Abstract
Replication protein A (RPA), the heterotrimeric single-stranded-DNA (ssDNA) binding protein (SSB) of eukaryotes, contains two homologous ssDNA binding domains (A and B) in its largest subunit, RPA1, and a third domain in its second-largest subunit, RPA2. Here we report that Saccharomyces cerevisiae RPA1 contains a previously undetected ssDNA binding domain (domain C) lying in tandem with domains A and B. The carboxy-terminal portion of domain C shows sequence similarity to domains A and B and to the region of RPA2 that binds ssDNA (domain D). The aromatic residues in domains A and B that are known to stack with the ssDNA bases are conserved in domain C, and as in domain A, one of these is required for viability in yeast. Interestingly, the amino-terminal portion of domain C contains a putative Cys4-type zinc-binding motif similar to that of another prokaryotic SSB, T4 gp32. We demonstrate that the ssDNA binding activity of domain C is uniquely sensitive to cysteine modification but that, as with gp32, ssDNA binding is not strictly dependent on zinc. The RPA heterotrimer is thus composed of at least four ssDNA binding domains and exhibits features of both bacterial and phage SSBs.
Single-stranded-DNA (ssDNA) binding proteins (SSBs) participate in almost every aspect of DNA metabolism. These proteins bind tightly to ssDNA with little or no sequence specificity and activate ssDNA by reducing its secondary structure and by stimulating DNA polymerases and DNA helicases. At first glance, the three well-studied SSBs encoded by phage T4 (gp32), Escherichia coli (Ecssb), and eukaryotes (replication protein A [RPA]) appear to have evolved very different ways of accomplishing this task. Gene 32 protein is a monomer of 33 kDa that binds ssDNA cooperatively (ω = 3,800), with a binding-site size of 8 nucleotides (nt) (19, 46). The structure of gp32 has been determined by X-ray crystallography and shown to contain a hydrophobic pocket composed of five β strands that contact the ssDNA bases and an electropositive cleft that contacts the phosphate backbone (52). While gp32 is a zinc metalloprotein, zinc is not required for ssDNA binding but is required for cooperativity and stabilization of the structure of the core fragment (19, 20, 43). The prototype bacterial SSB, Ecssb, is a tetramer of four identical 19-kDa subunits, or protomers, with at least two well-characterized DNA binding modes (38). At low salt concentrations (<10 mM NaCl) two protomers bind 35 nt of ssDNA with unlimited cooperativity (ω = 105) (18, 50), and at high salt concentrations (≥0.2 M NaCl) four protomers bind 65 nt of ssDNA with limited cooperativity (ω = 420) (9, 38, 39). This larger binding mode results in compaction of the ssDNA due to higher-order binding or wrapping about the tetramer (10, 12, 23).
The nuclear SSB of eukaryotes, RPA, is a heterotrimeric complex that was originally identified as a Homo sapiens protein (hsRPA) required for simian virus 40 (SV40) DNA replication in vitro (17, 59–61). RPA has been identified in numerous species, including the yeast Saccharomyces cerevisiae (scRPA), where it is a complex of subunits of 69 (RPA1), 36 (RPA2), and 13 (RPA3) kDa (8). Each subunit of RPA is required for SV40 DNA replication (16, 30) and for viability in yeast (7, 26).
RPA1 is known to bind ssDNA on its own and for some time was thought to be the only subunit that binds ssDNA (8, 16, 30, 62). Structure-function analysis revealed that the N-terminal 18 kDa of RPA1 is dispensable for SV40 DNA replication (21) but that the C terminus, which contains a putative Cys4-type zinc-binding domain, is required for RPA2 binding (21, 35, 36). The central portion of hsRPA1 (residues 168 to 442) contains a major ssDNA binding domain (21, 22, 35, 44). The notion that RPA might contain additional ssDNA binding domains was suggested by the fact that this domain has a 20-fold lower affinity for ssDNA than does the heterotrimer (21) and by the fact that different binding-site sizes have been reported for RPA. These include an 8-nt binding mode that is dependent on glutaraldehyde cross-linking (3), a 30-nt binding mode that is obtained by fluorescence quenching and electrophoretic mobility shift assay (EMSA) (2, 32–34, 41, 55), and a 90-nt binding mode that is obtained by fluorescence quenching and electron microscopy (EM) (1). These various sizes might be explained if RPA has multiple ssDNA binding domains and alternative binding modes like Ecssb (45).
Genetic and biochemical analysis in yeast revealed that the major ssDNA binding domain of yeast RPA1 is composed of two homologous subdomains, A and B, with weak sequence similarity to Ecssb (45). Both domains are required for viability, and domain A can functionally replace domain B. Based on a model of ssDNA binding by Ecssb (11), a pair of conserved aromatic residues in each domain was identified by amino acid sequence alignment and proposed to stack with the ssDNA. One of these, F238, is likely to be important for ssDNA binding since it was the only residue in scRPA1 that was individually required for viability. Portions of this model have been confirmed by the crystal structure of residues 181 to 422 of hsRPA1 (RPA1181–422) bound to ssDNA (5). Bochkarev and colleagues (5) showed that the central ssDNA binding domain of hsRPA1 is composed of two structurally homologous subdomains, or “OB folds” (42) (for oligonucleotide oligosaccharide binding folds), and that binding by each domain involves a number of hydrogen bonds as well as the stacking of ssDNA bases with a pair of aromatic residues, one of which is F238. To closely resemble Ecssb, however, two additional ssDNA binding domains must exist in the RPA heterotrimer. One of these is in the middle subunit, since it was demonstrated that RPA2 can bind ssDNA as part of the heterotrimer (45) or as part of the RPA2-RPA3 subcomplex of hsRPA (6). Binding by the RPA2-RPA3 subcomplex of hsRPA is stimulated by the C terminus of RPA1, suggesting that this domain might contain another ssDNA binding activity (6). RPA3 was proposed to serve as a binding domain; however, no direct evidence has yet been obtained to support this idea (45).
Here we show that the fourth ssDNA binding domain of RPA lies in the C terminus of RPA1, adjacent to domains A and B. This result confirms that both cellular SSBs, Ecssb and RPA, have at least four ssDNA binding domains. Curiously, the new domain in the C terminus of RPA1 has several features in common with the phage SSB gp32. This suggests that these very different SSBs are likely to have a common mechanism of function, if not a common origin.
MATERIALS AND METHODS
DNA constructions.
Fragments of the RFA1 gene encoding scRPA1 were isolated on NdeI/BamHI cassettes by 12 rounds of amplification with Vent DNA polymerase and pJM136 (45) as the template. Fragments were ligated into pET11a to produce the following E. coli expression plasmids: pJM163 (M1 to K180), pJM158 (M, T181 to N294), pJM159 (M, V295 to D415), pJM165 (M, S416 to A485), pJM164 (M, S416 to N511), pJM157 (M, F481 to S601), pJM155 (M, F481 to A621), and pJM185 (M, S416 to A621). Point mutations were introduced by two rounds of oligonucleotide-directed PCR mutagenesis, and the mutated sequences were ligated into pET11a for E. coli expression or into a yeast shuttle vector to test genetic complementation. The following domain C mutation expression plasmids were identical to pJM185 except for the indicated mutations: pJM174 (M, S416 to S601, with a C-terminal truncation of 20 amino acids in domain C) and pJM186 (C505A and C508A). To test whether an RFA1 gene with point mutations could complement an RFA1 deletion, we placed the mutations in the context of an intact RFA1 gene by using the “domain swap” vector pDS1 (45) to produce pDS1.13 (F238A), pDS1.28 (F269A), pDS1.14 (W360A), and pDS1.29 (F385A). Plasmid pDS1C was constructed for placing domain C mutations into RFA1 by introducing a SalI site just downstream of the Cys4 motif (resulting in the amino acid changes T510V and N511D) and a BglII site near the end of domain C (resulting in the amino acid change Y594S). Plasmid pDS1C was shown to confer wild-type activity by a complementation test and was used to place mutations in RFA1, which resulted in the following plasmids: pJM183 (C505A and C508A), pDS1C.9 (F537A), and pDS1C.10 (F563A). Complementation with the RPA2 F143A mutation was tested with pJM243 (45), and complementation with the RPA2 W101A mutation was tested with pJM254 (W101A), which was constructed in pDS2 (45). Maltose binding protein (MBP) fusions were constructed in the vector pMAL-c2 (New England Biolabs) by inserting BamHI fragments that were generated by PCR as described above into the unique BamHI site of the vector. Plasmid pJM410 expresses the MBP-A domain fusion (T181 to N294), pJM411 expresses the MBP-B fusion (V295-D415), and pJM412 expresses the MBP-C fusion (G418-A621). Genetic complementation of the rfa1-1::TRP1 and rfa2-1::TRP1 null mutations was performed as described previously (45).
Protein expression, purification, and ssDNA binding assay.
Recombinant RPA1 proteins were expressed in strain BL21 (DE3) by using the T7 expression system (54). Cells were grown in Luria-Bertani medium at 37°C in the presence of ampicillin and induced for 16 h by the addition of 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested and lysed as described previously (45), and the insoluble pellet from 6 ml of culture was resuspended in 0.3 ml of 10 M urea. The sample was then diluted with 0.3 ml of 2× buffer A (25 mM Tris [pH 7.5], 1 mM EDTA, 0.01% Nonidet P-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT]) containing 200 mM NaCl, and the sample was centrifuged 15 min at 4°C. The soluble portion was then sequentially dialyzed to 2, 1, 0.5, and 0 M urea in dialysis buffer consisting of buffer A and 100 mM NaCl, 10 mM MgCl2, and 20 μM ZnSO4. Each dialysis step was carried out for 8 to 16 h. Precipitates were removed by centrifugation, and the soluble sample, typically 0.3 mg of recombinant protein per ml, was stored at −80°C. Protein concentrations were determined by comparison to known standards on Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels.
MBP fusion proteins were expressed in strain XL1-Blue grown in Luria-Bertani medium with 0.2% glucose and 0.1 mg of ampicillin per ml. A 1-liter culture was induced for 2 h with 0.3 mM IPTG, and the cells were collected by centrifugation and resuspended in buffer A containing 50 mM NaCl and 1 mg of lysozyme per ml. All buffers for MBP-C also contained 20 μM ZnSO4 throughout the purification. All steps were performed either on ice or at 4°C. Following a 15-min incubation on ice, the samples were subjected to one freeze-thaw cycle and four sonication cycles (15 s each). The lysate was clarified by centrifugation, diluted fivefold with buffer A containing 50 mM NaCl, and applied to a 15-ml amylose-affinity resin. The resin was washed with 8 column volumes of buffer A containing 300 mM NaCl, and the MBP fusion proteins were eluted in a step with buffer A containing 50 mM NaCl and 10 mM maltose. Peak fractions were identified by the Bradford assay, pooled, and loaded onto a Mono Q column. This column was washed with 3 ml of buffer A containing 50 mM NaCl and eluted with an 8-ml gradient from 50 mM to 1 M NaCl. The peak fractions (0.15 M NaCl) were identified by SDS-polyacrylamide gel electrophoresis (PAGE), pooled, and dialyzed to buffer A containing 50 mM NaCl.
Typically, 10 μl of ssDNA binding reaction mixtures containing protein extracts from E. coli were incubated with a 10,000 cpm (2 fmol) of 32P-labeled 17-base oligonucleotide (universal sequencing primer) in the following solution: 10 mM HEPES (pH 7.5)–0.5 mM DTT–10 μg of sheared salmon sperm DNA per ml. Reaction mixtures were cross-linked and analyzed as described previously (45). Binding reactions for MBP fusion proteins were carried out under identical conditions except that they were UV cross-linked at 500 J/m2 to minimize the time of UV exposure. Gels were analyzed with a phosphorimager, and the intensities of the bound and free probes were quantitated with IP-Lab Gel software. The Kd for each binding reaction was then determined by fitting the data to the Langmuir equation.
Amino acid sequence analysis.
The amino acid sequences of the four domains shown in Fig. 3 were initially aligned with the PILEUP program. This alignment identified the absolutely conserved aspartic acid as well as the invariant N-terminal aromatic residue in all four domains. An aromatic residue corresponding to the C-terminal stacking residue was found to be conserved within each of the four domains but was not aligned between domains. The residues at this position in domains A and B were aligned with each other based on the structural alignment of hsRPA1181–422 (5), which consisted of shifting the stacking residue of domain B three residues upstream. The aromatic residue in domain D had aligned with this residue in domain B and was similarly shifted three residues. The aromatic residue of domain C was shifted three residues downstream. The PHD structure prediction program has been described previously (47–49).
FIG. 3.
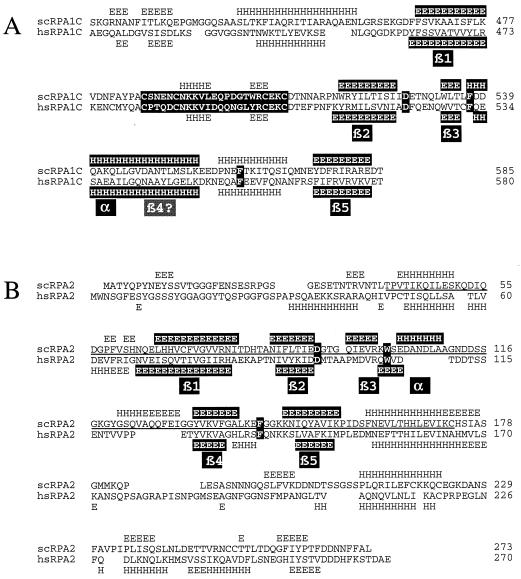
Amino acid sequence alignment of the four ssDNA binding domains of RPA. The amino acid sequences of the indicated proteins were aligned with the PILEUP program and optimized as described in Materials and Methods. The secondary structure of hsRPA1181–422, presented at the top of the sequences, was taken from Bochkarev et al. (5). Highly conserved residues are indicated in white type on a black background, and the putative zinc-binding domain in domain C is shaded. Numbers to the right of the protein designations refer to amino acid numbers. Note that 14 amino acids are deleted from the scRPA2 sequence at residue 115. Hs, H. sapiens; Xl, Xenopus laevis; Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe; Os, Oryza sativa; Cf, Crithidia fasiculata; Ce, Caenorhabditis elegans.
PMPS inactivation and reactivation.
p-Hydroxymercuriphenylsulfonate (PMPS) treatment was performed essentially as described previously (15, 20). All buffers and water were first passed over 30 ml of Chelex-100 resin (Bio-Rad), and the pH was adjusted with NaOH. Extracts were prepared without zinc and dialyzed twice against 1,000 volumes of buffer A lacking DTT and containing 10 mM EDTA. Concentrations of samples were made to be 2.5 mM in PMPS (Sigma) for 1 h on ice and adjusted to 50 mM in EDTA. Sensitivity to PMPS was determined by assaying the sample as usual, except that the assay buffer did not contain DTT. Reactivation was performed by dialyzing the PMPS-treated sample against TNG buffer (25 mM Tris [pH 7.5], 0.2 M NaCl, 5% glycerol) containing 1 mM EDTA to remove unreacted PMPS. The samples were then incubated with 0.1 M β-mercaptoethanol and increasing concentrations of zinc sulfate for 30 min on ice. Samples were assayed in the absence of DTT.
RESULTS
Identification of a new ssDNA binding domain in RPA1.
A UV-cross-linking EMSA was previously used to characterize the ssDNA binding activity of domains A and B of scRPA1 (45). In this method, an oligonucleotide 32P labeled at its 5′ end is incubated with an extract made from E. coli expressing a defined fragment of RPA protein and the mixture is irradiated with UV light to covalently link bound protein to the oligonucleotide. Activity is then detected by gel shift assay on a denaturing polyacrylamide gel. The experiments described here use the 17-nt universal sequencing primer to minimize the number of multiple protein-ssDNA interactions, but similar results were obtained with oligo(dT)40.
To search for additional ssDNA binding domains within the RPA1 subunit, fragments of the yeast gene encoding RPA1 were cloned into an E. coli expression vector such that the entire RPA1 protein could be expressed as several protein fragments (Fig. 1). Upon induction and cell lysis, each RPA1 fragment was found in the insoluble fraction. These proteins were solubilized in urea and refolded by stepwise dialysis. SDS-PAGE revealed that all RPA1 fragments were soluble and present in nearly equal amounts except for the smallest one (fragment 4), which was about fourfold less concentrated (data not shown). Portions of each extract were then subjected to the UV EMSA, and as was shown previously (45), fragments encompassing domains A and B resulted in a clear mobility shift (Fig. 1). At least two background bands were observed with all extracts, including extracts of E. coli expressing the vector alone (lane V). A strong mobility shift was also seen in assays of fragment 8, which encompasses residues 416 to 621. Assays of smaller fragments within this region (fragments 4 to 7) did not result in binding; however, we cannot rule out the possibility of a signal comigrating with the background bands. It should be noted that the RPA binding activities in this assay resulted in a corresponding decrease in the signal of the major background band (Fig. 1). This effect was due to limiting amounts of probe used in this experiment and was not observed when the ratio of probe to extract was increased (data not shown, but see Fig. 2). We also note that the signal obtained in the UV EMSA depends on both a protein’s ssDNA binding affinity and its efficiency of cross-linking, which may differ between proteins. The new binding domain located in the 206 amino acids C terminal to domain B is hereafter referred to as domain C. The ssDNA binding domain in RPA2 (6, 45) is here renamed domain D.
FIG. 1.
Identification of a new ssDNA binding domain in RPA1. Fragments of the scRPA1 protein were expressed in E. coli and tested for ssDNA binding activity by the UV EMSA. At the top is a schematic of the scRPA1 protein illustrating the locations of the three ssDNA binding domains and the eight protein fragments (numbered horizontal black bars) that were tested in this assay. Numbers above the schematic refer to the amino acid numbers of the scRPA1 protein. For each protein fragment, a portion of extract containing approximately 0.3 μg of recombinant protein was incubated with a 17-nt oligonucleotide 32P labeled at its 5′ end (P) and cross-linked with UV light. Products were resolved on a 6% denaturing polyacrylamide gel. V, extract made from E. coli expressing the vector alone; A*, B*, and C*, RPA1 domains A, B, and C; bkg, background band.
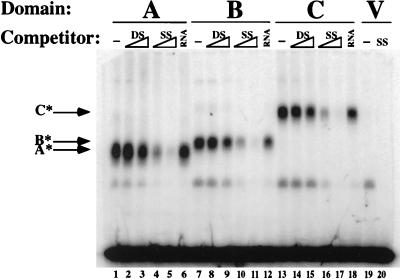
FIG. 2.
RPA1 domain C binds ssDNA specifically. Extracts of E. coli containing approximately 0.3 μg of recombinant protein were assayed in the presence of the indicated competitor, namely, 0.5 or 3 μg of unlabled lambda DNA before (double stranded [DS]) or after (single stranded [SS]) boiling or 3 μg of yeast tRNA (RNA). V, extract of E. coli expressing the vector alone or competed with 3 μg of ssDNA. A*, B*, and C*, RPA1 domains A, B, and C.
To determine the specificity of domain C binding, UV EMSAs were performed on extracts of E. coli expressing domains A, B, and C in the presence of competitor nucleic acid. As shown previously, binding by domains A and B is competed by the presence of unlabeled ssDNA but not by the presence of unlabeled double-stranded DNA (dsDNA) (Fig. 2; e.g., compare lanes 2 and 3 to lanes 4 and 5). The ssDNA binding activity of these two domains is also resistant to added RNA as the competitor (e.g., compare lanes 1 and 6). Domain C shows an identical pattern: it is specifically competed by ssDNA but not by dsDNA or RNA (lanes 13 to 18).
Amino acid sequence analysis of domain C.
To identify residues of domain C that might contribute to its ssDNA binding activity, the amino acid sequences of domains A, B, C, and D (RPA2) from a number of species were compared (Fig. 3). The alignment of domains A and B was anchored by using the structural comparison of hsRPA1181–422 described by Bochkarev et al. (5), and presented at the top of the alignment is a schematic of the corresponding secondary structure of this region. As described previously, this structure is an OB fold which consists of five strands of β-sheet with an α-helix connecting the third and fourth strands (42). A pair of aromatic residues in hsRPA1 domains A and B (one at the end of strand 3 and one in the loop between strands 4 and 5) make stacking interactions with the DNA bases. Several nonconserved residues in β-strands 1 and 2 (a structure termed the β-hairpin) make hydrogen bonds with the phosphate backbone and the DNA bases. A very similar structure in gp32 indicated that the ssDNA binding pocket of gp32 is also an OB fold (5, 52).
Highlighted in Fig. 3 are three residues that are conserved in all four ssDNA binding domains. One residue is an absolutely conserved aspartic acid that lies at the end of the β2 strand. This residue was previously noted for being conserved in domains A and B (45), and mutation of this residue produces phenotypes in yeast when it is mutated in domains A (53), B (45), and D (RPA2) (40, 51). Approximately 10 residues downstream of the conserved aspartic acid is an invariant aromatic residue that lies at the end of the β3 strand and is homologous to the N-terminal stacking residue, F238, of hsRPA1A. Approximately 30 residues further downstream is another aromatic residue which is conserved in all domains except RPA4 (31). This aromatic residue lies in the loop between strands β4 and β5 and is homologous to the C-terminal stacking residue F269 of hsRPA1A.
While domains A and B of all species probably consist of OB folds like their human homologs, the alignment in Fig. 3 implies that domains C and D (RPA2) are also OB folds. With domain C, however, it cannot be so simple, since the β1 strand is replaced by the C-terminal portion of the Cys4 motif (Fig. 3). But the position of the Cys4 motif relative to the positions of the remaining secondary structures is similar to that of the zinc-binding protein gp32. Specifically, the β-hairpin of gp32 was shown to be interrupted by the zinc-binding domain (5, 52). This may be the case in domain C as well.
A secondary-structure prediction program was used to determine whether the sequences of domains C and D were consistent with that of an OB fold. This program accurately predicted the OB fold domains of gp32 as determined by its crystal structure (52). Further, the secondary structures of all species of RPA1 domain C were found to be similar to each other, and the results for the yeast and human proteins are presented in Fig. 4A. In both species, a β-sheet is predicted immediately upstream of the Cys4 motif and may be the β1 strand of an OB fold. Directly following the Cys4 motif is another β strand (β2) followed by a third shorter β strand (β3) and an α-helix. A β-sheet is predicted near residues 580 and is proposed to be β5. A discrepancy between this analysis and the OB fold of gp32 is that an α-helix is predicted to exist where β4 is expected. Thus, domain C may not be a typical OB fold. With domain D, the secondary-structure prediction contained all five β-sheets and the α-helix (Fig. 4B) (6). It is worth noting that all the components of the hypothetical OB fold in domain D lie within the minimal region of scRPA2 that is required for viability in yeast (residues 40 to 173) (45). What is remarkable about both secondary-structure predictions is that each of the conserved residues identified in Fig. 3 lies at the expected position relative to the secondary structures of the OB folds in hsRPA1: the invariant aspartic acid closely follows β2, the first aromatic closely follows a shorter β3, and the second aromatic lies in the loop between β4 and β5. Although this loop is predicted to be helical in domain C, so also were helices predicted in the β4-β5 loop of yeast and human domains A (data not shown).
FIG. 4.
Secondary-structure predictions of domains C and D are consistent with an OB fold. Amino acid sequences of domains C and D from yeast and humans were analyzed by the Predict Protein program, European Molecular Biology Laboratory, Heidelberg, Germany (48). The predicted secondary structures of domains C (A) and D (B) are displayed above the yeast (scRPA1C and scRPA2, respectively) and below the human (hsRPA1C and hsRPA2, respectively) sequences. The sequence of the Cys4 motif (A) and the three highly conserved residues within each amino acid sequence (A and B) are highlighted in white type on a black background. The proposed β strands of the OB fold are indicated. H, helix; E, extended β-sheet.
Mutational analysis of domain C.
The sequence analysis described above makes predictions regarding the roles of the Cys4 motif and potential stacking residues in domain C function. To test these predictions, mutations were generated in domain C and the mutant proteins were expressed in E. coli and assayed for ssDNA binding activity. Wild-type domain C and domain C with a C-terminal truncation of 20 amino acids are active in ssDNA binding, indicating that the extreme C terminus is not required for binding (Fig. 5). But, changing cysteines 505 and 508 to alanine resulted in a recombinant protein that could no longer bind ssDNA. Similar mutations in the Cys4 motif of hsRPA are known to reduce or eliminate RPA activity in SV40 DNA replication (35, 36) and mismatch repair (37). Consistent with the role of RPA in these essential processes, complementation analysis revealed that the C505A-C508A double mutation is lethal in yeast (Table 1).
FIG. 5.
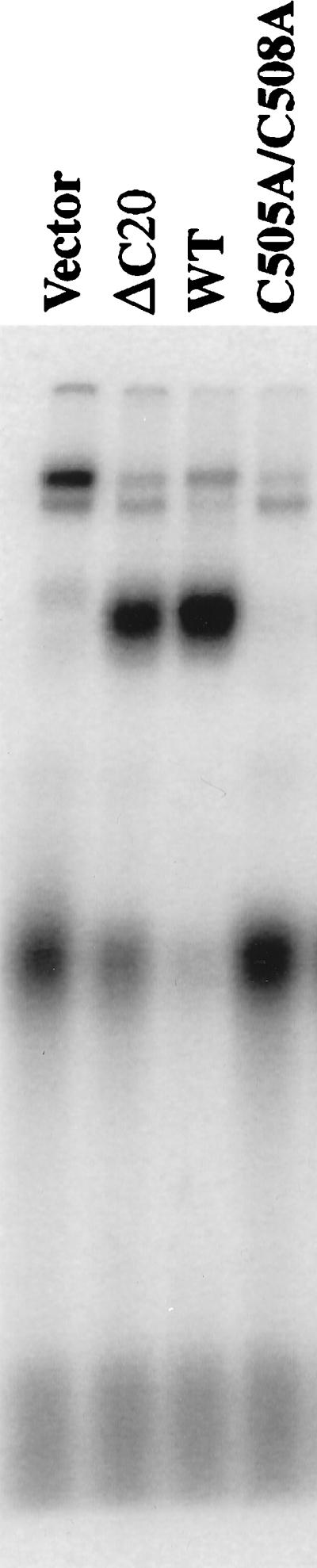
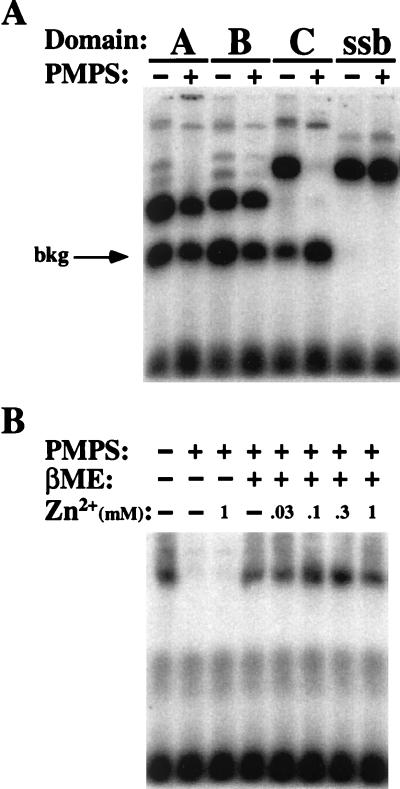
ssDNA binding activity of domain C requires the Cys4 motif. Extracts of E. coli expressing domain C of scRPA1 with the indicated mutations were assayed by UV EMSA. Vector is an extract of E. coli expressing the vector alone. ΔC20 is domain C with a C-terminal truncation of 20 amino acids. WT, wild type.
TABLE 1.
Mutational analysis of conserved residues in RPA
| Subunit | Domain | Mutation at interval β3–α-helix | Viabilitya | Mutation at interval β4–β5 | Viability | Mutations at interval β1 | Viability |
|---|---|---|---|---|---|---|---|
| RPA1 | A | F238A | − | F269A | + | NAb | NA |
| RPA1 | B | W360A | + | F385A | + | NA | NA |
| RPA1 | C | F537A | − | F563A | + | C505A and C508A | − |
| RPA2 | D | W101A | + | F143A | + | NA | NA |
Ability of strain SBY102 (domains A, B, and C) or SBY105 (domain D) bearing a single-copy plasmid with the indicated mutation in RPA1 or RPA2 to form colonies on 5-fluoro-orotic acid as described previously (45).
NA, not assayed.
To further analyze the role of the Cys4 motif in ssDNA binding, domain C activity was tested for sensitivity to the reversible cysteine-modifying reagent PMPS (15, 20, 27). Unlike domains A and B, treatment of domain C with PMPS abolished ssDNA binding activity (Fig. 6A). Ecssb was resistant to PMPS treatment, as was the background binding present in the extract. This result is consistent with genetic and biochemical evidence supporting a role for this domain in RPA function (35–37).
FIG. 6.
Domain C binding activity is sensitive to cysteine modification. (A) Extracts of E. coli expressing the indicated domains of scRPA1 or purified Ecssb (ssb) were incubated in the presence or absence of 2.5 mM PMPS for 1 h, made 50 mM in EDTA, and assayed for ssDNA binding activity by UV EMSA. (B) An extract of E. coli expressing domain C was treated with 2.5 mM PMPS for 1 h, made 50 mM in EDTA, and dialyzed against Chelex-100-treated TNG buffer containing 1 mM EDTA. The modified protein was then treated with the indicated reagents for 30 min before assay by UV EMSA. βME, β-mercaptoethanol; bkg, background.
To test whether a divalent metal was required for binding by domain C, the PMPS-modified protein was treated with high concentrations of EDTA and extensively dialyzed to remove all metals. The protein modification was then reversed with the reducing agent β-mercaptoethanol. As before, the PMPS-modified protein was inactive and could not be stimulated by the presence of 1 mM zinc sulfate (Fig. 6B). However, reversal of the modification with β-mercaptoethanol restored ssDNA binding activity even in the absence of added zinc. Titration of zinc sulfate into the reaction mixture had only a slight stimulating effect on binding. These results indicate that while recombinant domain C protein is sensitive to cysteine modification, zinc is not strictly required for ssDNA binding.
The role of the proposed stacking residues in RPA function in vivo was tested by mutation and complementation analysis in yeast. The N-terminal stacking residue of each domain, analogous to F238 in the β3–α-helix interval of hsRPA1A, was mutated to alanine. This mutation was found to be lethal in domains A and C but not in domains B and D (Table 1). The proposed C-terminal stacking residues were similarly tested. None of these C-terminal mutations had any noticeable effect on viability or growth rate when they were tested for complementation in yeast (Table 1). Thus, with respect to viability, the phenotypes of corresponding mutations in domains A and C are identical. Remarkably, of over 60 point mutations that we have made in scRPA1, only F238 and F537 were individually essential for viability. The fact that these residues are conserved by amino acid sequence alignment suggests that they perform the same function in ssDNA binding.
Quantitation of ssDNA binding affinity.
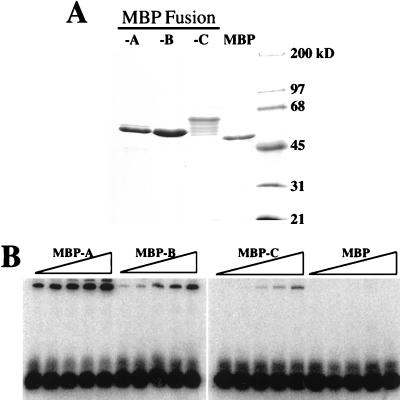
In order to measure the affinity of ssDNA binding by domains A, B, and C, we attempted to purify them from the E. coli extract. Unfortunately, fractionation of the refolded proteins caused them to precipitate quantitatively. To facilitate the recovery of soluble protein, we fused each of the RPA domains to MBP and purified them by affinity and ion-exchange chromatography. Figure 7A shows an SDS-PAGE analysis of the purified proteins which were estimated to be 95% pure. The MBP-C fusion appears as a ladder of bands. This pattern was obtained on multiple purifications and was insensitive to the amount of zinc sulfate present during purification. We assume that these bands arise from limited proteolytic degradation.
FIG. 7.
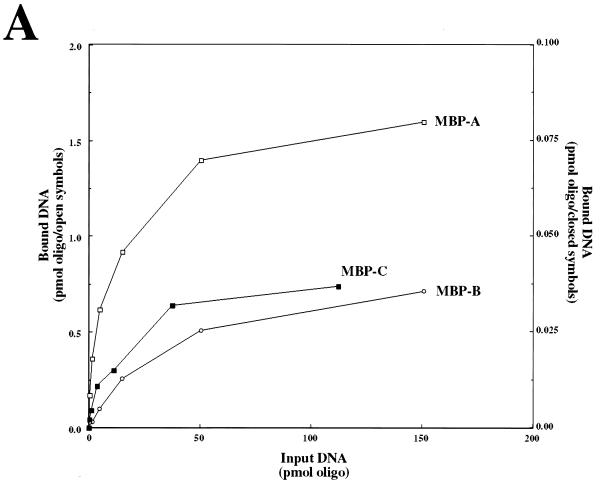
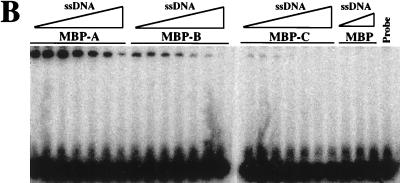
ssDNA binding activity of MBP-RPA fusion proteins. (A) Two micrograms of each purified MBP fusion protein was resolved by SDS–10% PAGE and stained with Coommassie blue. (B) Increasing amounts of the indicated MBP fusion protein or native MBP were incubated with a fixed amount of probe, UV cross-linked, and resolved by denaturing gel electrophoresis. Protein titrations are 0.1, 0.3, 1, 3, and 10 μg in 15-μl binding reaction mixtures.
Increasing amounts of each MBP fusion protein were incubated with a fixed amount of probe and UV cross-linked, and the products were resolved by denaturing gel electrophoresis. As expected, the MBP-A, MBP-B, and MBP-C fusion proteins possessed a ssDNA binding activity that was not present in purified MBP alone (Fig. 7B). Note that at high levels of MBP-A a slower-moving band which may represent multiple proteins binding to a single oligonucleotide is observed. To confirm that the binding of these proteins to ssDNA was specific, we performed two additional experiments. First, a fixed amount of each MBP fusion protein was incubated with increasing amounts of probe to test whether the binding could be saturated with ssDNA substrate. As shown in Fig. 8A, increasing amounts of probe resulted in increasing amounts of DNA binding until the signal reached a plateau at 50 pmol of input oligonucleotide (3- to 10-fold molar excess). Second, we tested that the binding of MBP fusion proteins to ssDNA was in equilibrium by reversing the probe-binding with unlabeled ssDNA. Following a standard incubation of MBP fusion protein with probe, unlabeled oligonucleotide was added and a second incubation was performed. The products were then subjected to UV cross-linking and gel electrophoresis. The presence of increasing amounts of competitor oligonucleotide resulted in decreased levels of probe binding by each MBP fusion protein, indicating that the binding of all MBP fusion proteins was specific (Fig. 8B). Again, no binding was seen by MBP alone. The amounts of free and bound probes present in protein titrations of MBP fusion proteins were quantitated by phosphorimaging, and dissociation constants were calculated. Based on these calculations we find that the binding affinities of MBP-A and MBP-B are approximately equal and two to three times greater than that of MBP-C.
FIG. 8.
ssDNA binding activities of MBP-RPA fusion proteins are saturable and reversible. (A) Increasing amounts of input DNA were incubated with 0.3 μg of MBP-A (5.4 pmol), 0.3 μg of MBP-B (5.4 pmol), or 1 μg of MBP-C (15 pmol) and assayed by UV EMSA. MPB-C values correspond to the 1/20th scale on the right. oligo, oligonucleotide. (B) One microgram of each MBP fusion protein was preincubated with probe (10 fmol) and then incubated with 0, 1, 3, 10, 30, 100, and 300 pmol of unlabeled oligonucleotide. MBP alone was incubated with 0, 100, and 300 pmol unlabeled oligonucleotide. The last lane is a control lacking protein.
DISCUSSION
RPA, like its prokaryotic homologs, is involved in multiple aspects of DNA metabolism (58, 60), including roles in DNA replication (17, 59, 61), DNA repair (13, 24), and genetic recombination (25, 53). While the role of the RPA complex in these processes is well documented, the functions of RPA’s individual subunits are not well understood. It has been noted that RPA resembles Ecssb in that both cellular SSBs are multimeric complexes; RPA is a heterotrimer and Ecssb is a homotetramer. The identification of ssDNA binding domains A and B in RPA1 and a third domain in RPA2 (5, 6, 45) suggested that RPA might contain a fourth ssDNA binding domain analogous to that in Ecssb. The identification of a new ssDNA binding domain in the C terminus of RPA1 supports the idea that cellular SSBs require multiple ssDNA binding domains to carry out their essential function.
The putative zinc-binding domain in the C terminus of RPA1 has long been recognized and compared to that in gp32 (16, 26), but its role in RPA function was not obvious. C-terminal truncations of RPA1 are known to eliminate SV40 DNA replication and the RPA1-RPA2 interaction but to only slightly reduce the ssDNA binding activity of the 70-kDa subunit (21, 22, 35, 36). The essential role of the RPA1 C terminus in SV40 DNA replication may therefore be explained by its effect on trimer formation rather than on ssDNA binding activity. However, the fact that point mutations in the Cys4 motif do not disrupt the RPA1-RPA2 interaction but still eliminate SV40 DNA replication in vitro (35, 36) suggests that it is the ssDNA binding activity of this domain that is important for function. Indeed, the role of the Cys4 motif in the ssDNA binding activity of domain C was confirmed both with point mutations and by chemical modification (Fig. 5 and 6). We conclude that the ssDNA binding activity of this domain is essential for RPA function and that the C-terminal region of RPA1 is required for both ssDNA binding and complex formation. The dual role of the RPA1 C terminus might explain why the secondary structure of this domain was predicted to have an α-helix where the β4 strand of the OB fold was expected. Experiments to test whether this helix contributes to the additional activity of binding RPA2 are in progress.
Having demonstrated that the Cys4 motif is essential for domain C activity, we were surprised to find that zinc is not required for ssDNA binding. While the possibility of trace metal contamination cannot be ruled out in this experiment, precautions were taken to avoid such impurities. For example, all buffers were treated with Chelex-100 resin, which removes divalent metals (15, 27), and different preparations of water and β-mercaptoethanol gave identical results. The Cys4 motif of domain C may therefore function like the zinc-binding domain of gp32, which does not require zinc for ssDNA binding but for cooperativity of binding (19, 43). It has been shown that apo-gp32, which lacks zinc, binds a single-site substrate with the same affinity as metallo-gp32 (19). In contrast, the binding affinity of metallo-gp32 was higher than that of apo-gp32 on larger substrates due to an increase in cooperativity. The binding-site size of the 206-amino-acid domain C protein has not been determined, but it is possible that the small stimulation of binding by zinc seen in Fig. 6B is due to enhanced cooperativity of binding to the 17-nt substrate used in this study. Confirmation of a role for zinc in this process awaits further experiments with larger substrates.
The data presented here lead to a revised model of RPA (45). This model proposes that the RPA heterotrimer consists of at least four ssDNA binding domains that are each essential for RPA activity and likely to serve distinct roles in RPA function. Domains A, B, and C lie within RPA1, and domain D lies within RPA2. The three domains of RPA1 constitute a very strong binding site and may bind ssDNA simultaneously at low concentrations of salt. At 250 mM NaCl, domain D is capable of binding ssDNA as well (45). Since domain D lies within another subunit, it is possible that higher-order binding or wrapping results from the interaction of ssDNA with domain D, leading to compaction of the ssDNA. While EM has provided some evidence for ssDNA wrapping by yeast RPA (1) and salt-dependent compaction of ssDNA by human RPA (56), investigators in the latter study suspected that factors other than wrapping were the cause of compaction.
This model may explain the discrepancy in ssDNA binding-site sizes that have been reported for RPA. Given that RPA is a complex of at least four different binding domains, it is reasonable to expect multiple binding-site sizes, perhaps similar to that of Ecssb. EM studies of hsRPA bound to oligonucleotides suggested that the 8-nt binding mode, which is identified by glutaraldehyde cross-linking (3), is an initial event that subsequently resolves into a stable 30-nt mode (4). Based on the known interaction of domains A and B with 8 nt of ssDNA (5), it is likely that this weak initial mode arises by cross-linking of ssDNA to these two domains (28). The current model then suggests that interaction of ssDNA with domains A, B, and C leads to the stable 30-nt mode, which was observed to have a distinctly elongated appearance by EM (4, 28). As described above, interactions between ssDNA and all four domains may account for the 90-nt mode in the presence of high concentrations of salt (1). Alternatively, it is possible that the 8- and 30-nt binding modes represent interactions with domain A and the A-B pair, respectively. The 90-nt mode might then arise from the binding of the C-D pair. Further experiments will be needed to distinguish between the multiple possibilities arising from these redundant domains. These studies will also need to address the role of metals in binding-site size and cooperativity of ssDNA binding by RPA.
If RPA is truly a structural and functional homolog of Ecssb, then the analysis of RPA provides a unique opportunity to study the role of higher-order binding in DNA metabolism. Since each of the four protomers of Ecssb are equivalent, it is not possible to ask if each of them is essential to SSB function, or whether they perform specific functions in the homotetramer. With RPA, each of the four protomers is distinct and each has now been shown to be required for yeast viability (45) and for SV40 DNA replication in vitro (22, 30, 35, 36). While there is some evidence for functional redundancy in RPA (domain A can substitute for domain B), each domain may have a specific function since domain B cannot substitute for domain A (45) and preliminary experiments in this lab indicate that domains A, B, and D cannot function in place of domain C in yeast. Recently, a detailed mutagenesis of the RPA1 subunit in yeast revealed that point mutations placed throughout its length can differentially affect its multiple functions in DNA metabolism (58). Such a result would be expected if the domains of RPA1 have specific functions. Other mutant studies indicate that conditional-lethal alleles of RPA2 are defective in replication fork movement at the nonpermissive temperature (40, 51). The ssDNA binding activity of RPA2 may therefore be critical to RPA function in the elongation phase of DNA replication. Further in vitro experiments are needed to reconcile the different results in studies of higher-order binding by RPA and to test whether domains C and D play a role in wrapping ssDNA.
What advantage could a multimeric SSB have over a monomeric one? A clue may be found by comparing the efficiencies of various SSBs in T-antigen-catalyzed unwinding (29) and unwinding of a pseudo-origin template (28). It is curious that most replicative SSBs that work in these assays (RPA, Ecssb, adenovirus DBP, and herpesvirus ICP8) are known to bind ssDNA in a multimerized fashion. RPA and Ecssb are now known to be multimers, and it has been shown that DBP uses a C-terminal hook to multimerize (57) and drive strand displacement synthesis (14). While the mechanism of ssDNA binding by ICP8 has not been determined, it is possible that this large protein of 128 kDa also contains multiple ssDNA binding domains. gp32 and T7 gp2.5, which bind ssDNA with high affinity, being monomers, do not function in these unwinding assays (28, 29), and mutant RPA complexes lacking one or more ssDNA binding domains are capable of only minor levels of unwinding (21). These findings suggest that the multiple domains of RPA play an important role in denaturing double-stranded DNA. It will be of interest to test this idea and determine whether RPA domains C and D regulate this process in eukaryotic cells.
ACKNOWLEDGMENTS
Thanks go to Alexey Bochkarev and Aled Edwards for suggesting the use of the secondary-structure prediction program, to John Diffley for advice on PMPS treatment, and to David Norris and Jan Mullen for comments on the manuscript.
This work was supported by NIH grant GM55583.
REFERENCES
- 1.Alani E, Thresher R, Griffith J D, Kolodner R D. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 2.Atrazhev A, Zhang S, Grosse F. Single-stranded DNA binding protein from calf thymus. Purification, properties, and stimulation of the homologous DNA-polymerase-alpha-primase complex. Eur J Biochem. 1992;210:855–865. doi: 10.1111/j.1432-1033.1992.tb17489.x. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell L J, Borowiec J A. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell L J, Borowiec J A, Mastrangelo I A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol Cell Biol. 1996;19:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 6.Bochkareva E, Frappier L, Edwards A M, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 7.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 8.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 9.Bujalowski W, Lohman T M. Limited co-operativity in protein-nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single-stranded nucleic acids in the “beaded,” (SSB)65 mode. J Mol Biol. 1987;195:897–907. doi: 10.1016/0022-2836(87)90493-1. [DOI] [PubMed] [Google Scholar]
- 10.Bujalowski W, Overman L B, Lohman T M. Binding mode transitions of Escherichia coli single-strand binding protein–single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 11.Casas-Finet J R, Khamis M I, Maki A H, Chase J W. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Lett. 1987;220:347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- 12.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coverley D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 14.Dekker J, Kanellopoulos P N, Loonstra A K, van Oosterhout J A, Leonard K, Tucker P A, van der Vliet P C. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 1997;16:1455–1463. doi: 10.1093/emboj/16.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diffley J F X, Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989;246:1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- 16.Erdile L F, Heyer W D, Kolodner R, Kelly T J. Characterization of a cDNA encoding the 70-kDa single-stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J Biol Chem. 1991;266:12090–12098. [PubMed] [Google Scholar]
- 17.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari M E, Bujalowski W, Lohman T M. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 19.Giedroc D P, Keating K M, Williams K R, Coleman J E. The function of zinc in gene 32 protein from T4. Biochemistry. 1987;26:5251–5259. doi: 10.1021/bi00391a007. [DOI] [PubMed] [Google Scholar]
- 20.Giedroc D P, Keating K M, Williams K R, Konigsberg W H, Coleman J E. Gene 32 protein, the single-stranded DNA binding protein from bacteriophage T4, is a zinc metalloprotein. Proc Natl Acad Sci USA. 1986;83:8452–8456. doi: 10.1073/pnas.83.22.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes X V, Wold M S. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 22.Gomes X V, Wold M S. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 23.Griffith J D, Harris L D, Register J D. Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harbor Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 24.He Z, Wong J M S, Maniar H S, Brill S J, Ingles C J. Assessing the requirements for nucleotide excision repair proteins of Saccharomyces cerevisiae in an in vitro system. J Biol Chem. 1996;271:28243–28249. doi: 10.1074/jbc.271.45.28243. [DOI] [PubMed] [Google Scholar]
- 25.Heyer W D, Kolodner R D. Purification and characterization of a protein from Saccharomyces cerevisiae that binds tightly to single-stranded DNA and stimulates a cognate strand exchange protein. Biochemistry. 1989;28:2856–2862. doi: 10.1021/bi00433a017. [DOI] [PubMed] [Google Scholar]
- 26.Heyer W D, Rao M R, Erdile L F, Kelly T J, Kolodner R D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt J B, Neece S H, Schachman H K, Ginsburg A. Mercurial-promoted Zn2+ release from Escherichia coli aspartate transcarbamoylase. J Biol Chem. 1984;259:14793–14803. [PubMed] [Google Scholar]
- 28.Iftode C, Borowiec J A. Denaturation of the simian virus 40 origin of replication mediated by human replication protein A. Mol Cell Biol. 1997;17:3876–3883. doi: 10.1128/mcb.17.7.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny M K, Lee S-H, Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases α and δ. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 31.Keshav K F, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C, Paulus B F, Wold M S. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 33.Kim C, Snyder R O, Wold M S. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C, Wold M S. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 35.Kim D K, Stigger E, Lee S H. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y L, Shivji M K, Chen C, Kolodner R, Wood R D, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 38.Lohman T M, Ferrari M E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 39.Lohman T M, Overman L B. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 40.Maniar H S, Wilson R, Brill S J. Roles of replication protein-A subunits 2 and 3 in DNA replication fork movement in Saccharomyces cerevisiae. Genetics. 1997;145:891–902. doi: 10.1093/genetics/145.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitsis P G, Kowalczykowski S C, Lehman I R. A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry. 1993;32:5257–5266. doi: 10.1021/bi00070a038. [DOI] [PubMed] [Google Scholar]
- 42.Murzin A G. OB (oligonucleotide oligosaccharide binding) fold—common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadler S G, Roberts W J, Shamoo Y, Williams K R. A novel function for zinc(II) in a nucleic acid-binding protein. Contribution of zinc(II) toward the cooperativity of bacteriophage T4 gene 32 protein binding. J Biol Chem. 1990;265:10389–10394. [PubMed] [Google Scholar]
- 44.Pfuetzner R A, Alexey Bochkarev A, Frappier L, Edwards A M. Replication protein A—characterization and crystallization of the DNA binding domain. J Biol Chem. 1997;272:430–434. doi: 10.1074/jbc.272.1.430. [DOI] [PubMed] [Google Scholar]
- 45.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. A hierarchy of SSB protomers in replication protein-A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 46.Prigodich R V, Casas-Finet J, Williams K R, Konigsberg W, Coleman J E. 1H NMR (500 MHz) of gene 32 protein-oligonucleotide complexes. Biochemistry. 1984;23:522–529. doi: 10.1021/bi00298a019. [DOI] [PubMed] [Google Scholar]
- 47.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 48.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 49.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 50.Ruyechan W T, Wetmur J G. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single-stranded DNA. Biochemistry. 1975;14:5529–5534. doi: 10.1021/bi00696a023. [DOI] [PubMed] [Google Scholar]
- 51.Santocanale C, Neecke H, Longhese M P, Lucchini G, Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J Mol Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- 52.Shamoo Y, Friedman A M, Parsons M R, Konigsberg W H, Steitz T A. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 53.Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 55.Sugiyama T, Zaitseva E M, Kowalczykowski S C. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 56.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 57.Tucker P A, Tsernoglou D, Tucker A D, Coenjaerts F E, Leenders H, van der Vliet P C. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 1994;13:2994–3002. doi: 10.1002/j.1460-2075.1994.tb06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umezu K, Sugawara N, Chen C, Haber J E, Kolodner R D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wold M S. RPA: a heterotrimeric, single-stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 61.Wold M S, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wold M S, Weinberg D H, Virshup D M, Li J J, Kelly T J. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]