Abstract
The human double-stranded RNA (dsRNA)-dependent protein kinase PKR inhibits protein synthesis by phosphorylating translation initiation factor 2α (eIF2α). Vaccinia virus E3L encodes a dsRNA binding protein that inhibits PKR in virus-infected cells, presumably by sequestering dsRNA activators. Expression of PKR in Saccharomyces cerevisiae inhibits protein synthesis by phosphorylation of eIF2α, dependent on its two dsRNA binding motifs (DRBMs). We found that expression of E3 in yeast overcomes the lethal effect of PKR in a manner requiring key residues (Lys-167 and Arg-168) needed for dsRNA binding by E3 in vitro. Unexpectedly, the N-terminal half of E3, and residue Trp-66 in particular, also is required for anti-PKR function. Because the E3 N-terminal region does not contribute to dsRNA binding in vitro, it appears that sequestering dsRNA is not the sole function of E3 needed for inhibition of PKR. This conclusion was supported by the fact that E3 activity was antagonized, not augmented, by overexpressing the catalytically defective PKR-K296R protein containing functional DRBMs. Coimmunoprecipitation experiments showed that a majority of PKR in yeast extracts was in a complex with E3, whose formation was completely dependent on the dsRNA binding activity of E3 and enhanced by the N-terminal half of E3. In yeast two-hybrid assays and in vitro protein binding experiments, segments of E3 and PKR containing their respective DRBMs interacted in a manner requiring E3 residues Lys-167 and Arg-168. We also detected interactions between PKR and the N-terminal half of E3 in the yeast two-hybrid and λ repressor dimerization assays. In the latter case, the N-terminal half of E3 interacted with the kinase domain of PKR, dependent on E3 residue Trp-66. We propose that effective inhibition of PKR in yeast requires formation of an E3-PKR-dsRNA complex, in which the N-terminal half of E3 physically interacts with the protein kinase domain of PKR.
Mammalian PKR is a double-stranded RNA (dsRNA)-dependent protein kinase that is transcriptionally induced by interferon and becomes activated in virus-infected cells by dsRNAs produced during the virus life cycle. PKR interferes with virus propagation by phosphorylating the α subunit of translation initiation factor 2 (eIF2α), converting eIF2 from a substrate to an inhibitor of its guanine nucleotide exchange factor, eIF2B. This leads to inhibition of viral protein synthesis at the translation initiation step. PKR is activated by dsRNA in vitro, and the N-terminal half of the protein contains two copies of a dsRNA binding motif (DRBM) also present in other dsRNA binding proteins (reviewed in references 11 and 33). When expressed in yeast (Saccharomyces cerevisiae) cells, PKR inhibits general translation initiation and cell growth due to hyperphosphorylation of eIF2α. Mutations in the DRBMs which impair dsRNA binding by PKR in vitro also reduce the ability of PKR to phosphorylate eIF2α in yeast, consistent with the idea that the DRBMs mediate the stimulatory effect of dsRNA on PKR kinase activity (10, 40). The importance of dsRNA binding for kinase activation in vivo is also shown by the fact that viruses encode negative regulators of PKR that interfere with the binding of dsRNA activators to the enzyme. VA RNAI, encoded by adenovirus, binds to the DRBMs but fails to activate the enzyme. The vaccinia virus E3L product is a dsRNA-binding protein capable of inhibiting PKR by sequestering dsRNA activators (3, 7, 26, 49).
Vaccinia virus E3 contains one copy of the conserved DRBM in the C-terminal half of the protein that is necessary and sufficient for dsRNA binding in vitro (8) and also to provide the host range (9), replication efficiency, and interferon resistance in cultured cells characteristic of wild-type vaccinia virus (45). The ability of E3 to inhibit PKR in cell extracts can be partially reversed by adding large amounts of dsRNA (1, 14, 26), leading to the conclusion that E3 inhibits PKR by sequestering dsRNA activators. Consistent with this view, an E3L-deleted virus can be functionally complemented by expression of heterologous dsRNA binding proteins, including rotavirus NSP3 (32), reovirus S4 (2), the cellular human immunodeficiency virus transactivation response (TAR) RNA binding protein (TRBP) (36), and Escherichia coli RNase III (44). Additionally, E3 was found to suppress the dsRNA-dependent 2′,5′-oligoadenylate synthetase/RNase L portion of the interferon response pathway (2).
The C-terminal portion of E3 containing the DRBM exists as a dimer in solution and appears to bind dsRNA cooperatively; thus, protein-protein interactions contribute to the dsRNA binding affinity of E3. Removal of the N-terminal half of the protein had no significant effect on the affinity of E3 for dsRNA; however, it reduced the formation of higher-order oligomers in solution (22). Although the N-terminal domain was not required for replication or interferon sensitivity of vaccinia virus during infections of cultured cells, it remains possible that it confers a replication advantage in infected whole animals (45).
There is evidence that PKR acts as a dimer and that the N-terminal region containing the DRBMs mediates dimerization in addition to dsRNA binding. PKR activation exhibits second-order kinetics with respect to protein concentration (28), and the enzyme was purified as a phosphorylated dimer from L cells (31). In addition, two mutant PKR alleles containing deletions in DRBM-1 or DRBM-2 functionally complemented when coexpressed in the same yeast cells (40). It was also shown that an N-terminal segment of PKR containing both DRBMs was coimmunoprecipitated with full-length PKR from transfected COS cells (50). Similarly, N-terminal segments were shown to interact with themselves and with full-length PKR in various in vivo and in vitro protein interaction assays (12, 37, 38, 48).
The observation that PKR activation is inhibited by very high concentrations of dsRNAs (24) suggested that dimer formation is promoted by binding of two PKR molecules to the same dsRNA molecule. Accordingly, high concentrations of dsRNAs would favor dissociation of dimers into monomers bound to different dsRNA molecules (28). In agreement with this view, binding of full-length PKR to an N-terminal fragment containing the DRBMs in vitro was dependent on dsRNA (12). Moreover, chemical cross-linking and gel filtration experiments indicated that TAR RNA promotes PKR dimerization (4a). Other studies indicated that PKR dimerization is relatively unaffected by point mutations in the DRBMs which impair dsRNA binding in vitro (12, 35, 37, 38, 50), suggesting that dimerization can occur through protein-protein interactions in the absence of dsRNA binding. Cooperative binding to dsRNA of PKR N-terminal fragments (41) confirms that the DRBMs are capable of protein-protein interactions regardless of whether these are sufficient for dimerization in the absence of dsRNA binding.
Interestingly, mutations that reduced dsRNA binding in vitro impaired dimerization in vivo by full-length PKR but not by the isolated N-terminal domain of the protein. To account for these last observations, it was proposed that the dimerization domain in the N-terminal region of PKR is masked by interactions between the N-terminal and C-terminal halves of the protein and that dsRNA binding stimulates a conformational change that exposes the N-terminal dimerization surface, leading to autophosphorylation and PKR activation (51). The idea that the N-terminal region functions negatively was originally prompted by observations that its removal leads to constitutively active PKR function (50, 54). A recent study using the yeast two-hybrid assay confirmed that the N-terminal and C-terminal halves of PKR can physically interact (42).
Previously, we obtained genetic evidence that high-level PKR activity requires self-interactions between the protein kinase domains in a PKR dimer. Expression of PKR-K296R (and several other kinase domain point mutants) did not detectably interfere with wild-type PKR in yeast cells, whereas mutant PKR alleles with deletions in the kinase domain did interfere with wild-type PKR function in yeast (40). Since PKR mutants with point mutations or deletions in the kinase domain were comparably expressed and were presumed to bind dsRNA equally well, the dominant interference by the deletion alleles was difficult to explain by sequestering of dsRNA activators. Accordingly, we proposed that kinase domain deletion mutants formed heterodimers with wild-type PKR that were less active than those formed with PKR-K296R because the deletions eliminated important interactions between the kinase domains in the dimer (40). In vitro studies with recombinant PKR have provided independent evidence for self-interactions in the kinase domain and the relative inactivity of heterodimers formed between wild-type and kinase domain deletion mutants (37). In addition, a recent study using in vivo protein interaction assays confirmed the existence of a second dimerization domain in PKR located between residues 244 and 296 (48).
The fact that overexpression of PKR-K296R protein did not detectably interfere with PKR function in yeast (40) suggested that dsRNA activators may be too abundant in yeast cells to allow inhibition of PKR by sequestering of dsRNA. Accordingly, we reasoned that if expression of E3 inhibited PKR in yeast, this might indicate that E3 does not rely exclusively on sequestering dsRNA. In this report, we show that E3 can indeed inhibit PKR in yeast and that this inhibition requires the nonconserved N-terminal half of E3 in addition to its DRBM. In addition, we provide several lines of evidence indicating that E3 inhibits PKR by forming inactive E3-PKR-dsRNA complexes. Our results lead us to propose that E3 inhibits PKR by a novel mechanism in which the inhibitory E3 N-terminal domain is tethered to PKR by interactions between the DRBMs of the two proteins.
MATERIALS AND METHODS
Plasmids and yeast strains.
Construction of the plasmids containing PKR-K296R (p1470) and PKR-ΔK (p1766) under the control of the GAL-CYC1 promoter was described previously (40). Plasmid p1545, from which wild-type PKR was produced in strains coexpressing various E3 proteins, contains a 2.8-kbp ApaI-PstI fragment from p1420 (15) inserted into the high-copy-number TRP1 yeast plasmid pRS424 (46). p2521 containing PKR-ΔK was constructed by inserting a 2.8-kbp ApaI-BamHI fragment from p1766 into the high-copy-number LEU2 plasmid pRS425 (46). Plasmid pHY26, containing the vaccinia virus E3L coding sequence on an EcoRI-BamHI fragment in vector pSG5, has been previously described (53). An EcoRI fragment containing E3L was isolated from pHY26, blunted ended, and further digested with BamHI. The resulting fragment was inserted between the SmaI and BamHI sites of pEMBLyex4 (6), creating plasmid pC178.
To facilitate subcloning and mutagenesis of the E3L coding sequence, an SstI-BamHI fragment from plasmid pC178 was inserted into SstI-, BamHI-digested pUC19, creating p2286. A PCR fusion technique (52) was used to insert the nine codons encoding the hemagglutinin (HA) epitope (TAC CCA TAC GAC GTA CCA GAC TAT GCA) immediately following the E3L start codon in plasmid p2286, creating plasmid pR124. p2245 was created by inserting an SstI-BamHI fragment from pR124 containing the HA-tagged E3L coding sequence into pEMBLyex4. p2558 was created by inserting an SstI-BamHI fragment from p2245 into p2444, a modified pEMBLyex4 vector in which the URA3 marker is replaced by TRP1 (39). Silent mutations which generated ClaI restriction sites within the E3L coding sequence were introduced by PCR fusion. ClaI sites were created by altering the codons for Asp-7 (GAC to GAT) and Ile-85 (ATA to ATC) of plasmid p2286 to create pR49. pR49 was digested with ClaI and religated to produce the E3LΔ7-86 allele in pR50. The HA epitope was inserted immediately after the E3L start codon in pR50 as described above, and the sequence containing the HA-tagged version of E3LΔ7-86 (HA-E3LΔ7-86) was inserted as an SstI-BamHI fragment into pEMBLyex4, creating plasmid p2446. PCR fusion was used to introduce a BsrGI site at the codon for Ser-81 (TCG to TAC) and Asp-82 (GAC to AGC) of pR124 to create pR141. Digestion with BsrGI followed by religation generated pR144. The HA-E3LΔ60-82 allele was isolated from pR144 as an SstI-XbaI fragment and ligated with SstI-, XbaI-digested pEMBLyex4 to create p2604. A BsrGI site was introduced by PCR fusion at the Val-15 codon (GTG to GTA) and Cys-16 codon (TGT to CAT) in pR124, and digestion of pR124 with BsrGI followed by religation generated pR143 containing HA-E3LΔ15-56. This tagged allele was inserted into pEMBLyex4 as an SstI-BamHI fragment, creating p2602. PCR fusion was used to construct E3L-K167A,R168A by changing the codon for Lys-167 to GCA and the codon for Arg-168 to GCA in p2245, creating pR187. PCR fusion was used to construct E3L-W66A,F67A,M68A by changing the three relevant codons to GCG, GCT, and GCG, respectively, in p2245, creating p2601. Similarly each codon was individually changed to GCG, GCT, or GCG, respectively, using the same PCR fusion technique, creating p2603 (W66A), p2638 (F67A), and p2639 (M68A).
The plasmid vectors for yeast two-hybrid analysis, pGBT10 and pGAD425, and the constructs encoding the fusion proteins containing the GAL4 activation domain (AD) and various PKR segments, pAD-PKR-K296R, pAD-PKRΔ243-451, pAD-PKRΔ297-551, pAD-PKRΔ1-366, and pAD-PKRΔ1-243, have been described previously (19). Plasmids encoding fusion proteins between the GAL4 DNA binding domain (BD) (pGBT10) and wild-type E3 (pR162), E3-K167A,R168A (pL194-4), E3-W66A (pL181-10), E3Δ60-82 (pL182-3), E3Δ7-86 (pL165-2), E3Δ7-86,K167A,R168A (PL199-1), E3Δ105-190,W66A (pL168-3), and E3Δ105-190 (PL166-4), were constructed by PCR using as primers oligonucleotides that introduce NdeI and BamHI sites at the beginning and end, respectively, of the E3L coding sequence. Where necessary, a stop codon was included in the 3′ oligonucleotide. Following amplification, the PCR products were digested with NdeI and BamHI and ligated with NdeI-, BamHI-digested pGBT10.
Plasmids encoding GST-E3 fusions were constructed by using p2645, a derivative of vector pGEX-5X-3 (Pharmacia) produced by inserting a pair of annealed oligonucleotides, RO97 (5′ GATCGTCCATATGCTCG) and RO98 (5′ GATCCGAGCATATGGAC), which contain an NdeI site at the BamHI site of pGEX-5X-3. In p2645, the ATG initiation codon is within the NdeI site, and a BamHI site was reintroduced downstream. Plasmids encoding GST-E3 WT (wild type) (pR194), GST–E3-W66A (pR195), GST–E3-Δ7-86 (pR196), GST–E3-Δ7-86,K167A,R168A (pL201-2), GST–E3-Δ105-190 (pR198), and GST–E3-Δ105-190,W66A (pR199) were made by inserting into p2645 the appropriate NdeI-SalI fragment obtained from the plasmids described above encoding fusions between the GAL4 BD and various E3 segments.
Plasmids used for in vitro transcription and translation of PKR mutant proteins, pcDNA1/Neo, pcDNA1/Neo-PKR WT, pcDNA1/Neo-PKR-K296R, pcDNA1/Neo-PKR-Δ297-551, and pcDNA1/Neo-PKR-Δ1-243, were described previously (19). pcDNA-PKR-Δ1-367 was made by synthesizing a HindIII-BamHI fragment by PCR using primers FZP29 (5′ CCGGAAGCTTGCCGCCACCATGTTCTGTGATAAAGG 3′) and P68-3′ (5′ TATCAGAAGCAGGATCCCGGGGATCCCTAACATGTGTGTCGTTCA 3′), with HindIII and BamHI sites shown in italics and start and stop codons shown in boldface, and inserting it between the HindIII and BamHI sites of pcDNA1/Neo. The inserted fragment contains an ATG codon in the optimum Kozak sequence context (29) in frame with the PKR-Δ1-367 coding sequence. The sequence of the inserted fragment was verified by using a Sequenase version 2.0 DNA sequencing kit from United States Biochemical/Amersham Life Sciences.
Construction of the isogenic S. cerevisiae strains RY1-1 and RY1-12 (a ura3-52 leu2-3 leu2-112 gcn2Δ trp1-Δ63), containing two copies and a single copy, respectively, of the wild-type human PKR coding sequence under control of the GAL-CYC1 promoter integrated at the LEU2 locus, was described previously (40). S. cerevisiae J82 (a ura3-52 leu2-3 leu2-112 gcn2Δ sui2Δ trp1-Δ63, p1098 [SUI2-S51A, LEU2]) and H1894 (a ura3-52 leu2-3 leu2-112 gcn2Δ trp1-63Δ) (15) are derivatives of strain H1645 (16). Strain Hf7c (a ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::[GAL 17-mers]3-CYC1-lacZ) (18) was used for yeast two-hybrid analysis. Media used to culture yeast strains and to conduct growth tests for phenotypic analysis of PKR alleles were described previously (40).
Immunoblot analysis of E3 and PKR protein expression.
Whole-cell extracts were prepared, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to nitrocellulose membranes, and the membranes were treated with blocking solution exactly as described previously (40) except for the addition of complete protease inhibitor (CPI) cocktail (Boehringer Mannheim) to the lysis buffer and the use of 8 to 16% gradient gels. Immunodetection of E3 protein was conducted with monoclonal or polyclonal antibodies against the HA epitope, HA-12CA5 (Boehringer Mannheim) or HA.11 (BabCo), respectively, used at a dilution of 1:500 in blocking solution. Immunodetection of PKR was conducted with a PKR-specific monoclonal antibody (71/10; Ribogene) at a dilution of 1:1,000 in blocking solution. Visualization of immune complexes by using the enhanced chemiluminescence (ECL) detection system (Amersham) was conducted as specified by the manufacturer.
Poly(I-C) binding assays.
Whole-cell extracts were prepared by breaking cells with glass beads in KR lysis buffer (20 mM Tris-HCl [pH 8.0], 50 mM KCl, 400 mM NaCl, 20% glycerol, 0.1% Triton X-100, 0.5 mM EDTA) containing CPI cocktail; 100 μg of whole-cell extract was incubated with poly(I-C)-agarose in 200 μl of buffer A (150 mM KCl, 20 mM HEPES, 10% glycerol, 5 mM magnesium acetate) plus CPI cocktail for 1 h at 4°C. The agarose beads were collected and washed three times with KR lysis buffer, and the pellet was resuspended in 30 μl of 2× Laemmli sample buffer (30). Proteins in the supernatant were precipitated with trichloroacetic acid, washed once with 80% ethanol, dried, resuspended in 30 μl of 2× Laemmli sample buffer, and boiled for 5 min. The proteins were resolved by SDS-PAGE on 8 to 16% polyacrylamide gradient gels. Immunodetection of E3 proteins was conducted as described above.
Coimmunoprecipitation of PKR and E3.
Whole-cell extracts were prepared by breaking the cells with glass beads in immunoprecipitation lysis buffer (20 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM magnesium acetate, 1 mM dithiothreitol) containing CPI cocktail (Boehringer Mannheim). To immunoprecipitate PKR and E3, protein samples (500 μg) were diluted to a final volume of 0.05 ml in nondenaturing binding buffer (20 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM magnesium acetate, 0.1% Triton X-100) containing CPI cocktail (Boehringer Mannheim). Three microliters of monoclonal antibodies against the HA epitope (HA-12CA5; Boehringer Mannheim) was added, and samples were incubated at 4°C for 2 h with rocking. Immune complexes were collected using GammaBind Plus protein G-Sepharose beads and washed three times with 0.10 ml of nondenaturing binding buffer. The beads were resuspended in 30 μl of 2× Laemmli sample buffer (30), boiled for 3 min, resolved by SDS-PAGE on 8 to 16% gradient gels, and subjected to immunoblot analysis for detection of PKR and E3 proteins as described above. The efficiencies of immunoprecipitating E3 proteins and PKR were estimated by video image densitometry of the resulting films, using the NIH Image software (version 1.61).
In vivo protein interaction assays.
For yeast two-hybrid analysis, GAL4 fusion constructs were introduced into yeast strain Y190 (obtained from S. Elledge, Baylor College of Medicine) by standard techniques (25). Transformants were selected on synthetic complete dextrose (SC) medium (43) lacking tryptophan and leucine. The strength of the protein-protein interactions was measured by stimulation of the HIS3 reporter present in this strain as assayed by growth on SC medium lacking histidine, tryptophan, and leucine and containing 24 or 30 mM 3-aminotriazole. The procedures followed in performing λ repressor dimerization assays and the λN-PKR-Δ1-243 and λN-PKR-K296R constructs used in these experiments were described previously (48). Plasmids encoding glutathione S-transferase (GST)–E3 fusions were described above. PC168-derived plasmids encoding the λ repressor N-terminal DNA-binding domain (λN) fused with the indicated PKR proteins and p2645-derived plasmids encoding GST alone or GST fused with the indicated E3 proteins were cotransformed into E. coli AG1688 (obtained from J. C. Hu, Texas A&M University). Cotransformants were selected on B medium containing 50 μg of ampicillin and 20 μg of chloramphenicol per ml. Cultures grown overnight (30°C) in LB supplemented with the antibiotics, 10 mM MgSO4, and 0.2% maltose were used to create bacterial lawns containing 100 nM isopropylthio-β-d-galactoside (IPTG). Lawns were then spotted with 5-μl aliquots of serial dilutions of a λKH54 phage lysate (109 PFU) at 10-fold intervals. Infected lawns were incubated overnight at 30°C, and the inhibition of dimerization mediated by λN-PKR fusions was scored by appearance of dot plaques on the lawns.
GST binding assays.
Transformants of E. coli BL21(DE3) bearing plasmids encoding GST or different GST-E3 fusion proteins were grown to an optical density at 600 nm (OD600) of 0.6 to 0.7, and IPTG was added to 1 mM to induce expression of the GST proteins for 2 h. Total proteins were extracted by sonicating cells in 1× phosphate-buffered saline containing CPI cocktail (Boehringer Mannheim), followed by centrifugation at 12,000 × g for 30 min at 4°C. 35S-labeled proteins PKR-K296R, PKR-Δ243-551, PKR-Δ297-551, PKR-Δ1-243, and PKR-Δ1-367 were produced by in vitro transcription and translation using plasmids pcDNA1/Neo-PKR K296R, pcDNA1/Neo-PKR WT linearized with BanI, pcDNA1/Neo-PKR-Δ297-551, pcDNA1/Neo-PKR-Δ1-234, and pcDNA1/Neo-PKR-Δ1-367, respectively, [35S]-Pro Mix (mixture of [35S]methionine and [35S]cysteine; Amersham), and the TNT T7 coupled reticulocyte lysate system (Promega) as instructed by the manufacturer. GST binding assays were conducted essentially as described previously (19), with the following modifications: binding buffer (1× phosphate-buffered saline) contained CPI cocktail (Boehringer Mannheim) as the only source of protease inhibitors; 15 μl of glutathione-Sepharose beads (50% slurry) instead of 50 μl was added to each reaction mixture; after washing, the glutathione-Sepharose beads were resuspended in 15 μl of 2× SDS sample buffer, boiled, and resolved by SDS-PAGE on 10 to 20% polyacrylamide gradient gels.
Assays of ribosome binding by E3 and PKR in yeast cell extracts.
Detection of ribosome association in whole-cell extracts was conducted essentially as described previously (54). The distribution of proteins in the gradients was analyzed by separating 25 μl from each 600-μl fraction by SDS-PAGE on 10 to 20% gradient gels followed by immunoblot analysis of PKR and HA-E3 proteins conducted as described above.
RESULTS
Genetic evidence that E3 inhibits PKR in yeast through complex formation.
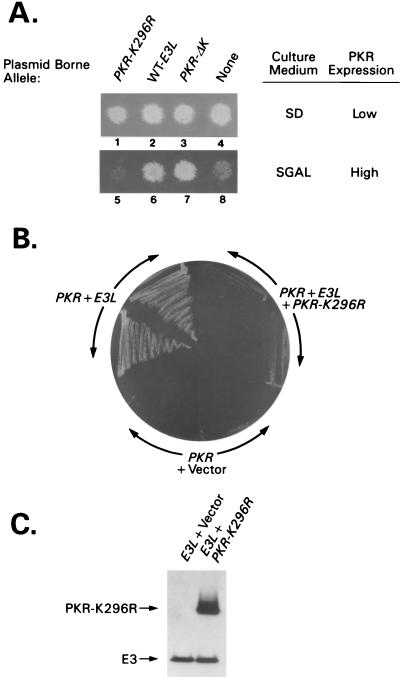
S. cerevisiae contains a protein kinase known as GCN2 that phosphorylates eIF2α on serine-51 (16), the same reaction catalyzed by PKR in mammalian cells. To test whether E3 can inhibit PKR activity in yeast, we used a gcn2Δ strain (RY1-1) expressing wild-type PKR cDNA under the control of a galactose-inducible (GAL) promoter from a plasmid integrated in the genome. When RY1-1 is cultured on medium containing galactose as the carbon source (SGal medium [10% galactose, 2% raffinose]), PKR is expressed at levels high enough to cause extensive phosphorylation of eIF2α, with attendant inhibition of protein synthesis and cell growth (15) (Fig. 1A, lane 8). When PKR is expressed at low levels on medium with glucose as the carbon source (SD medium), the amount of eIF2α phosphorylation is not great enough to reduce growth rate (lane 4). A multicopy plasmid containing E3L cDNA under the control of the same GAL promoter was introduced into strain RY1-1 and assayed for inhibition of PKR function in cells grown on SGal medium. In parallel, two PKR alleles encoding catalytically defective proteins with wild-type DRBMs were compared with E3L for the ability to inhibit wild-type PKR function: (i) PKR-K296R, which is recessive to wild-type PKR in yeast, and (ii) PKR-ΔK, completely lacking the kinase domain, which is dominant negative to wild-type PKR (40). When expressed at high levels, the recessive PKR-K296R allele had no effect on wild-type PKR function (compare lanes 5 and 8), whereas the dominant-negative PKR-ΔK allele inhibited wild-type PKR and restored growth on SGal medium (lane 7), as reported previously (40). The E3L construct resembled PKR-ΔK and reversed the growth inhibition associated with PKR overexpression (lane 6), indicating that E3 interferes with PKR function in yeast cells.
FIG. 1.
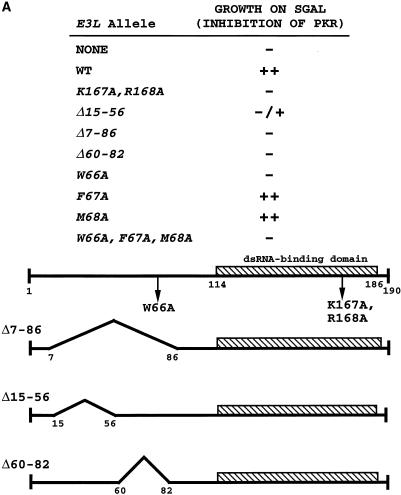
Genetic evidence that E3 inhibits PKR in yeast through heterocomplex formation. (A) A plasmid containing the PKR-K296R (p1470), PKR-ΔK (p1766), or wild-type E3L (p2245) cDNA, all under the control of the CYC1-GAL promoter, or empty vector p1079 (none) was introduced into the gcn2Δ yeast strain RY1-1 containing two copies of the wild-type PKR allele (also under CYC1-GAL control) integrated at the LEU2 locus. Patches of transformants were grown on SD medium, replica plated to SD or SGal medium, and incubated for 3 to 4 days at 30°C. (B) Plasmids containing E3L (p2558) and PKR K296R (p1470) or the corresponding empty vectors (p2444 and p1079) were introduced into strain RY1-1, and transformants were streaked for single colonies on SGal medium and incubated for 5 to 6 days at 30°C. (C) The parental strain of RY1-1 lacking the integrated copies of PKR, H1894, was transformed with plasmids containing E3L (p2558), plus either PKR-K296R (p1470) or empty vector (pEMBLyex4), and grown on SGal medium for ∼12 h. Whole-cell extracts were prepared, and 20 μg of total cell protein was fractionated by SDS-PAGE and subjected to immunoblot analysis using monoclonal antibodies against PKR and against HA to detect HA-tagged E3. ECL (Amersham) was used to visualize immune complexes. Transformants of strain H1894 rather than the RY1-1 transformants described for panel B were analyzed to eliminate differential effects of wild-type PKR on expression of E3.
The fact that E3 inhibited wild-type PKR in yeast whereas PKR-K296R did not, even though both proteins contain DRBMs, could be explained by proposing that E3 is expressed at higher levels and thus can sequester dsRNA activators more effectively than PKR-K296R. Alternatively, E3 could inhibit PKR by forming defective heteromeric complexes, in the manner proposed previously to account for the dominant-negative phenotype of PKR-ΔK (40). If the former explanation is correct, then coexpressing PKR-K296R and E3 in the same cells should enhance down-regulation of wild-type PKR by increasing the abundance of functional DRBMs without changing the number of active kinase domains. Alternatively, if E3 must form complexes with PKR to inhibit kinase activity, then coexpressing PKR-K296R might interfere with E3 inhibitory function by displacing it from the inhibited PKR-E3 complexes and forming PKR–PKR-K296R heterodimers of relatively higher activity plus inactive PKR-K296R–E3 complexes.
To test these predictions, we introduced the PKR-K296R construct into RY1-1 transformants containing integrated wild-type PKR and plasmid-borne E3L. As described above, cells overexpressing wild-type PKR on galactose medium failed to grow, whereas coexpressing E3 suppressed the toxicity of PKR and rescued growth on galactose medium (Fig. 1B, PKR + Vector versus PKR + E3L). Coexpressing the inactive PKR-K296R protein together with E3 neutralized the effect of E3 and restored the toxicity of the wild-type PKR (Fig. 1B, PKR + E3L + PKR-K296R). Immunoblot analysis showed that E3 levels were unaffected by coexpressing PKR-K296R in the same cells (Fig. 1C); thus, the ability of PKR-K296R to reverse E3 inhibitory function is unlikely to result from a reduction in E3 expression. Our results can be explained by proposing that E3 inactivates PKR by forming a complex with it and that the defective PKR-K296R protein rescues the wild-type kinase by sequestering E3 in nonfunctional PKR-K296R–E3 complexes.
Identification of residues in the N-terminal domain of E3 required for inhibition of PKR.
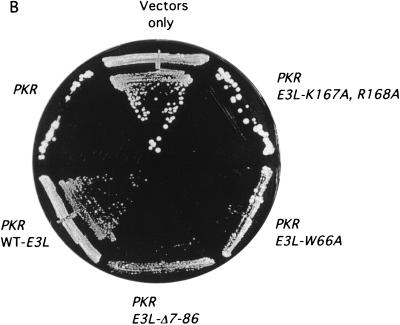
To determine the importance of dsRNA binding by E3 for its ability to inhibit PKR in yeast, we introduced alanine substitutions at Lys-167 and Arg-168 in the E3 DRBM. These residues are conserved in the DRBMs of other dsRNA binding proteins (47), and it was shown that alanine substitutions in the corresponding residues of PKR DRBM-1 abolished or weakened dsRNA binding in vitro (34, 38). Moreover, a single threonine substitution in E3 at Lys-167 was shown previously to abolish dsRNA binding in vitro (8). We also constructed several deletion and point mutations in the N-terminal half of E3 to determine whether this uncharacterized part of the protein was required for E3 inhibitory function (Fig. 2A). These mutations were introduced into a plasmid-borne E3L construct identical to that described above except that coding sequences for the influenza virus HA epitope were added to the beginning of the coding region to facilitate immunodetection of E3 proteins. Addition of the HA epitope had no effect on E3 function, as judged by growth assays of the type represented in Fig. 1A (data not shown).
FIG. 2.
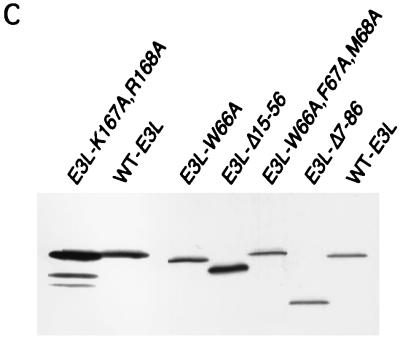
Genetic evidence that the N-terminal domain of E3 is critically required for inhibition of PKR. (A) Summary of growth phenotypes on SGal medium conferred by different E3L alleles in strain RY1-1, indicating their ability to inhibit PKR function in yeast. The mutant E3 proteins are represented schematically at the bottom, from amino acids 1 to 190. Transformants of RY1-1 bearing the indicated plasmid-borne E3L alleles were analyzed for growth as described for Fig. 1 and scored relative to transformants containing wild-type E3L or vector alone. The E3L alleles were introduced into RY1-1 on the follow ing plasmids: E3L (wild type [WT]), p2445; E3-K167A,R168A, p2612; E3-Δ15-56, p2602; E3-Δ7-86, p2446; E3-Δ60-82, p2604; E3-W66A, p2603; E3-F67A, p2638; E3-M68A, p2639; E3-W66A,F67A,M68A, p2601; NONE, empty vector p1079. (B) Transformants of RY1-1 bearing the indicated plasmid-borne alleles or vector alone, described above, were streaked for single colonies on SGal medium and incubated for 7 to 10 days at 30°C. (C) The gcn2Δ eIF-2α-S51A yeast strain J82 expressing wild-type PKR from plasmid p1545 was transformed with plasmids containing the indicated E3L alleles described above, grown in SD medium at 30°C for ∼30 h, and shifted to SGal medium for ∼12 h. Whole-cell extracts were prepared and 20 μg of total protein was fractionated by SDS-PAGE and subjected to immunoblot analysis using monoclonal antibodies against the HA epitope to detect the HA-tagged E3. The ECL system was used to detect the immune complexes. Transformants of strain J82 rather than the RY1-1 transformants described for panel A were analyzed to eliminate the differential effects of wild-type PKR on the expression of the various mutant E3 proteins. The blot was stripped and probed with antibodies against poly(A) binding protein (PAB1) to verify that equal amounts of whole-cell protein were loaded in all lanes (data not shown). Lanes 1 and 2 and lanes 3 to 7 derived from different experiments.
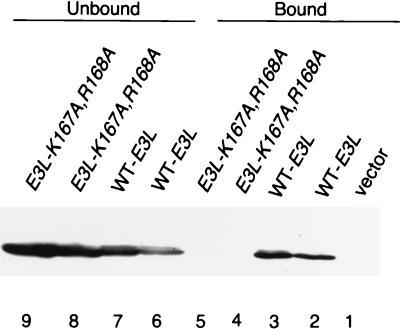
The K167A,R168A double substitution in the DRBM abolished E3 function, restoring the lethal effect of high-level PKR expression on SGal medium (Fig. 2A and B). To verify that this mutant protein was incapable of binding dsRNA, we compared it to wild-type E3 for the ability to bind to poly(I-C)-agarose beads. As shown in Fig. 3, under conditions where ca. 50% of wild-type E3 in the extract bound to the resin (lanes 6 and 7 versus 2 and 3), none of the E3-K167A,R168A mutant protein was recovered in the bound fraction (lanes 4 and 5 versus 8 and 9). Interestingly, the Δ7-86 and Δ60-82 deletions and the W66A substitution also abolished the anti-PKR function of E3 (Fig. 2A and B), whereas Ala substitutions at Phe-67 and Met-68, flanking Trp-66, had no effect on E3 function (Fig. 2A). It was shown previously that the N-terminal half of E3 makes no contribution to the binding affinity for dsRNA, as purified full-length E3 and the C-terminal half of E3 containing the DRBM had indistinguishable dissociation constants (22). Thus, the requirement for the N-terminal half of E3 for inhibition of PKR cannot be explained by a reduction in dsRNA binding.
FIG. 3.
The K167A and R168A mutations impair dsRNA binding by E3 in vitro. Transformants of strain J82 containing the indicated HA-tagged E3L alleles or vector alone (p1079) were grown in SD medium for ∼30 h and then shifted to inducing conditions (SGal medium) for ∼12 h, and whole-cell extracts were prepared. Aliquots containing 100 μg of total protein were incubated with poly(I-C)-agarose for 1 h at 4°C. Proteins which bound to poly(I-C)-agarose were collected by centrifugation and eluted by boiling in 2× Laemmli sample buffer. Unbound proteins in the supernatant were trichloroacetic acid precipitated, washed with ethanol, and resuspended by boiling in 2× Laemmli sample buffer. Proteins were resolved by SDS-PAGE using 8 to 16% gradient gels and subjected to immunoblot analysis using monoclonal antibodies against the HA epitope (HA-12CA5) and ECL to detect immune complexes. Lanes 1 to 5 contain the fraction of E3 proteins which bound to poly(I-C)-agarose; lanes 6 to 9 contain the fraction of E3 proteins which did not bind to poly(I-C)-agarose.
We conducted immunoblot analysis of mutant and wild-type E3 proteins in whole-cell extracts to determine whether the impaired functions of the K167A R168A, Δ15-56, Δ7-86, and W66A E3L alleles resulted from instability of the mutant proteins. For this experiment, the E3L constructs were expressed in a strain containing a nonphosphorylatable form of eIF2α (eIF2α-S51A) to eliminate the inhibitory effect of PKR function on protein synthesis in strains containing defective E3 proteins. The results shown in Fig. 2C indicated that all of the mutant proteins were present in amounts similar or even greater than that of wild-type E3. Thus, the mutations seem to impair E3 function, not its expression or stability.
PKR down-regulates its own expression in yeast by a mechanism that depends on phosphorylation of eIF2α, such that PKR abundance is inversely proportional to its eIF2α kinase activity (15, 27, 40). Accordingly, the levels of PKR in strains coexpressing E3 proteins should be inversely proportional to the anti-PKR function of the E3 proteins. This expectation was borne out by immunoblot analysis of PKR levels in strains expressing wild-type eIF2α, where we found that PKR accumulated to higher levels in transformants expressing wild-type E3 than in those containing the inactive E3 protein E3-Δ7-86, E3-W66A, or E3-K167A,R168A (data not shown).
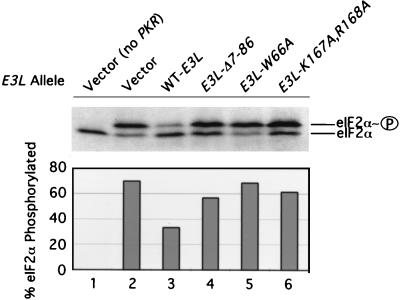
To demonstrate more directly that E3 inhibits PKR function in yeast, we measured the relative amounts of eIF2α phosphorylated and unphosphorylated on serine-51 in extracts from yeast strains expressing PKR and various E3 proteins. As shown in Fig. 4, the presence of wild-type E3 decreased the proportion of phosphorylated eIF2α from ca. 70% to ca. 35%. In contrast, the E3-Δ7-86, E3-W66A, and E3-K167A,R168A proteins produced little or no reduction in the extent of eIF2α phosphorylation. Taken together, the results in Fig. 2 to 4 indicate that both the dsRNA binding activity of E3 and residues in its N-terminal domain between amino acids 60 and 82 are critically required for the inhibition of PKR function in yeast cells.
FIG. 4.
Expression of E3 reduces eIF2α phosphorylation by PKR in yeast cells. Transformants of strain RY1-12 (containing the single-copy chromosomal PKR construct) bearing the indicated E3L alleles or vector alone (lanes 2 to 6) and the vector transformant of strain H1894 lacking the PKR construct (lane 1) were grown to saturation in SD medium containing the necessary supplements for 2 days, diluted 1:50 into 50 ml of fresh SD medium, and grown to an OD600 of 0.5 to 1. Cells were harvested, resuspended in 50 ml of SGal medium, and grown overnight. Whole-cell extracts were prepared, and 20 μg of total protein was resolved by isoelectric focusing PAGE and then subjected to immunoblot analysis using polyclonal eIF2α antibodies as described previously (16). The positions of basally phosphorylated (eIF2α) and eIF2α phosphorylated on Ser-51 (eIF2α∼P) are indicated at the right. The intensities of both signals were quantitated with a scanner (Silverscanner III) and NIH Image software (version 1.61), and the resulting data are shown graphically below the blot expressed as the percentage of total eIF2α phosphorylated on Ser-51.
E3 and PKR interact in vivo in a manner dependent on both the DRBM and the N-terminal domain of E3.
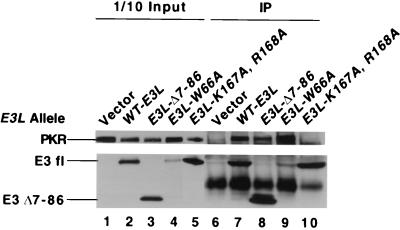
To investigate whether E3 and PKR physically interact in yeast cells, we asked whether PKR could be coimmunoprecipitated with E3 from extracts prepared from strains coexpressing PKR and the HA-tagged mutant or wild-type E3 proteins. These experiments were carried out with the eIF2α-S51A strain to eliminate autoregulation of PKR expression. As shown in Fig. 5, PKR was immunoprecipitated with anti-HA antibodies from the extract containing wild-type HA-tagged E3 but not from one lacking E3 (lanes 1, 2, 6, and 7), indicating that PKR is physically associated with wild-type E3 in cell extracts. In these experiments, 15 to 20% of the total PKR in the cell extracts was coimmunoprecipitated with 20 to 40% of the wild-type E3. After correction for the efficiency of immunoprecipitating E3, these data suggest that 50 to 70% of the PKR in the cell is physically associated with E3.
FIG. 5.
Coimmunoprecipitation of wild-type PKR with mutant and wild-type E3 proteins from yeast cell extracts. The gcn2Δ eIF-2α-S51A strain J82 bearing wild-type PKR on plasmid p1545 was transformed with plasmids encoding the indicated HA-tagged E3L alleles (see the legend to Fig. 2A) or with the vector p1079 alone. Transformants were grown under inducing conditions in SGal medium for ∼12 h, and whole-cell extracts were prepared. Aliquots containing 500 μg of total protein were immunoprecipitated (IP) with monoclonal antibodies against the HA epitope. Immune complexes were fractionated by SDS-PAGE on 8 to 16% polyacrylamide gradient gels and subjected to immunoblot analysis using a monoclonal antibody against PKR (71/10) and a polyclonal antibody against the HA epitope (HA.11). Lanes 6 to 10 contain immunoprecipitated proteins; lanes 1 to 5 contain 50 μg of the starting extracts used for the immunoprecipitations. WT-E3L, wild-type E3L allele.
The K167A,R168A mutation abolished complex formation between E3 and PKR (Fig. 5, lane 10), suggesting that the dsRNA binding activity of E3 is required for a stable interaction with PKR in vivo. This conclusion implies that PKR-E3 complexes also contain dsRNA. The Δ7-86 and Δ15-56 mutations reproducibly decreased the yield of PKR in immune complexes compared to that seen with wild-type E3 (lanes 7 and 8 and data not shown). After correcting for the efficiencies of immunoprecipitating wild-type E3 and E3Δ7-86, we found that Δ7-86 reduced complex formation with PKR to 60% of the level seen for wild-type E3. By contrast, E3-W66A complexed with PKR as efficiently as did wild-type E3 (lanes 7 and 9). (The amount of E3-W66A protein was atypically reduced relative to that of wild-type E3 in this particular extract; nevertheless, a similar amount of PKR was coimmunoprecipitated with wild-type E3 and E3-W66A.) This last result indicates that Trp-66 in the N-terminal half of E3 does not function in the inhibition of PKR by mediating complex formation between the two proteins.
Evidence for multiple interactions involving different segments of E3 and PKR.
We used the yeast two-hybrid assay to investigate physical interactions between different segments of E3 and PKR. Protein fusions between the GAL4 AD and various PKR segments were tested for interactions with GAL4 BD fusions bearing various E3 segments (Fig. 6). The full-length AD-PKR fusion contains the K296R mutation because the corresponding fusion containing wild-type PKR is toxic in yeast (19). It was shown previously that these AD-PKR fusion proteins were expressed at comparable levels in yeast cells (19). Immunoblot analysis of whole-cell extracts using anti-HA antibodies showed that the different BD-E3 fusion proteins also were expressed at similar levels (data not shown).
FIG. 6.
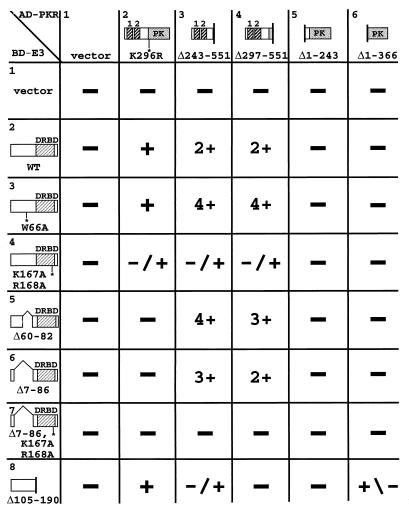
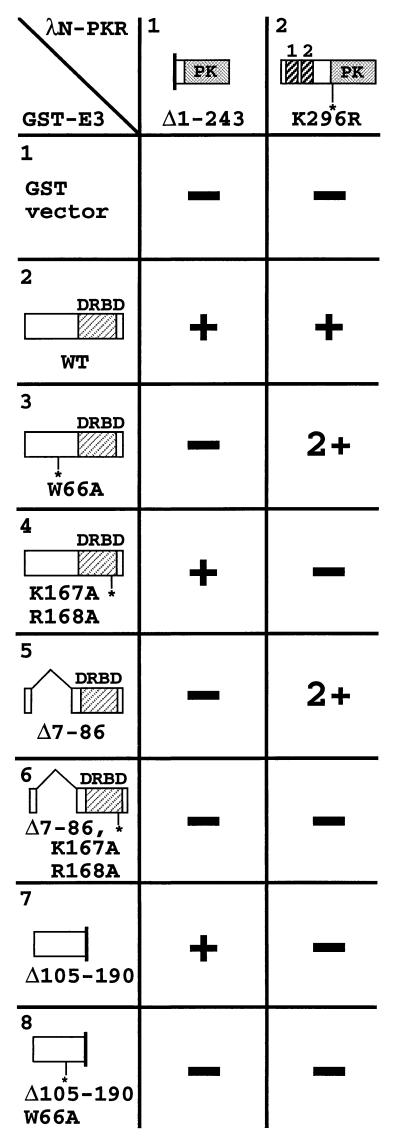
Summary of interactions between PKR and E3 segments in the yeast two-hybrid assay. Strain Y190 was cotransformed with TRP1 plasmids encoding the GAL4 BD alone (vector) or fused with wild-type E3 protein (WT) or the indicated E3 mutant proteins (BD-E3 fusions) and with a LEU2 plasmid encoding the GAL4 AD alone (vector) or fused with the indicated PKR proteins (AD-PKR fusions). Transformants were streaked for single colonies on SC medium containing 30 mM 3-AT and lacking histidine, leucine, and tryptophan. The colony size was scored for each transformant and ranked as −, −/+, +/−, +, 2+, 3+, or 4+. DRBD, dsRNA binding domain; PK, protein kinase domain.
The two PKR fusions bearing the DRBMs but lacking the protein kinase domain, Δ243-551 and Δ297-551, interacted strongly with all of the E3 fusions containing the wild-type DRBM but not with those containing the K167A,R168A mutation or with the E3-Δ105-190 fusion which lacks the DRBM entirely (Fig. 6, columns 3 and 4). The full-length PKR fusion showed a moderate interaction with wild-type E3 but little or no interaction with the E3-K167A,R168A fusion (column 2, rows 2 and 4). These data suggest that E3 interacts with PKR through its DRBM and that the dsRNA binding activity of this domain is required for E3-PKR complex formation.
Interestingly, the E3 N-terminal deletions Δ60-82 and Δ7-86 showed no interaction with full-length PKR (Fig. 6, column 2, rows 5 and 6). These data suggest that the E3 N-terminal domain contributes to the stability of complexes containing full-length PKR and E3 proteins, in accordance with results of the coimmunoprecipitation experiments described above. In contrast, the Δ60-82 and Δ7-86 E3 fusions interacted strongly with the truncated PKR fusions containing DRBMs but lacking the kinase domain (columns 3 and 4, rows 5 and 6). Thus, it appears that the E3 N-terminal half is required for interaction with full-length PKR but dispensable for interaction with PKR segments containing DRBMs but lacking the kinase domain. This seemingly paradoxical result could be explained by proposing that the PKR DRBMs are masked by interactions with the kinase domain, as suggested recently (42, 51). If the N-terminal half of E3 interacts with the kinase domain, this could free the DRBM-half of PKR for interaction with the DRBM-half of E3.
The N-terminal half of E3 (Δ105-190) interacted with full-length PKR (Fig. 6, column 2, row 8); however, this E3 segment interacted weakly or not at all with two kinase domain segments (columns 5 and 6, row 8). We could not test the effect of the W66A mutation on interaction between the N-terminal half of E3 and full-length PKR because the BD–E3-Δ105-190,W66A fusion activated transcription in the absence of any AD fusion (data not shown). Full-length E3 failed to interact with the kinase domain segments; however, while this work was in progress, Sharp et al. found that full-length E3 fused to the GAL4 AD interacted with the PKR kinase domain segment Δ1–366 fused to the GAL4 BD (42). Thus, the two-hybrid assay provides some evidence that the N-terminal half of E3 can interact with PKR.
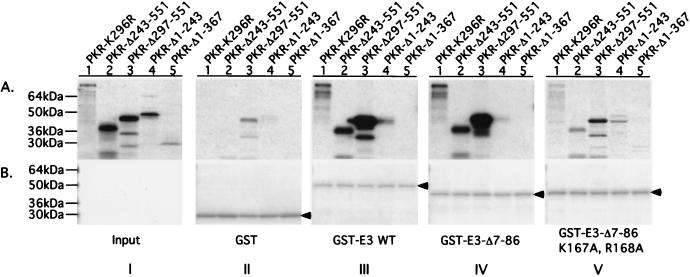
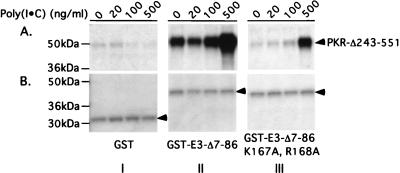
We sought to confirm the two-hybrid interactions between different segments of PKR and E3 by in vitro binding assays using recombinant proteins. Selected E3 segments were expressed in E. coli as GST fusion proteins, and bacterial cell extracts containing the fusions were incubated with radiolabeled PKR polypeptides synthesized by in vitro translation. The GST-E3 proteins were precipitated with glutathione-Sepharose beads, and the bound PKR proteins were analyzed by SDS-PAGE and autoradiography (GST pull-down assays). Full-length PKR and PKR peptides containing the DRBMs bound to GST fusions containing either full-length E3 or the C-terminal half of the protein bearing the DRBM (Fig. 7, III and IV, lanes 1 to 3). Binding of the PKR polypeptides to GST–E3-Δ7-86 was greatly reduced by the K167A and R168A substitutions in the E3 DRBM (IV and V, lanes 1 to 3). The two PKR fragments containing only the protein kinase domain (PKR-Δ1-243 and PKR-Δ1-367) showed little or no binding to these three GST-E3 fusions (I to V, lanes 4 and 5). Although binding of the PKR-Δ243-551 polypeptide occurred without addition of dsRNA to the reactions, we found that its interaction with GST–E3-Δ7-86 (and even with the E3 derivative containing the K167A and R168A substitutions) was stimulated by addition of poly(I-C) at 500 ng/ml (Fig. 8). Apparently, the K167A and R168A substitutions greatly diminish but do not completely abolish E3 dsRNA binding activity. These last results support the idea that complex formation between segments of PKR and E3 containing their DRBMs is dependent on dsRNA binding by E3.
FIG. 7.
Analysis of in vitro interactions between segments of PKR and E3 containing their respective DRBMs. Aliquots of bacterial extracts containing 20 to 100 μg of total protein predetermined to contain similar amounts of GST or GST-E3 fusion proteins (as indicated at bottom of panel B, sections II to V) were combined with an extract prepared from the parental bacterial strain devoid of GST proteins to achieve 200 μg of total bacterial protein. These mixtures were incubated with the 35S-labeled PKR proteins indicated at the top of panel A, section I, and the GST or GST-E3 fusion proteins, along with any bound 35S-labeled PKR proteins, were precipitated by using glutathione-agarose beads and resolved by SDS-PAGE. The 35S-labeled proteins were visualized by autoradiography (A, sections II to V), and the GST or GST-E3 fusion proteins were visualized by Coomassie blue staining (B, sections II to V). Section I in panel A, shows 1/20 (lanes 1 to 4) or 1/10 (lane 5) of the input amounts of 35S-labeled PKR proteins used in the binding assays depicted in sections II to V. Arrowheads in panel B identify GST or the relevant GST-E3 fusion protein.
FIG. 8.
dsRNA binding is required for strong interaction between the DRBM-containing segments of E3 and PKR in vitro. Binding reactions between 35S-labeled PKR-Δ243-551 and either GST (I), GST–E3-Δ7-86 (II), or GST–E3-Δ7-86,K167A,R168A (III) were carried out exactly as described for Fig. 7 except that poly(I-C) was added to the reactions at the final concentrations shown above the lanes in panel A. The 35S-labeled PKR-Δ243-551 proteins precipitated with GST or the GST-E3 fusions were visualized by autoradiography (A), and the precipitated GST or GST-E3 fusion proteins were visualized by Coomassie blue staining (B).
In contrast with both the two-hybrid and coimmunoprecipitation results, removing the N-terminal domain of E3 (in GST–E3-Δ7-86) did not detectably impair its interaction with full-length PKR (Fig. 7, III and IV, lanes 1 to 3). Moreover, we did not observe a significant interaction between the N-terminal half of E3 and full-length PKR in these in vitro binding experiments (data not shown). To explain this last result, it could be proposed that the E3 N-terminal fragment does not fold properly or lacks an important posttranslational modification when expressed in bacterial cells, or that the GST moiety interfered with its PKR binding activity. Alternatively, the rate of dissociation of this particular PKR-E3 complex may too great to allow its detection by the GST pull-down technique, which involves washing immobilized complexes with large volumes of buffer.
To obtain corroborative evidence for interaction between the N-terminal half of E3 and PKR, we turned to the λ repressor dimerization assay for protein-protein interactions (23). In this system, the protein segments of interest are expressed in E. coli as fusions to the N-terminal domain of λ cI repressor, which contains the DNA binding domain but lacks the dimerization domain of cI. Interaction between the protein segments under study mediates dimerization of the λN fusion proteins, leading to repression of the λ pR promoter that can be assayed in several ways. With this technique, it was shown that PKR contains a dimerization domain located between the DRBMs and kinase subdomain II (residues 244 to 296) in addition to the previously identified dimerization domain in the N-terminal half of PKR (residues 1 to 167) (48). Moreover, dimerization of full-length PKR in this assay was disrupted by coexpression of a GST fusion to P58IPK protein, a cellular inhibitor of PKR active during influenza virus infections that binds PKR residues 244 to 296 (19). These findings provided evidence that P58IPK inhibits PKR function, at least partly, by interfering with PKR dimerization through residues 244 to 296 (48).
We used the same approach to determine whether the N-terminal half of E3 can interact with the kinase domain of PKR. PKR residues 244 to 551, containing the complete kinase domain, can mediate dimerization in the λ repressor dimerization assay (48). We found that coexpressing full-length GST-E3 or the GST fusion containing the N-terminal half of E3 (GST–E3-Δ105-190), but not the W66A derivatives of these GST-E3 proteins, blocked dimerization by the λN-PKR fusion containing kinase domain residues 244 to 551 (λN–PKR-Δ1-243) (Fig. 9, column 1, rows 2, 3, 7, and 8). As expected, inactivating the dsRNA binding activity of full-length GST-E3 by the K167A and R168A mutations had no effect on its ability to disrupt λN–PKR-Δ1-243 dimerization. Moreover, the DRBM half of E3 (GST–E3-Δ7-86) had no effect on dimerization of this λN-PKR kinase domain fusion (column 1, rows 4, 5, and 6). These findings suggest that the N-terminal half of E3 can interact with the C-terminal half of PKR and prevent dimerization via PKR residues 244 to 296, independent of dsRNA binding by the E3 protein.
FIG. 9.
Summary of inhibition of PKR dimerization by E3 segments in the λ repressor dimerization assay. E. coli AG1688 was cotransformed with pC168-derived plasmids encoding λN fused with the indicated PKR proteins and with p2645-derived plasmids encoding GST alone or GST fused with the indicated E3 proteins. Cotransformants were tested for immunity to superinfection by λKH54, a mutant lacking the ability to synthesize its own repressor, by using a dot plaque assay. GST alone had no or minimal effect on λN-PKR fusion dimerization-induced resistance to λ superinfection (−). Coexpression of a GST-E3 fusion that reduced resistance to λ superinfection by at least 10-fold (+) or 100-fold (2+) was scored as disruption of λN-PKR fusion dimerization. DRBD, dsRNA binding domain; PK, protein kinase domain.
Quite different results were obtained with the full-length λN–PKR-K296R fusion in the dimerization assay. In this case, the GST fusions containing full-length E3 or the DRBM half, but not the N-terminal half, of E3 blocked dimerization by λN–PKR-K296R in a manner dependent on residues K167 and R168 in the E3 DRBM (Fig. 9, column 2). In addition, the W66A mutation had no effect on the ability of full-length GST-E3 to interfere with λN–PKR-K296R dimerization. These findings suggest that the DRBM-bearing portion of E3 is required to disrupt dimerization mediated by the DRBM-containing segment of PKR and, as concluded above, that the interaction between DRBM-bearing segments of E3 and PKR is dependent on dsRNA binding by E3.
The fact that the N-terminal half of E3 is not necessary or sufficient to disrupt dimerization by full-length PKR in this assay could indicate that the DRBM-containing region of PKR can mediate dimerization even when the self-interaction of PKR residues 244 to 296 is blocked by binding of the N-terminal half of E3 to the kinase domain. This contrasts with the finding that binding of P58IPK to residues 244 to 296 is sufficient to disrupt dimerization by full-length PKR (48). It is possible, however, that the isolated E3 N-terminal half can bind to PKR residues 244 to 551 without impeding self-interactions in the N-terminal region of PKR, whereas binding of P58IPK to the 244 to 296 region impairs self-interactions by both PKR dimerization domains. Our results also suggest that self-interactions by PKR residues 244 to 296 cannot mediate dimerization by full-length PKR when the DRBM half of E3 is bound to the N-terminal region of PKR containing the DRBMs, despite the fact that residues 244 to 296 are sufficient for dimerization of the isolated C-terminal half of PKR. Perhaps binding of the E3 DRBM to the N-terminal region of PKR sterically blocks self-interactions between PKR residues 244 to 296. Finally, none of the GST-E3 proteins could prevent dimerization by a λN-PKR fusion containing only residues 244 to 296 (data not shown), indicating that the N-terminal half of E3 requires residues in the kinase domain, located C terminally to position 296, in order to bind to PKR and block dimerization by residues 244 to 296. This last finding is in agreement with recent results discussed below indicating that E3 makes contact with the C-terminal lobe of the PKR kinase domain (42).
E3 blocks ribosome binding by PKR, but this activity is insufficient to inhibit PKR function.
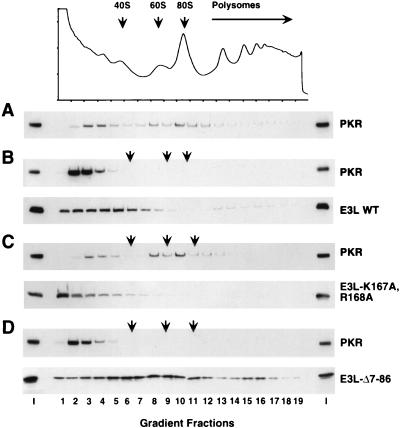
It was reported that the DRBMs in PKR mediate a stable interaction with yeast ribosomes and that the ribosome targeting of PKR could be an important aspect of the requirement for DRBMs to achieve high-level phosphorylation of eIF2α in yeast cells (54). Accordingly, we asked whether coexpression of E3 would interfere with ribosome binding by PKR. In the absence of E3 expression, a substantial fraction of PKR in the cell extract cosedimented through sucrose gradients with 40S and 60S ribosomal subunits, 80S ribosomes, and polysomes (Fig. 10A), in agreement with previous findings (54). Coexpression of wild-type E3 shifted the distribution of PKR from the ribosomal particles to the top of the gradient. A minor fraction of the total E3 cosedimented with 40S subunits and polysomes (Fig. 10B). Interestingly, coexpression of E3-K167A,R168A did not alter the ribosome association of PKR, and a smaller proportion of this mutant E3 protein than of wild-type E3 cosedimented with 40S subunits (Fig. 10C). These findings suggest that displacement of PKR from ribosomes is dependent on the dsRNA binding activity of E3. This could be accounted for by proposing that E3 competes with PKR for dsRNA binding sites in rRNA. Alternatively, since complex formation between E3 and PKR requires the dsRNA binding activity of E3, displacement of PKR from the ribosomes could depend on complex formation with E3.
FIG. 10.
Expression of E3 displaces PKR from ribosomes in a manner that depends on the dsRNA binding activity of E3 but not its N-terminal domain. Transformants of strain J82 (expressing eIF2α-S51A) containing PKR plasmid p1545 and either a plasmid encoding wild-type E3 (p2245), E3-K167A,R168A (p2612), or E3-Δ7-86 (p2246) (B to D) or the empty vector p1079 (A) were grown in SGal medium to an OD600 of ≈1.5. Whole-cell extracts prepared in the presence of 50 mg of cycloheximide per ml and 10 mM MgCl2 were resolved by velocity sedimentation on 5 to 47% sucrose gradients. The gradients were fractionated, and absorbance at 254 nm was recorded to determine the position of the free 40S and 60S subunits, 80S monosomes, and polysomes (indicated by arrows). The OD254 absorbance profile is shown for the gradient analyzed in panel A; the OD254 profiles for panels B to D were essentially identical to that shown in panel A. The distribution of proteins along the gradients was visualized by SDS-PAGE and immunoblot analysis. The first and last lanes in each panel were loaded with 1/20 of the input (I) extracts applied to the gradients.
Expression of the E3-Δ7-86 mutant led to complete displacement of PKR from the ribosomes and showed even greater association with ribosomes than did wild-type E3 (Fig. 10D). It has been shown that the N-terminal domain of E3 promotes oligomerization of E3 in high-molecular-weight complexes (22). Perhaps in lacking the ability to oligomerize, E3-Δ7-86 can interact more stably with ribosomes. The fact that expression of E3-Δ-7-86 displaced PKR from ribosomes, but did not detectably decrease eIF2α phosphorylation by PKR, suggests that ribosome binding is not essential for high-level phosphorylation of eIF2α by PKR in yeast and that displacement of PKR from ribosomes is not the sole function of E3 required for inhibiting PKR. We propose that binding of the N-terminal half of E3 to the kinase domain of PKR is also crucial for preventing eIF2α phosphorylation by PKR.
DISCUSSION
It is generally thought that E3 inhibits PKR solely by sequestering dsRNA molecules required for activation of PKR function. We questioned whether this was its only mode of action after finding that E3 could inhibit PKR in yeast, because previous observations with dominant-negative PKR alleles had suggested that dsRNA activators are very abundant in yeast cells (40). We considered an alternative possibility in which the complete inhibition of PKR by E3 would additionally require complex formation by the two proteins. Consistent with this possibility, E3 inhibitory activity was reversed by coexpressing the mutant PKR-K296R protein along with wild-type PKR (Fig. 1B and C). If E3 inhibited PKR in yeast solely by sequestering dsRNA, then overexpression of PKR-K296R should enhance rather than antagonize E3 by further reducing the levels of free dsRNA. Instead, we propose that overexpression of PKR-K296R displaced E3 from wild-type PKR, forming inactive PKR-K296R-E3 complexes and partially functional PKR/PKR-K296R dimers.
Additional evidence for a second mode of E3 function came from the fact that mutations in the N-terminal half of E3, including a single alanine substitution at Trp-66, led to complete loss of its anti-PKR function in yeast. Chang and Jacobs reported that deletion of the N-terminal 83 residues of E3 did not affect its ability to bind dsRNA in vitro (8). Ho and Shuman showed that the apparent dissociation constants for dsRNA bound by full-length E3 and a fragment containing only the C-terminal 90 residues were virtually identical (22), indicating that the N-terminal half of E3 makes no contribution to the binding affinity of E3 for dsRNA in vitro. We found that the K167A,R168A double mutation in the E3 DRBM abolished in vitro binding of GST–E3-Δ7-86 to PKR, indicating that dsRNA binding by E3 is required for its interaction with PKR. In contrast, the Δ7-86 mutation in the N-terminal half of E3 had no effect on GST-E3 binding to PKR (Fig. 7), implying that the Δ7-86 mutation does not reduce dsRNA binding by E3 in vitro. The same conclusion can be drawn from the fact that coimmunoprecipitation of PKR with E3 was abolished by the K167A and R168A mutations but unaffected by the W66A mutation in E3, indicating that the latter mutation did not impair dsRNA binding by E3 in vivo (Fig. 5). Similarly, the W66A, Δ60-82, and Δ7-86 mutations did not impair the in vivo interaction of E3 with PKR-Δ243-551 in the two-hybrid assay, whereas E3-K167A,R168A failed to interact with this PKR segment. Combined with the previous results of Ho and Shuman, these observations indicate that the N-terminal half of E3, and Trp-66 in particular, does not contribute to dsRNA binding in vitro or in vivo and thus is involved in a distinct aspect of E3 anti-PKR function.
Using four different assays for protein-protein interactions, we obtained strong evidence for heterocomplex formation by PKR and E3. The assay of greatest physiological relevance, coimmunoprecipitation from yeast extracts, suggested that a majority of the PKR molecules in yeast were physically associated with E3 in a manner dependent on the dsRNA binding activity of E3. The yeast two-hybrid and GST pull-down assays revealed complex formation between the C-terminal half of E3 and the N-terminal half of PKR, the segments containing their DRBMs, and this interaction also depended on dsRNA binding by the E3 partner. These observations, plus the fact that binding between PKR-Δ243-551 and E3-Δ7-86,K167A,R168A was rescued by high concentrations of dsRNA, indicated that complex formation is critically dependent on the DRBM-containing segments of both proteins and dsRNA binding by E3. Because the N-terminal half of E3 was not essential for coimmunoprecipitation of PKR with E3, we suggest that protein-protein contacts involving the DRBM-containing segments, plus mutual binding to the same dsRNA molecules, make the most important contributions to the stability of these E3-PKR-dsRNA complexes in vivo (Fig. 11). Similar protein contacts via DRBMs have been proposed to explain dimerization by PKR N-terminal segments, and heterocomplex formation by PKR and TRBP, in cases where the DRBMs contained point mutations that abolish dsRNA binding in vitro (4, 12, 35, 37, 38, 50). In addition, the purified E3 DRBM dimerizes in solution (22), and purified segments containing the DRBMs of PKR (41) or E3 (22) each show cooperative binding to dsRNA in vitro, indicating the existence of protein-protein interactions by DRBMs bound to the same dsRNA molecules.
FIG. 11.
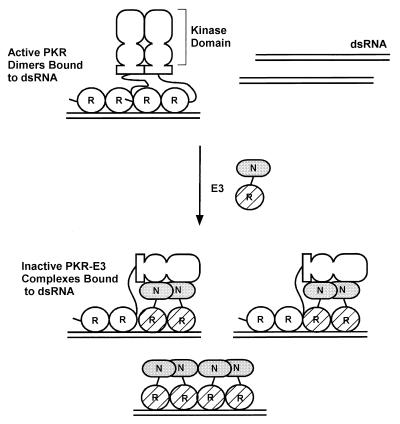
Hypothetical model for inhibition of PKR function by E3 through the formation of inactive heteromeric complexes. PKR is shown schematically with its two DRBMs (R) connected by a linker to the dimerization domain located between residues 244 and 296 (shown as a rectangle) and the N-terminal and C-terminal lobes of the kinase domain (depicted as two ovals). E3 is depicted with its single DRBM (R) hatched and the N-terminal domain (N) shaded. The active form of PKR is depicted as a dimer bound to dsRNA (28, 31, 40), with dimerization mediated by interactions involving the N-terminal region containing the DRBMs (12, 37, 48, 50), the kinase domain (37, 40), and the region from residues 244 to 296 (48) and by binding to the same dsRNA molecule (12, 37, 51). E3 is shown inhibiting PKR by forming inactive heterocomplexes, disrupting PKR homodimers. In addition, the N-terminal domain of E3 is shown interacting with the kinase domain of PKR, interfering with some aspect of kinase function. Binding to dsRNA by E3 greatly contributes to the stability of the PKR-E3 complex. E3 can also inhibit kinase activation by sequestering dsRNA molecules. See text for details.
The fact that coimmunoprecipitation of PKR with E3 was dependent on Lys-167 and Arg-168 in the E3 DRBM suggests that PKR resides in heteromeric complexes containing both E3 and dsRNA. This seems at odds with the dsRNA sequestration model, in which E3 prevents PKR from interacting with dsRNA, and more consistent with the notion that E3 inhibits PKR via heterocomplex formation. It could be argued that E3 and PKR do not directly interact with one another in these complexes but simply bind independently to the same dsRNA molecules. To explain our coimmunoprecipitation results by this hypothesis, the numbers of dsRNA and E3 molecules would have to be nearly equivalent in yeast cells. If dsRNA molecules were in large molar excess of E3, then PKR would most frequently bind to dsRNA molecules lacking E3. If E3 was in large molar excess of dsRNA, it would compete with PKR for limited dsRNA binding sites (as suggested by the dsRNA sequestration model). In either case, most of the PKR would not be physically linked with E3. It is conceivable that the dsRNAs are long enough to accommodate multiple protein molecules and that the prevailing E3/PKR ratio is such that most of the PKR is bound to dsRNAs containing at least one molecule of E3 without postulating protein-protein contacts between E3 and PKR. Even if this were true, however, it would still be necessary to propose a second function for E3 besides sequestration of dsRNA to explain why the PKR molecules bound to dsRNA in heteromeric complexes with E3 are catalytically inactive.
Another argument in favor of protein-protein contacts between E3 and PKR comes from the fact that expression of E3 displaced PKR from ribosomes even though it appeared that most of the wild-type E3 was not stably bound to ribosomes (Fig. 10B). If E3 was competing with PKR for dsRNA binding sites in rRNA, one would expect to find a large fraction of E3 associated with ribosomes. A possible objection to this interpretation could be that E3 was produced in large excess of the ribosomes, such that all dsRNA binding sites in rRNA would be bound by E3 even though most of the E3 was nonribosomal. This seems unlikely considering that a greater fraction of E3-Δ7-86 than of wild-type E3 was bound to ribosomes (Fig. 10B and D), with no differences in the overall levels of these two proteins (Fig. 2C). We suggest that E3-PKR heterocomplexes interact with ribosomes less efficiently than do PKR monomers or homodimers, although the molecular explanation for this difference in binding remains to be elucidated.
Results from coimmunoprecipitation and two-hybrid assays indicating that deletion of the N-terminal half of E3 decreased the yield of E3-PKR complexes provided evidence for E3-PKR interactions involving the N-terminal half of E3 and PKR. Direct interaction of the isolated N-terminal half of E3 with PKR was detected in both the yeast two-hybrid and λ dimerization assays. Results of the latter experiments localized the interaction to the kinase domain of PKR and revealed a dependence on Trp-66, the E3 residue critically required for its anti-PKR function in yeast. Based on these findings, we suggest that the N-terminal half of E3 binds to the PKR kinase domain in a manner that interferes with kinase activation or catalysis and is tethered to PKR through interactions between their DRBMs and mutual binding to the same dsRNA molecules (Fig. 11). From the results of the λ dimerization assays, it could be proposed that the N-terminal half of E3 impedes dimerization of PKR via segment 244–296, as suggested previously for P58IPK (48). Interestingly, Trp-66 resides within the largest stretch of amino acids shared between E3 and the parapoxvirus orf virus OV20.0L gene product, a DRBM-containing PKR inhibitor that is 31% identical to E3 (21). Thus, the importance of the N-terminal half of E3 for inhibition of PKR may be a conserved feature of these related proteins. Our model for E3 action (Fig. 11) combines mechanisms of PKR inhibitors which bind to the kinase domain, including hepatitis c virus NS5A (20), baculovirus Autographa californica PK2 (17), P58IPK, and vaccinia virus K3 protein, with that of TRBP, which forms heterodimers with PKR through interactions between their respective DRBMs (4). K3, also encoded by vaccinia virus, functions as a pseudosubstrate inhibitor by binding to PKR (5, 13, 27) between kinase subdomains VI and XI (12, 13, 19).
We could not confirm a physical interaction between the N-terminal half of E3 and PKR in the GST pull-down assays. Moreover, the N-terminal half of E3 interacted with full-length PKR but not with the isolated kinase domain in the yeast two-hybrid experiments. Finally, in the λ dimerization assay, the N-terminal half of E3 was dispensable for the interaction between E3 and full-length PKR even though it interacted with the isolated kinase domain in this assay. These discrepancies concerning interactions of the E3 N-terminal half and the PKR kinase domain could reflect a high off rate for this interaction; alternatively, it might be impaired by juxtaposition of E3 with GST or the GAL4 BD in certain protein fusions where it failed to interact with PKR. In agreement with the latter suggestion, Sharp et al. observed a significant interaction between full-length E3 and PKR kinase domain segment 367–551 in the two-hybrid assay when these segments were fused to the GAL4 AD and BD, respectively, but not when the fusions were constructed vice versa (42). They also reported that radiolabeled full-length E3 bound to the PKR kinase domain segment 242–551, but not to the larger PKR segment 99–551. Interestingly, the interaction between E3 and PKR segment 242–551 was competed with unlabeled E3, eIF2α, or K3 proteins (42). These last findings imply that E3 and K3 have overlapping binding sites in the C-terminal lobe of the PKR kinase domain.
E3 is localized in both the nucleus and the cytoplasm of vaccinia virus-infected cells (53), and nuclear localization is dependent on its N-terminal domain (9). This finding led us to consider that the N-terminal half of E3 might function by sequestration of E3-PKR heterodimers in the nucleus. Using indirect immunofluorescence and monoclonal antibodies against PKR and HA-tagged E3, we found that both proteins were localized in the cytoplasm whether expressed individually or coexpressed in the same yeast cells (data not shown). Thus, we have no evidence that E3 inhibits the ability of PKR to phosphorylate eIF2α by sequestering it in the nucleus. It is conceivable, however, that the N-terminal half of E3 leads to sequestration of PKR in cytoplasmic aggregates where it cannot interact efficiently with eIF2.
The previous finding that the ability of E3 to inhibit PKR in cell extracts could be partially reversed by adding large amounts of dsRNA (1, 14, 26) was an important observation indicating that E3 inhibits PKR by sequestering dsRNA activators. Can this result be reconciled with our proposal that E3 inhibits PKR by heterocomplex formation? One possibility is that an E3 dimer is required to interact with each PKR monomer to prevent PKR dimerization (Fig. 11), and that E3 dimers would be dissociated (through binding to separate dsRNA molecules) at dsRNA concentrations lower than required to dissociate PKR dimers. In accordance with the latter stipulation, certain mutations in the PKR DRBMs that abolish dsRNA binding activity do not eliminate dimer formation (12, 35, 37, 38, 50), whereas we showed that E3 must bind dsRNA to interact stably with PKR. Thus, the protein-protein contacts may be stronger or more extensive in a PKR dimer than in an E3 dimer.
Deletion of the N-terminal 83 amino acids of E3 did not affect its ability to confer upon vaccinia virus interferon resistance during infections of RK-13 cells and the ability to replicate in HeLa cells (45). These findings suggest that inhibition of PKR by E3 in these cells requires only sequestration of dsRNA activators. There is evidence, cited above, that E3 can prevent PKR activation in vitro by sequestering dsRNA activators. Because the DRBMs in E3 and PKR bind dsRNA with comparable affinities (22, 41), the dsRNA sequestration mechanism requires that E3 be produced in large molar excess of both PKR and dsRNA activators. Presumably, these conditions were satisfied in RK-13 and HeLa cells, where a vaccinia virus mutant lacking the N-terminal domain of E3 could prevent PKR activation (45). It is possible that these conditions were not met in our yeast strains because they contain endogenous dsRNAs in considerable excess of the E3 protein being produced. Consequently, the E3 DRBM alone could not prevent PKR activation, and the N-terminal domain was additionally required for full inhibition of PKR function in the context of E3-PKR heterocomplexes. This latter mechanism could be important during viral infections when the concentration of dsRNA is very high, allowing E3 to block PKR activation even when it cannot completely prevent the occurrence of some free dsRNA. Another possibility is that the N-terminal half of E3 is required to prevent PKR activation by a dsRNA-independent mechanism that might operate in yeast and, presumably, in certain mammalian cells. At least in yeast, this would require that the PKRDRBMs are needed primarily for dimerization rather than dsRNA binding. Based on the findings in this paper, it will be interesting to examine the effects of N-terminal mutations in E3 on virus propagation in different cell types and during a systemic infection of the natural animal host.
ACKNOWLEDGMENTS
We thank Tyson Sharp and Rosemary Jagus for extensive discussion of results prior to publication and Bobbie Felix for help in preparation of the manuscript.
REFERENCES
- 1.Akkaraju G R, Whitaker-Dowling P, Younger I S, Jagus R. Vaccinia specific kinase inhibitory factor prevents translational inhibition by double-stranded RNA in rabbit reticulocyte lysate. J Biol Chem. 1989;264:10321–10325. [PubMed] [Google Scholar]
- 2.Beattie E, Denzler K L, Tartaglia J, Perkus M E, Paoletti E, Jacobs B L. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie E, Paoletti E, Tartaglia J. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology. 1995;210:254–263. doi: 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- 4.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Carpick B W, Graziano V, Schneider D, Maitra R K, Lee X, Williams B R G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-1 trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 5.Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2α-specific protein kinase. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- 6.Cesareni G, Murray J A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow J K, Hollaender A, editors. Genetic engineering: principals and methods. Vol. 9. New York, N.Y: Plenum Press; 1987. pp. 135–154. [Google Scholar]
- 7.Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang H-W, Jacobs B L. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 9.Chang H-W, Uribe L H, Jacobs B L. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J Virol. 1995;69:6605–6608. doi: 10.1128/jvi.69.10.6605-6608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong K L, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R G. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 139–172. [Google Scholar]
- 12.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig A W B, Cosentino G P, Donze O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 14.Davies M V, Chang H W, Jacobs B L, Kaufman R J. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dever T E, Chen J J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T D, Hinnebusch A G. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 17.Dever T E, Sripriya R, McLachlin J R, Lu J, Fabian J R, Kimball S R, Miller L K. Disruption of cellular translational control by a viral truncated eukaryotic translation initiation factor 2α kinase homolog. Proc Natl Acad Sci USA. 1998;95:4164–4169. doi: 10.1073/pnas.95.8.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale M, Tan S-L, Wambach M, Katze M G. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins P58IPK and vaccinia virus K3L is mediated by unique domains: implications for kinase regulation. Mol Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale M J, Korth M J, Tang N M, Tab S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 21.Haig D M, McInnes C J, Thomson J, Wood A, Bunyan K, Mercer A. The orf virus OV20.0L gene product is involved in interferon resistance and inhibits an interferon-inducible, double-stranded RNA-dependent kinase. Immunology. 1998;93:335–340. doi: 10.1046/j.1365-2567.1998.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho C K, Shuman S. Physical and functional characterization of the double-stranded RNA binding protein encoded by the vaccinia virus E3 gene. Virology. 1996;217:272–284. doi: 10.1006/viro.1996.0114. [DOI] [PubMed] [Google Scholar]
- 23.Hu J C, O’Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 24.Hunter T, Hunt T, Jackson R J, Robertson H D. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 25.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jagus R, Gray M M. Proteins that interact with PKR. Biochimie. 1994;76:779–791. doi: 10.1016/0300-9084(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 27.Kawagishi-Kobayashi M, Silverman J B, Ung T K, Dever T E. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostura M, Mathews M B. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Langland J O, Jacobs B L. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr = 66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 32.Langland J O, Pettiford S M, Jiang B, Jacobs B L. Products of the porcine N5P3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase, PKR. J Virol. 1994;68:3821–3829. doi: 10.1128/jvi.68.6.3821-3829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews M B. Viral evasion of cellular defense mechanisms: regulation of the protein kinase DAI by RNA effectors. Semin Virol. 1993;4:247–257. [Google Scholar]
- 34.McMillan N A, Carpick B W, Hollis B, Toone W M, Zamanian-Daryoush M, Williams B R G. Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J Biol Chem. 1995;270:2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- 35.Ortega L G, McCotter M D, Henry G L, McCormack S J, Thomis D C, Samuel C E. Mechanism of interferon action. Biochemical and genetic evidence for the intermolecular association of the RNA-dependent protein kinase PkR from human cells. Virology. 1996;215:31–39. doi: 10.1006/viro.1996.0004. [DOI] [PubMed] [Google Scholar]
- 36.Park H, Davies M V, Langland J O, Chang H W, Nam Y S, Tartaglia J, Paoletti E, Jacobs B L, Kaufman R J, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel R C, Stanton P, McMillan N M J, Williams B R G, Sen G C. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 39.Pavitt G D, Yang W, Hinnebusch A G. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romano P R, Green S R, Barber G N, Mathews M B, Hinnebusch A G. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmedt C, Green S R, Manche L, Taylor D R, Ma Y, Mathews M B. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 42.Sharp, T. V., A. Romashko, F. Moonan, B. Joshi, G. N. Barber, and R. Jagus. The vaccinia virus E3L gene product interacts with both the regulatory and substrate binding regions of PKR: implications for PKR autoregulation. Virology, in press. [DOI] [PubMed]
- 43.Sherman F, Fink G R, Lawrence C W. Methods of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 44.Shors T, Jacobs B L. Complementation of deletion of the vaccinia virus E3L gene by the Escherichia coli RNase III gene. Virology. 1997;227:77–87. doi: 10.1006/viro.1996.8319. [DOI] [PubMed] [Google Scholar]
- 45.Shors T, Kibler K V, Perkins K B, Seidler-Wulff R, Banaszak M P, Jacobs B L. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology. 1997;239:269–272. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan S L, Gale M J, Katze M G. Double-stranded RNA-independent dimerization of the interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson J C, Chang H W, Jacobs B L. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- 50.Wu S, Kaufman R J. Double-stranded (ds) RNA binding and not dimerization correlates with the activation of the dsRNA-dependent protein kinase (PKR) J Biol Chem. 1996;271:1756–1763. doi: 10.1074/jbc.271.3.1756. [DOI] [PubMed] [Google Scholar]
- 51.Wu S, Kaufman R J. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 52.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1988;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuwen H, Cox J H, Yewdell J W, Bennink J R, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 54.Zhu S, Romano P R, Wek R C. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J Biol Chem. 1997;272:14434–14441. doi: 10.1074/jbc.272.22.14434. [DOI] [PubMed] [Google Scholar]