Abstract
Sorting nexin 1 (SNX1) is a protein that binds to the epidermal growth factor (EGF) receptor and is proposed to play a role in directing EGF receptors to lysosomes for degradation (R. C. Kurten, D. L. Cadena, and G. N. Gill, Science 272:1008–1010, 1996). We have obtained full-length cDNAs and deduced the amino acid sequences of three novel homologous proteins, which were denoted human sorting nexins (SNX2, SNX3, and SNX4). In addition, we identified a presumed splice variant isoform of SNX1 (SNX1A). These molecules contain a conserved domain of ∼100 amino acids, which was termed the phox homology (PX) domain. Human SNX1 (522 amino acids), SNX1A (457 amino acids), SNX2 (519 amino acids), SNX3 (162 amino acids), and SNX4 (450 amino acids) are part of a larger family of hydrophilic molecules including proteins identified in Caenorhabditis elegans and Saccharomyces cerevisiae. Despite their hydrophilic nature, the sorting nexins are found partially associated with cellular membranes. They are widely expressed, although the tissue distribution of each sorting nexin mRNA varies. When expressed in COS7 cells, epitope-tagged sorting nexins SNX1, SNX1A, SNX2, and SNX4 coimmunoprecipitated with receptor tyrosine kinases for EGF, platelet-derived growth factor, and insulin. These sorting nexins also associated with the long isoform of the leptin receptor but not with the short and medium isoforms. Interestingly, endogenous COS7 transferrin receptors associated exclusively with SNX1 and SNX1A, while SNX3 was not found to associate with any of the receptors studied. Our demonstration of a large conserved family of sorting nexins that interact with a variety of receptor types suggests that these proteins may be involved in several stages of intracellular trafficking in mammalian cells.
Cell surface receptors mediate a wide variety of biological processes, including transmembrane signalling in response to extracellular ligands (7, 19) and receptor-mediated endocytosis of macromolecules (12, 16, 27). In addition to participating in events initiated at the plasma membrane, cell surface receptors traverse both the secretory pathway during their biosynthesis and the endocytic pathway following internalization from the cell surface (8, 10). Many different proteins are required to form the complex molecular machinery used to direct cellular trafficking of receptors and to mediate signalling downstream of these molecules (6, 36). A variety of interaction cloning techniques have been used by investigators to identify some of the proteins involved in these processes (3, 25). Recently, Kurten et al. (21) used the yeast two-hybrid system to clone a cDNA encoding a protein, sorting nexin 1 (SNX1), that bound to the cytoplasmic domain of the epidermal growth factor (EGF) receptor. Based primarily on its homology to a protein (Mvp1p) known to be involved in targeting hydrolases to the vacuole in yeast, it was hypothesized that SNX1 may be involved in EGF receptor degradation in lysosomes (15). Moreover, it was reported that SNX1 promoted degradation of the EGF receptor but did not exert the same effect on other related tyrosine kinases (21).
We now report the cloning of three novel proteins that are homologous to human SNX1, which were designated SNX2, SNX3, and SNX4. All four sorting nexins contain an ∼100-amino-acid region, which was termed the phox homology (PX) domain, that is also contained in at least three Saccharomyces cerevisiae proteins (32). Two of these proteins, Mvp1p and Vps5p, are required for the proper sorting of carboxypeptidase Y to the vacuole (15, 18, 30). A third yeast protein, Grd19p, has recently been shown to be required to maintain two late-Golgi enzymes (dipeptidyl amino peptidase A and Kex2) in their proper locations by retrieving mislocalized molecules from the prevacuolar compartment (41). Interestingly, Vps5p exhibits the greatest sequence similarity with SNX1 and SNX2 described here, while Grd19p is most closely related to SNX3.
In addition, we report that SNX1, SNX2, and SNX4 associate with a variety of receptors, including receptors for insulin, EGF, platelet-derived growth factor (PDGF), and leptin. In contrast, we did not detect binding of SNX3 to these different receptor types. Taken together, these findings suggest that mammalian cells possess multiple sorting nexins that, by analogy to their yeast homologs, are likely to play an important role in protein trafficking among various organelles.
MATERIALS AND METHODS
Cells.
COS7 cells were obtained from the American Type Culture Collection (ATCC, Rockville, Md.). NIH 3T3 cells overexpressing human EGF receptors were kindly provided by J. Schlessinger (17). The cells were maintained in Dulbecco’s modified medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (Life Technologies, Inc., Gaithersburg, Md.), 100 U of penicillin and 100 μg of streptomycin per ml.
Antibodies.
Anti-insulin receptor β-subunit antibody (C19), anti-EGF receptor antibody (1005), anti-PDGF receptor, β-isoform antibody (958), and anti-c-myc antibodies (A-14 and 9E10) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Anti-hemagglutinin (HA) antibody (12CA5) was from Boehringer Mannheim Corp. (Indianapolis, Ind.). Anti-transferrin receptor antibody (6F11) was from Advanced Immuno Chemical Corp. (Long Beach, Calif.). Anti-leptin receptor antibody directed against the common extracellular domain of the receptor isoforms (N-term OB-R) was from Research Diagnostics, Inc. (Flanders, N.J.). Donkey anti-rabbit immunoglobulin G (IgG)-peroxidase and sheep anti-mouse IgG-peroxidase were obtained from Amersham Life Science, Inc. (Arlington Heights, Ill.), and mouse anti-goat IgG-peroxidase was from Jackson ImmunoResearch Laboratories (West Grove, Pa.).
ESTs related to SNX1.
Using the published amino acid sequence for human SNX1 (GenBank accession no. U53225), we searched the expressed sequence tag (EST) nucleotide sequence database of the National Center for Biotechnology Information for molecules related to SNX1 at the amino acid level. Four classes of human ESTs were identified by using the BLAST algorithm that had a high degree of amino acid homology with SNX1 (1). EST 236890, which was obtained from an infant brain cDNA library (GenBank accession no. T17214), showed 100% nucleotide identity with SNX1 over 95 amino acids. EST 309337, which was obtained from a human fetal lung cDNA library (GenBank accession no. W40457), showed 71% amino acid identity with and 81% amino acid similarity to SNX1 over 167 amino acids, while the translated sequence of EST 324810, which was obtained from a senescent fibroblast cDNA library (GenBank accession no. W49550), showed 29% amino acid identity with and 61% amino acid similarity to SNX1 over 54 amino acids. In addition, EST 78509, obtained from a human adult liver library (GenBank accession no. T60550) showed 30% identity with and 50% similarity to SNX1 over 105 amino acids. The high degree of amino acid homology between these human ESTs and SNX1 suggested that three additional human sorting nexin molecules existed in the human genome. Using these and other overlapping clones, we obtained full-length cDNA molecules and determined the sequences of human SNX2, SNX3, and SNX4 (see Fig. 1 and below).
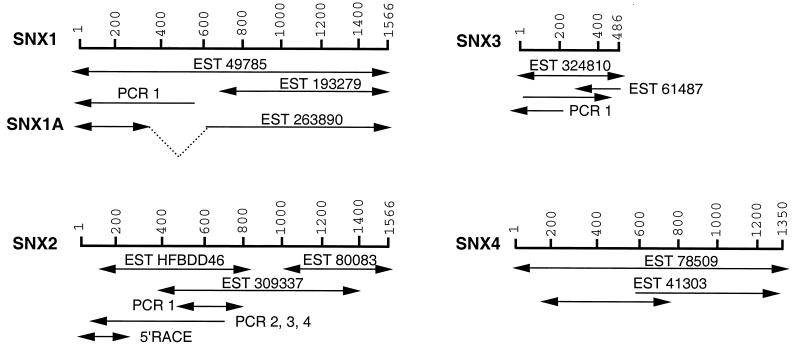
FIG. 1.
Construction and characterization of SNX1, SNX1A, SNX2, SNX3, and SNX4 cDNAs. Five human sorting nexins (SNX1, SNX1A, SNX2, SNX3, and SNX4) are depicted. For each full-length cDNA, the nucleotide numbering is shown. Listed below each molecule are the fragments (ESTs, PCR, or 5′-RACE products) used to determine the nucleotide sequences of the cDNAs and to construct full-length cDNAs as detailed in Materials and Methods. One or both strands of each cDNA fragment were sequenced as indicated by the directions of the arrowheads. In every case, both strands of DNA were sequenced from at least one clone.
Cloning of SNX1, SNX1A, SNX2, SNX3, and SNX4 cDNAs.
I.M.A.G.E. Consortium cDNA clones (23) were obtained from Research Genetics (Huntsville, Ala.) or the ATCC. Plasmid DNA was isolated with reagents supplied by QIAGEN, Inc. (Chatsworth, Calif.), and sequenced on one or both strands starting with vector-specific primers, followed by a series of sequence-specific primers as indicated (see Fig. 1). PCR was performed with the Expand high-fidelity PCR kit (Boehringer Mannheim Corp.) and 5′-Stretch Plus human pancreas and skeletal muscle cDNA libraries (Clontech, Palo Alto, Calif.) to obtain additional sequence information for SNX1, SNX2, and SNX3. A 5′-rapid amplification of cDNA ends (RACE) strategy (Life Technologies, Inc., Gaithersburg, Md.) with total human skeletal muscle as a template was used to complete the 5′ end of the SNX2 cDNA (Clontech). When necessary, standard molecular biology techniques were used to construct full-length cDNAs. All sequencing was performed with the ABI PRISM dye terminator cycle sequencing kit and an ABI automated sequencer (model 373A; Perkin-Elmer Corp., Applied Biosystems Division, Foster City, Calif.).
Construction of epitope-tagged sorting nexins and receptor expression vectors.
Epitope tags were introduced at the 5′ end of each sorting nexin molecule and associated mutant proteins by PCR with the Expand high-fidelity PCR kit (Boehringer Mannheim Corp.). SNX1, SNX1A, SNX2, SNX3, and SNX4 were myc tagged, and SNX2 was also HA tagged. Sequences of the primers used are available upon request. The epitope-tagged molecules were then cloned into either pCDNA(3.1±) (Invitrogen Corp., Carlsbad, Calif.) or pCMV5myc-1 expression vectors (2) by standard techniques. Full-length human insulin receptor (33), PDGF receptor (40), and EGF receptor (28) were cloned into pCDNA(3.1±) (Invitrogen Corp.) or pCIS vectors as previously described (33, 34). Full-length human leptin receptor short and long isoforms (11, 22, 37) were cloned into pCIneo vector (Promega Corp., Madison, Wis.) by standard techniques.
Tissue distribution of sorting nexins.
Multiple-tissue Northern blots containing approximately 2 μg of purified poly(A)+ RNA from human tissues and a multiple tissue dot blot were obtained from Clontech. The dot blot RNAs were normalized to the mRNA levels of eight different housekeeping genes, thus allowing the relative levels of the various sorting nexin mRNAs to be determined by densitometry of autoradiographs. To obtain probes for the Northern blot analyses, we digested SNX1, -2, and -3 cDNAs with EcoRI and NotI, while SNX4 cDNA was digested with NcoI and XhoI. The resulting 1,000- to 2,000-nucleotide fragments were gel purified, 32P-labeled with a randomly primed DNA labeling kit (Boehringer Mannheim Corp.), diluted in hybridization buffer (50% [vol/vol] formamide, 8× Denhardt’s solution, 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2 PO4, and 1 mM EDTA; pH 7.7], 0.5% (wt/vol) sodium dodecyl sulfate [SDS], 1-mg/ml salmon sperm DNA), and hybridized for 18 to 24 h at 42°C. The blots were then washed according to the manufacturer’s instructions.
Transient transfection of sorting nexins and various receptors.
Totals of 2 × 105 to 3 × 105 COS7 cells were plated into each well of a six-well dish at 16 to 20 h before the start of the transfection. The cells were washed twice with serum-free DMEM; DNA (2 μg of total DNA per well) was transfected into the cells with Lipofectamine (6 μl per well) according to the manufacturer (Life Technologies, Inc.). The cells were incubated for 5 to 6 h with the DNA-Lipofectamine complexes in serum-free DMEM and then incubated overnight in 10% (vol/vol) fetal bovine serum. The cells were harvested 28 to 30 h after the start of the transfection.
Coimmunoprecipitation experiments.
Transfected COS7 cells were washed in ice-cold phosphate-buffered saline (PBS) and scraped into 250 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 0.5% [vol/vol], Triton X-100, 0.3 M NaCl plus protease inhibitor tablet [Boehringer Mannheim Corp.]). The cells were then solubilized for 30 min on ice, and insoluble debris was removed by centrifugation at 14,000 × g for 20 min. The proteins were then detected by immunoblotting with anti-HA, anti-myc, anti-insulin receptor, anti-PDGF receptor, and anti-EGF receptor antibodies or by immunoprecipitation of extract (400 μl) with receptor-specific antibodies (1/100 dilution; see Antibody section above), followed by immunoblotting with anti-HA or anti-myc antibodies (24, 35).
Subcellular localization of sorting nexins.
NIH 3T3 cells overexpressing human EGF receptors (17) were transiently transfected with the various epitope-tagged sorting nexin molecules as described above. At 24 to 26 h posttransfection, the cells (∼5 × 106 to 6 × 106 per well) were placed on ice and washed twice with ice-cold PBS. Subsequently, the cells were scraped and lysed in 250 μl of ice-cold homogenization buffer (10 mM HEPES [pH 7], 0.25 M sucrose, 0.5 mM MgCl2, and protease inhibitor tablet [Boehringer Mannheim Corp.]) by 15 passages through a 25-gauge syringe. To obtain total membrane and cytosolic fractions, the lysates were centrifuged at 240,000 × g for 45 min at 4°C in a Beckman TLA 120.2 rotor (Beckman Instruments, Palo Alto, Calif.). Following the centrifugation step, the membrane pellet was resuspended in 250 μl of homogenization buffer with eight strokes with a Teflon pestle and a Potter homogenizer. Aliquots of the total lysates and cytosolic and membrane fractions were then solubilized in Laemmli buffer and run on 7.5% (vol/vol) SDS gels and EGF receptors, and the various epitope-tagged sorting nexins were detected by Western blotting (35).
Nucleotide sequence accession numbers.
The predicted amino sequences for human SNX1, SNX1A, SNX2, SNX3, and SNX4 cDNAs have been deposited in the National Center for Biotechnology Information database under accession no. 3152940, 3152942, 3152938, 3127053, and AF065485, respectively.
RESULTS
Identification of human SNX1 homologs.
In an effort to identify homologs of human SNX1, we searched the dbEST database of the National Center for Biotechnology Information with the entire amino acid sequence of human SNX1 or its PX domain (amino acids 161 to 272) (21, 32). This led to identification of four groups of human EST sequences which, when translated, showed a high degree of homology to SNX1 as judged by the BLAST algorithm (1). For each group, we obtained several EST clones, determined their nucleotide sequences, and constructed full-length cDNAs (Fig. 1).
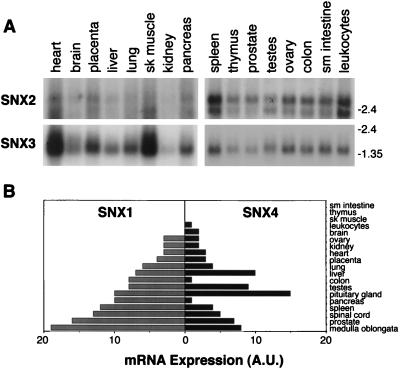
Group 1 consisted of ESTs that encode all or part of SNX1 and included a putative splice variant isoform of SNX1 that we have designated SNX1A. The deduced amino acid sequence of our SNX1 cDNA clone showed complete agreement with the published sequence of SNX1, except for three amino acid differences (Fig. 2). There was one major difference between the translated sequence of SNX1A and SNX1, i.e., a deletion of 195 nucleotides that resulted in an in-frame deletion of 65 amino acids (Fig. 2, underlined region). Thus, SNX1A cDNA is predicted to encode a 457-amino-acid hydrophilic protein with a pI of 5.3. PCR was used to confirm the presence of both SNX1 and SNX1A mRNAs in multiple tissues (data not shown). In addition, Northern blot analyses showed that human SNX1/1A mRNAs are ubiquitously expressed, with the highest levels in the medulla oblongata, prostate, spinal cord, spleen, pancreas, and pituitary gland (Fig. 3B).
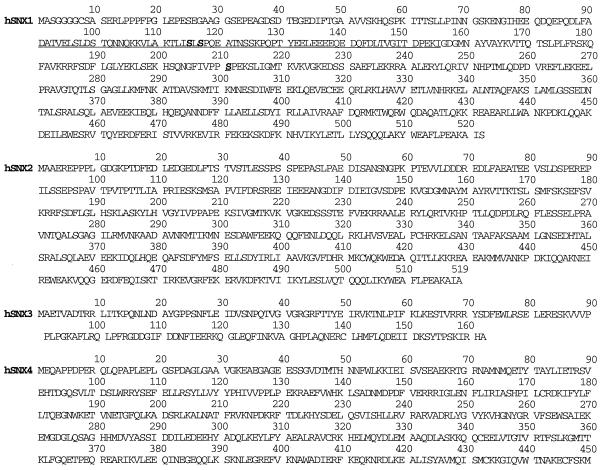
FIG. 2.
Predicted amino acid sequences of SNX1, SNX1A, SNX2, SNX3, and SNX4. The predicted amino acid sequences for human SNX1 (hSNX1), SNX1A, SNX2, SNX3, and SNX4 cDNAs are shown. The SNX1 amino acid sequence was identical to the published SNX1 sequence (21), except for the three residues indicated by boldface and double underlining. We found serine at positions 115 and 117. At position 211, we found proline (codon CCC) in one clone and serine (TCC) in two other clones, suggesting that there is a polymorphic amino acid. The entire coding sequence of SNX1A was shown to be identical to that of SNX1, except for an in-frame deletion of 195 nucleotides. The 65 amino acid residues (91 to 115) deleted from SNX1A are underlined.
FIG. 3.
Tissue distributions of SNX1, SNX2, SNX3, and SNX4 mRNAs. Filters containing poly(A)+ mRNA from the indicated human tissues were hybridized with radiolabeled human SNX2 or SNX3 probes (A). A multiple-tissue dot blot was hybridized with radiolabeled human SNX1 or SNX4 probes (B) as described in Materials and Methods. Autoradiographs of the dot blots were scanned with a laser densitometer, and the individual spots were quantified with ImageQuant software from Molecular Dynamics. The numbers on the x axis denote arbitrary units (AU).
The second group of ESTs encodes pieces of a SNX1 homolog that we have designated human SNX2. This novel cDNA is predicted to encode a protein of 519 amino acids with a pI of 4.8 (Fig. 2). SNX1 and SNX2 are 63% identical and 73% similar over much of their lengths (Fig. 4A). As shown by Northern blot analyses, SNX2 mRNA is ubiquitously expressed, with transcripts of approximately 3.1 and 2.4 kb detected in all human tissues examined. However, the ratio of the two SNX2 mRNAs and their absolute amounts varied among the tissues studied. The highest levels of SNX2 mRNA were found in spleen, heart, skeletal muscle, and peripheral leukocytes, while very little SNX2 mRNA was detected in the kidney, liver, or brain (Fig. 3A).
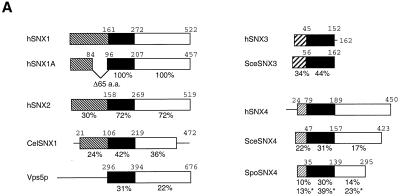
FIG. 4.
Sequence identity between human sorting nexins and related molecules from other species. (A) Schematic illustrations of the domain architecture of human (h) SNX1, SNX1A, SNX2, SNX3, and SNX4 and related proteins from C. elegans, S. cerevisiae, and S. pombe. The protein labeled CelSNX1 is an ORF found in the nematode sequencing project database (GenBank accession no. Z69384). The protein labeled SceSNX4 is an S. cerevisiae ORF (accession no. S56808), and the protein labeled SpoSNX4 is an S. pombe ORF (accession no. 2389002). SceSNX3 (accession no. AF016101) is the newly described S. cerevisiae protein known as Grd19p (41). Vps5p and Mvp1p are two additional S. cerevisiae molecules involved in sorting to the yeast vacuole (15, 18, 30). The cross-hatched boxes represent conserved N-terminal domains of variable lengths. The black boxes represent a highly conserved domain of ∼100 amino acids which was termed the PX domain (32). The white boxes represent a conserved domain C terminal to the PX domain. The thin lines are regions with limited homology. The percentages of amino acid identity between each human sorting nexin and related proteins from other species as determined by pairwise alignment with the ALIGN program (13) are shown below the various domains. The percentages of amino acid identity between S. cerevisiae and S. pombe SNX4 are indicated by asterisks. The carboxyl-terminal regions of the SNX1-like group and the SNX4 group may be distantly related, but this is not illustrated. (B) Multiple sequence alignment of the PX domains from each human sorting nexin and related molecules from other species. The sequences were aligned with CLUSTAL W (38), followed by manual refinement. Amino acid residues conserved in at least 6 of 10 proteins of the sorting nexin-like aligned sequences (top group) or other PX-containing proteins (bottom group) are highlighted in black. The amino acids that were predicted to serve as binding sites for SH3 and/or WW domain-containing proteins are indicated by asterisks (9, 31). Cons, consensus.
The third and fourth groups of ESTs were identified via their high degree of sequence homology with a portion of SNX1 called the PX domain (amino acids 161 to 272 [Fig. 4B]). The novel cDNA that we have designated human SNX4 is predicted to encode a protein of 450 amino acids with a pI of 5.6 (Fig. 2). SNX4 mRNA is also widely expressed, with its highest levels in the pituitary gland, testes, and liver (Fig. 3B). In contrast, the protein that we have designated SNX3 is a molecule of only 162 amino acids and a pI of 9.2 (Fig. 2). A single transcript of ∼1.9 kb was detected in all human tissues hybridized with an SNX3 probe. The tissue distribution of SNX3 closely parallels that seen for SNX2, with the highest levels of mRNA detected in the peripheral leukocytes, spleen, heart, and skeletal muscle, and greatly reduced levels in the kidney (Fig. 3A).
Sorting nexin homologs in yeasts and Caenorhabditis elegans.
To examine the evolutionary conservation of the sorting nexins, we searched the complete genome of the budding yeast S. cerevisiae and the known open reading frames (ORFs) from C. elegans and Schizosaccharomyces pombe for sequences similar to those for SNX1, SNX2, SNX3, and SNX4. The best match to human SNX3 was the recently described S. cerevisiae protein, Grd19p (ORF YOR357c) (41). Likewise, a search of Grd19p against libraries of human ESTs showed no closer homolog than SNX3. Thus, the two appear to be orthologs, and we designate YOR375c as SceSNX3. Figure 4A shows the percentage of amino acid identity between SNX3 and SceSNX3 as determined with the ALIGN program (13). Similar results suggest that human SNX4 and yeast YJL036w (GenBank accession no. Z49311) may be orthologs; we designate YJL036w as SceSNX4. Likewise, the S. pombe hypothetical protein that we designate SpoSNX4 (GenBank accession no. 239002) probably represents an SNX4 ortholog, although SpoSNX4 has a shorter C-terminal domain than either the human or S. cerevisiae SNX4 molecule (Fig. 4A). Finally, the two closely related human proteins, SNX1 and SNX2, showed a high degree of sequence homology along most of their lengths with the recently described S. cerevisiae protein, Vps5p (18, 30), and the hypothetical C. elegans protein that we have designated CelSNX1 (GenBank accession no. 1439669). The best matches from searching yeast Vps5p against all human ESTs were the SNX1 and SNX2 groups, again suggesting orthology.
A common feature of the human sorting nexins is the presence of an ∼100-amino-acid region that was termed the PX domain. This domain was first identified as a conserved sequence in the 40- and 47-kDa subunits of NADPH oxidase and has subsequently been identified in a variety of proteins with varied functions (14, 26, 32). Figure 4B shows a multiple sequence alignment of the PX domains from the sorting nexins and their predicted orthologs, as well as other PX domain-containing proteins. The alignment was created with Clustal W (38) and was edited by hand. A tree constructed from the PX domain alignment by using the unweighted pair group method with arithmetic mean (29) grouped all of the proteins that we have designated human sorting nexins or their orthologs into a single branch of the tree along with one additional protein, Mvp1p (data not shown). Inspection of the PX domain alignment shows general features of the PX domain but also some possible signatures for the sorting nexin-like subset. The strongest motif in the alignment, [VIL] X R R [FY] S [DE] F, is matched well in most of the members of the alignment, and not just in the human sorting nexins and their orthologs (Fig. 4B). In contrast, the PX domains of all candidate sorting nexins (including Mvp1p), but few other proteins, matched, with one mismatch or fewer, to the motif [VIM] [VI] P P [LP] P X K. Toward the C terminus of the PX domain, the dipeptide motif HP is found in the human sorting nexins and Mvp1p, while DP appears in Vps5p, its binding partner Vps17p (18, 20), and a homolog of Vps5p that was designated YKR078w. No other proteins in the alignment matched the HP or DP sequence at that position.
Sorting nexins exist in both cytosolic and membrane-bound pools.
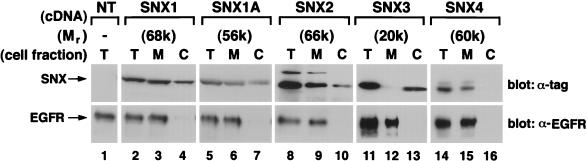
To study the functions of the sorting nexin proteins, each full-length cDNA containing an epitope tag at its 5′ end was cloned into a mammalian expression vector and transiently transfected into either COS7 or NIH 3T3 cells. Western blotting of cell extracts with anti-tag antibodies showed that each sorting nexin was expressed. However, the apparent molecular mass of each sorting nexin protein differed from its calculated mass by ∼8,000 to 12,000 kilodaltons. We then inquired whether the sorting nexins were located in the cytosol or whether they were associated with cellular membranes. For these studies, NIH 3T3 cells were transiently transfected with the various sorting nexin cDNAs. After homogenization of the cells in sucrose-containing buffer, we carried out ultracentrifugation to generate cytosolic and membrane fractions. As judged by Western blotting of the cell fractions, all of the sorting nexins partitioned into both membrane and cytosolic fractions (Fig. 5). SNX1, SNX1A, SNX2, and SNX4 were associated predominantly with membranes, whereas SNX3 was found mainly in the cytosol. Similar results were found with endogenous mouse SNX1 and SNX2 when the appropriate SNX-specific polyclonal antibodies were used (data not shown). In addition, the membrane-associated forms of the various sorting nexins could be partially extracted with 1 M NaCl, suggesting that they behave as peripheral membrane proteins (data not shown).
FIG. 5.
Analysis of the association of sorting nexins with membranes. NIH 3T3 cells overexpressing EGF receptors were transiently transfected with epitope-tagged sorting nexin cDNAs. After 26 to 30 h, total-cell extracts (T), and total membrane (M) and (C) cytosolic fractions were prepared as described in Materials and Methods. The distributions of the EGF receptors and the epitope-tagged sorting nexins were determined by Western blotting with an anti-EGF receptor antibody and anti-epitope antibodies, respectively.
Oligomerization of the sorting nexin molecules.
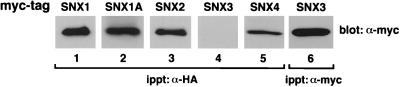
Given that the yeast protein Vps5p, which is a member of the SNX1/2 subgroup of sorting nexins, forms a heteromeric complex with another PX domain-containing protein, Vps17p (20), we examined whether SNX2 was able to associate with any of the other human sorting nexins. COS7 cells were transfected with HA-tagged SNX2 alone or in combination with myc-tagged versions of the other sorting nexins. SNX2 forms heteromeric complexes with SNX1, SNX1A, and SNX4 but not with SNX3 (Fig. 6, lanes 1, 2, and 5). Likewise, SNX2 was able to oligomerize with itself (lane 3). SNX2 did not associate with SNX3, although SNX3 was readily expressed (lanes 4 and 6).
FIG. 6.
Oligomerization of the sorting nexin molecules. COS7 cells were transiently transfected with expression vectors encoding HA-tagged SNX2 in combination with myc-tagged versions of SNX1, SNX1A, SNX2, SNX3, and SNX4. Total-cell extracts were immunoprecipitated (ippt) with an anti-myc antibody and electrophoresed, and the resulting blots were probed with either anti-myc (lane 6) or anti-HA (lanes 1 to 5) antibodies to detect the various immunoprecipitated epitope-tagged sorting nexin molecules.
Association with receptor tyrosine kinases.
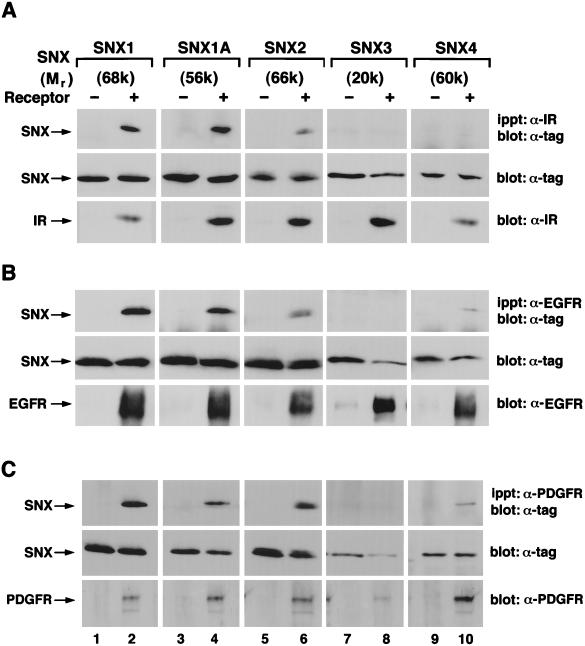
To test for the ability of the various sorting nexins to associate with growth factor receptors, COS7 cells were transiently transfected with expression plasmids encoding epitope-tagged sorting nexin cDNAs (Fig. 7, lanes 1 to 10). In addition, some cells were also cotransfected with an expression plasmid for the insulin receptor (Fig. 7A). Each epitope-tagged sorting nexin molecule was readily detected in cell extracts made from the transfected cells (Fig. 7A, middle panel). Likewise, overexpression of the insulin receptor was also detected in the appropriate extracts (Fig. 7A, lower panel). After incubation of cell extracts with an insulin receptor β-subunit antibody followed by Western blotting of the precipitates with antibody to the epitope tag, we found that SNX1, SNX1A, and SNX2 and small amounts of SNX4 coimmunoprecipitated with the insulin receptor (Fig. 7A, upper panel). Kinase-inactive insulin receptors coimmunoprecipitated amounts of SNX1, SNX2, and SNX4 similar to that immunoprecipitated with the wild-type receptor (data not shown). We did not detect association between SNX3 and overexpressed insulin receptors (lane 8). Neither did we detect association between the transfected sorting nexin molecules and the small amount of endogenous COS7 insulin receptors (lanes 1, 3, 5, 7, and 9). However, we consistently observed that overexpression of insulin receptor decreased the level of expression of epitope-tagged SNX3 (Fig. 7A, lanes 7 and 8). When these experiments were performed with extracts from cells overexpressing EGF receptors and the various sorting nexins (Fig. 7B), we found that SNX1 and SNX1A were readily detected in EGF receptor immunoprecipitates. SNX2 and SNX4 were detected, but to a lesser degree, and SNX3 was undetectable (Fig. 7B, upper panel). Likewise, SNX1, SNX1A, and SNX2 were readily coimmunoprecipitated with PDGF receptors (Fig. 7C, lanes 2, 4, and 6), while SNX4 was coimmunoprecipitated to a lesser extent (lane 10). As with the insulin and EGF receptors, no association was detectable between the PDGF receptor and SNX3 (Fig. 7C, lane 8). Furthermore, previous work suggested that the last 66 amino acids of SNX1 are required for this protein, when overexpressed, to accelerate the turnover of EGF receptors (21). However, we found that deletion of 66 C-terminal amino acids of SNX1 did not appreciably disrupt the association of the truncated SNX1 molecule with the receptor tyrosine kinases for EGF and insulin (data not shown). Nevertheless, because these C-terminal 66 amino acids bound the EGF receptor in the yeast two-hybrid system, our findings suggest that the receptor tyrosine kinases for EGF and insulin have multiple binding sites for SNX1. In addition, we found that stimulation of cells with either insulin or EGF for 5 to 60 min did not change the amounts of the various sorting nexins associated with the respective receptor tyrosine kinases (data not shown).
FIG. 7.
Differential association of SNX1, SNX1A, SNX2, SNX3, and SNX4 with the insulin receptor (IR), EGF receptor, and PDGF receptor. COS7 cells were transiently transfected with expression vectors encoding epitope-tagged SNX1 (lanes 1 and 2), SNX1A (lanes 3 and 4), SNX2 (lanes 5 and 6), SNX3 (lanes 7 and 8), or SNX4 (lanes 9 and 10). Some cells were also transfected with expression vector encoding the human insulin receptor (A), EGF receptor (B), and PDGF receptor (C) (lanes 2, 4, 6, 8, and 10). Total-cell extracts were immunoprecipitated (ippt) with the appropriate anti-receptor antibody and electrophoresed, and resulting blots were probed with either anti-myc or anti-HA antibodies to detect coimmunoprecipitated epitope-tagged sorting nexin molecules (upper panel). To check for expression of the various receptors and each of the sorting nexin molecules, each sample was also immunoblotted with an anti-tag antibody (middle panel) or an anti-receptor antibody (lower panel). Shown above each lane is the approximate molecular weight corresponding to the electrophoretic mobility of each sorting nexin protein. This experiment was repeated three times with similar results.
Association with endogenous transferrin receptors.
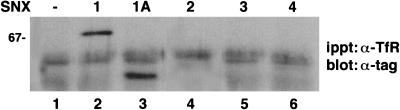
Having demonstrated the association of several sorting nexins with receptor tyrosine kinases, we inquired whether the various sorting nexins also associated with receptors that constitutively internalize and recycle. To test this, COS7 cells were transiently transfected with expression plasmids encoding epitope-tagged sorting nexin cDNAs. Western blotting of cell extracts made from the transfected cells confirmed that each epitope-tagged sorting nexin molecule was readily expressed (data not shown). After incubation of cell extracts with a transferrin receptor antibody followed by Western blotting of the precipitates with an anti-tag antibody, we found that only SNX1 and SNX1A coimmunoprecipitated with endogenous transferrin receptors (Fig. 8, lanes 2 and 3). We did not detect association between endogenous transferrin receptors and SNX2, SNX3, or SNX4 (lanes 4 to 6).
FIG. 8.
Differential association of SNX1, SNX1A, SNX2, SNX3, and SNX4 with endogenous transferrin receptors. COS7 cells were transiently transfected with expression vectors encoding epitope-tagged SNX1, SNX1A, SNX2, SNX3, or SNX4. Total-cell extracts were immunoprecipitated (ippt) with an anti-transferrin receptor antibody and electrophoresed, and the resulting blots were probed with either anti-myc or anti-HA antibodies to detect coimmunoprecipitated epitope-tagged sorting nexin molecules. This experiment was repeated three times with similar results.
Association with leptin receptor isoforms.
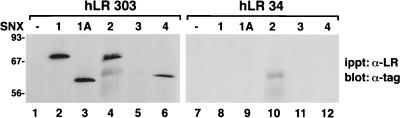
Then, we asked if the various sorting nexins associated with a third type of cellular receptor, cytokine receptors. COS7 cells were transiently transfected with expression plasmids encoding epitope-tagged sorting nexin cDNAs. In addition, some cells were also cotransfected with expression plasmids encoding the two most abundant isoforms of the human leptin receptor (22, 37). These isoforms share a large ligand binding domain and transmembrane domain and 29 cytoplasmic domain amino acids. Following this conserved region, the short form of the human leptin receptor (hLR34) contains 5 unique carboxyl-terminal amino acids, and the long form of the human leptin receptor (hLR303) contains 274 unique carboxyl-terminal amino acids. Western blotting of cell extracts made from transfected cells confirmed that each epitope-tagged sorting nexin molecule was readily expressed. Likewise, we found that each leptin receptor isoform was similarly expressed, as shown by 125I-leptin binding to solubilized cell extracts (data not shown). After incubation of cell extracts with a leptin receptor antibody directed against the extracellular domain of the receptor isoforms, followed by Western blotting of the precipitates with an antitag antibody, we found that SNX1, SNX1A, SNX2, and SNX4 coimmunoprecipitated with the long form of the leptin receptor (Fig. 9, lanes 2, 3, 4, and 6). We did not detect association between SNX3 and hLR303 (lane 5). In contrast, the short form of the receptor showed little if any association with the sorting nexin molecules (Fig. 9, lanes 8 to 12). Two other isoforms of the human leptin receptor that possess 15 and 67 unique amino acids following the carboxyl-terminal splice site (4, 12) also showed little if any association with the sorting nexins (data not shown).
FIG. 9.
Differential association of SNX1, SNX1A, SNX2, SNX3, and SNX4 with the short and long isoforms of the human leptin receptor. COS7 cells were transiently transfected with expression vectors encoding epitope-tagged SNX1, SNX1A, SNX2, SNX3, or SNX4. The cells were also transfected with an expression vector encoding either the long form (hLR303) or the short form (hLR34) of the human leptin receptor. Total-cell extracts were immunoprecipitated (ippt) with an anti-leptin receptor antibody made against the common extracellular domain of the receptor isoforms. The samples were then electrophoresed, and the resulting blots were probed with either anti-myc or anti-HA antibodies to detect coimmunoprecipitated epitope-tagged sorting nexin molecules. This experiment was repeated three times with similar results.
DISCUSSION
In this report, we have identified three novel human sorting nexin molecules (SNX2, SNX3, and SNX4) and SNX1A, a presumed splice variant isoform of SNX1. These mammalian proteins appear to be closely related to proteins in yeasts and C. elegans, indicating that they are part of an evolutionarily important family. The sorting nexins are all hydrophilic proteins that lack obvious hydrophobic regions capable of serving as membrane anchors, and yet they are found partially associated with cellular membranes. They also contain a conserved domain of ∼100 amino acids termed the PX domain that may be involved in protein-protein interactions. The sorting nexins also associate with each other and with a variety of other cellular proteins, suggesting that they exist as part of multisubunit complexes. Based on the functions of their yeast homologs, it is likely that the mammalian sorting nexins described here serve similar functions related to intracellular trafficking of proteins to various organelles.
Sorting nexin subgroups.
Human SNX1, SNX1A, and SNX2 are part of a subgroup of sorting nexins that include a related protein from C. elegans and the recently described S. cerevisiae protein, Vps5p (18, 30). These proteins share a high degree of amino acid similarity over their lengths. These findings suggest that this subclass of sorting nexin proteins may have evolved from a common ancestral precursor and thus may perform related functions in humans, yeasts, and invertebrates. Human SNX3, SNX4, and their respective orthologs make up two additional subgroups of sorting nexins. The SNX4 subgroup appears to be more closely related to the SNX1/2 subgroup than the SNX3 subgroup. The SNX4 subgroup contains amino acids in the PX domains and in the region just C terminal to the PX domains that are also conserved in the SNX1/2 subgroup. In contrast, the SNX3 subgroup consists of shorter proteins and has no sequence similarity to the other two groups outside the PX domain. It is interesting to note that yeast has at least five sorting nexin-like proteins. Additional searches of the EST databases suggest that several additional human sorting nexin proteins also exist.
Functions of yeast sorting nexin-like proteins.
Vps5p, which is a member of the SNX1/2 subgroup of sorting nexins, is a highly charged phosphoprotein that forms a heteromeric complex with another PX domain-containing protein, Vps17p (20). Vps5p is peripherally associated with a dense membrane fraction distinct from Golgi, endosomal, and vacuolar membranes. Cells that lack either Vps5p or Vps17p possess vacuoles of altered morphology and exhibit defects in the sorting of the vacuolar hydrolase, carboxypeptidase Y, due to mislocalization of the carboxypeptidase Y sorting receptor, Vps10p. They also missort late-Golgi membrane proteins to the vacuole. It is thought that Vps5p and Vps17p and perhaps other proteins may form a complex on transport vesicles that are responsible for recycling receptors such as Vps10p from endosomes back to the Golgi protein, where they then mediate another round of delivery of newly synthesized vacuolar hydrolases (18, 30). Interestingly, SNX2 is also able to form homomeric complexes as well as heteromeric complexes with SNX1, SNX1A, and SNX4. In addition, the sorting nexins are able to associate with a number of different cell surface receptors (see below). The regions of the sorting nexin molecules required for oligomerization, the specificity and degree of oligomerization, and the functional role of the oligomerization process will require further investigation.
Mvp1p is another S. cerevisiae protein that contains a sorting nexin-like PX domain. Its PX domain is more similar to the PX domains found in the sorting nexins described in this study than to other PX domain-containing proteins. Mvp1p is known to interact with Vps1p, a high-molecular-weight GTPase that associates with Golgi membranes. Vps1p is part of a family of proteins that includes the mammalian protein dynamin, which is involved in the early stages of clathrin-mediated endocytosis (15). Thus, in yeast cells, correct sorting of acid hydrolases to the vacuole appears to require several multiprotein complexes. One contains at least a sorting nexin (Vps5p), another PX domain-containing protein (Vps17p), and a sorting receptor (Vps10p), while the second contains a sorting nexin-like protein (Mvp1p) and a dynamin-like GTPase (Vps1p).
Grd19p is a third S. cerevisiae sorting nexin-like protein that appears to be the yeast ortholog of SNX3. Grd19p has recently been shown to interact with the cytoplasmic domain of dipeptidyl aminopeptidase A, a resident late-Golgi enzyme. Unlike the other yeast sorting nexin proteins described to date, Grd19p is not required for vacuolar protein sorting or the recycling of the carboxypeptidase Y sorting receptor Vps10p. Instead, Grd19p is required to maintain resident proteins such as Kex2p and dipeptidyl aminopeptidase A in the late-Golgi membrane by retrieving escaped molecules from the prevacuolar compartment (41). The molecular mechanisms used by these yeast sorting nexin-like molecules to direct proteins to specific locations and to maintain them there are presently unknown. Further studies will be required to determine if Grd19p and/or SNX3 is also part of a multiprotein complex involved in trafficking to and from the late-Golgi membrane.
Association of sorting nexins with receptors.
SNX1 was cloned by using a two-hybrid screen with a portion of the cytoplasmic domain of the EGF receptor as bait (21). In the present study, we inquired whether SNX1 and/or other sorting nexins could bind to EGF receptors, as well as to other cell surface receptors. In contrast to the work reported by Kurten et al. (21), our studies performed with mammalian cells showed that SNX1 binding is not specific for the EGF receptor. Rather, SNX1 bound to receptors for PDGF, EGF, and insulin. SNX2 and SNX4 also associated with these three classes of receptor tyrosine kinases. Interestingly, the SNX-receptor interactions were not affected by activation of the receptors by ligand. Both active and inactive receptors associated similarly with the receptor tyrosine kinases. The various sorting nexins also showed differential association with several leptin receptor isoforms. SNX1, SNX1A, SNX2, and SNX4 associated with the long isoform of the leptin receptor but not with the short and medium isoforms. Genetic evidence suggests that the long isoform of the leptin receptor mediates most of the biologically important signalling induced by leptin binding. However, recent studies have suggested that the short form is also activated following ligand addition (4, 5). Given that the short and medium isoforms lack a JAK box found in the long isoform, it is possible that the sorting nexins associate with the leptin receptor long form indirectly, possibly through JAKs. Lastly, the transferrin receptor, which internalizes and recycles constitutively, also shows interactions with a sorting nexin. However, association of transferrin receptors is restricted to SNX1 and SNX1A. The specific binding of transferrin receptors to SNX1, but not SNX2 and SNX4, may be important for determining the fate of the receptor (i.e., recycled versus degraded) (39).
There are several potential reasons why we did not detect association of SNX3 with the receptors studied. First, SNX3 may bind weakly or transiently to the receptors studied, and, thus, the amount of complex formed may be below the level of detection of our coimmunoprecipitation assay. Alternatively, SNX3 may bind to the receptors but be rapidly degraded after binding. In support of this idea, we find that overexpression of PDGF, EGF, or insulin receptors consistently diminished the SNX3 overexpression in cell extracts, whereas the amounts of SNX1, SNX2, and SNX4 were unchanged. Thus, it is possible that SNX3 might associate with the receptors studied here but then be rapidly degraded. Finally, SNX3 may bind to molecules other than the plasma membrane receptors studied here. In support of this idea, Kurten et al. (21) showed that SNX1 contained an EGF receptor binding site in its C-terminal 66 amino acids. SNX3 possesses only a PX domain and 45 amino acids N terminal to the PX domain. It does not contain a large C-terminal domain like SNX1, SNX2, and SNX4 that may be important for receptor recognition. In addition, SNX3 appears to be the human ortholog of Grd19p. Thus, it is likely that SNX3 may associate with late-Golgi resident proteins rather than cell surface receptors.
Function of sorting nexins in mammalian cells.
At present, the functions of SNX2, SNX3, and SNX4 in mammalian cells are not known. Kurten et al. (21) reported that overexpression of SNX1 resulted in accelerated degradation of EGF receptors. However, we have been unable to demonstrate that overexpression of SNX1 or any of the other sorting nexins increased the turnover of insulin or EGF receptors (unpublished observations). One possible reason for the discrepancy may be that the sorting nexins appear to exist as subunits of a multiprotein complex. Therefore, it may be difficult to see accelerated degradation of receptors in our system because we did not overexpress all of the components of the complex. Nevertheless, the conclusions of Kurten et al. (21) are entirely consistent with the role that Vps5p, the yeast protein most closely related to SNX1 and SNX2, is thought to play in vacuolar sorting. Further studies are necessary to elucidate the functions of mammalian sorting nexins.
Conclusion.
The sorting nexins described here, as well as related proteins from yeast, appear to be a large family of proteins involved in membrane trafficking. Further studies will be required to identify the proteins with which the human sorting nexins associate, to determine how sorting nexins associate with cellular membranes and with each other, and to elucidate the role that these proteins play in receptor trafficking. Possible roles include involvement in the recycling of receptors from endosomes back to the plasma membrane, targeting of receptors from endosomes to lysosomes for degradation, sorting of receptors from the trans-Golgi network to the plasma membrane, and retrieval of late-Golgi proteins from endocytic compartments.
ACKNOWLEDGMENTS
We are grateful to Axel Ullrich for the generous gift of the insulin receptor cDNA. In addition, we thank Marc Reitman for helpful discussions and for critical reading of the manuscript. Finally, we thank Rachel Kulansky, Neinke Grossman, and Jill Sherman for contributions to this project.
ADDENDUM IN PROOF
While this paper was under review, Seaman et al. reported that Vps5p, the yeast ortholog of SNX1 and SNX2, is part of a multiprotein membrane-associated complex. This may serve as a novel coat involved in vesicle transport from endosomes back to the late-Golgi membrane (M. N. J. Seaman, J. M. McCaffery, and S. D. Emi, J. Cell Biol. 142:665–681, 1998).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;125:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ansersson S, Davis D L, Dahlback H, Jornall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Benmerah A, Gagnon J, Beque B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N. The tyrosine kinase substrate Eps15 is constitutively associated with the plasma membrane adaptor, AP-2. J Cell Biol. 1995;131:1831–1838. doi: 10.1083/jcb.131.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett B D, Solar G P, Yaun J Q, Mathias J, Thomas G R, Matthews W. A role for leptin and its cognate receptor in hematopoiesis. Curr Biol. 1996;6:1170–1180. doi: 10.1016/s0960-9822(02)70684-2. [DOI] [PubMed] [Google Scholar]
- 5.Bjorbaek C, Uotani S, da Silva B, Flier J S. Divergent signaling capacities of the long and short isoforms of the leptin receptor. J Biol Chem. 1997;272:32686–32695. doi: 10.1074/jbc.272.51.32686. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino J S, Marks M, Ohno H, Kirchhausen T. Mechanisms of signal-mediated protein sorting in the endocytic and secretory pathways. Proc Am Assoc Physicians. 1996;108:285–295. [PubMed] [Google Scholar]
- 7.Carpentier J-L, Paccaud J-P, Gordon P, Rutter W J, Orci L. Insulin-induced surface redistribution regulates internalization of the insulin receptor and requires its autophosphorylation. Proc Natl Acad Sci USA. 1992;89:162–166. doi: 10.1073/pnas.89.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpentier J-L, Paccaud J-P, Backer J, Gilbert A, Orci L, Kahn C R. Two steps of insulin receptor internalization depend on different domains of the β-subunit. J Cell Biol. 1993;122:1243–1245. doi: 10.1083/jcb.122.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan D C, Bedford M T, Lender P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides that functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C P, Lazar C S, Walsh B J, Kumuro M, Collawn J F, Kuhn L A, Tainer J A, Trowbridge I S, Farquhar M G, Rosenfeld M G, Wiley H S, Gill G N. Ligand-induced internalization of the epidermal growth factor receptor is mediated by multiple endocytic codes analogous to the tyrosine motif found in constitutively internalized receptors. J Biol Chem. 1993;268:19312–19320. [PubMed] [Google Scholar]
- 11.Cioffi J A, Shafer A W, Zupancic T J, Smith-Gbur J, Mikhail A, Platika D, Snodgrass H R. Novel B2219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat Med. 1996;2:585–589. doi: 10.1038/nm0596-585. [DOI] [PubMed] [Google Scholar]
- 12.Davis C G, Van Driel G I R, Russel D W, Brown M S, Goldstein J L. The low density lipoprotein receptor: identification of amino acids in the cytoplasmic domain required for rapid endocytosis. J Biol Chem. 1987;262:4075–4082. [PubMed] [Google Scholar]
- 13.Dayhoff M O, Barker W C, Hunt L T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Vlahos C, Liu R, Brown R F, Badwey J A. Antagonists of phosphatidylinositol 3-kinase block activation of several novel protein kinases in neutrophils. J Biol Chem. 1995;270:11684–11691. doi: 10.1074/jbc.270.19.11684. [DOI] [PubMed] [Google Scholar]
- 15.Ekena K, Stevens T H. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol Cell Biol. 1995;15:1671–1678. doi: 10.1128/mcb.15.3.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein J L, Brown M S, Anderson R G, Russell D W, Schneider W J. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 17.Honegger A M, Dull T J, Felder S, Van Obberghen E, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell. 1987;23:199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- 18.Horazdovsky B F, Davies B A, Seaman M N N, McLaughlin S A, Yoon S H, Emr S D. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasuga M, Kahn C R, Hedo J A, Van Obberghen E, Yamada K M. Insulin-induced receptor loss in cultured human lymphocytes is due to accelerated receptor degradation. Proc Natl Acad Sci USA. 1978;78:6917–6921. doi: 10.1073/pnas.78.11.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohrer K, Emr S D. The yeast VPS17 gene encodes a membrane-associated protein required for the sorting of soluble vacuolar hydrolases. J Biol Chem. 1993;268:559–569. [PubMed] [Google Scholar]
- 21.Kurten R C, Cadena D L, Gill G N. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 22.Lee G H, Proenca R, Montez J M, Carroll K M, Darvishzadeh J G, Lee J I, Friedman J M. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 23.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 24.Levy-Toledano R, Caro L H, Hindman N, Taylor S I. Streptavidin blotting: a sensitive technique to study cell surface proteins; application to investigate autophosphorylation and endocytosis of biotin-labeled insulin receptors. Endocrinol. 1993;113:1803–1808. doi: 10.1210/endo.133.4.8404622. [DOI] [PubMed] [Google Scholar]
- 25.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signalling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 26.Lyons D M, Mahanty S K, Choi K-Y, Manandhar M, Elion E A. The SH3 domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1997;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 28.Merlino G T, Stahle C, Jhappan C, Linton R, Mahon K A, Willingham M C. Inactivation of a sperm motility gene by insertion of an epidermal growth factor receptor transgene whose product is overexpressed and compartmentalized during spermatogenesis. Genes Dev. 1991;5:1395–1406. doi: 10.1101/gad.5.8.1395. [DOI] [PubMed] [Google Scholar]
- 29.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. pp. 293–298. [Google Scholar]
- 30.Northwehr S F, Hindes A E. A yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- 31.Pawson T. Protein modules and signalling networks. Nature. 1955;373:573–779. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 32.Ponting C P. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Prot Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quon M J, Zarnowski M J, Guerre-Millo M, de la Luz Sierra M, Taylor S I, Cushman S W. Transfection of DNA into isolated rat adipose cells by electroporation: evaluation of promoter activity in transfected adipose cells which are highly responsive to insulin after one day in culture. Biochem Biophys Res Commun. 1993;194:338–346. doi: 10.1006/bbrc.1993.1825. [DOI] [PubMed] [Google Scholar]
- 34.Quon M J, Chen H, Lin C H, Zhou L, Ing B L, Zarnowski M J, Klinghoffer R, Kazlauskas A, Cushman S W, Taylor S I. Effects of overexpressing wild-type and mutant PDGF receptors on translocation of Glut 4 in transfected rat adipose cells. Biochem Biophys Res Commun. 1996;226:587–594. doi: 10.1006/bbrc.1996.1400. [DOI] [PubMed] [Google Scholar]
- 35.Renfrew Haft C, Klausner R D, Taylor S I. Involvement of dileucine motifs in the internalization and degradation of the insulin receptor. J Biol Chem. 1994;269:26286–26294. [PubMed] [Google Scholar]
- 36.Seaman M N J, Burd C G, Emr S D. Receptor signalling and the regulation of endocytic membrane transport. Curr Opin Cell Biol. 1996;8:549–556. doi: 10.1016/s0955-0674(96)80034-2. [DOI] [PubMed] [Google Scholar]
- 37.Tartaglia L S, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL-W—Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trowbridge I S, Collawn J F, Hopkins C R. Signal-dependent membrane trafficking in the endoctyic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 40.Valius M, Kazlauskas A. Phospholipase C-gamma 1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell. 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- 41.Voos W, Stevens T H. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–589. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]