Abstract
Cell type control of meiotic gene regulation in the budding yeast Saccharomyces cerevisiae is mediated by a cascade of transcriptional repressors, a1-α2 and Rme1. Here, we investigate the analogous regulatory pathway in the fission yeast Schizosaccharomyces pombe by analyzing the promoter of mei3, the single gene whose expression is sufficient to trigger meiosis. The mei3 promoter does not appear to contain a negative regulatory element that represses transcription in haploid cells. Instead, correct regulation of mei3 transcription depends on a complex promoter that contains at least five positive elements upstream of the TATA sequence. These elements synergistically activate mei3 transcription, thereby constituting an on-off switch for the meiosis pathway. Element C is a large region containing multiple sequences that resemble binding sites for Mc, an HMG domain protein encoded by the mating-type locus. The function of element C is extremely sensitive to spacing changes but not to linker-scanning mutations, suggesting the possibility that Mc functions as an architectural transcription factor. Altered-specificity experiments indicate that element D interacts with Pm, a homeodomain protein encoded by the mating-type locus. This indicates that Pm functions as a direct activator of the meiosis pathway, whereas the homologous mating-type protein in S. cerevisiae (α2) functions as a repressor. Thus, despite the strong similarities between the mating-type loci of S. cerevisiae and S. pombe, the regulatory logic that governs the tight control of the key meiosis-inducing genes in these organisms is completely different.
The differentiation of a diploid somatic cell into haploid sex cells via a meiotic cell cycle is a hallmark of eukaryotic organisms. Gene regulation in haploid and diploid cells is tightly regulated, although in distinct manners. Haploids need tight control of the intracellular signals and genes that trigger a meiotic cell cycle, because meiosis is a lethal event. In contrast, diploids induce a meiotic cell cycle only upon an appropriate environmental signal(s) or a developmental program.
The meiotic regulatory cascade has been well studied in the budding and fission yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. In both organisms, there is a complex pathway that starts from the mating-type loci and involves meiosis-specific genes and other genes that have multiple cellular roles (20, 29). Both organisms contain three copies of mating-type cassettes; one of these is active and two are silent. There are two mating-type alleles (a and α for S. cerevisiae and P and M for S. pombe), and each cassette expresses two distinct proteins. Finally, two of the mating-type proteins, S. cerevisiae α2 and S. pombe Pm, contain homeodomains that are 41% identical and are likely to recognize similar DNA sequences (15, 28).
In S. cerevisiae, cell type control of meiotic gene regulation is mediated by transcriptional repression (20). Diploid a/α cells specifically express the a1-α2 heterodimer that directly represses transcription of RME1 by binding to its promoter (5). Rme1 directly represses transcription of IME1, the key gene whose product directly activates a set of genes that are required for meiosis (6). Although IME1 transcription is necessary for meiosis, it is not sufficient to trigger a meiotic cell cycle without other cellular signals (13, 23). In starved haploid cells, IME1 transcription is toxic but not lethal (21). Thus, the S. cerevisiae mating-type loci activate IME1 transcription and the meiotic pathway by an indirect mechanism, i.e., by repressing a repressor.
Meiotic regulation in S. pombe can be reduced to the transcriptional status of just one gene, mei3 (28). Transcription of mei3 is required for meiosis and is sufficient to trigger meiosis independently of ploidy and nutritional stress (19). In wild-type cells, mei3 is transcribed only transiently in nitrogen-starved diploids (19). Mei3 is not homologous to Ime1 (23), and it mediates its effect on meiosis by a different mechanism. Mei3 binds to the Pat1 (Ran1) kinase and blocks its autophosphorylation and the phosphorylation of Ste11 and Mei2, the physiological substrates (17, 25). Pat1 kinase activity is required for a mitotic cell cycle; the lack of Pat1 kinase activity leads to a meiotic cell cycle independently of ploidy and nutritional stress. Because Mei3 is necessary and sufficient to induce meiosis, it is critical that mei3 transcription is tightly controlled in haploid cells.
Genetic experiments indicate that mei3 is regulated by the coexpression of the four mating-type proteins (29). Two of the mating-type proteins, Mc and Pc, are expressed in vegetatively growing cells, whereas the other two proteins, Mm and Pm, are induced specifically by nitrogen starvation (15). Thus, in wild-type haploids, mei3 is not expressed, due to the lack of either Mm or Pm. Nitrogen-starved diploids in which the pheromone receptor is bound by pheromone are the only cells in which both Mm and Pm are expressed (2). In principle, the mating-type proteins could bind directly to the mei3 promoter and activate transcription. Alternatively, these proteins could indirectly activate mei3 transcription via another, as of yet unidentified, gene(s). By analogy with meiotic regulation in S. cerevisiae, one possibility is that the mating-type proteins directly repress transcription of a currently unidentified repressor which directly represses mei3 transcription.
To understand the molecular mechanisms of mei3 transcriptional regulation, we performed a detailed mutational analysis of the mei3 promoter. We define five positive regulatory elements that synergistically activate and correctly regulate mei3 transcription but obtain no evidence for negative control elements. We demonstrate that one of the critical mei3 promoter elements is a binding site for Pm and that Pm functions as a direct activator of mei3 transcription in vivo. In addition, we suggest that another mei3 promoter element may contain multiple binding sites for Mc. These observations indicate that the underlying logic of meiotic transcriptional regulation in S. pombe is very different from that found in S. cerevisiae. In addition, the analysis here represents the first detailed dissection of a promoter from S. pombe.
MATERIALS AND METHODS
DNA molecules.
The starting molecule, pWH4, is a derivative of pBSKS+ (Stratagene) that has a mutated SacI site and contains the 1.78-kb HindIII ura4 fragment cloned between the PstI and SmaI sites and a 1.95-kb HindIII-EcoRI mei3 fragment with a translationally silent mutation of the KpnI site in the coding region. pWH6 is a derivative of pWH4 in which the coding and 3′ untranslated regions of mei3 are replaced by a 1.9-kb SacI-SpeI ade6 fragment containing the coding region (with an engineered SacI site at the ATG initiation codon) and 3′ untranslated region. pWH8 is a derivative of pWH4 in which the mei3 coding region was replaced by a 1.69-kb SacI-PstI fragment containing the firefly luciferase coding region (7) with an engineered SacI site at the ATG initiation codon.
mei3 deletion mutant DNAs were generated by standard methods with exonuclease III or Bal31 nuclease digestion or by PCR mutagenesis. In general, promoter fragments contain KpnI sites at the 5′ end, SacI sites at the 3′ end, and SalI (in most cases) or MluI sites (in some cases) at the junction point. Linker-scanning and base pair substitution mutations were created by PCR-mediated site-directed mutagenesis with appropriate oligonucleotides. Insertion-scanning mutations were created by linearizing linker-scanning derivatives with the linker restriction site, blunting the ends, and religating or by inserting oligonucleotide linkers of appropriate size into the linker restriction site. All mutations were created in pWH4 and were confirmed by DNA sequencing. To generate ade6 and luciferase fusions, Asp718-SacI mei3 promoter fragments were transferred into pWH6 and pWH8, respectively.
To generate altered-specificity derivatives of Pm, an 830-bp DNA fragment that encodes Pm and extends from the endogenous BamHI site 140 bp upstream of the transcription start site to an engineered XbaI site 140 bp downstream of the translational stop codon was generated by PCR amplification of S. pombe genomic DNA and cloned between the BamHI and XbaI sites of pBS-KS. Derivatives of this plasmid containing desired mutations in the putative DNA recognition helix of Pm were generated by site-directed two-step PCR mutagenesis. Wild-type and mutant Pm fragments (generated by cleavage with PstI and Ecl136II) were subcloned between the PstI and SmaI polylinker sites of pAD2 (1), a high-copy-number plasmid with an ade6 selectable marker.
Construction of yeast strains.
S. pombe strains used in this study are listed in Table 1. DNA molecules containing the mutant mei3 promoter derivatives were linearized with StuI and introduced into appropriate ura4 mutant strains, as described previously (4). To confirm proper integration of these DNAs into the ura4 locus, Ura+ strains were analyzed on Southern blots in which SmaI-digested genomic DNA (10 μg) was hybridized with a ura4 probe (32P-labeled 1.78-kb HindIII fragment). The parental strain (or gene convertants) yields a 5.3-kb fragment, whereas correct integrants yield a single fragment of 12 kb (size varies slightly depending on whether the DNA was derived from pWH4, pWH6, or pWH8). Therefore, proper integration could be assayed on a Southern blot by observing a shift of a single 5.3-kb SmaI ura4 fragment to a single SmaI ura4 fragment of approximately 12 kb. Incorrect or multiple integrants contain multiple ura4 fragments of various sizes.
TABLE 1.
S. pombe strains
| Strain | Genotype | Source |
|---|---|---|
| WP3 (SP67) | h90 ade6-210 leu1-32 | S. pombe strain bank |
| WP16 | h90 ade6-210 ura4-595 leu1-32 mei3::LEU2+ | This work |
| WP17 | h+ ade6-D1 ura4-595 leu1-32 | This work |
| WP20 | h90 ade6-210 leu1-32 mei3::LEU2+ ura4-595::mei3+::ura4+ | This work |
| WP45 | h− ade6-D1 ura4-595 pro2-1 | This work |
| WP54 | h+/h− ade6-210/ade6-216 leu1+/leu1-32 (diploid) | This work |
| WP62 | h− ade6-D1 ura4-595::mei3::ade6::ura4+ pro2-1 | This work |
| WP64 | h+ ade6-D1 ura4-595::mei3::ade6::ura4+ leu1-32 | This work |
| WP66 | h− ade6-D1 ura4-595::mei3::ade6::ura4+ leu1-32 mei3::LEU2+ | This work |
| WP151 | h+ pro2-1 ade6-D1 leu1-32 mei3::LEU2+ ura4-595 | This work |
| WP162 | h− ade6-D1 leu1-32 mei3::LEU2+ ura4-(595 or 294) lys1-131 | This work |
| WP356 | h− ade6-D1 leu1-32 lys1-131 mei3::LEU2+ ura4-(595 or 294)::mei3::LUC::ura4+ | This work |
| WP367 | h−/h+ diploid (WP356 × WP151) | This work |
| WP434 | h90 ade6-210 leu1-32 mei3::LEU2+ ura4-595::mei3-d1::ura4+ | This work |
| WP495 | h90 ade6-210 leu1-32 mei3::LEU2+ ura4-595::mei3-d2::ura4+ | This work |
| WP496 | h90 ade6-210 leu1-32 mei3::LEU2+ ura4-595::mei3-d3::ura4+ | This work |
| WP499 | h90 ade6-210 leu1-32 mei3::LEU2+ ura4-595::mei3-d4::ura4+ | This work |
Phenotypic assays.
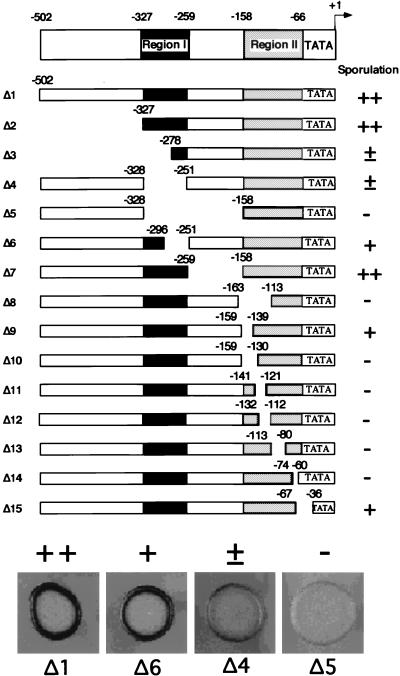
To assay for sporulation, WP16 (h90 leu1-32 ade6-210 ura4-595 mei3::LEU2+) cells containing pWH4 derivatives were grown overnight at 30°C in 2 ml of YEL plus Ade medium, washed once in H2O and once in 150 mM NaCl, and resuspended in the 20-μl residual volume of 150 mM NaCl. Twenty microliters of this cell suspension was spotted on SPA sporulation plates, incubated for 3 days at room temperature, and exposed to iodine vapors for 3 min. Iodine stains the polysaccharides that are found in the ascus walls and in the cell walls of ascospores but not in vegetative cells. Wild-type cells appear as a dark ring at the edges of the spots, representing cells that are in contact with the surface of the plate and which have mated and sporulated. Cells that are not in contact with the plate surface do not receive enough nutrients to mate and sporulate; hence, they are unstained. Sporulation phenotypes are defined as follows: ++, sporulation indistinguishable from the wild type; +, noticeably less sporulation than the wild type; ±, barely detectable sporulation; −, no detectable sporulation (see Fig. 1). Sporulation phenotypes are consistent among independent Ura+ transformants containing the same mei3 promoter construct.
FIG. 1.
Definition of two positive regulatory regions by deletion analysis of the mei3 promoter. For each mei3 deletion mutant, solid bars indicate regions that are present and blank spaces indicate regions that are deleted; nucleotide positions at the boundaries indicate the last residue that is present and are defined with respect to the +1 transcriptional initiation site. Sporulation phenotypes (assessed by iodine staining) conferred by these mei3 derivatives are defined as follows: ++, indistinguishable from a wild-type strain; +, noticeably reduced from wild-type levels; ±, barely detectable; −, no detectable sporulation. Sporulation assays that indicate the phenotypic range are shown for selected mei3 derivatives (Δ1, Δ6, Δ4, and Δ5). The approximate locations of regions I and II are indicated (see text).
The activity of mei3 promoter mutants in nitrogen-sated haploids was assayed by using WP17, WP45, or WP162 (heterothallic ura4 ade6Δ) strains containing pWH6 derivatives (mei3-ade6 fusions). Ade+ phenotypes were tested by the ability of these strains to grow on glucose minimal medium lacking adenine and by their color on rich medium (Ade+ strains are white, whereas Ade− strains are red).
For altered-specificity experiments, pAD2 plasmids expressing wild-type and mutant derivatives of Pm were introduced into WP16 derivatives carrying wild-type or mutant D alleles by selecting for Ade+ colonies. Plasmid-borne Pm derivatives were expressed from the natural promoter and hence presumably only under conditions in which the endogenous Pm was expressed.
Quantitation of mei3 promoter activity by luciferase assays.
pWH8 derivatives integrated into the ura4 locus of WP162 (h− mei3 ura4 lys1) were mated to WP151 (h+ mei3 ura4 pro2) on a sporulation plate overnight, and diploids were subsequently selected on minimal NBA plus 0.01% Ade plates. Diploid (or haploid control) cultures (50 ml) were grown at 30°C in EMM-2 minimal medium lacking lysine and proline to mid-log phase (A600 = 0.7) and then divided into two portions. One portion (35 ml) was washed twice with water, resuspended in 30 ml of EMM-2 containing amino acids but lacking NH4Cl, and incubated at 22°C for 4 h with shaking. The remaining portion was diluted into 45 ml of EMM-2 lacking lysine and proline and grown for 3.5 h at 30°C. After measuring the A600, cultures were washed once with ice-cold H2O, resuspended in 0.9 ml of buffer (100 mM KPO4 [pH 7.8], 1 mM dithiothreitol) on ice, and divided into three samples (approximately 0.4 ml) to which 0.2 ml of acid-washed glass beads were added. Cells were disrupted by vortexing in a multitube vortexer on high at 4°C for 12 min, and insoluble material was removed by microcentrifugation for 5 min at 4°C and placed on ice. Luciferase activity was measured by adding 100 μl of each sample to a glass vial containing 200 μl of assay buffer (25 mM Tricine, 15 mM MgCl2, 5 mM ATP, 500 μg of bovine serum albumin per ml) and 100 μl of 0.5 μM luciferin that had been placed in a luminometer. Photon emission was detected for 10 s, and luciferase activity was normalized to the number of cells. To minimize experimental variation, luciferase activities of the various strains were normalized to that of the strain containing the intact mei3 promoter (502 nucleotides of mei3 promoter sequence) by using samples that were prepared and analyzed in parallel, and assays were carried out on independent occasions. The experimental error is ±30% (except for strains that express very low luciferase activity, where the error is higher).
RESULTS
Defining the minimal mei3 promoter by sequential 5′ deletions.
The mei3 promoter region (defined here as sequences 502 bp upstream of the mRNA initiation site) contains a putative TATA element (TATAAG) located between −33 and −28 with respect to the mapped transcription start site (19). This location is in excellent accord with in vitro transcription reactions reconstituted with S. pombe components (9, 18), and TATAAG is a moderately functional TATA element (27). In the vicinity of the mRNA initiation site, the mei3 promoter contains a sequence (GCATCCA, located between −2 and +5) that weakly resembles a eukaryotic initiator element (consensus PyPyAN[T/A]PyPy) (14).
To assess whether the isolated mei3 promoter (to −502) is sufficient to confer nitrogen-starvation-dependent transcriptional induction, a mei3 segment (−502 to +94) was fused translationally to the luciferase structural gene. A stable diploid strain containing this DNA integrated at the ura4 locus was grown to mid-log phase in nitrogen-rich medium and shifted to medium lacking nitrogen. A peak of luciferase activity was observed 3 h after nitrogen starvation (data not shown), a result in accord with the observation that mei3 mRNA levels are maximal between 2.5 and 4 h after nitrogen starvation (19). Thus, the information necessary for proper mei3 transcription resides in the 502 nucleotides of mei3 promoter and 5′ untranslated region.
To define the minimal mei3 promoter region, mei3 DNAs successively deleted for sequences upstream of the mRNA initiation site were integrated in single copy at the ura4 locus of a mei3 deletion strain (WP16), and the resulting transformants were tested for sporulation efficiency. Deletions that contain 502 and 327 bp upstream of the mRNA initiation site (Δ1 and Δ2, respectively) have sporulation phenotypes indistinguishable from a wild-type strain, whereas a deletion retaining 278 bp (Δ3) has markedly reduced sporulation efficiency (Fig. 1). These results suggest that sequences downstream of −328 are necessary and sufficient to confer proper mei3 expression and that the region between −502 and −327 is not essential.
Crude internal deletions define two positive regulatory regions.
To define elements necessary for mei3 transcription, we analyzed the sporulation phenotypes conferred by a set of internal deletions within the mei3 promoter region. Two separable promoter regions are identified from the crude deletion analysis shown in Fig. 1. Region I (−327 to −259) is defined by the observation that strains containing Δ7 sporulate normally whereas strains containing the more extensive Δ5 fail to sporulate. A deletion that removes much of region I (Δ6) is partially defective for sporulation. For region II (−158 to −66), the distal end is defined by Δ7, which sporulates normally, and the proximal end is loosely defined by Δ15, which sporulates with a reduced efficiency. Most internal deletions within region II are completely unable to sporulate. However, a small deletion at the distal end, Δ9, sporulates with reduced efficiency, suggesting that this derivative only partially disrupts a positive element in region II.
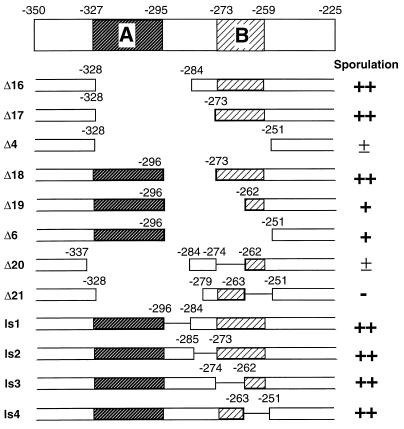
Dissection of region I into elements A and B.
Region I was dissected by smaller internal deletions and linker-scanning mutations (Fig. 2). Elements A and B are defined by nonoverlapping portions of region I that confer increased sporulation over that conferred by Δ4, which is completely deleted for region I. Element A (−327 to −295) is defined by Δ6 and Δ19, which confer partial sporulation activity in the absence of element B. This location is consistent with the phenotypic distinction between Δ2 and Δ3 (Fig. 1). However, deletion of element A has no significant phenotypic effect if element B is present (e.g., Δ16 and Δ17). Element B (−273 to −256) is defined by the comparison of Δ4, which has an extremely weak sporulation phenotype, and Δ17, which confers wild-type sporulation. Sequences between −262 and −273 are important for element B, because their removal leads to reduced promoter activity (compare Δ18 and Δ19). Consistent with this observation, linker-scanning mutations in element B significantly reduce (Δ20) or eliminate (Δ21) sporulation under conditions where element A is deleted. However, linker-scanning mutations in element B (ls1 to ls4) do not affect sporulation in the context of the intact mei3 promoter region, presumably due to compensation by element A and perhaps by sequences between −273 and −295. In this regard, the downstream endpoint of element A is poorly defined and may extend beyond −295. Thus, region I contains at least two partially redundant positive elements that contribute to mei3 transcription.
FIG. 2.
Definition of elements A and B by deletion analysis of region I. The structures and sporulation phenotypes of the indicated mei3 derivatives are shown as described in the legend to Fig. 1, with the addition that solid lines indicate the location of linker-scanning mutations. The approximate locations of elements A and B are indicated (see text).
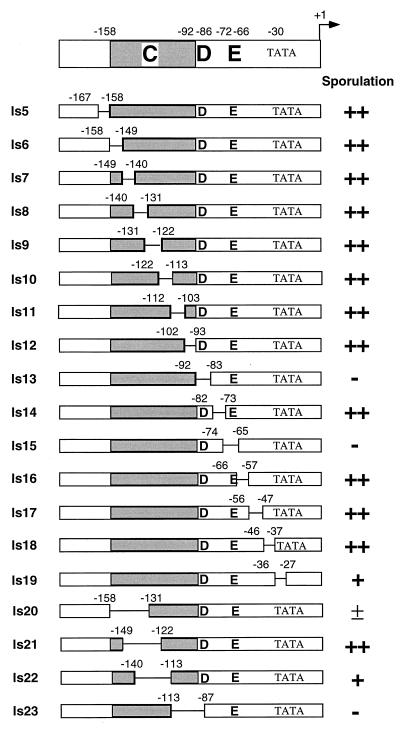
Identification of elements D and E in region II.
Region II was initially dissected by insertion-scanning mutations that replace 10 bp of the mei3 promoter region with 12 or 14 bp of heterologous sequence (Table 2). At least one positive element is located between −158 and −86, because all insertion-scanning mutations in this region are severely or completely defective for sporulation whereas Δ22, Δ51, and Δ52 are fully functional. In addition, there is a distinct element (termed E) that is defined by a small internal deletion (Δ14 [Fig. 1]) which removes residues −73 to −60. Element E is clearly separable from the further upstream elements by virtue of insertion-scanning mutations Δ51 and Δ52, which do not detectably affect sporulation; these derivatives also establish the upstream boundary of element E at −72.
TABLE 2.
Insertion-scanning mutations
| mei3 allele | Endpoint
|
Spacing change | Sporulation phenotype | |
|---|---|---|---|---|
| Upstream | Downstream | |||
| Δ22 | −167 | −158 | +4 | ++ |
| Δ23 | −158 | −149 | +4 | + |
| Δ24 | −158 | −149 | +20 | ± |
| Δ25 | −149 | −140 | +2 | − |
| Δ26 | −149 | −140 | +4 | − |
| Δ27 | −149 | −140 | +8 | − |
| Δ28 | −149 | −140 | +20 | − |
| Δ29 | −140 | −131 | +4 | − |
| Δ30 | −140 | −131 | +10 | − |
| Δ31 | −140 | −131 | +12 | − |
| Δ32 | −140 | −131 | +14 | − |
| Δ33 | −140 | −122 | +1 | ± |
| Δ34 | −131 | −122 | +4 | − |
| Δ35 | −131 | −122 | +10 | − |
| Δ36 | −131 | −122 | +12 | − |
| Δ37 | −131 | −122 | +14 | − |
| Δ38 | −131 | −113 | +1 | − |
| Δ39 | −122 | −113 | +4 | − |
| Δ40 | −122 | −113 | +4 | − |
| Δ41 | −122 | −113 | +10 | − |
| Δ42 | −122 | −113 | +12 | − |
| Δ43 | −122 | −113 | +14 | − |
| Δ44 | −112 | −113 | +8 | − |
| Δ45 | −112 | −103 | +4 | − |
| Δ46 | −112 | −103 | −1 | − |
| Δ47 | −102 | −93 | +4 | ± |
| Δ48 | −102 | −93 | +10 | − |
| Δ49 | −102 | −93 | +12 | − |
| Δ50 | −102 | −93 | +14 | − |
| Δ51 | −86 | −83 | +2 | ++ |
| Δ52 | −82 | −73 | +4 | ++ |
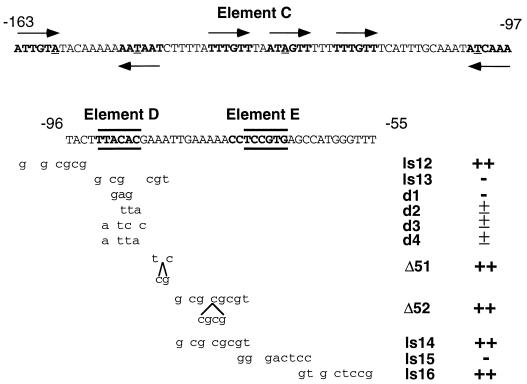
Because insertion-scanning mutations affect both nucleotide sequence and spacing, we further dissected region II with 10-bp linker-scanning mutations covering sequences from −167 to −27 (Fig. 3). In striking contrast to the result with the comparable insertion-scanning mutations, nearly all of the linker-scanning mutations confer wild-type sporulation. The two exceptions, ls13 (−92 to −83) and ls15 (−74 to −65), are completely defective for mei3 promoter function, respectively defining two positive elements, D and E. Sporulation is reduced, but not eliminated, by ls19, a linker-scanning mutation that destroys the putative TATA element. The fact that the ls15 mutation eliminates sporulation indicates that element E is defined by a specific sequence, not by an effect on spacing. Further, residues −72 to −67 (TCCGTG) are particularly important for element E function because linker-scanning mutations on either side of this region, ls14 (−82 to −73) and ls16 (−66 to −57), confer normal sporulation (Fig. 4).
FIG. 3.
Definition of elements D and E by deletion analysis of linker-scanning mutations. The structures and sporulation phenotypes of the indicated mei3 derivatives are shown as described in the legend to Fig. 1, except that linker-scanning mutations are defined by the regions that are mutated; e.g., ls13 substitutes 10 bp for nucleotides −92 to −83 inclusive. The locations of elements D, E, and C, which is defined primarily by insertion-scanning mutations (see Table 3), are also indicated.
FIG. 4.
Nucleotide sequences of elements C, D, and E and additional alleles of element D. Element C, which is defined primarily by insertion-scanning mutations (see Table 3), is a large region that contains six sequences (boldface letters with arrows indicating orientation) that strongly resemble consensus recognition sites for Mc and other DNA-binding domains that contain the HMG structural motif (8, 10); underlined bases deviate from the consensus. The most critical bases of elements D and E are shown in boldface letters and are bracketed by horizontal lines. Shown below are the structures (mutated base pairs are indicated in small letters) and phenotypes of the relevant mei3 derivatives used to define these elements. Derivatives d1 to d4 were designed to assess whether element D is recognized by Pm in vivo (see Fig. 6).
Element D is defined by the −92 to −83 linker-scanning mutation (ls13), and residues −92 to −87 (TTACAC) are particularly important because a disruption of sequences further downstream (Δ51) does not affect sporulation (Fig. 4). The spacing between elements D and E is not critical because insertion-scanning mutations (Δ51 and Δ52) between them have minimal phenotypic effects. Element D coincides with a sequence (CTTTACACG, located between −86 and −94) that has eight of nine matches with the consensus half-site, A(A/T)NTACAPyPu, recognized by the S. cerevisiae α2 homeodomain (28).
Mating-type protein Pm interacts with element D in vivo.
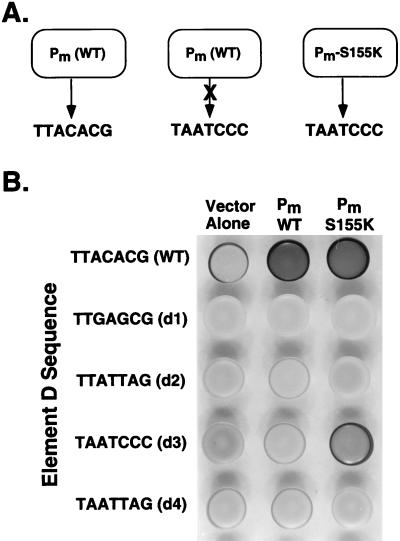
Given the element D sequence and the strong similarity between the α2 and Pm homeodomains (28), we hypothesized that Pm might activate mei3 transcription in vivo by directly binding to element D of the promoter. This hypothesis predicts the existence of altered-specificity derivatives of Pm that function at mei3 promoters containing specifically mutated versions of element D (Fig. 5A). Based on DNA-binding specificity experiments involving the bicoid and antennapedia homeodomains (11, 12), we generated a bicoid-like derivative of Pm (S155K) and the following four alleles of element D: a bicoid-like site (TAATCCC [d3]), antennapedia-like binding sites (TTATTAG [d2] and TAATTAG [d4]), and a mutant site (TTGAGCG [d1]) that presumptively would not interact with homeodomains (underlining indicates differences from wild-type mei3 element D).
FIG. 5.
Pm interacts with element D in vivo. (A) Wild-type Pm interacts with wild-type element D (TTACACG) but not with the bicoid-like d3 allele (TAATCCC), indicating that the Pm-element D interaction is critical for sporulation. An altered-specificity derivative of Pm (S155K) that interacts with the d3 allele should effectively recreate the Pm-element D interaction and permit sporulation. (B) Sporulation phenotypes (assayed by iodine staining) for strains containing the indicated alleles of element D (WP20, WP434, WP495, WP496, and WP499) and plasmids overexpressing the wild type or S155K derivative of Pm. All strains, including the control containing the pAD2 vector, contain the chromosomal copy of Pm.
Multicopy plasmids expressing the Pm derivatives from the natural promoter were introduced into wild-type strains containing single integrated copies of the mei3 alleles containing element D mutations (Fig. 5B). As expected, promoters containing the element D mutations are extremely defective in the ability to support sporulation in the presence of normal amounts of Pm (vector only). However, the bicoid-like S155K derivative confers efficient (and perhaps greater than wild-type) levels of sporulation (and hence mei3 expression) in combination with the bicoid-like d3 allele but not with any of the other mutations of element D. In addition, sporulation is observed when the S155K derivative is introduced into a strain containing the wild-type promoter but lacking Pm (data not shown). Thus, the S155K derivative has relaxed DNA-binding specificity in that it interacts with wild-type and bicoid-like versions of element D but not with antennapedia-like or mutated forms of this element. The ability of the S155K derivative of Pm to suppress a specific mutation of element D in a manner consistent with the known DNA-binding specificity of bicoid (11, 12) indicates that Pm interacts with element D in vivo. Consistent with this conclusion, overexpression of wild-type Pm weakly suppresses the sporulation defect conferred by the d2, d3, and d4 alleles.
Unusual properties of element C.
Element C is defined by the observation that sporulation is severely or completely defective in strains containing insertion-scanning mutations that locally disrupt any small region between −158 and −92 (Table 2). Although the boundaries of elements C and D are not precisely defined (and may overlap), element C is distinct from element D because its function cannot be eliminated by linker scanning. Mutations that disrupt the distal (Δ9, Δ23, Δ24) or proximal (Δ47) end of element C confer some mei3 promoter function, whereas centrally located mutations are generally nonfunctional. This observation might reflect disruption of redundant subelements at the ends of element C or the importance of centrally located residues for element C function.
The unusual feature of element C is that 10-bp linker-scanning mutations anywhere within this large region do not significantly affect sporulation (Fig. 3), whereas insertion-scanning mutations at the same positions drastically reduce mei3 promoter function (Table 2). It is unlikely that the linker sequences themselves encode positive elements that compensate for the disruption of element C. In all cases tested, changing the linker sequence does not affect the sporulation phenotype. In addition, several of the internal deletions that disrupt element C function have the same junction sequences as the linker-scanning mutations that have no phenotypic effect. Finally, in all cases tested, mei3 function is disrupted by small deletions within element C that decrease spacing by 1, 5, 9, and 18 bp or by insertion mutations that increase spacing by 1, 2, 4, 8, 9, 10, 12, 14, and 20 bp (Table 2). In the most dramatic example, ls11 and Δ46 differ only by the deletion of a single base pair yet have completely different sporulation phenotypes. Taken together, these observations suggest that any change in spacing drastically affects element C function whereas a change in the nucleotide sequence has much less effect.
We also analyzed very long linker-scanning mutations in which 27- or 28-bp regions were replaced with heterologous DNA (Fig. 3). Two such mutations (ls20 and ls22) significantly reduce, but do not completely eliminate, mei3 expression. However, the −149 to −121 mutation (ls21) has substantial overlap with the above mutations yet confers wild-type levels of sporulation. These apparently contradictory observations are suggestive of redundancy in element C (see Discussion). As expected, a large linker-scanning mutation that also disrupts element D (ls23) completely eliminates mei3 promoter function.
Quantitation of mei3 promoter activity in mutant strains.
To quantitate mei3 promoter activity, mei3 derivatives with a range of sporulation phenotypes were fused to the luciferase reporter gene and integrated into the ura4 locus. Luciferase activity was assayed in uninduced (nitrogen sated) and induced (nitrogen starved for 4 h) mei3 diploids (Table 3). Interestingly, all derivatives with noticeable defects in sporulation (i.e., +, ±, and − phenotypes) show <10% of the wild-type levels of luciferase activity. This observation suggests that a limited amount of mei3 transcription is sufficient to confer significant (perhaps even wild-type) levels of sporulation. Importantly, although experimental errors in the luciferase measurements make it difficult to discriminate among the classes of sporulation-defective mutants, it is clear that any visible reduction of sporulation reflects a severe decrease in mei3 promoter function. For this reason, we assayed only a limited number of derivatives with non-wild-type sporulation properties.
TABLE 3.
Luciferase assays
| mei3 allele | Endpoint
|
Phenotype
|
||
|---|---|---|---|---|
| Upstream | Downstream | Sporulation | Luciferase activity (%) | |
| Δ1 | None | −502 | ++ | 100 |
| Δ7 | −258 | −159 | ++ | 32 ± 8 |
| Δ16 | −327 | −285 | ++ | 62 ± 8 |
| Δ18 | −295 | −274 | ++ | 37 ± 5 |
| Δ22 | −167 | −158 | ++ | 90 ± 30 |
| Δ51 | −86 | −83 | ++ | 40 ± 7 |
| Δ52 | −82 | −73 | ++ | 180 ± 51 |
| ls1 | −295 | −285 | ++ | 63 ± 25 |
| ls3 | −273 | −263 | ++ | 109 ± 24 |
| ls5 | −167 | −158 | ++ | 53 ± 6 |
| ls6 | −158 | −149 | ++ | 47 ± 7 |
| ls7 | −149 | −140 | ++ | 30 ± 6 |
| ls8 | −140 | −131 | ++ | 34 ± 6 |
| ls9 | −131 | −122 | ++ | 44 ± 11 |
| ls10 | −122 | −113 | ++ | 41 ± 5 |
| ls11 | −112 | −103 | ++ | 32 ± 3 |
| ls12 | −102 | −93 | ++ | 47 ± 14 |
| ls14 | −82 | −73 | ++ | 109 ± 26 |
| ls16 | −66 | −57 | ++ | 138 ± 50 |
| ls17 | −56 | −47 | ++ | 81 ± 10 |
| ls18 | −46 | −37 | ++ | 97 ± 24 |
| Δ9 | −158 | −140 | + | 5 ± 2 |
| ls22 | −140 | −113 | + | 2 ± 1 |
| Δ11 | −140 | −122 | ± | 2 ± 1 |
| Δ5 | −327 | −159 | − | 2 ± 1 |
| Δ10 | −158 | −131 | − | 3 ± 1 |
| Δ34 | −131 | −122 | − | 1 ± 1 |
| Δ45 | −112 | −103 | − | 4 ± 1 |
| Δ46 | −112 | −103 | − | 3 ± 1 |
| ls13 | −92 | −83 | − | 6 ± 1 |
| dl | −90 | −88 | − | 9 ± 4 |
However, we examined most of the derivatives that displayed wild-type sporulation phenotypes, given that a low level of mei3 expression might be sufficient for sporulation. In many cases, these derivatives express luciferase at levels comparable (within a factor of 2) to that of the wild-type control. However, some of these derivatives are modestly affected in mei3 promoter function, because they express luciferase at a level of approximately 30 to 50% of the wild type; these include a subset of the insertion-scanning mutants within element C, the large deletion between regions I and II (Δ7), the deletion between elements A and B (Δ18), and the insertion-scanning mutation at the downstream edge of element D (Δ51). Although these partially defective mei3 derivatives affect our description of the mei3 promoter (see Discussion), elements A to E are defined by derivatives with much more severe transcriptional and phenotypic effects.
Evidence against negative elements in the mei3 promoter region.
Expression of mei3 in haploids is sufficient to trigger meiosis and initiation of the sporulation pathway, even when cells are grown in nitrogen-rich medium (19). In this regard, when vector sequences are fused directly to positions −99 and −20, respectively, the resulting strains are extremely unstable and exhibit iodine staining in nitrogen-rich medium that is characteristic of constitutive sporulation and mei3 expression (data not shown). In contrast, for all of the mei3 derivatives tested in this study, we have never observed iodine staining in nitrogen-rich medium or strain instability characteristic of haploid strains undergoing inappropriate meiosis. This observation suggests that the mei3 promoter does not contain negative elements that are required to repress transcription in haploid cells.
As an independent assay for mei3 expression in haploid cells, we fused most of our mei3 promoter derivatives to the ade6 reporter gene and assayed the resulting DNAs for the ability to support growth on medium lacking adenine. In principle, elimination of a negative element that represses transcription in haploids should result in an Ade+ phenotype. However, all of the strains tested were Ade−, indicating that the mei3 promoter derivatives were inactive in nitrogen-rich medium. In contrast, mei3::ade6 derivatives in which vector sequences were fused to positions −99 or −20 were Ade+. Thus, despite analyzing numerous derivatives, we have been unable to generate promoter mutants that express mei3 in nitrogen-rich medium. This observation argues against negative elements in the mei3 promoter region, and it strongly suggests that meiotic regulation of mei3 expression is under positive control.
DISCUSSION
Evidence that S. pombe does not utilize a repression mechanism to prevent meiosis in haploids.
It is essential that eukaryotic organisms have a mechanism to prevent meiosis from occurring in haploid cells. In S. cerevisiae, this occurs by an active repression mechanism involving direct binding of the Rme1 repressor to the key meiosis-inducing gene, IME1 (6, 22). Loss of Rme1 function, either by mutation or by a1-α2 repression, leads to inappropriate meiosis in starved haploid cells. In contrast, our detailed mutational analysis of the mei3 promoter provides strong evidence that the failure of haploid S. pombe cells to express mei3 and trigger an inappropriate meiotic cycle is not due to a transcriptional repression mechanism. In particular, inappropriate mei3 transcription in haploid cells was not observed with any of the large number and wide variety of mutations throughout the mei3 promoter region. This result is significant because promoter analyses of the type described here have uncovered negative regulatory elements in a wide variety of eukaryotic promoters. Indeed, in the case of meiotic regulation in S. cerevisiae, negative elements were easily identified in the promoters of IME1 (6, 22) and IME2, a gene that acts downstream of IME1 (3).
It is impossible to prove that a given promoter does not contain a negative element. It is always possible that a negative element in a promoter might be obscured by redundant negative elements or by overlapping positive elements that are required for expression. Thus, the validity of the argument that a given promoter lacks a negative element depends on the number and variety of mutations tested; in this regard, our analysis is extensive. Furthermore, negative elements are generally easy to uncover, because even a partial loss of repression can lead to significant levels of transcription (e.g., 50% repression results in transcription only twofold below wild-type levels). Because our assays for inappropriate transcription in haploid cells (iodine staining and Ade+ phenotype) are very sensitive, it is likely that the various mei3 promoter derivatives are inactive in haploid cells and that our analysis would uncover mutants that only partially relieve repression.
The mei3 promoter is complex and contains at least five positive elements.
The mei3 region between −327 and the ATG initiation codon is necessary and sufficient to mediate proper mei3 expression. Within this region, we have defined five promoter elements by the following criteria. Element E is defined by a linker-scanning mutation (ls15) and a small deletion (Δ14) that eliminate sporulation. Element D is defined by linker-scanning mutations and multiple base pair substitutions that eliminate sporulation. Element C is defined by insertion-scanning mutations that map to a large region (−158 to −93) and greatly reduce or eliminate mei3 expression. Element B is defined by Δ19, a moderately sized deletion (−295 to −262) that significantly reduces mei3 expression, and by two linker-scanning mutations that eliminate sporulation when element A is deleted (derivatives Δ21 and Δ22). However, linker-scanning mutations in element B (ls3 and ls4) have no phenotypic effects in an otherwise intact mei3 promoter. Element A is defined loosely as a region (−327 to −295) that is important for mei3 expression when element B is deleted.
The five promoter elements are separable because regions between them can be heavily mutated or deleted with no detectable effect on sporulation and modest or no reduction of mei3 expression (as assayed by luciferase fusions). Thus, elements A and B are separated by Δ18 (−295 to −273), elements B and C are separated by Δ7 (−256 to −158) and the insertion-scanning mutation Δ22, and elements D and E are separated by linker-scanning (ls14) and insertion-scanning (Δ51 and Δ52) mutations. Elements C and D have not been physically separated, but they are functionally distinct because element D can be inactivated by linker-scanning and base pair substitution mutations whereas element C can be inactivated only by insertion-scanning mutations. With the notable exception of element C (see below), the mei3 promoter elements do not require precise spacing relationships with each other.
Several promoter derivatives are modestly affected in mei3 expression, despite displaying a wild-type sporulation phenotype (Table 3). Such partially defective mei3 derivatives can be interpreted in a variety of ways. First, these derivatives could inactivate a promoter element whose function is partially redundant with another element; e.g., the modest effects of linker-scanning mutations in element C might reflect redundant functions within this element (see below). Second, these derivatives might partially affect the function of one of the five defined elements by altering residues at the element boundaries. This explanation is likely for Δ51, which affects the downstream edge of element D, and for Δ18, which affects the poorly defined boundary between elements A and B. Third, in the case of deletion mutants (particularly the large Δ7 between regions I and II), transcriptional defects might arise from altered spacing between elements. While recognizing that these partially defective mutants affect our description of the mei3 promoter, we have defined the five positive elements based on the properties of mutants with severe phenotypic and transcriptional effects.
It is important to note that our view of the mei3 promoter region represents a formal description that does not presuppose a particular molecular mechanism. Furthermore, our description represents the simplest interpretation that is consistent with all the data. As is always the case with promoter dissections, we cannot exclude more complex interpretations. Finally, although we have identified and localized five elements within the mei3 promoter region, the precise boundaries of some of these elements (particularly A, B, and C) have not been defined and the existence of additional positive elements has not been excluded.
Evidence that Mc binds to element C and acts as an architectural transcription factor.
Element C is very large (60 bp) and has the unusual property of being inactivated by insertion-scanning mutations but not by linker-scanning mutations. We doubt that element C is simply a spacer sequence that maintains the correct distance between other promoter elements, because insertion-scanning or deletion mutations that change the spacing relationship of elements A, B, and E to element C do not have phenotypic effects. Instead, we suggest that element C contains subelements that have a precise spacing relationship to each other and/or to element D. However, the failure of 10-bp linker-scanning mutations to inactivate element C implies that such putative subelements would be functionally redundant. Consistent with this possibility, some linker-scanning mutations within element C reduce mei3 expression (Table 3), and a large linker-scanning mutation (ls20) can virtually inactivate element C.
Interestingly, element C contains six sequences (ATTGTA, ATTATT, TTTGTT, ATAGTT, TTTGTT, and TTTGAT) that resemble the consensus recognition sequence, (A/T)TTGTT, for DNA-binding proteins with an HMG structural motif (10). One of the mating-type proteins, Mc (15), contains a region that strongly resembles (32% identity, 64% similarity over 76 amino acids) the HMG domain of SRY, a protein that determines male sex in mammals. Furthermore, Mc behaves like a typical HMG protein; it binds the HMG consensus sequence with high affinity primarily through interactions with the minor groove, and it severely bends DNA (8). For these reasons, we speculate that element C consists of multiple Mc interaction sites and that Mc binds and bends DNA to form a highly specific nucleoprotein structure that is important for mei3 transcription (Fig. 6). This model, in which Mc functions as an “architectural transcription factor” (26), could account for the apparent paradox of functional redundancy and severe spacing constraints, particularly if protein-protein interactions between Mc molecules energetically compensate for weak protein-DNA interactions due to linker-scanning mutations.
FIG. 6.
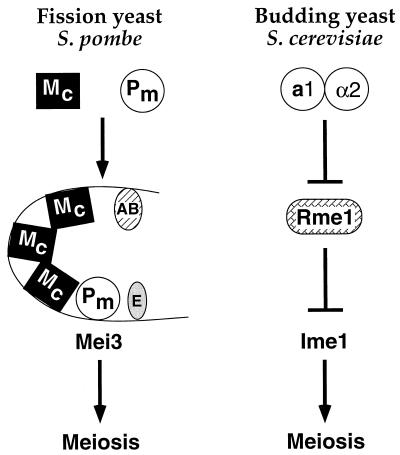
Different regulatory logic for cell type control of the key meiosis-inducing genes in fission and budding yeasts. In S. pombe, diploids express two distinct DNA-binding proteins, Mc, which contains an HMG domain, and Pm, which contains a homeodomain homologous to α2. Pm binds to element D of the mei3 promoter, and Mc is likely (but not directly shown) to bind multiple sites within element C. These proteins, together with putative proteins interacting with elements E and A or B, stimulate transcription of mei3, the trigger for meiosis. Haploids do not undergo meiosis because Pm and perhaps these other proteins are not present or active. Given the unusual properties of element C, we speculate that mei3 transcription requires the formation of an enhancesome that depends on DNA-binding and bending properties of the HMG protein Mc (see text). In S. cerevisiae, haploids do not undergo meiosis because they express Rme1, a repressor that binds the promoter of IME1, the key meiosis-inducing gene. Diploids can undergo meiosis because they specifically express the a1 and α2 homeodomain proteins, which form a heterodimeric repressor that directly binds the RME1 promoter and blocks production of Rme1. Hence, a1-α2 indirectly cause the expression of Ime1 by repressing a repressor, whereas Pm and presumably Mc directly bind the mei3 promoter and activate transcription. Furthermore, α2 functions as a repressor whereas the homologous Pm functions as an activator. The key meiosis-inducing proteins, Mei3 and Ime1, have unrelated molecular functions, and the pathways downstream of these proteins are completely different.
Pm interacts with element D and directly activates mei3 transcription.
Several lines of evidence indicate that element D interacts with Pm in vivo. First, element D strongly resembles an α2 binding site, and the Pm homeodomain is 41% identical (70% similar) to the α2 homeodomain. Although the recognition helix of Pm is five residues shorter than that of α2, all of the residues that contact bases or phosphates (28) are conserved between α2 and Pm, including homeodomain residue 50 (S155 in Pm), which plays an important role in DNA-binding specificity (11). Second, mutational analysis of element D indicates that the critical base pairs for sequence recognition by α2 are essential for mei3 expression. Third, overexpression of Pm partially suppresses the sporulation-defective phenotype conferred by several element D mutations. Fourth, and most convincing, the S155K derivative of Pm strongly suppresses a specific mutation of element D; it permits mei3 expression from the d3 allele but not from other mutant forms of element D. Furthermore, this allele-specific suppression reflects a direct protein-DNA interaction between Pm and element D because the combination of compensating mutations is consistent with the known DNA-binding specificity of bicoid (11, 12). Given that Pm interacts with element D in vivo and that element D is positively required for mei3 transcription, we conclude that Pm functions as a transcriptional activator at the mei3 promoter.
The mei3 promoter acts as an on-off developmental switch because transcription requires synergistic activation by multiple elements.
It is commonly observed that efficient transcription of eukaryotic genes requires the combinatorial and synergistic action of distinct elements in the promoter region. Wild-type levels of mei3 transcription require the synergistic activation of elements C, D, and E and region I (which contains elements A and B). Mutations in any of these four components drastically reduces mei3 expression.
In the context of wild-type S. pombe cells, the requirement for at least four independent elements restricts mei3 expression to circumstances when the proteins bound to these elements are functionally active. At a minimum, these circumstances include the presence of the M and P mating-type loci, the pheromone signal, and the environmental condition of nitrogen starvation. Given that the mei3 promoter elements appear to be unrelated in DNA sequence, we presume that Pm, Mc, and two or three other proteins must be present and active under the specific conditions that are appropriate for mei3 transcription (Fig. 6). Under other conditions, we presume that one or more of these proteins are absent or inactive. Thus, the mei3 promoter constitutes an on-off developmental switch that is responsible for the tight regulation of mei3 expression that is critical in the S. pombe life cycle.
In several respects, the complex mei3 promoter resembles the human beta interferon enhancer. Transcription of the beta interferon gene is restricted to the specific circumstance of virus infection, and this restriction reflects the requirement for the binding of multiple and distinct proteins to their cognate elements in the enhancer (16, 24). In this sense, the beta interferon enhancer functions as an on-off environmental switch. The function of the beta interferon enhancer depends on the architectural transcription factor HMG-I, and it is exquisitely sensitive to spacing changes between individual elements. Thus, transcription of the beta interferon gene requires the assembly of a precisely structured multiprotein-DNA complex termed an “enhancesome” (24). By analogy, we speculate that developmentally regulated expression of mei3 might involve the assembly of an enhancesome (Fig. 6), a structure not previously described for unicellular eukaryotes. However, the observation that elements A, B, E, and perhaps D do not require precise spacing relationships suggests that a putative mei3 enhancesome might be restricted to element C and Mc. In contrast, the beta interferon enhancesome is composed of distinct elements that interact with different proteins.
The regulatory logic for cell type control of the key meiosis-inducing gene differs between fission and budding yeasts.
In many respects, mating-type control of sporulation is similar in S. cerevisiae and S. pombe. Both organisms contain one active and two silent mating-type cassettes, each of the two mating-type alleles expresses two proteins, and S. cerevisiae α2 and S. pombe Pm contain homologous homeodomains. Nevertheless, in contrast to the S. cerevisiae mating-type proteins, which directly repress the Rme1 repressor of the key meiotic control gene (IME1), the S. pombe mating-type proteins (Pm and probably Mc) directly activate mei3, the gene that triggers meiosis (Fig. 6). Furthermore, the homologous homeodomain proteins regulate transcription of the direct target genes in opposite manners; i.e., α2 functions as a repressor whereas Pm functions as an activator. Finally, to deal with the critical issue of preventing expression of meiosis-inducing genes in haploid cells, S. cerevisiae uses an active repression mechanism whereas S. pombe does not appear to do so. Thus, our results demonstrate that the underlying logic of cell type control of the key meiosis-inducing gene in fission yeast is fundamentally different from that in budding yeast.
ACKNOWLEDGMENTS
We are indebted to Vicki Chandler and Jo Ann Wise for permitting W.J.v.H. to carry out some of these experiments in their laboratories. We thank Charles Hoffman, Amar Klar, Maureen McLeod, Fred Ponticelli, Fred Winston, and Jo Ann Wise for plasmids and/or yeast strains. We thank Fred Ponticelli for technical advice in the early stages of this work and Brendan Cormack, Mark Lee, and Jo Ann Wise for comments on the manuscript. This work was supported by a predoctoral fellowship to W.J.V.H. from the Howard Hughes Medical Institute, a National Institutes of Health (NIH) postdoctoral fellowship to D.R.D., and research grants to K.S. from NIH (GM30186 and GM53720).
REFERENCES
- 1.Althoff S M, Stevens S W, Wise J A. The Srp54 GTPase is essential for protein export in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:7839–7854. doi: 10.1128/mcb.14.12.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono T, Yanai H, Miki F, Davey J, Shimoda C. Mating pheromone-induced expression of the mat1-Pm gene of Schizosaccharomyces pombe: identification of signalling components and characterization of upstream controlling elements. Yeast. 1994;10:757–770. doi: 10.1002/yea.320100607. [DOI] [PubMed] [Google Scholar]
- 3.Bowdish K S, Mitchell A P. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broker M. Transformation of intact S. pombe cells with plasmid DNA. BioTechniques. 1987;5:516–517. [Google Scholar]
- 5.Covitz P A, Herskowitz I, Mitchell A P. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-α2. Genes Dev. 1991;5:1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- 6.Covitz P A, Mitchell A P. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 7.de Wet J R, Wood J V, DeLuca M, Helinski D R, Subramani S. The firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dooijes D, van de Wetering M, Knippels L, Clevers H. The Schizosaccharomyces pombe mating-type gene mat-Mc encodes a sequence-specific DNA-binding high mobility group box protein. J Biol Chem. 1993;268:24813–24817. [PubMed] [Google Scholar]
- 9.Flanagan P M, Kelleher R J, Tschochner H, Sayre M H, Kornberg R D. Simple derivation of TFIID-dependent RNA polymerase II transcription systems from Schizosaccharomyces pombe and other organisms, and factors required for transcriptional activation. Proc Natl Acad Sci USA. 1992;89:7659–7663. doi: 10.1073/pnas.89.16.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 11.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 12.Hanes S D, Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991;251:426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- 13.Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann J, Smale S T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 15.Kelly M, Burke J, Smith M, Klar A, Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhancesome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 17.Li P, McLeod M. Molecular mimicry in development—identification of Ste11(+) as a substrate and Mei3(+) as a pseudosubstrate inhibitor of Ran1(+) kinase. Cell. 1996;87:869–880. doi: 10.1016/s0092-8674(00)81994-7. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Flanagan P M, Tschochner H, Kornberg R D. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263:805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- 19.McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell A P, Bowdish K S. Selection for early meiotic mutants in yeast. Genetics. 1992;131:65–72. doi: 10.1093/genetics/131.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagee S, Sherman A, Shenhar G, Robzyk K, Ben-Doy N, Simchen G, Kassir Y. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1985–1995. doi: 10.1128/mcb.18.4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith H E, Su S S Y, Neigeborn L, Driscoll S E, Mitchell A P. Role of IME1 expression in control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6110. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhancesome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- 26.Werner M H, Burley S K. Architectural transcription factors: proteins that remodel DNA. Cell. 1997;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 27.Wobbe C R, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Crystal structure of a MATα2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto M. The molecular mechanisms of meiosis in fission yeast. Trends Biochem Sci. 1996;21:18–22. [PubMed] [Google Scholar]