Abstract
Members of the C/EBP (CCAAT/enhancer binding protein) family of transcription factors play important roles in mediating the acute-phase response (APR), an inflammatory process resulting from infection and/or tissue damage. Among the C/EBP family of proteins, C/EBPβ and -δ were thought to be the primary mediators of the APR. The function of C/EBPα in the APR has not been fully characterized to date. Here, we investigate the role of C/EBPα in the APR by using neonatal mice that lack C/EBPα expression. Northern blot analysis of acute-phase protein gene expression in neonatal mice treated with purified bacterial lipopolysaccharide or recombinant interleukin 1β as an inflammation stimulus showed a strong APR in wild-type mice, but a response in C/EBPα null animals was completely lacking. The C/EBPα knockout and wild-type mice demonstrated elevations in C/EBPβ and -δ mRNA expression and DNA binding as well as increased DNA binding of NF-κB, all of which are known to be important in the APR. Null mice, however, failed to activate STAT3 binding in response to lipopolysaccharide. Our results provide the first evidence that C/EBPα is absolutely required for the APR in neonatal mice, is involved in STAT3 regulation, and cannot be compensated for by other C/EBP family members.
The acute-phase response (APR) is an evolutionarily conserved reaction to a wide range of inflammation stimuli. Cytokines and signaling molecules are produced and secreted by macrophages, fibroblasts, and epithelial cells at the site of trauma. Interleukin 1 (IL-1) and IL-6 are two of the primary cytokine mediators of the APR. Hepatocytes have a high density of cytokine receptors per cell, and the liver has the largest number of cells with receptors, making it a primary organ involved in the APR. In response to cytokines, changes in expression of various acute-phase protein (APP) genes occur, with an up regulation of positive APP genes (those genes whose expression increases during the APR) and a down regulation of negative APP genes (genes whose expression decreases during the APR). The protein products of these genes are secreted from hepatocytes and, in combination with the effects of the cytokines themselves, bring about the systemic and metabolic changes seen in response to inflammation (31).
There are two categories of APP genes based on their cytokine responsiveness. Type 1 genes respond to IL-1, IL-6, tumor necrosis factor alpha (TNF-α), and glucocorticoids, while type 2 genes respond to IL-6 and glucocorticoids but not IL-1 or TNF-α (31). Several families of transcription factors have been shown to play important roles in mediating the changes in APP gene expression seen in the liver, including the C/EBP (CCAAT/enhancer binding protein) (23, 25, 43), NF-κB (50, 55), and STAT (signal transducers and activators of transcription) (45, 46) protein families. Expression of C/EBPβ and -δ is up regulated by IL-1 and IL-6 (1, 25, 28, 42), while IL-1 and TNF-α increase NF-κB activation (50, 55). NF-κB, C/EBPβ, and C/EBPδ all increase IL-6 expression (1, 28, 38). IL-6, in turn, activates STAT proteins through the IL-6 receptor or via other pathways (32, 58, 65). The STAT family members STAT1, STAT3, and STAT5b have been identified as binding to the IL-6-responsive elements in the promoters of several type 2 APP genes and are known to be important for regulation of these genes (30, 45, 46, 62). STAT3 has also been implicated in the regulation of C/EBPδ expression (60). Members of the NF-κB family play important roles in regulating type 1 APP genes. There are several instances of adjacent NF-κB and C/EBP binding sites in the promoters of APP and cytokine genes (2), specifically in the C3 (15), serum amyloid A (SAA) (36, 48), alpha 1-acid glycoprotein (AGP) (33), IL-6 (2), IL-8 (38), and angiotensinogen (47) promoters. Additionally, the basic leucine zipper domain (bZIP) of C/EBP proteins has been shown to be able to directly interact with the Rel homology domain of NF-κB proteins (49, 56), which may have some bearing on the expression of APP genes containing adjacent C/EBP and NF-κB binding sites.
During the APR, both C/EBPβ and -δ show rapid and dramatic increases in mRNA expression, while C/EBPα exhibits a moderate decrease (4). Consensus C/EBP binding sites have been identified in the promoters of a number of APP genes, including those for hemopexin, C-reactive protein, and haptoglobin (Hp) (42), AGP (3), albumin (20), transferrin (51), the third component of complement (25), T-kininogen (13), SAA (23, 35), and transthyretin (14). C/EBPβ and -δ have been identified as binding to the C/EBP binding sites in the promoters of several of the above mentioned APP genes in response to inflammatory stimuli (23, 25, 43). C/EBPα is thought to play a role in homeostatic APP gene regulation, but its role in the APR has been thought to be minimal, in that its binding activity decreases (4). A good model system in which to study the role of C/EBPα in the APR is the C/EBPα knockout mouse. However, the mutant homozygous C/EBPα animals do not survive beyond 24 h after birth due to metabolic complications (57), necessitating the use of neonatal mice in these studies. Although there have been many studies of the APR in adult mice, and a few in neonatal and fetal animals of various species, no extensive molecular studies of APP gene regulation in neonatal mice have been performed. The data we present here demonstrates that normal neonatal mice mount an APR to inflammation, yet C/EBPα null mice fail to up regulate APP genes. C/EBPβ and -δ expression levels are elevated and NF-κB binding is activated in the C/EBPα knockout mice in response to either lipopolysaccharide (LPS) or IL-1β. An elevation in STAT3 binding is lacking in the null mice in response to LPS, highlighting the essential role for STAT3 in the inflammatory response and linking STAT3 regulation to C/EBPα expression. This indicates that C/EBPα is not a “bystander” but plays an important role in the APR independently of the essential transcription factors, C/EBPβ, C/EBPδ, and NF-κB.
MATERIALS AND METHODS
APR induction.
Newborn (0- to 3-h old) mice were injected with 5 mg of LPS per kg of body weight intraperitoneally to induce a generalized inflammatory response. To test the specific IL-1 response, 10 μg of recombinant human IL-1β per kg was injected intraperitoneally into neonatal mice. All neonatal mice were injected periodically with 10% glucose subcutaneously to counteract the hypoglycemia seen in C/EBPα−/− pups.
Northern analysis.
Northern blot analyses were performed on livers harvested at various time points after LPS treatment, including 0, 1, 4, 6, and 12 h, on lungs harvested at 12 h post-LPS treatment, and on livers harvested at 6 h after IL-1 treatment. Total RNA was isolated by using RNA Stat-60 in accordance with the manufacturer’s directions (Tel-Test, Inc.). Twenty-five micrograms of total RNA was loaded on an 0.8% agarose–2.2 M formaldehyde gel, transferred onto a Zeta probe nylon membrane (Bio-Rad), and hybridized sequentially with specific probes as described in the supplier’s manual. Quantification was achieved by using the Molecular Dynamics phosphorimaging system and Image Quant software. The values obtained for each probe were normalized relative to the 18S rRNA control values. The statistical significance of differences between the genotypes was assessed by using Student’s t test.
EMSA.
Preparation of nuclear extracts (NE) from liver tissue was carried out as described previously (53). Briefly, liver tissue was homogenized, centrifuged to pellet nuclei, and washed. The nuclei were lysed with high-salt buffer on ice for 30 min and then centrifuged to remove debris. Electrophoretic mobility shift assays (EMSA) were performed as previously described (52), with minor modifications. For C/EBP proteins, the bZIP1 canonical C/EBP binding site was used, with the sequence as reported previously (25). Samples were run on 8% nondenaturing polyacrylamide gels in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA). For NF-κB binding, the consensus oligonucleotide used, from Promega, had the sequence 5′-AGTTGAGGGGACTTTCCCAGGC-3′. Binding reaction mixtures containing 10 μg of NE and antibodies in binding buffer were preincubated on ice for 20 min prior to the addition of radiolabeled NF-κB oligonucleotide. Samples were then incubated on ice for 40 min and then run on a 6% nondenaturing polyacrylamide gel in 0.5× TBE at 4°C. Competition experiments were carried out with a 100-fold excess of unlabeled oligonucleotide for NF-κB or AP-1 (from Santa Cruz Biotechnology, Inc.; with the sequence 5′-CGCTTGATGACTCAGCCGGAA-3′) in the binding reaction mixture prior to the addition of labeled NF-κB. For STAT3 EMSA, binding reactions were carried out in binding buffer containing 10 mM HEPES (pH 7.6), 50 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 10% glycerol, and 5 mM dithiothreitol, with 10 μg of NE and antibodies. After preincubation on ice for 1 h, radiolabeled STAT3 oligonucleotide (from Santa Cruz Biotechnology, Inc.; with the sequence 5′-GATCCTTCTGGGAATTCCTAGATC-3′) was added, followed by incubation at room temperature for 20 min. Samples were run on a 6% nondenaturing polyacrylamide gel at 4°C. All gels were dried and exposed to X-ray film at −80°C with intensifying screens. The following antibodies were used: for C/EBPα, 14AAX antibody; for C/EBPβ, C-19X; for C/EBPδ, C-22X; and for STAT3, C-20X (all from Santa Cruz Biotechnology, Inc.). Polyclonal antibodies against NF-κB p50 and p65 were generously provided by Rebecca Taub.
Western analysis.
Forty micrograms of liver NE was electrophoresed on a 0.1% sodium dodecyl sulfate–12% polyacrylamide gel, and Western immunoblotting was performed as described previously (53), with sequential hybridization with polyclonal STAT3 antibodies (Santa Cruz Biotechnology, Inc.; C-20X, 1:2,000 dilution), antibodies specific to STAT3 phosphorylated on tyrosine 705 (New England Biolabs; 1:1,500 dilution, overnight incubation with primary antibody), and β-actin (Sigma; 1:5,000 dilution) antibodies as a loading control. As a control for the location and phosphorylation of STAT3, 10 μl of control extracts from NIH 3T3 cells either untreated or treated with IL-6 (New England Biolabs) was run on the same blot as the mouse liver NE. Samples were quantified by using a Molecular Dynamics personal densitometer. The values obtained for each probe were normalized relative to the β-actin values.
RESULTS
LPS induces the APR in neonatal mice.
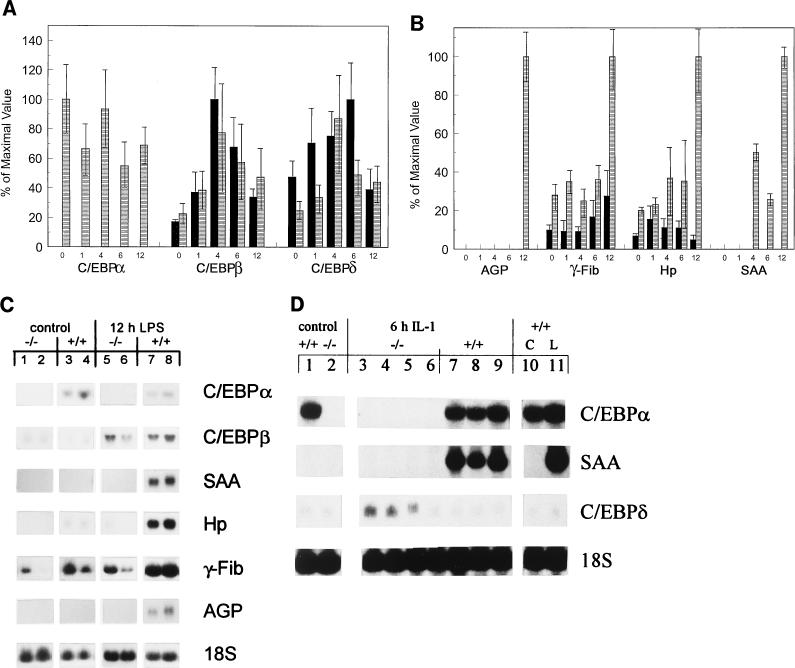
To determine the feasibility of studying the APR in neonatal mice, newborn C/EBPα wild-type (C/EBPα+/+) pups were assayed for responsiveness to LPS. Northern blot analyses of livers harvested at various time points after LPS administration showed that the wild-type neonatal pups (n ≥ 3 per time point) were able to mount a response to LPS treatment, through elevation of both the immediate-early genes, those of C/EBPβ and -δ (Fig. 1A), as well as several classical APP genes, those of AGP, γ-fibrinogen (γ-Fib), Hp, and SAA (Fig. 1B). Representative Northern blots, showing results for RNA from both untreated control and 12-h-post-LPS-treated C/EBPα−/− (Fig. 1C, lanes 1 and 2 and lanes 5 and 6, respectively) and C/EBPα+/+ (Fig. 1C, lanes 3 and 4 and lanes 7 and 8, respectively) mouse livers were probed sequentially with the indicated APP gene probes.
FIG. 1.
Lack of APP gene induction in C/EBPα−/− mice in response to LPS or IL-1. Northern analysis of 25 μg of total RNA from newborn C/EBPα−/− (−/−) and C/EBPα+/+ (+/+) mice at 0, 1, 4, 6, and 12 h post-LPS treatment. (A and B) Expression of C/EBP gene family (A) and APP genes (B). Blots were quantified, and the phosphorimaging value of each probe was normalized to the value of the 18S rRNA probe used as a loading control. The mean value for each genotype at each time point was determined. The largest normalized value for a given gene probe was set at 100%, and the normalized values of the other samples were compared to this maximal value. For each probe (indicated below the appropriate LPS time course), the data was expressed as the percentage of the maximal value ± standard deviation obtained for that probe. Each bar represents the results for three animals, except those for null and wild-type mice at 12 h post-LPS treatment (n = 5 and 4, respectively) and for null mice at 1 h post-LPS treatment (n = 4). ■, null mice; ▤, wild-type mice. (C) Representative Northern blots showing results for RNA from the same blot for two control (glucose-treated) −/− (lanes 1 and 2) and +/+ (lanes 3 and 4) pups and for RNA from the same blot for two −/− (lanes 5 and 6) and +/+ (lanes 7 and 8) mice at 12 h post-LPS treatment. The blots were hybridized with each individual probe sequentially, with the probe used indicated to the right of the panel. (D) Northern blot results for 25 μg of total liver RNA from control (glucose-treated) −/− (lane 2) and +/+ (lane 1) mice, 6-h-IL-1-treated −/− (lanes 3 to 6) and +/+ (lanes 7 to 9) animals, and +/+ control (C) (lane 10) and 6-h-LPS-treated (L) (lane 11) mice, hybridized sequentially with the probes indicated to the right of the panel.
Newborn C/EBPα+/+ mice had measurable basal levels of C/EBPδ, γ-Fib, C/EBPβ, and Hp gene expression but not detectable expression of the SAA or AGP gene (Fig. 1A and B). In response to LPS, there was no significant difference in the expression of the C/EBPα gene through 12 h of treatment. As expected, there were elevations in the expression levels of the C/EBPβ and -δ genes, both peaking at 4 h post-LPS treatment, with over a threefold increase above basal levels in each case (Fig. 1A). The C/EBPα+/+ pups demonstrated a dramatic induction of both the AGP and SAA genes by 12 h after LPS administration. Additionally, there was over a threefold increase in γ-Fib gene expression and a fivefold elevation in Hp gene expression in the C/EBPα+/+ neonates by 12 h post-LPS treatment (Fig. 1B). All of these increases were statistically significant as measured by Student’s t test (P = 0.001 for the γ-Fib gene and P < 0.0001 for the SAA, AGP, and Hp genes). The results demonstrate that LPS elicits an APR in wild-type neonatal mice and that this process can be studied in C/EBPα null mice in order to discriminate the role of C/EBPα from those of other C/EBP family members.
C/EBPα null mice fail to mount an APR in response to LPS or IL-1β.
In contrast to the observations made with wild-type (C/EBPα+/+) neonates, C/EBPα null (C/EBPα−/−) mice showed no increase in APP gene expression in response to LPS in the liver (Fig. 1B and C). Basal levels of γ-Fib and Hp gene expression were present but reduced by about threefold in C/EBPα−/− mice relative to their C/EBPα+/+ littermates, and neither gene showed a statistically significant increase in expression in response to LPS, in contrast to the elevation seen in wild-type mice. Both SAA and AGP gene expression levels were undetectable in LPS-treated C/EBPα−/− pups, in contrast to the large increases seen in C/EBPα+/+ mice (Fig. 1B and C). As expected, the C/EBPα gene was not expressed in the livers of C/EBPα−/− animals under any conditions. The lack of an APR gene response was not due to a failure to activate the C/EBPβ or C/EBPδ gene, as the induction of these genes over control levels was statistically significant (for the C/EBPβ gene, P < 0.05 at all times of LPS treatment, and for the C/EBPδ gene, P = 0.035 at the 6-h time point) (Fig. 1A). The expression levels of the C/EBPβ gene were not significantly different between the mouse genotypes in control or LPS-treated livers, while the expression of the C/EBPδ gene was actually higher in C/EBPα−/− pups than in C/EBPα+/+ mice and was statistically significant in both the control mice (P = 0.027) and at 6 h after LPS administration (P = 0.04). These results demonstrate that C/EBPα−/− mice were able to elevate the expression levels of both the C/EBPβ and -δ genes in response to LPS, indicating that the C/EBPα null animals responded to the LPS treatment and that the C/EBPα gene was not necessary for the acute-phase induction of expression of these C/EBP gene family members.
IL-1 was administered to mice to bypass the requirement for activation of and cytokine release by macrophages. At 6 h post-IL-1 treatment, C/EBPα+/+ mice demonstrated an APR, as measured by mRNA induction of the SAA gene (Fig. 1D, lanes 7 to 9). Basal levels of SAA gene expression were undetectable in the C/EBPα+/+ mice (Fig. 1D, lanes 1 and 10). The C/EBPα−/− animals exhibited no SAA gene response to IL-1, with no detectable SAA gene mRNA in either treated or untreated livers (Fig. 1D, lanes 2 to 6). However, the C/EBPα−/− mice up regulated expression of the C/EBPδ gene (Fig. 1D, lanes 3 to 6) to levels threefold higher on average than C/EBPα+/+ mice did (Fig. 1D, lanes 7 to 9). The C/EBPβ gene expression levels were also elevated in response to IL-1 in C/EBPα−/− mouse livers, showing a moderate increase relative to that in livers from C/EBPα+/+ animals and levels three times higher than that in livers from control untreated mice (data not shown). These results indicate that the IL-1 signaling pathway is functioning to elevate expression of C/EBP gene family members but not SAA gene expression in the C/EBPα−/− mouse livers.
The defect in null mice is liver specific.
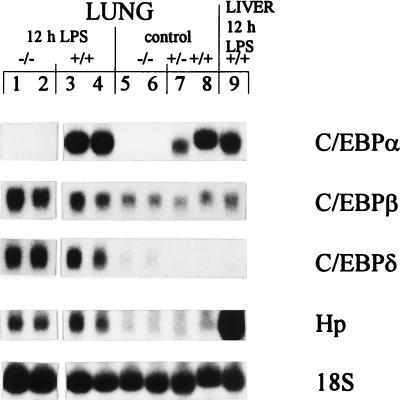
In addition to the liver, the lung is also able to mount an APR to LPS (26). An analysis of APP gene expression in the lung at 12 h post-LPS treatment was performed to determine whether the lack of a response in C/EBPα−/− mice was liver specific (Fig. 2). Quantitation by phosphorimaging showed that there was no difference between the mouse genotypes in C/EBPβ gene mRNA levels in the lungs of LPS-treated mice (Fig. 2, lanes 1 to 4) or untreated animals (lanes 5 to 8). Basal C/EBPδ gene expression was detectable in the lung, with that in C/EBPα−/− animals (Fig. 2, lanes 5 and 6) two times higher than that in their wild-type and heterozygous littermates (Fig. 2, lanes 7 and 8); these levels increased in all mice in response to LPS (Fig. 2, lanes 1 to 4). In contrast to the results with the liver, in which case C/EBPα−/− mice failed to elevate APR gene expression levels, Hp was induced by LPS in the lungs of mice of both genotypes (Fig. 2, lanes 1 to 4), although levels were lower than those seen in the liver (Fig. 2, lane 9). Additionally, similar to the results with the liver, lung samples showed no basal expression of the SAA gene. However, both C/EBPα+/+ and C/EBPα−/− mice showed minor elevations in SAA gene expression in the lung at 12 h after LPS treatment (data not shown). The AGP gene was not expressed in the lung in response to LPS in animals of any genotype (data not shown). Thus, all mice appeared to have the ability to mount an APR in the lung in a subset of APP genes normally elevated in the liver, suggesting that the cytokine signals were activated in C/EBPα−/− mice and that the requirement for C/EBPα expression was liver specific.
FIG. 2.
LPS response in neonatal C/EBPα−/− mouse lung. Northern blot analysis of 25 μg of total lung RNA from 12-h-LPS-treated C/EBPα−/− (−/−) (lanes 1 and 2) and C/EBPα+/+ (+/+) (lanes 3 and 4) mice and control (glucose-treated) −/− (lanes 5 and 6), +/− (lane 7), and +/+ (lane 8) mice, with results for 25 μg of RNA from 12-h-LPS-treated mouse liver (lane 9) shown for comparison, hybridized sequentially with the indicated probes.
Null mice properly induce other C/EBP family members in response to LPS.
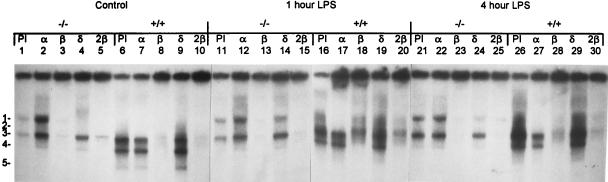
In the liver, there is an induction of both C/EBPβ and -δ that accompanies the APR (4). EMSA were performed on liver NE to verify the nature of the C/EBP proteins that were responsive to LPS in the mice (Fig. 3). Since there are many combinations of the different C/EBP proteins in the neonatal mouse liver, the picture is quite complicated. Four complexes, numbered 2, 3, 4, and 5, were seen binding to a canonical C/EBP site in C/EBPα+/+ mice in the presence of preimmune (PI) serum (Fig. 3, lane 6). With the addition of antibodies to C/EBPα, complex 2 was shifted completely, indicating the presence of C/EBPα in the DNA binding complex (Fig. 3, lanes 7, 17, and 27). Antibodies to C/EBPβ shifted or neutralized complexes 4 and 5 completely and partially diminished complexes 2 and 3 (Fig. 3, lanes 8, 18, and 28). An addition of twice the amount of C/EBPβ antibody also failed to completely shift this complex (Fig. 3, lanes 10, 20, and 30). This demonstrated that complexes 4 and 5 contained C/EBPβ isoforms, complex 2 included both C/EBPα and -β, and complex 3 likely contained C/EBPβ heterodimerized with another protein. In the C/EBPα+/+ mice, LPS treatment resulted in an increase in complexes 3 and 4 (containing C/EBPβ) (Fig. 3, lanes 6, 16, and 26). Antibodies to C/EBPδ did not alter any of the complexes in the C/EBPα+/+ mice, indicating that C/EBPδ was not binding to DNA in these neonatal pups (Fig. 3, lanes 9, 19, and 29). Western blot analyses of C/EBPδ protein levels showed a greatly reduced quantity of C/EBPδ protein in the wild-type mouse livers relative to that in the null mice (data not shown). There was a significant decline in the C/EBPα-containing complexes by 12 h of LPS treatment (data not shown), corresponding to the reported reduction in C/EBPα protein binding during the APR (4). Although C/EBPα protein levels were reduced at this time, there was still some C/EBPα protein present, indicating the possibility that it may play a direct role in the APR in wild-type animals.
FIG. 3.
C/EBPβ and -δ are present in C/EBPα−/− mice and respond to LPS treatment. EMSA of C/EBP binding activity in C/EBPα−/− (−/−) and C/EBPα+/+ (+/+) mice treated with glucose (control) (lanes 1 to 10) or LPS for 1 (lanes 11 to 20) or 4 (lanes 21 to 30) h are shown. Complexes are numbered to the left of the figure. Ten micrograms of liver NE was preincubated with 1 μl of antisera against C/EBPα (α), C/EBPβ (β), C/EBPδ (δ), or PI serum as a control or with 2 μl of C/EBPβ (2β) antiserum to determine the nature of the complexes. The C/EBPα and C/EBPδ antisera showed a stabilizing effect on the NE proteins. The antisera used are indicated above the lanes.
In untreated C/EBPα−/− mice, only two major complexes, 1 and 3, were observed in the presence of PI serum (Fig. 3, lane 1). Complex 3 appeared to be the same as that seen in the C/EBPα+/+ animals, but complex 1 was unique to C/EBPα−/− mice. Both complexes 1 and 3 were mostly neutralized by antibodies to C/EBPβ (Fig. 3, lanes 3, 13, and 23). Doubling the quantity of C/EBPβ antibody did not lead to complete neutralization of these complexes (Fig. 3, lanes 5, 15, and 25), indicating that other proteins may also be present. Antibodies to C/EBPδ also shifted complex 1 (Fig. 3, lanes 4, 14, and 24), demonstrating the presence of C/EBPβ/δ heterodimers. The binding activities of both C/EBPβ and C/EBPδ increased in response to LPS (Fig. 3, lanes 3, 4, 13, 14, 23, and 24).
In mice of both genotypes, the different C/EBPβ-containing complexes showed an overall increase in C/EBPβ protein levels, as expected, and likely correspond to the proportions of the different C/EBPβ protein isoforms that are observed during the APR (of sizes 38, 35, 21, and 14 kDa) (7, 17). Interestingly, Western blot analyses of C/EBPβ protein levels in the liver showed significant differences between the mouse genotypes in the C/EBPβ protein isoform composition, with the null pups having a greatly increased proportion of the full-length C/EBPβ isoforms (of sizes 38 and 35 kDa) to that of the smaller, 21-kDa, isoform, while the wild-type animals primarily expressed the small 21-kDa and 14-kDa isoforms, with little full-length C/EBPβ protein (11). These results correlate with the differences in complex mobility seen in the C/EBP EMSA of the present study, with the null livers possessing C/EBPβ-containing complexes with decreased electrophoretical mobility and wild-type livers having faster-migrating complexes (Fig. 3). In the C/EBPα−/− mice, both C/EBPβ and -δ, important regulators of the inflammatory response, were induced in response to LPS, although the differences seen in these two proteins between the mouse genotypes may impact the APR.
NF-κB is induced in null mice.
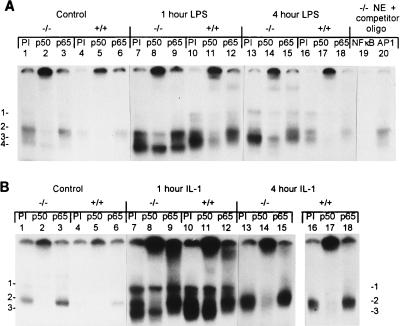
To determine whether the lack of an APR in C/EBPα−/− mice was due to defective regulation of NF-κB activity, known to be important for the APR (50, 55), EMSA were performed on both LPS- and IL-1-treated and control liver NE (Fig. 4). In control C/EBPα+/+ mouse livers, the p50 subunit (complex 2) of NF-κB was detected in the presence of PI serum, as shown by supershift analysis using antibodies to p50 and p65 (Fig. 4A, lane 4 to 6). In untreated neonatal C/EBPα−/− mouse livers, three complexes, 1, 2, and 4, were detected as binding to the NF-κB oligonucleotide in the presence of PI serum (Fig. 4A, lane 1). By using specific antisera, complex 2 was shown to contain p50 (Fig. 4A, lanes 2 and 8), while complex 4 was not significantly altered by either antibody (Fig. 4A, lanes 1 and 9). As tested with competing oligonucleotides, the complexes were specific to NF-κB and were not competed by an AP1 oligonucleotide (Fig. 4A, lanes 19 and 20). At 1 h post-LPS treatment, C/EBPα+/+ mouse livers had an increased level of complex 2, containing p50, and showed the appearance of complex 3, which contained p50 and p65 subunits, as determined by supershift analysis (Fig. 4A, lanes 10 to 12). C/EBPα−/− mouse livers at 1 h post-LPS treatment also showed an elevation of complex 2 (p50) and complex 4 (Fig. 4A, lanes 7 to 9). At the 4-h time point, NF-κB binding was declining in the C/EBPα+/+ mouse livers yet was still elevated above control levels (Fig. 4A, lanes 16 to 18). By 4 h after LPS administration, C/EBPα−/− mouse livers had increased levels of both complex 1 (p50) and complex 3, containing both p50 and p65 (Fig. 4A, lanes 13 to 15). Although the C/EBPα−/− mice had higher basal levels of NF-κB binding, there was a clear induction of p50 and p65 in response to LPS. Therefore, NF-κB DNA binding complexes were available for participation in APR gene regulation. However, the differences seen in complex composition may have an impact on the APR.
FIG. 4.
Induction of NF-κB family members by LPS and IL-1 in all genotypes. EMSA analysis of NF-κB binding activity on 10 μg of liver NE from C/EBPα−/− (−/−) and C/EBPα+/+ (+/+) mice, with preincubation with 1 μl of PI serum used as a control, or of NF-κB p50 or p65 antisera to identify proteins in the complexes is shown. Genotypes and treatments are indicated above the lanes. The antiserum used is listed above each lane, and complexes are numbered on the left (A and B) and/or right (B) of the panel. (A) NF-κB response to LPS. Mice were treated with glucose (control) (lanes 1 to 6) or LPS for 1 (lanes 7 to 12) or 4 (lanes 13 to 18) h and analyzed for protein binding. Competition using 100-fold excess of either unlabeled NF-κB or AP1 (from Santa Cruz Biotechnology, Inc.) oligonucleotides (oligo) and 10 μg of liver NE from C/EBPα−/− mice demonstrated that the complexes were specific to NF-κB; the competitor is specified above the lanes (lanes 19 and 20). (B) NF-κB response to IL-1. Mice were treated with glucose (control) or IL-1 for 1 or 4 h and analyzed for protein binding. Samples in lanes 16 to 18 were run on a separate gel.
To determine whether the increased binding to the NF-κB oligonucleotide in response to LPS was the result of a functional IL-1 signaling pathway, NE from mouse livers treated for various times with IL-1 were analyzed by EMSA (Fig. 4B). As with LPS treatment, IL-1 induced NF-κB binding in both C/EBPα+/+ and C/EBPα−/− mice by 1 h (Fig. 4B, lanes 7 and 10). Complexes containing p50 (number 2) and both p50 and p65 (number 3) were induced in C/EBPα−/− (Fig. 4B, lanes 7 to 9) and C/EBPα+/+ (Fig. 4B, lanes 10 to 12) mouse livers at the 1-h time point. By 4 h after IL-1 administration, NF-κB binding was decreasing in all animals, was composed primarily of complex 2 (p50-containing) binding (Fig. 4B, lanes 13 to 18), and returned to basal levels by 6 h (data not shown). This demonstrates that the IL-1 signaling pathway is functioning in C/EBPα−/− livers and that NF-κB p50 and p65 proteins are present and inducible in response to both IL-1 and LPS yet this induction is not sufficient to mediate an up regulation of APP gene expression in the C/EBPα−/− mice.
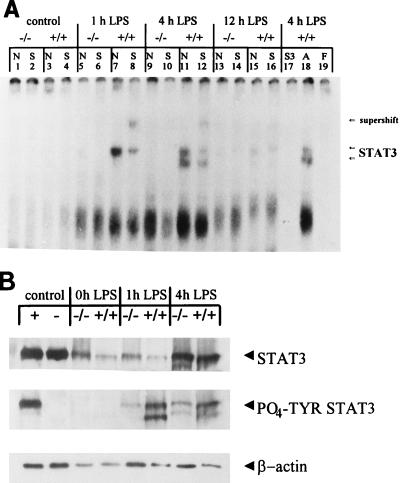
STAT3 binding is not induced in null mice in response to LPS.
EMSA were also performed to study STAT3 binding in LPS-treated mice as an indicator of the IL-6 signaling pathway (45, 46) (Fig. 5A). In response to LPS, the C/EBPα+/+ mice demonstrated induction of complexes binding to the STAT3 oligonucleotide at the 1- and 4-h time points that were shown to contain STAT3 by using specific antibodies (Fig. 5A, lanes 7, 8, 11, and 12). These complexes were reduced by 12 h after LPS administration (Fig. 5A, lanes 15 and 16). In C/EBPα−/− mouse livers, however, these complexes were virtually undetectable at any time (Fig. 5A, lanes 1, 2, 5, 6, 9, 10, 13, and 14). All complexes were competed by cold STAT3 oligonucleotide (Fig. 5A, lane 17) but not by an AP1 oligonucleotide (Fig. 5A, lane 18). The lower-mobility, LPS-inducible complex present in all mice was not shifted by antibodies to STAT1, STAT3, or STAT5b (data not shown), and the proteins contained in this complex remain unknown but were equally well expressed in mice of both genotypes. Western blot analysis (n = 2 for each genotype at each time point) was performed to assess the STAT3 protein levels and phosphorylation status in liver NE in LPS-treated and control mice (Fig. 5B). Tyrosine phosphorylation of STAT3 has been shown to be important for appropriate functioning (reviewed in references 16 and 24). Densitometry was used to quantify the amount of STAT3 and tyrosine-phosphorylated STAT3 present relative to the β-actin loading control. STAT3 was detectable in control mice of both genotypes. In response to LPS, STAT3 protein levels were increased in all mice to various degrees. On average, there were increases of 5-fold in C/EBPα−/− mouse livers and 10-fold in C/EBPα+/+ mouse livers after 4 h of LPS treatment. Interestingly, the C/EBPα−/− mice had reduced levels of tyrosine-phosphorylated STAT3, with levels around sixfold lower than those in C/EBPα+/+ animals at the 4-h LPS treatment time point (Fig. 5B). The fact that tyrosine phosphorylation has been demonstrated to be critical for DNA binding (16, 24) may help explain the absence of detectable STAT3 binding in the null mice and may impact the APR in these animals.
FIG. 5.
No STAT3 binding in response to LPS treatment in C/EBPα−/− mouse livers. (A) EMSA analysis of STAT3 binding activity by using 10 μg of liver NE from C/EBPα−/− (−/−) and C/EBPα+/+ (+/+) mice treated with glucose (control) (lanes 1 to 4) or LPS for 1 (lanes 5 to 8), 4 (lanes 9 to 12), or 12 (lanes 13 to 16) h, with preincubation with either no antiserum (N) or 1 μl of STAT3 antiserum (S) to identify proteins in the complexes. Genotypes and treatments are indicated above the lanes. The antisera used are listed above the lanes, and the positions of STAT3-containing complexes and antibody supershift are shown to the right of the panel. Competition using a 100-fold excess of either unlabeled STAT3 (S3) or AP1 (A) (from Santa Cruz Biotechnology, Inc.) oligonucleotides and 10 μg of liver NE from 4-h LPS-treated +/+ mice demonstrated that the complexes were specific to STAT3 (competitors are specified above the lanes [lanes 17 and 18]). Free probe (F) was run without proteins present (lane 19). (B) Western blot analysis of STAT3 protein levels in 40 μg of liver NE. The mouse genotypes and treatments are indicated above the lanes, and the antisera used as probes are indicated on the right, with the polyclonal STAT3 antibodies denoted as STAT3 and the tyrosine-phosphorylated STAT3 antibodies designated PO4-TYR STAT3. “Control” refers to the NIH 3T3 cell extracts (New England Biolabs) that were used to control for the position of STAT3 and the phosphorylation status of STAT3. The positive control extracts (+) contain tyrosine-phosphorylated STAT3, while the negative control extracts (−) contain nonphosphorylated STAT3. The untreated control mouse liver extracts are designated as 0h LPS.
DISCUSSION
Although neonatal pups have not been studied extensively, they are capable of mounting an APR to the inflammatory stimuli LPS and IL-1 and permit the analysis of molecular pathways leading to the APR that are relevant to this stage of development. The results demonstrate that C/EBPα null mice fail to generate an appropriate response to LPS or IL-1, despite the induction and activation of several other transcription factors (C/EBPβ, C/EBPδ, and NF-κB) critical for the APR. The fact that IL-1 treatment led to an elevation of C/EBPβ and -δ expression as well as an increase in the DNA binding ability of these two proteins and members of the NF-κB family suggests that the IL-1 signaling pathway is functional in the C/EBPα−/− mice. Yet the failure to elevate the type 1, IL-1-responsive, SAA gene expression in C/EBPα−/− mice receiving IL-1 shows that C/EBPα is required for some aspect of the hepatic response to IL-1 and that IL-1 signaling alone is not sufficient for the APR induction of the SAA gene in the neonate. Nevertheless, it is possible that the differences seen between the mouse genotypes in the levels of induction of C/EBPδ and NF-κB transcription factors may have an influence on the APR in the null mice.
Furthermore, the data show a reduction in the basal expression levels of both the γ-Fib and Hp genes in C/EBPα−/− mouse livers. This is likely a direct result of the absence of C/EBPα, since C/EBPα is thought to be a basal regulator of these genes (8, 40). The question remains, however, whether C/EBPα has additional direct or indirect roles in mediating the APR. In the case of a direct role, C/EBPα may bind directly to the C/EBP sites in the promoters of the APP genes, either as homodimers or as heterodimers with other family members or with NF-κB family members, to directly activate gene expression. However, studies of DNA binding proteins stimulated by IL-1 and IL-6 treatment of cultured hepatoma cells showed reduced binding of C/EBPα compared to that of untreated control cells, with concomitant increases in binding of C/EBPβ and/or C/EBPδ (23, 25). Consistent with the cell culture results, studies of mouse livers demonstrated that C/EBPα bound to the AGP promoter under normal conditions and was replaced by C/EBPβ after LPS treatment (3). These studies suggested that C/EBPβ and -δ were the predominant C/EBP proteins active in DNA binding during the inflammatory response. C/EBPα may have more of an indirect rather than a direct role in the APR. The potential indirect roles of C/EBPα are numerous. As described above, multiple signaling pathways may be affected by C/EBPα. C/EBPα could also have effects on the chromatin conformation in the APP gene promoters, maintaining an open conformation of use during the APR. p300, a protein known to have roles in chromatin remodeling, has been shown to increase the transactivational capability of both C/EBPα and -β. p300 also directly interacts with C/EBPβ in a region conserved in the C/EBP family (39). Therefore, it is possible that C/EBPα may interact with p300 to alter chromatin conformation. Alternatively, C/EBPα might be important for recruitment of proteins, such as other C/EBP and/or NF-κB family members, to the APP gene promoters during the APR.
STAT3 is thought to act primarily through the IL-6 signaling pathway to elevate transcription of many of the APP genes. Binding sites for STAT3 have been identified in several of the APP genes studied here, including the γ-Fib (64), AGP (59), and Hp (27) genes. The absence of STAT3 binding in C/EBPα−/− mouse livers in response to LPS is likely to play a role in the failure to elevate the transcription of these genes. Although there was an elevation in the level of nuclear STAT3 protein in C/EBPα−/− mouse livers in response to LPS, STAT3 did not bind to DNA in these animals and, therefore, was apt to be nonfunctional during the APR. Analysis of the phosphorylation status of STAT3 in the livers showed a reduced level of tyrosine-phosphorylated STAT3 present in the null animals. Since tyrosine phosphorylation is known to be critical for STAT3 DNA binding (reviewed in references 16 and 24), the decreased level present in the null animals may be responsible for the lack of STAT3 binding in the C/EBPα−/− mice and may explain, at least in part, why the APR is compromised in the C/EBPα-deficient mice. Additionally, C/EBPα−/− mice have been shown to have significantly reduced levels of the IL-6 receptor on hepatocytes (63), which may play a role in the reduced STAT3 tyrosine phosphorylation in these animals.
While the failure to properly activate STAT3 in the C/EBPα−/− mouse liver in response to LPS is an important component of the alterations seen in the APR, it is likely that there are other pathways that are affected that also impact the APR. For example, mice deficient in IL-1β (18), IL-1 receptor type I (34), TNF (37), or IL-6 (19, 29) all demonstrate a relatively normal LPS response, providing evidence for significant redundancy of function in cytokine signaling. More evidence for this redundancy comes from a study of six different cytokines sharing gp130 as a receptor subunit (CNTF, IL-6, LIF, OSM, IL-11, and cardiotrophin-1) that were all able to induce SAA gene expression when administered individually (9). Given the multiple induction pathways of SAA gene expression, it is likely that there are additional defects in the C/EBPα−/− mice that account for the impaired LPS response. Additionally, the IL-6-deficient animals are able to activate STAT3 in response to LPS (6), suggesting that the IL-6 receptor may be used by other cytokines in these mice to activate STAT3 and the APP genes. C/EBPα may broadly disrupt all pathways or may be specifically required for induction of SAA gene expression, regardless of its activation by multiple signaling pathways.
The SAA gene is thought to be primarily regulated by C/EBPβ and -δ and NF-κB in the APR (36, 48), yet these proteins are active and available in the C/EBPα−/− mice. However, each of these proteins demonstrates some difference in the level and timing of induction between the genotypes, and these differences, taken together, may be a component of the altered APR in the null mice. The role of STAT3 in SAA gene induction is not fully understood, since a STAT3 binding site has not been identified in the characterizations of the SAA gene promoter to date. IL-6 induction of SAA gene expression has been reported, including the identification of an IL-6-responsive element (44), which may well bind STAT3. In addition, in cell culture studies, antibodies to IL-6 interfere with the ability of IL-1 to induce SAA gene expression in hepatocytes (61). The lack of STAT3 binding ability in the C/EBPα−/− animals may well have some effect on the absence of the SAA gene in these mice. It is likely, however, that additional defects in the C/EBPα−/− mouse livers account for the total lack of APP gene induction by LPS or IL-1. For example, both TNF-α (61) and okadaic acid (21) have been shown to inhibit the ability of IL-1 and IL-6 to elevate SAA gene expression. The pathways affected by these substances may require C/EBPα actively. Additionally, a new STAT-like factor, LIL-STAT, has been identified as both LPS and IL-1 inducible (54), leading to the possibility that additional STAT family members are adversely affected in the C/EBPα−/− mice.
Studies of the APR in adult C/EBPβ knockout mice, using turpentine as an inducer of inflammation, showed a reduction in the magnitude of, and an earlier peak in, SAA gene mRNA expression as compared to that in wild-type mice, while the expression of several other APP genes was not adversely affected (12). The composition of the C/EBPβ-containing complexes binding to DNA were somewhat altered in the C/EBPα−/− mice. To determine whether these differences in the C/EBPα−/− mouse livers played a role in the defective APR, newborn C/EBPβ knockout mice were analyzed for a response to LPS. By 6 h after LPS treatment, SAA gene expression was induced in neonatal C/EBPβ null and wild-type mice (data not shown). The presence of a response to LPS in the C/EBPβ null mice suggests that although C/EBPβ may be important for the APR, other factors are able to compensate for its absence in the C/EBPβ knockout animals, in striking contrast to the C/EBPα-deficient mice. C/EBPβ protein isoforms are known to have differing transactivational abilities (17, 22, 41) and have been shown to change in composition during the APR (7). The variation in the composition of C/EBPβ complexes in the C/EBPα knockout mice could influence the regulation of APP genes. Additionally, it is possible that the elevated basal levels of C/EBPδ protein in the C/EBPα−/− mouse liver may interfere with the proper functioning of the APR.
These results demonstrate an indispensable, and heretofore unknown, role for C/EBPα in the hepatic response to inflammation in vivo. In addition to supporting the prevailing view that C/EBPα is involved in the homeostatic maintenance of APP gene expression, our data clearly show that C/EBPα is critical for the induction of the APR in neonatal mice and cannot be replaced by other C/EBP family members. The effects of treating the mice with LPS and IL-1 indicate that the IL-1 and LPS signaling pathways can function in the null animals to activate C/EBPβ, C/EBPδ, and NF-κB. The failure of STAT3 to be properly activated in the C/EBPα−/− mouse liver is likely to be one factor in the loss of the inflammatory response seen in these animals and may have far-reaching implications for the APR. Since SAA gene expression has not been definitively associated with STAT3 to date, it appears that other C/EBPα-dependent factors are required to increase the expression of the SAA gene and potentially the other classic APR genes studied. Although we cannot completely eliminate the differences in the complex composition of the NF-κB proteins as causal, it appears that the complexes that are induced in the C/EBPα+/+ mice are also induced in the C/EBPα knockout animals. These findings may be of significance to infants born prematurely, prior to the full expression of C/EBPα, which occurs late in gestation (10). Such infants have severe metabolic complications (5), which would be exacerbated in a situation in which inflammatory responses to infection or wound healing were required.
ACKNOWLEDGMENTS
We thank the members of the Darlington lab for their support and encouragement throughout these studies. Recombinant human IL-1β was a gift from the National Cancer Institute. C/EBPβ knockout and heterozygous breeding mice were generously provided by Valeria Poli.
This work was supported by the National Institutes of Health (grants DK45285, AG13663, DK53045, T32-DK07664, and T32-GM08307) and the Moran Foundation.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev. 1992;127:25–50. doi: 10.1111/j.1600-065x.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 3.Alam T, An M R, Mifflin R C, Hsieh C C, Ge X, Papaconstantinou J. Trans-activation of the alpha 1-acid glycoprotein gene acute phase responsive element by multiple isoforms of C/EBP and glucocorticoid receptor. J Biol Chem. 1993;268:15681–15688. [PubMed] [Google Scholar]
- 4.Alam T, An M R, Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J Biol Chem. 1992;267:5021–5024. [PubMed] [Google Scholar]
- 5.Allen M C. An overview of long-term outcome. In: Witter F R, Keith L G, editors. Textbook of prematurity: antecedents, treatment, and outcome. Boston, Mass: Little, Brown & Co.; 1993. pp. 371–384. [Google Scholar]
- 6.Alonzi T, Fattori E, Cappelletti M, Ciliberto G, Poli V. Impaired stat3 activation following localized inflammatory stimulus in IL-6- deficient mice. Cytokine. 1998;10:13–18. doi: 10.1006/cyto.1997.0250. [DOI] [PubMed] [Google Scholar]
- 7.An M R, Hsieh C C, Reisner P D, Rabek J P, Scott S G, Kuninger D T, Papaconstantinou J. Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol Cell Biol. 1996;16:2295–2306. doi: 10.1128/mcb.16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann H, Jahreis G P, Morella K K, Won K, Pruitt S C, Jones V E, Prowse K R. Transcriptional regulation through cytokine and glucocorticoid response elements of rat acute phase plasma protein genes by C/EBP and JunB. J Biol Chem. 1991;266:20390–20399. [PubMed] [Google Scholar]
- 9.Benigni F, Fantuzzi G, Sacco S, Sironi M, Pozzi P, Dinarello C A, Sipe J D, Poli V, Cappelletti M, Paonessa G, Pennica D, Panayotatos N, Ghezzi P. Six different cytokines that share GP130 as a receptor subunit, induce serum amyloid A and potentiate the induction of interleukin-6 and the activation of the hypothalamus-pituitary-adrenal axis by interleukin-1. Blood. 1996;87:1851–1854. [PubMed] [Google Scholar]
- 10.Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 11.Burgess-Beusse, B. L., N. A. Timchenko, and G. J. Darlington. C/EBPα is an important mediator of C/EBPβ protein isoform production. Submitted for publication. [DOI] [PubMed]
- 12.Cappelletti M, Alonzi T, Fattori E, Libert C, Poli V. C/EBP beta is required for the late phases of acute phase gene induction in the liver and for tumour necrosis factor-alpha, but not interleukin-6, regulation. Cell Death Differ. 1996;3:29–35. [PubMed] [Google Scholar]
- 13.Chen H M, Liao W S. Differential acute-phase response of rat kininogen genes involves type I and type II interleukin-6 response elements. J Biol Chem. 1993;268:25311–25319. [PubMed] [Google Scholar]
- 14.Costa R H, Grayson D, Darnell J E., Jr Multiple hepatocyte enriched nuclear factors function in the regulation of transthyretin and α1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darlington G J, Wilson D R, Juan T S-C. Transcriptional regulation of the human C3 gene. In: Mackiewicz A, Kushner I, Baumann H, editors. Acute phase proteins: molecular biology, biochemistry, and clinical applications. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 425–442. [Google Scholar]
- 16.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 17.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 18.Fantuzzi G, Zheng H, Faggioni R, Benigni F, Ghezzi P, Sipe J D, Shaw A R, Dinarello C A. Effect of endotoxin in IL-1 beta-deficient mice. J Immunol. 1996;157:291–296. [PubMed] [Google Scholar]
- 19.Fattori E, Cappelletti M, Costa P, Sellitto C, Cantoni L, Carelli M, Faggioni R, Fantuzzi G, Ghezzi P, Poli V. Defective inflammatory response in interleukin 6-deficient mice. J Exp Med. 1994;180:1243–1250. doi: 10.1084/jem.180.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 21.Ganapathi M K. Okadaic acid, an inhibitor of protein phosphatases 1 and 2A, inhibits induction of acute-phase proteins by interleukin-6 alone or in combination with interleukin-1 in human hepatoma cell lines. Biochem J. 1992;284:645–648. doi: 10.1042/bj2840645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh C-C, Xiong W, Xie Q, Rabek J P, Scott S G, An M A, Reisner P D, Kuninger D T, Papaconstantinou J. Effects of age on the posttranscriptional regulation of CCAAT/enhancer binding protein alpha and CCAAT/enhancer binding protein beta isoform synthesis in control and LSP-treated livers. Mol Biol Cell. 1998;9:1479–1494. doi: 10.1091/mbc.9.6.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J H, Liao W S. Induction of the mouse serum amyloid A3 gene by cytokines requires both C/EBP family proteins and a novel constitutive nuclear factor. Mol Cell Biol. 1994;14:4475–4484. doi: 10.1128/mcb.14.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 25.Juan T S, Wilson D R, Wilde M D, Darlington G J. Participation of the transcription factor C/EBP delta in the acute phase regulation of the human gene for complement component C3. Proc Natl Acad Sci USA. 1993;90:2584–2588. doi: 10.1073/pnas.90.7.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalmovarin N, Friedrichs W E, O’Brien H V, Linehan L A, Bowman B H, Yang F. Extrahepatic expression of plasma protein genes during inflammation. Inflammation. 1991;15:369–379. doi: 10.1007/BF00917353. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Baumann H. The carboxyl-terminal region of STAT3 controls gene induction by the mouse haptoglobin promoter. J Biol Chem. 1997;272:14571–14579. doi: 10.1074/jbc.272.23.14571. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita S, Akira S, Kishimoto T. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Kohler G. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 30.Kordula T, Travis J. The role of Stat and C/EBP transcription factors in the synergistic activation of rat serine protease inhibitor-3 gene by interleukin-6 and dexamethasone. Biochem J. 1996;313:1019–1027. doi: 10.1042/bj3131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kushner I, Mackiewicz A. The acute phase response: an overview. In: Mackiewicz A, Kushner I, Baumann H, editors. Acute phase proteins: molecular biology, biochemistry, and clinical applications. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 4–14. [Google Scholar]
- 32.Leaman D W, Leung S, Li X, Stark R G. Regulation of STAT-dependent pathways by growth factors and cytokines. FASEB J. 1996;10:1578–1588. [PubMed] [Google Scholar]
- 33.Lee Y-M, Miau L-H, Chang C-J, Lee S-C. Transcriptional induction of the alpha-1 acid glycoprotein (AGP) gene by synergistic interaction of two alternative activator forms of AGP/enhancer-binding protein (C/EBPβ) and NF-κB or Nopp140. Mol Cell Biol. 1996;16:4257–4263. doi: 10.1128/mcb.16.8.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leon L R, Conn C A, Glaccum M, Kluger M J. IL-1 type I receptor mediates acute phase response to turpentine, but not lipopolysaccharide in mice. Am J Physiol. 1996;271:R1668–R1675. doi: 10.1152/ajpregu.1996.271.6.R1668. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Liao W. Expression of rat serum amyloid A1 gene involves both C/EBP-like and NFkb-like transcription factors. J Biol Chem. 1991;266:15192–15201. [PubMed] [Google Scholar]
- 36.Li X, Liao W S. Cooperative effects of C/EBP-like and NF kappa B-like binding sites on rat serum amyloid A1 gene expression in liver cells. Nucleic Acids Res. 1992;20:4765–4772. doi: 10.1093/nar/20.18.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marino M W, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, Basu S, Old L J. Characterization of tumor necrosis factor-deficient mice. Proc Natl Acad Sci USA. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsusaki T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mink S, Haenig B, Klempnauer K-H. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan J G, Courtois G, Fourel G, Chodosh L A, Campbell L, Evans E, Crabtree G R. Sp1, a CAAT-binding factor, and the adenovirus major late promoter transcription factor interact with functional regions of the gamma-fibronogen promoter. Mol Cell Biol. 1988;8:2628–2637. doi: 10.1128/mcb.8.6.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramji D P, Vitelli A, Tronche F, Cortese R, Ciliberto G. The two C/EBP isoforms, IL-6DBP/NFIL6 and C/EBP delta/NF-IL6 beta, are induced by IL-6 to promote acute phase gene transcription via different mechanisms. Nucleic Acids Res. 1993;21:289–294. doi: 10.1093/nar/21.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray A, Ray B K. Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-β and C/EBPδ and their activation by phosphorylation. Mol Cell Biol. 1994;14:4324–4332. doi: 10.1128/mcb.14.6.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray A, Ray B K. A novel cis-acting element is essential for cytokine-mediated transcriptional induction of the serum amyloid A gene in nonhepatic cells. Mol Cell Biol. 1996;16:1584–1594. doi: 10.1128/mcb.16.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ripperger J, Fritz S, Richter K, Dreier B, Schneider K, Lochner K, Marschalek R, Hocke G, Lottspeich F, Fey G H. Isolation of two interleukin-6 response element binding proteins from acute phase rat livers. Ann NY Acad Sci. 1995;762:252–260. doi: 10.1111/j.1749-6632.1995.tb32330.x. [DOI] [PubMed] [Google Scholar]
- 46.Ripperger J, Fritz S, Richter K, Hocke G M, Lottspeich F, Fey G H. Transcription factors Stat3 and Stat5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- 47.Ron D, Brasier A R, Habener J F. Transcriptional regulation of hepatic angiotensinogen gene expression by the acute-phase response. Mol Cell Endocrinol. 1990;74:C97–C104. doi: 10.1016/0303-7207(90)90221-s. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu H, Yamamoto K. NF-kappa B and C/EBP transcription factor families synergistically function in mouse serum amyloid A gene expression induced by inflammatory cytokines. Gene. 1994;149:305–310. doi: 10.1016/0378-1119(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 49.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 51.Theisen M, Behringer R R, Cadd G G, Brinster R L, McKnight G S. A C/EBP-binding site in the transferrin promoter is essential for expression in the liver but not the brain of transgenic mice. Mol Cell Biol. 1993;13:7666–7676. doi: 10.1128/mcb.13.12.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timchenko N, Wilson D R, Taylor L R, Wilde A M, Abdelsayed S, Sawadogo M, Darlington G J. Autoregulation of the human C/EBPα gene by stimulation of upstream stimulatory factor binding. Mol Cell Biol. 1995;15:1192–1202. doi: 10.1128/mcb.15.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timchenko N A, Harris T E, Wilde M, Bilyeu T A, Burgess-Beusse B L, Finegold M J, Darlington G J. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukada J, Waterman W, Koyama Y, Webb A C, Auron P E. A novel STAT-like factor mediates lipopolysaccharide, interleukin 1 (IL-1), and IL-6 signaling and recognizes a gamma interferon activation site-like element in the IL1B gene. Mol Cell Biol. 1996;16:2183–2194. doi: 10.1128/mcb.16.5.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 56.Vietor I, Oliveira I C, Vilcek J. CCAAT box enhancer binding protein alpha (C/EBP-alpha) stimulates kappaB element-mediated transcription in transfected cells. J Biol Chem. 1996;271:5595–5602. doi: 10.1074/jbc.271.10.5595. [DOI] [PubMed] [Google Scholar]
- 57.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Impaired energy homeostasis in C/EBP alpha knock-out mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe S, Arai K. JAK-STAT system in signal transduction via cytokine receptors. Curr Opin Genet Dev. 1996;6:587–596. doi: 10.1016/s0959-437x(96)80088-8. [DOI] [PubMed] [Google Scholar]
- 59.Won K-A, Baumann H. The cytokine response element of the rat α1-acid glycoprotein gene is a complex of several interacting regulatory sequences. Mol Cell Biol. 1990;10:3965–3978. doi: 10.1128/mcb.10.8.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada T, Tobita K, Osada S, Nishihara T, Imagawa M. CCAAT/enhancer-binding protein delta gene expression is mediated by APRF/STAT3. J Biochem. 1997;121:731–738. doi: 10.1093/oxfordjournals.jbchem.a021647. [DOI] [PubMed] [Google Scholar]
- 61.Yap S H, Moshage H J, Hazenberg R P, Roelofs H M, Bijzet J, Limburg P C, Aarde L A, van Rijswijk M H. Tumor necrosis factor (TNF) inhibits interleukin (IL)-1 and/or IL-6 stimulated synthesis of C-reactive protein (CRP) and serum amyloid A (SAA) in primary cultures of human hepatocytes. Biochim Biophys Acta. 1991;1091:405–408. doi: 10.1016/0167-4889(91)90207-e. [DOI] [PubMed] [Google Scholar]
- 62.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9509. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 63.Zhang P, Iwama A, Datta M W, Darlington G J, Link D C, Tenen D G. Upregulation of interleukin 6 and granulocyte colony-stimulating factor receptors by transcription factor CCATT enhancer binding protein α (C/EBPα) is critical for granulopoiesis. J Exp Med. 1998;188:1–12. doi: 10.1084/jem.188.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, Fuentes N L, Fuller G M. Characterization of the IL-6 responsive elements in the gamma fibrinogen gene promoter. J Biol Chem. 1995;270:24287–24291. doi: 10.1074/jbc.270.41.24287. [DOI] [PubMed] [Google Scholar]
- 65.Zhong Z, Wen Z, Darnell J E., Jr STAT3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]