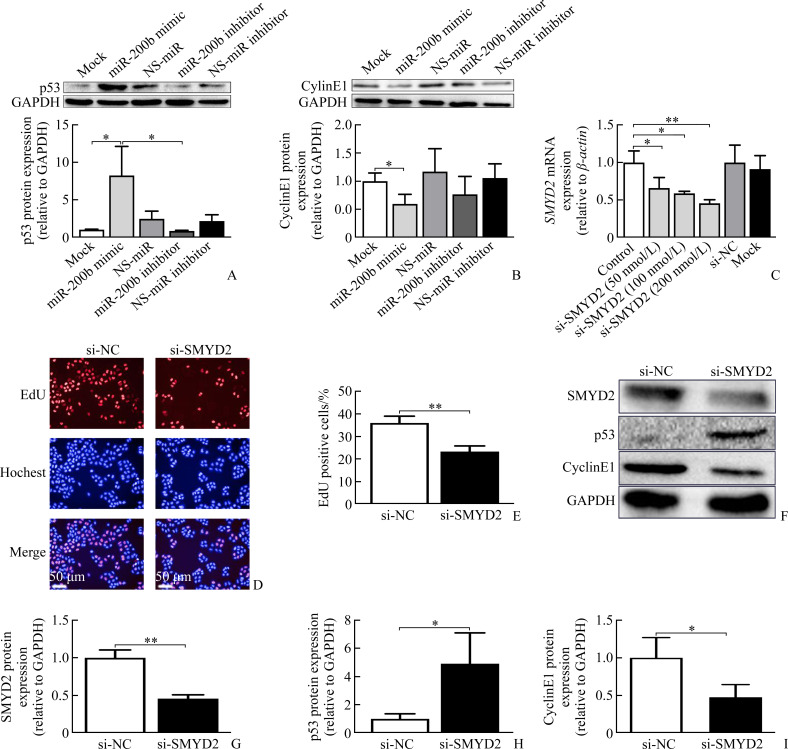
The protein expressions of p53 were obviously increased by treatment with miR-200b mimic compared with mock or NS-miR. Similarly, p53 expression was significantly reduced after administration of miR-200b inhibitor (P<0.05, Figure 3A). Furthermore, we also tested CyclinE1 to evaluate whether miR-200b influences the cell cycle. The results showed that CyclinE1 expression was significantly reduced in the miR-200b mimic group (all P<0.05), while no significant difference was found in the miR-200b inhibitor group (Figure 3B).
Figure 3. SMYD2 is essential for miR-200b mediated p53-CyclinE1 pathway activation.
A-B: Expressions of p53 and CyclinE1 protein were measured with Western blotting in MHCC-97L cell line. C: Expression of SMYD2 mRNA was measured with RT-PCR in MHCC-97L cells transfected with si-SMYD2 (50, 100, 200 nmol/L) or si-NC for 36 h. D-E: MHCC-97L cells were cultured till cell density was about 50% and transfected with si-SMYD2 (200 nmol/L) or si-NC for 36 h. MHCC-97L cell proliferation was measured by EdU assay in 96-well plates. F-I: Expressions of SMYD2, p53 and CyclinE1 protein were measured with Western blotting in the MHCC-97L cells transfected with si-SMYD2 (200 nmol/L) or si-NC for 36 h (n=3). *P<0.05, **P<0.01. SMYD2: SET and MYND domaincontaining protein 2.
2.4. SMYD2 is essential for miR-200b mediated p53-CyclinE1 pathway activation
MHCC-97L cells were transfected with SMYD2-specific siRNA in different doses and si-NC (Figure 3C). We found that SMYD2-specific siRNA dose-dependently reduced the SMYD2 mRNA levels (50, 100, 200 nmol/L) and si-SMYD2 (200 nmol/L) significantly inhibited SMYD2 protein expressions. Moreover, the suppression of SMYD2 by siRNA knockdown promoted p53 protein expression and reduced CyclinE1 expression (Figure 3F-3I). Further, we performed EdU incorporation assay to confirm that SMYD2 knockdown obviously inhibited cell proliferation (Figure 3D-3E).
3. Discussion
HCC remains one of the most lethal tumors despite current advances in testing and diagnosis[31]. More advanced therapeutic measures are not to be neglected to improve prognosis and reduce mortality in HCC patients. Therefore, it is of great importance to explore effective therapeutic targets, which may improve the efficacy of molecular-targeted drugs for HCC. Previous studies[19] have reported that the positive expression of SMYD2 is associated with poor prognosis in patients with primary HCC. Highly expressed SMYD2 is correlated with the tumor size, vascular invasion, differentiation, and TNM stage, and these results indicate that SMYD2 may be responsible for the cell aggressive phenotype.
Indeed, our present study has observed significant up-regulation of SMYD2 mRNA levels in tumor tissues compared with matched adjacent normal tissues. The suppression of SMYD2 by siRNA knockdown obviously inhibited the cell proliferation by performing EdU incorporation assay. These findings are somewhat in agreement with the previous notion. In view of the vital role of SMYD2 in HCC, further investigations are therefore needed to determine how SMYD2 is regulated and the potential of targeting SMYD2 as a treatment for HCC.
Current researches[32-36] have shown that miRNAs are involved in the tumor development, diagnosis and treatment. MiR-200b is a member of the miR-200 family and has been reported to be down-regulated in many cancers, such as ovary, breast, bladder, pancreas, prostate and esophagus[37-39]. These studies prompt us to explore the role that miR-200b might play in HCC and to provide mechanistic insights into earlier observations that implicated regulation in contributing to cell proliferation. In this study, we have found significant down-regulation of miR-200b in HCC tissues compared with matched non-cancerous tissues, which is contrary to the observed level of SMYD2 expression. To further determine the relationship between miR-200b and SMYD2, dual-luciferase reporter assay was performed after bioinformatics predictions and the results show that miR-200b can combine with 3'-UTR of SMYD2 to inhibit its transcription. These findings indicate that the high expression of SMYD2 in HCC may be due to inhibition of miR-200b. The further study confirmes this hypothesis that SMYD2 protein expressions are suppressed by miR-200b mimic, while SMYD2 is promoted by miR-200b inhibitor. This study provides intriguing insights into the role of miR-200b inhibition as the major reason for the HCC development and draw for the first time the link between the miR-200b-SMYD2 pathway and cell proliferation.
The results of this study have suggested a close tie between the miR-200b/SMYD2 and cell proliferation in HCC. A large number of studies[40-41] have shown that the cell proliferation is closely related to the cell cycle. It is not clear that whether miR-200b/SMYD2 regulates the cell proliferation by modulating protein expressions of cell cycle. It is well known that CyclinE1 is a vital factor to regulate the cell cycle. Amplification of CyclinE1 is associated with the poor outcome in breast, lung, and other solid cancers, and CyclinE1/CDK2 serves as an important therapeutic target in high-grade serous ovarian cancer[40]. Another study[41] has found that CyclinE1 expression level is useful as a biomarker that allows identification of cervical lesions with a higher risk for progression to cervical cancer with high sensitivity and precision. The reduced expressions of CyclinE1 are closely related to inhibited the cell proliferation[42]. CyclinE1 even participates in deteriorate endothelial cell senescence and vascular aging[43]. Indeed, in this study, miR-200b mimic significantly inhibits the CyclinE1 expression in MHCC-97L cells. However, to our surprise, miR-200b inhibitor has no effect on the increase of the CyclinE1 expression and cell proliferation. Since MHCC-97L cells are always in a state of excessive proliferation and the expression of miR-200b is commonly low in HCC cell lines. Therefore, miR-200b inhibitor probably means lower miRNA effects on the cell proliferation. These results may be reasonable because the effect of miRNA inhibitors on the cell proliferations according to many already published papers[43-44].
Previous studies[45-46] have showed that the reduced CyclinE1 expression is always accompanied by the increased p53 expression, and vice versa. Underscored by phylogenetic conservation, a primordial function of p53 depends on DNA damage-imposed cell cycle arrest and subsequent, conditional apoptosis, that is to say, p53 is also implicated in the cell cycle regulation. Thus, we speculate that miR-200b may affect the p53 expression, similar as CylinE1. According to our results, the protein expressions of p53 are obviously increased by treatment with miR-200b mimic compared with mock or NS-miR. More importantly, a significant reduced p53 expression is noted after administration of miR-200b inhibitor. This result suggests that miR-200b may have a more direct regulatory effect on p53 than cyclinE1.
SMYD2 is reported to be an oncogenic methyltransferase that represses the functional activity of the tumor suppressor proteins p53 and RB[47]. Inhibition of the SMYD2 expression could enhance p53 pathway activity and induce cell apoptosis through regulating its target genes in non-small cell lung cancer[48]. On the basis of this, we knock down SMYD2 by siRNA in the MHCC-97L cells and find that the p53 protein expression is promoted. We also note a significant reduction in CyclinE1 expression, which is similar to the effect of miR-200b mimic, indicating that SMYD2 directly regulates p53-CyclinE1 pathway in HCC. It is worth noting that knockdown of SMYD2 dampened the cell proliferation of MHCC-97L cells, which may be partially through the up-regulation of p53 and the down-regulation of CyclinE1. However, at present, the exact mechanism for the modulation of SMYD2 on p53-CyclinE1 pathway is not clear, and further studies are required to clarify this point.
In summary, miR-200b is associated with HCC progression through targeting SMYD2 and regulating SMYD2/p53/CyclinE1 signaling pathway. MiR-200b potentially limites the SMYD2 expression, restores p53 contents and further arrests the cell cycle by inhibiting CyclinE1 expression. Mir-200b may be used as a potential target for HCC treatment.
Contributions:FANG Weijin, SONG Liying Research design, experimental operation, and paper writing; LI Zuojun Paper modification; MENG Peipei, ZUO Shanru Data collecting and analysis; LIU Shikun Paper supervision and revision. All authors have read and agreed to the final text.
Funding Statement
This work was supported by the Natural Science Foundation of Hunan Province (2018JJ3795), China.
Conflict of Interest
The authors declare that they have no conflicts of interest to disclose.
Note
http://xbyxb.csu.edu.cn/xbwk/fileup/PDF/2022101303.pdf
References
- 1. Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma[J]. Expert Rev Gastroenterol Hepatol, 2021, 15(11): 1295-1307. 10.1080/17474124.2021.1991792. [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604. 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asrani SK, Devarbhavi H, Eaton J, et al. Burden of liver diseases in the world[J]. J Hepatol, 2019, 70(1): 151-171. 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 4. Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification[J]. Adv Cancer Res, 2021, 149: 1-61. 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mattiuzzi C, Lippi G. Current cancer epidemiology[J]. J Epidemiol Glob Health, 2019, 9(4): 217-222. 10.2991/jegh.k.191008.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marmarelis M, Thompson JC, Aggarwal C, et al. Emerging uses of circulating tumor DNA in advanced stage non-small cell lung cancer[J]. Ann Transl Med, 2017, 5(18): 380. 10.21037/atm.2017.07.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin SB, Gregory RI. microRNA biogenesis pathways in cancer[J]. Nat Rev Cancer, 2015, 15(6): 321-333. 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Otmani K, Lewalle P. Tumor suppressor miRNA in cancer cells and the tumor microenvironment: Mechanism of deregulation and clinical implications[J]. Front Oncol, 2021, 11: 708765. 10.3389/fonc.2021.708765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou YL, Liu F, Ma CY, et al. Involvement of microRNAs and their potential diagnostic, therapeutic, and prognostic role in hepatocellular carcinoma[J/OL]. J Clin Lab Anal, 2022: e24673[2022-08-29]. 10.1002/jcla.24673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karbasforooshan H, Hayes AW, Mohammadzadeh N, et al. The possible role of Sirtuins and microRNAs in hepatocellular carcinoma therapy[J]. Cell Cycle, 2020, 19(23): 3209-3221. 10.1080/15384101.2020.1843813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown MA, Sims RJ, Gottlieb PD, et al. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex[J]. Mol Cancer, 2006, 5: 26. 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawamura S, Yoshigai E, Kuhara S, et al. smyd1 and smyd2 are expressed in muscle tissue in Xenopus laevis [J]. Cytotechnology, 2008, 57(2): 161-168. 10.1007/s10616-008-9128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan LB, Ding BC, Liu HR, et al. Inhibition of SMYD2 suppresses tumor progression by down-regulating microRNA-125b and attenuates multi-drug resistance in renal cell carcinoma[J]. Theranostics, 2019, 9(26): 8377-8391. 10.7150/thno.37628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu L, Kou F, Ji ZY, et al. SMYD2 promotes tumorigenesis and metastasis of lung adenocarcinoma through RPS7 [J]. Cell Death Dis, 2021, 12(5): 439. 10.1038/s41419-021-03720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Jin G, Guo YF, et al. SMYD2 suppresses p53 activity to promote glucose metabolism in cervical cancer[J]. Exp Cell Res, 2021, 404(2): 112649. 10.1016/j.yexcr.2021.112649. [DOI] [PubMed] [Google Scholar]
- 16. Sun JJ, Li HL, Ma H, et al. SMYD2 promotes cervical cancer growth by stimulating cell proliferation[J]. Cell Biosci, 2019, 9: 75. 10.1186/s13578-019-0340-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakakido M, Deng ZZ, Suzuki T, et al. Dysregulation of AKT pathway by SMYD2-mediated lysine methylation on PTEN[J]. Neoplasia, 2015, 17(4): 367-373. 10.1016/j.neo.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation[J]. Nature, 2006, 444(7119): 629-632. 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 19. Zuo SR, Zuo XC, He Y, et al. Positive expression of SMYD2 is associated with poor prognosis in patients with primary hepatocellular carcinoma[J]. J Cancer, 2018, 9(2): 321-330. 10.7150/jca.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho HS, Hayami S, Toyokawa G, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation[J]. Neoplasia, 2012, 14(6): 476-486. 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abu-Farha M, Lambert JP, Al-Madhoun AS, et al. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase[J]. Mol Cell Proteomics, 2008, 7(3): 560-572. 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 22. Siddique AB, Ebrahim HY, Tajmim A, et al. Oleocanthal attenuates metastatic castration-resistant prostate cancer progression and recurrence by targeting SMYD2[J]. Cancers (Basel), 2022, 14(14): 3542. 10.3390/cancers14143542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cavallari I, Ciccarese F, Sharova E, et al. The miR-200 family of microRNAs: fine tuners of epithelial-mesenchymal transition and circulating cancer biomarkers[J]. Cancers (Basel), 2021, 13(23): 5874. 10.3390/cancers13235874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L, Li P, Wang Q, et al. Diagnosis accuracy of the miR-200 family tumor marker series in ovarian cancer: a systematic review and meta-analysis[J]. Transl Cancer Res, 2022, 11(7): 2283-2290. 10.21037/tcr-22-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mekala JR, Naushad SM, Ponnusamy L, et al. Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer[J]. Gene, 2018, 641: 248-258. 10.1016/j.gene.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 26. Mei YH, Zheng JB, Xiang P, et al. Prognostic value of the miR-200 family in bladder cancer: a systematic review and meta-analysis[J/OL]. Medicine (Baltimore), 2020, 99(47): e22891[2021-08-20]. 10.1097/MD.0000000000022891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diaz-Riascos ZV, Ginesta MM, Fabregat J, et al. Expression and role of microRNAs from the miR-200 family in the tumor formation and metastatic propensity of pancreatic cancer[J]. Mol Ther Nucleic Acids, 2019, 17: 491-503. 10.1016/j.omtn.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park JW, Jeong JM, Cho KS, et al. miR-30a and miR-200c differentiate cholangiocarcinomas from gastrointestinal cancer liver metastases[J/OL]. PLoS One, 2021, 16(4): e0250083[2021-04-14]. 10.1371/journal.pone.0250083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moh-Moh-Aung A, Fujisawa M, Ito S, et al. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression[J]. Sci Rep, 2020, 10(1): 10418. 10.1038/s41598-020-67425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu XM, Chen D, Chen H, et al. YB1 regulates miR-205/200b-ZEB1 axis by inhibiting microRNA maturation in hepatocellular carcinoma[J]. Cancer Commun (Lond), 2021, 41(7): 576-595. 10.1002/cac2.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma[J]. BMJ, 2020, 371: m3544. 10.1136/bmj.m3544. [DOI] [PubMed] [Google Scholar]
- 32. Inoue J, Inazawa J. Cancer-associated miRNAs and their therapeutic potential[J]. J Hum Genet, 2021, 66(9): 937-945. 10.1038/s10038-021-00938-6. [DOI] [PubMed] [Google Scholar]
- 33. Shi Y, Liu ZH, Lin Q, et al. MiRNAs and cancer: key link in diagnosis and therapy[J]. Genes (Basel), 2021, 12(8): 1289. 10.3390/genes12081289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersen GB, Tost J. Circulating miRNAs as Biomarker in Cancer[J]. Recent Results Cancer Res, 2020, 215: 277-298. 10.1007/978-3-030-26439-0_15. [DOI] [PubMed] [Google Scholar]
- 35. Kabekkodu SP, Shukla V, Varghese VK, et al. Cluster miRNAs and cancer: Diagnostic, prognostic and therapeutic opportunities[J/OL]. Wiley Interdiscip Rev RNA, 2020, 11(2): e1563[2021-08-22]. 10.1002/wrna.1563. [DOI] [PubMed] [Google Scholar]
- 36. Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy[J]. J Pharm Pharmacol, 2020, 72(12): 1732-1749. 10.1111/jphp.13351. [DOI] [PubMed] [Google Scholar]
- 37. Gurbuz V, Sozen S, Bilen CY, et al. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression[J]. Oncol Lett, 2021, 22(5): 805. 10.3892/ol.2021.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu P, Liu K, Gao XX, et al. Transforming growth factor-β and bone morphogenetic protein 2 regulation of microRNA-200 family in chronic pancreatitis[J]. Pancreas, 2018, 47(2): 252-256. 10.1097/MPA.0000000000000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindner K, Borchardt C, Schöpp M, et al. Proton pump inhibitors (PPIs) impact on tumour cell survival, metastatic potential and chemotherapy resistance, and affect expression of resistance-relevant miRNAs in esophageal cancer[J]. J Exp Clin Cancer Res, 2014, 33(1): 73. 10.1186/s13046-014-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Etemadmoghadam D, Au-Yeung G, Wall M, et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer[J]. Clin Cancer Res, 2013, 19(21): 5960-5971. 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 41. del Moral-Hernández O, Hernández-Sotelo D, Alarcón-Romero LDC, et al. TOP2A/MCM2, p16INK4a, and CyclinE1 expression in liquid-based cytology: a biomarkers panel for progression risk of cervical premalignant lesions[J]. BMC Cancer, 2021, 21(1): 39. 10.1186/s12885-020-07740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu X, Tian J, Li QY. Downregulation of HOTTIP regulates insulin secretion and cell cycle in islet β cells via inhibiting MEK/ERK pathway[J]. Eur Rev Med Pharmacol Sci, 2018, 22(15): 4962-4968. 10.26355/eurrev_201808_15636. [DOI] [PubMed] [Google Scholar]
- 43. Ke YL, Li D, Zhao MM, et al. Gut flora-dependent metabolite Trimethylamine-N-oxide accelerates endothelial cell senescence and vascular aging through oxidative stress[J]. Free Radic Biol Med, 2018, 116: 88-100. 10.1016/j.freeradbiomed.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 44. Sun GS, Sun GQ, Chen X, et al. Meloxicam inhibits hepatocellular carcinoma progression and enhances the sensitivity of immunotherapy via the microRNA-200/PD-L1 pathway[J]. J Oncol, 2022. [2022-02-21]. 10.1155/2022/4598573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim KM, Park SH, Bae JS, et al. FAM83H is involved in the progression of hepatocellular carcinoma and is regulated by MYC[J]. Sci Rep, 2017, 7(1): 3274. 10.1038/s41598-017-03639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peng XY, Wen Y, Zha L, et al. TRIM45 suppresses the development of non-small cell lung cancer[J]. Curr Mol Med, 2020, 20(4): 299-306. 10.2174/1566524019666191017143833. [DOI] [PubMed] [Google Scholar]
- 47. Cowen SD, Russell D, Dakin LA, et al. Design, synthesis, and biological activity of substrate competitive SMYD2 inhibitors[J]. J Med Chem, 2016, 59(24): 11079-11097. 10.1021/acs.jmedchem.6b01303. [DOI] [PubMed] [Google Scholar]
- 48. Shang L, Wei MJ. Inhibition of SMYD2 sensitized cisplatin to resistant cells in NSCLC through activating p53 pathway[J]. Front Oncol, 2019, 9: 306. 10.3389/fonc.2019.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]