Abstract
Saccharomyces cerevisiae harbors two cyclophilin 40-type enzymes, Cpr6 and Cpr7, which are components of the Hsp90 molecular chaperone machinery. Cpr7 is required for normal growth and is required for maximal activity of heterologous Hsp90-dependent substrates, including glucocorticoid receptor (GR) and the oncogenic tyrosine kinase pp60v-src. In addition, it has recently been shown that Cpr7 plays a major role in negative regulation of the S. cerevisiae heat shock transcription factor (HSF). To better understand functions associated with Cpr7, a search was undertaken for multicopy suppressors of the cpr7Δ slow-growth phenotype. The screen identified a single gene, designated CNS1 (for cyclophilin seven suppressor), capable of suppressing the cpr7Δ growth defect. Overexpression of CNS1 in cpr7Δ cells also largely restored GR activity and negative regulation of HSF. In vitro protein retention experiments in which Hsp90 heterocomplexes were precipitated resulted in coprecipitation of Cns1. Interaction between Cns1 and the carboxy terminus of Hsp90 was also shown by two-hybrid analysis. The functional consequences of CNS1 overexpression and its physical association with the Hsp90 machinery indicate that Cns1 is a previously unidentified component of molecular chaperone complexes. Thus far, Cns1 is the only tetratricopeptide repeat-containing component of Hsp90 heterocomplexes found to be essential for cell viability under all conditions tested.
Cells rely on molecular chaperones to facilitate the folding of nascent polypeptides, prevent protein aggregation, and enable proper inter- and intramolecular interactions. Indispensable in both Saccharomyces cerevisiae (5) and Drosophila melanogaster (13), Hsp90 is one of the most abundant and highly conserved chaperones. In vitro, Hsp90 displays general chaperone properties by preventing the aggregation of proteins such as citrate synthase (52) and casein kinase II (30) and by maintaining β-galactosidase (21) in a folding-competent state. In vivo, however, Hsp90 is not required for the folding of most proteins (33) but instead plays a key role in the maturation of a small subset of proteins typically involved in signal transduction (3, 32, 42). It is needed for the maturation of certain steroid hormone receptors (40, 43, 44), basic helix-loop-helix transcription factors (45), serine/threonine kinases (30, 41, 50, 51), and oncogenic tyrosine kinases (7). Depending on the particular substrate, the interactions with Hsp90 required for maturation can be transient or continuous (32).
In some cases, the role of the Hsp90 machinery is also regulatory. For example, while the chaperone is required to fold glucocorticoid receptor (GR) into its mature, ligand-responsive conformation, it also ensures that the receptor remains in an inactive state in the absence of ligand (3, 6, 8). Similarly, the association of Hsp90 with newly synthesized oncogenic tyrosine kinase pp60v-src keeps the kinase inactive until it dissociates from Hsp90 (7).
Recent discoveries have revealed additional regulatory roles for Hsp90 in signal transduction. For example, in endothelial cells, the induction of nitric oxide production elicits the binding of Hsp90 to nitric oxide synthase, increasing the activity of the enzyme (23). We have shown that Hsp90 and the Hsp90-associated cyclophilin 40 (Cyp40)-type enzyme Cpr7 are required for negative regulation of heat shock transcription factor (HSF) in S. cerevisiae (reference 17 and this report), revealing that some components of the Hsp90 machinery participate in regulation of the heat shock response.
Hsp90 is assisted in its various roles by a cohort of associated proteins including the chaperones Hsp70 and DnaJ, the immunophilins Cyp40, FKBP51, and FKBP52, and other, less well characterized proteins, including Hop (p60), Hip (p48), p23, and PP5, a protein phosphatase with immunophilin properties (4, 25, 38, 42, 46, 48). The Hsp90-associated proteins are highly conserved among eukaryotes, and the composition of Hsp90 heterocomplexes is also generally conserved (9). In vitro reconstitution studies of steroid receptor maturation have shown that the components of Hsp90 complexes are assembled in an ordered, multistep process (31, 38). This pathway involves a dynamic association of Hsp90, Hsp70, p60, and p48 at early or intermediate stages, while the final mature complex contains Hsp90, lower levels of Hsp70, p23, and an immunophilin (47). The mature complex enables the steroid receptor to achieve a conformation responsive to ligand stimulation.
The immunophilins associated with Hsp90 harbor peptidyl-prolyl isomerase domains and constitute two distinct families: FKBPs and cyclophilins, which are capable of binding the immunosuppressive drugs FK506 and cyclosporin A, respectively (22, 29, 38). In addition to an isomerase domain, the Hsp90-associated immunophilins also contain three tetratricopeptide repeat (TPR) motifs. TPRs are involved in protein-protein interactions (27), and the immunophilins interact with Hsp90 via these domains (38). Biochemical experiments have shown that only a single species of TPR-containing protein is found in association with a particular Hsp90 complex (27, 36). Furthermore, binding to Hsp90 by the immunophilins appears to be competitive, providing support for the hypothesis that Hsp90 dimers generate a single TPR-accepting pocket (27, 36, 39).
S. cerevisiae harbors two genes, CPR6 and CPR7, that encode Cyp40-type cyclophilins (18). Bolstered by biochemical evidence showing that conservation of Hsp90 heterocomplexes between mammals and S. cerevisiae includes the presence of Cyp40-type cyclophilins (9), we showed that both Cpr6 and Cpr7 can interact directly with Hsp90 (Hsp82) (16). Although both Cpr6 and Cpr7 are components of Hsp90 heterocomplexes, Cpr7 appears to be functionally distinct from Cpr6, as cells that harbor a null allele of CPR7 exhibit a slow-growth defect not observed with cpr6Δ cells (18). In fact, S. cerevisiae cells lacking all eight of the cyclophilin genes exhibit only the slow-growth phenotype conferred by the cpr7Δ mutation (15). Two lines of evidence indicate that the physical association between Cpr7 and the Hsp90 machinery reflects a functional relationship. First, cells devoid of Cpr7 exhibit decreased Hsp90-dependent functions including decreased activity of the heterologous Hsp90 substrates GR and the oncogenic kinase pp60v-src. Second, mutations such as hsc82Δ and sti1Δ, which decrease Hsp90 activity but do not by themselves impair growth, result in a severe growth defect in cells lacking CPR7 (16). Surprisingly, a derivative of Cpr7 lacking the isomerase domain retains the ability to interact with Hsp90 and is sufficient to restore Cpr7-dependent functions to cpr7Δ cells (16, 19).
To better understand the role(s) of Cpr7 in the function of the Hsp90 chaperone and the requirement for Cpr7 in normal growth, we pursued the analysis of suppressors of the cpr7Δ slow-growth phenotype. The isolation and analysis of spontaneous suppressors identified three unlinked genes (50a). Here we describe a multicopy suppressor encoded by the essential open reading frame (ORF) YBR155w, which we have designated CNS1 (for cyclophilin seven suppressor). We show that overexpression of CNS1 restores multiple Cpr7-dependent activities to cpr7Δ cells. Biochemical evidence indicating that Cns1 is a new component of the Hsp90 chaperone machinery is also presented.
MATERIALS AND METHODS
Strains and plasmids.
All S. cerevisiae strains used are derivatives of W303 background (leu2-112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 GAL SUC2) except for YBR155w/YBR12.05 (MATa/α cns1Δ::LEU2/CNS1 trp1/trp1 ura3/ura3 his3/his3 leu2/leu2; gift of T. Miosga), GPD::HSP82FP (MATa can1 ade2 leu2 his3 trp1 ura3 hsc82Δ::LEU2 hsp82Δ::LEU2 [pTGPD-H-Hsp82FP]; gift of S. Lindquist), and PJ69-4a (MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ; gift of P. James and E. Craig) (26). Strains harboring deletions for HSC82, STI1, and/or CPR7 have been described previously (16, 18).
CNS1 was cloned into centromeric and multicopy plasmids. pJM105 is a URA3-marked centromeric plasmid that contains an epitope-tagged derivative of Cns1 expressed under control of the CNS1 promoter. It was constructed as follows. CNS1 (lacking the 3′ termination codon but containing 0.6 kb of the 5′ untranslated sequence) was PCR amplified and cloned into XhoI/EcoRI sites of pRS316. A 99-bp fragment encoding three tandem copies of the influenza virus A hemagglutinin (HA) epitope was cloned in frame immediately downstream of CNS1 at the EcoRI/SpeI sites of pRS316. Expression of the Cns1-3HA fusion protein was confirmed by immunoblot analysis. The CNS1::3HA fusion was subcloned into pRS426, a multicopy URA3-marked vector (creating pJM107), into pRS314, a centromeric TRP1-marked vector (creating pJM110), and into pRS424, a multicopy TRP1-marked vector (creating pJM111). Expression of Cns1-3HA from pJM107, pJM110, and pJM111 was confirmed by immunoblot analysis. pJM110 was used as template to subclone the CNS1::3HA (beginning with the 5′ ATG of CNS1) fusion into the two-hybrid system bait vector pGBD-C2, creating pJM123. Plasmids used in assays for GR activity (p2A/GRGZ and pJM98), HSF activity (pHSE2-lacZ), and protein retention experiments (pTGPD-H-Hsp82FP, pJM5, and pGEX-2T) and plasmids overexpressing CPR7 (pAAD97), CPR6 (pJM90), and STI1 (pUSti1) have been described previously (9, 10, 16, 19, 32, 49).
Yeast transformations were carried out by the standard lithium acetate method, and bacterial transformations were performed by electroporation. Control vectors were introduced into wild-type cells as needed to maintain similar prototrophic backgrounds.
Screen for multicopy suppressors of cpr7Δ slow growth.
hsc82Δ cpr7Δ cells were transformed with plasmid DNA from a URA3-marked multicopy library of S. cerevisiae genomic inserts and plated to selective media. Rapidly growing transformant colonies were picked and, after streaking, assessed for growth rate by comparing colony sizes to those of control cells treated similarly. The rapid growth rate of the candidates was analyzed for plasmid dependency by plating to media containing 5-fluoro-orotic acid. Candidates that exhibited a return to the slow growth rate of hsc82Δ cpr7Δ cells on 5-fluoro-orotic acid were analyzed further. Plasmid DNA from positive candidates was rescued by transformation in Escherichia coli and reintroduced into cpr7Δ hsc82Δ and cpr7Δ recipients to confirm the ability to suppress the growth defect of these cells. Plasmids demonstrating an ability to suppress cpr7Δ slow-growth phenotypes were sequenced at the junction of the genomic insert and analyzed by using BLAST and the Saccharomyces genome database.
GR activity assay.
Wild-type and cpr7Δ cells were transformed with the GR expression plasmid p2A/GRGZ, which harbors the lacZ gene under the control of glucocorticoid response elements (32). A multicopy CNS1 plasmid, pJM107 or pJM25 (p2μ-CNS1; isolated from the genomic library described above), was introduced into these cells. Cells harboring either control plasmids or the multicopy CNS1 plasmids were assayed for β-galactosidase activity as described previously (16, 19).
HSF activity assay.
Wild-type and cpr7Δ cells harboring the HSF-dependent reporter construct pHSE2-lacZ were transformed with the multicopy CNS1 plasmid pJM111. Logarithmically growing cultures were lysed and assayed for β-galactosidase activity as previously described (17).
Protein retention assays.
Cell lysis for the protein retention assays was performed as follows. Logarithmically growing cultures of these cells were harvested, washed, and suspended in lysis buffer (LyB; 10 mM Tris [pH 7.3], 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 20% [wt/vol] glycerol, 1 mM dithiothreitol, aprotinin [0.4 μg/ml; Sigma], leupeptin [0.4 μg/ml; Sigma], antipain [0.5 μg/ml; Sigma], 2 mM phenylmethylsulfonyl fluoride). Cells were lysed by using acid-washed beads (B. Braun Biotech International) with six cycles of vortexing for 30 s followed by placement on ice for 60 s. Lysates were cleared by centrifugation, and protein concentrations were measured by using Bio-Rad protein assay reagents.
Precipitation using glutathione S-transferase (GST) fusion proteins expressed in E. coli was performed as described previously (16). Briefly, glutathione-Sepharose 4B beads (Pharmacia) were incubated with bacterial lysates containing either GST alone or GST-Cpr7 for 1 h at 4°C with mixing, and the beads were collected by centrifugation. After a brief wash, equal amounts of Sepharose-bound fusion protein were added to reaction mixtures containing 5× BB (1× BB consists of 20 mM Tris-HCl [pH 7.5], 150 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40), yeast lysate, and H2O for a final volume of 350 μl. After 1 h of incubation at 4°C, the Sepharose beads were collected and washed seven times in phosphate-buffered saline. Bound proteins were eluted in glutathione elution buffer (20 mM reduced glutathione [Sigma] in 50 mM Tris-HCl [pH 8.0]). The eluate was separated from the Sepharose beads by brief centrifugation, and aliquots of the supernatant were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
A plasmid expressing histidine-tagged Hsp82 (Hsp82FP) was used to analyze components of Hsp90 complexes by affinity chromatography on a nickel resin. Histidine-tagged Hsp82 expressed in S. cerevisiae cells as the sole source of Hsp90 was used to collect Hsp90 protein complexes on a nickel affinity matrix essentially as described previously (9). To detect Cns1 in Hsp90 complexes, wild-type cells or hsc82Δ hsp82Δ pTGPD-H-Hsp82FP cells were transformed with pJM111 (multicopy CNS1-3HA). A 50% slurry of Ni2+-nitriloacetic acid beads (100 μl; Qiagen) was washed three times with double-distilled H2O and equilibrated with LyB. Equal amounts of lysates were added to the beads and mixed for 30 min at 4°C. The beads were harvested and washed twice with wash buffer 1 (LyB, 5 mM imidazole, 1% Triton X-100) and twice with wash buffer 2 (LyB, 10 mM imidazole, 0.2% Triton X-100). Proteins were eluted by incubation in 150 mM imidazole (Sigma) for 10 min at 4°C with mixing. The imidazole-released proteins were precipitated with 10% (vol/vol) trichloroacetic acid in the presence of 8 μg of carrier protein (β-lactoglobulin; Sigma) per ml upon centrifugation for 10 min after 15 min on ice. The pellets were washed with 100% ethanol and air dried before suspension in 1× sample buffer for SDS-PAGE analysis.
Gels were stained with Coomassie blue or processed for immunoblot analysis. Antibodies with specificity for Hsp82 (5) (gift of S. Lindquist), Ssa (9) (gift of E. Craig), Sti1 (10) (from D. Toft; gift of S. Lindquist), and HA (monoclonal antibody 12CA5; BAbCo, Inc., Richmond, Calif.) were used. Enhanced chemiluminescence immunoblot detection reagents (Amersham) were used to detect the antibodies.
Two-hybrid analysis.
A modified two-hybrid system designed by James et al. (26) was used to investigate interactions between Cns1 and Hsp90. This system utilizes three reporter genes (LYS2::GAL1-HIS3 GAL2-ADE2, and met2::GAL7-lacZ) to detect protein-protein interactions. Plasmid pJM123 expressing the bait GAL4BD (GAL4 DNA binding domain)-Cns1-3HA was transformed into the two-hybrid reporter strains PJ69-4a and HKY140 (a cpr7Δ::TRP1 derivative of PJ69-4a). A prey construct expressing GAL4AD-Hsp82C (a fusion between the Gal4 activation domain and the last 182 amino acids of Hsp82) was isolated from a yeast genomic two-hybrid library and transformed into PJ69-4a and HKY140, both harboring pJM123. PJ69-4a and HKY140 cells containing both the Cns1 bait (pJM123) and Hsp82C prey plasmids exhibited the His+ Ade+ LacZ+ phenotype that resulted from activation of all three reporter genes. Cells harboring either bait or prey plasmids alone were His− Ade−, although they expressed low levels of β-galactosidase. Liquid β-galactosidase assays were conducted as described above.
RESULTS
Screen for multicopy suppressors of the cpr7Δ slow-growth phenotype.
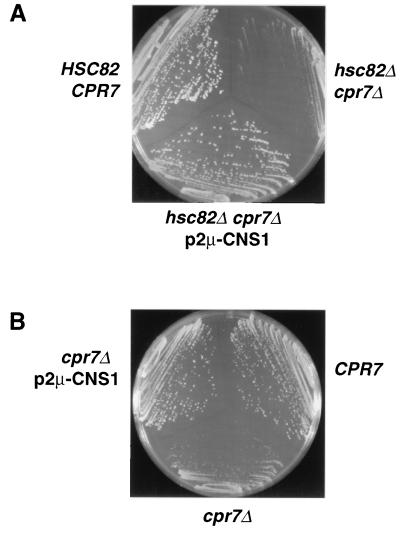
Cpr7 is a component of the Hsp90 machinery and is required for normal growth (16, 18). In contrast, deletion of HSC82, one of the two S. cerevisiae genes encoding Hsp90, does not affect the growth rate of cells at normal temperatures even though the level of Hsp90 is reduced by 90% (5). However, the cpr7Δ slow-growth phenotype is greatly exacerbated upon deletion of HSC82 (16). We took advantage of the severe growth phenotype of hsc82Δ cpr7Δ cells to identify genes whose overexpression could suppress the growth defect associated with the loss of Cpr7. hsc82Δ cpr7Δ cells were transformed with plasmid DNA from a multicopy S. cerevisiae library, and transformants that exhibited plasmid-dependent rapid growth (Fig. 1A) were retained for analysis. The plasmids harbored by the rapidly growing colonies were rescued, reintroduced into cpr7Δ hsc82Δ and cpr7Δ cells to confirm their ability to suppress the cpr7Δ-dependent growth phenotypes, and sequenced to identify the genomic region responsible for the suppression phenotype.
FIG. 1.
Overexpression of CNS1 suppresses the slow-growth phenotype of cpr7Δ hsc82Δ and cpr7Δ cells. cpr7Δ hsc82Δ and cpr7Δ cells harboring multicopy CNS1 plasmids were streaked to selective medium and incubated for 2 days at 30°C. (A) A multicopy plasmid containing the genomic region encompassing CNS1/YBR155w is able to suppress the slow growth of cpr7Δ hsc82Δ cells. Wild-type (HSC82 CPR7) and hsc82Δ cpr7Δ control cells harbor vectors only. (B) Suppression of slow-growth phenotype of cpr7Δ cells by a multicopy CNS1 plasmid (pJM111). Wild-type and cpr7Δ control cells harbor vectors only.
An exhaustive screen of the library revealed only two types of clones capable of suppressing the growth defect of cpr7Δ cells. One (11 isolates) contained genomic fragments encompassing the CPR7 gene. The other (nine isolates) contained fragments encompassing the ORF YBR155w, identified through the S. cerevisiae genome sequencing project (2); we designated this ORF CNS1. Although we expected to obtain HSC82 by this screen, it was not found among the suppressors, probably because hsc82Δ cpr7Δ pHSC82 cells grow as slowly as cpr7Δ cells. In contrast, when present on a multicopy plasmid, the genomic fragment harboring CNS1 conferred growth rates in both hsc82Δ cpr7Δ and cpr7Δ cells that were nearly equal to wild-type rates (Fig. 1).
To confirm that the CNS1/YBR155w ORF was responsible for the suppression of the cpr7Δ slow-growth phenotype, we constructed a TRP1-marked multicopy plasmid harboring the CNS1 ORF (pJM111) and tested its abilities to suppress the lethality of a cns1Δ (ybr155wΔ::LEU2) mutation (2) and increase the growth rate of a cpr7Δ recipient. Introduction of pJM111 into a diploid heterozygous for the ybr155wΔ::LEU2 mutation resulted in the occurrence of viable Leu+ spore colonies among the meiotic progeny, confirming that pJM111 expressed a functional CNS1 gene. In contrast, control diploids transformed with vector yielded only the expected 0:2 Leu+:Leu segregation pattern among the offspring (data not shown). The overexpression of CNS1 from pJM111 was able to suppress cpr7Δ-dependent slow-growth phenotypes. When introduced into cpr7Δ hsc82Δ and cpr7Δ recipient cells, pJM111 was able to increase the growth rate of these cells to nearly that of wild-type cells (Fig. 1B). Thus, the ORF corresponding to CNS1/YBR155w is sufficient for suppression of the slow-growth phenotype of cpr7Δ hsc82Δ and cpr7Δ cells.
The growth rate of cpr7Δ cells can also be exacerbated by deletion of STI1 (16), which encodes a nonessential component of Hsp90 heterocomplexes (34). To test further the ability of CNS1 overexpression to suppress growth defects conferred by deletion of CPR7, we introduced pJM111 into a sti1Δ cpr7Δ recipient strain and assessed the growth rates of the transformants by streaking for single colonies on plasmid-selective medium. Overexpression of CNS1 from pJM111 suppressed the severe slow-growth phenotype of sti1Δ cpr7Δ cells (data not shown). Thus, overexpression of CNS1 can functionally replace Cpr7 even in backgrounds in which Cpr7 is nearly essential for growth.
Although overexpression of CNS1 suppressed growth defects that resulted from loss of CPR7, the overexpression of two other Hsp90-associated proteins, Cpr6 and Sti1, failed to confer even partial suppression of the cpr7Δ growth phenotype (unpublished results).
Overexpression of CNS1 can suppress other cpr7Δ phenotypes.
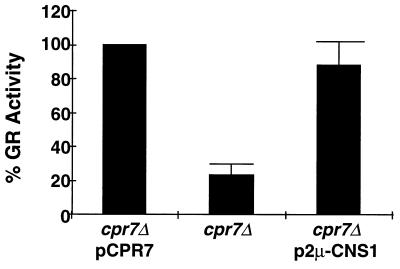
In addition to its role in normal growth, Cpr7 is required for full activity of heterologously expressed Hsp90 substrates such as GR (16). To test the breadth of Cns1 involvement in Cpr7-dependent functions, pJM107, a URA3-marked multicopy plasmid expressing CNS1 was introduced into cpr7Δ recipient cells that express the heterologous GR and harbor the glucocorticoid response element GRE-lacZ reporter construct p2A/GRGZ. GR-promoted lacZ expression was measured and compared to the activity in cpr7Δ cells and cpr7Δ cells expressing CPR7 from a centromeric plasmid. In these experiments GR activity was nearly fivefold lower in the cpr7Δ cells than in the CPR7 control cells. In contrast, GR activity was increased to nearly wild-type levels in cpr7Δ cells harboring the multicopy CNS1 plasmid (Fig. 2). Thus, increased gene dosage of CNS1 is sufficient to substantially restore Cpr7-dependent Hsp90 chaperoning of this heterologous substrate. Although overexpression of CNS1 restored GR activity to cpr7Δ cells, the overexpression of other Hsp90-associated proteins, namely, Cpr6 (19) and Sti1 (data not shown), could not do so.
FIG. 2.
Overexpression of CNS1 restores GR activity to cpr7Δ cells. cpr7Δ cells harbored a GR-dependent lacZ reporter gene and either the control vector (middle lane) or plasmids expressing CPR7 (pAAD97) or CNS1 (pJM107) as indicated. Cells were treated with hormone and assayed for GR activity as described in Materials and Methods. Each column is the mean ± standard deviation of at least four independent experiments expressed as a percentage of activity of cpr7Δ cells harboring pAAD97.
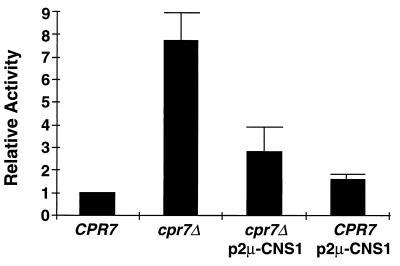
We recently discovered a specific endogenous role for Hsp90 in regulation of the heat shock response in S. cerevisiae (17). Hsp90 negatively regulates the transcriptional activity of HSF under both normal and stress conditions. Furthermore, Cpr7 plays a major role in this regulation. HSF activity is derepressed in cpr7Δ cells, resulting in a significant increase in steady-state HSF-dependent gene expression (17). Thus, as another test of the functional relationship between Cns1 and Cpr7, we assessed the ability of Cns1 to restore repression of HSF activity in cpr7Δ cells. Wild-type and cpr7Δ cells harboring the plasmid-borne HSF-dependent reporter HSE2::lacZ were transformed with the multicopy CNS1 plasmid pJM111 or with vector and assayed for β-galactosidase activity. In these experiments, HSE2::lacZ expression was eightfold higher in vector-harboring cpr7Δ cells than in the wild-type CPR7 control cells. Although overexpression of CNS1 in wild-type cells did not affect HSE2::lacZ expression, in cpr7Δ cells the increased CNS1 gene dosage largely restored repression of the reporter gene (Fig. 3). Thus, overexpression of CNS1 is able to substitute for Cpr7 in the down regulation of HSF.
FIG. 3.
Overexpression of CNS1 restores down regulation of HSF activity in cpr7Δ cells. Steady-state β-galactosidase activity in wild-type (CPR7) and cpr7Δ cells harboring the HSF-dependent reporter plasmid pHSE2-lacZ and either vector alone or the multicopy CNS1 plasmid pJM111 (p2μ-CNS1) was measured as described in Materials and Methods. Indicated activities are relative to those of wild-type cells. Each bar represents the mean ± standard deviation of at least six independent experiments.
Overexpression of Hsp90-associated TPR-containing proteins is unable to suppress the lethal phenotype of cns1Δ cells.
CNS1/YBR155w is an essential gene encoding a protein 385 amino acids in length that is weakly related to Sti1 (13% identity) (2), the S. cerevisiae homolog of the Hsp90-associated protein p60 (34). Cns1 harbors three TPR motifs in its amino-terminal half which share significant sequence identity with those of Cpr6 and Cpr7 (Fig. 4) and are also related to the TPR motifs in other Hsp90-associated proteins, including Sti1 and Ppt1, the S. cerevisiae homolog of PP5. A search of the databases failed to identify proteins that share significant sequence identity with the carboxy-terminal half of Cns1.
FIG. 4.
TPR motifs in Cns1. The location of each of the three TPR motifs in Cns1 is depicted by a hatched box. The TPR motifs of CN21 are compared with those of Cpr6 and Cpr7. Sites containing identical amino acids are shaded. Underlined amino acids correspond to residues that fit the TPR consensus motif *--*G-*Y/F-----*--A*--Y/F--A*-*-P----- (24), where ∗ represents any large hydrophobic amino acid and - represents any amino acid.
To investigate further the relationship between Cns1 and the Hsp90 machinery, other TPR-containing proteins known to associate with the chaperone were tested for the ability, when overexpressed, to suppress the lethality of cns1Δ cells. Plasmids overexpressing Cpr6, Cpr7, or Sti1 were introduced into a CNS1/cns1Δ heterozygous diploid, transformants were sporulated, and the meiotic progeny were analyzed. Overexpression of CPR7, CPR6, or STI1 failed to suppress the cns1Δ lethal phenotype (data not shown) indicating that the essential function performed by Cns1 cannot be replaced by these TPR-containing proteins.
Cns1 is a component of the Hsp90 chaperone machinery.
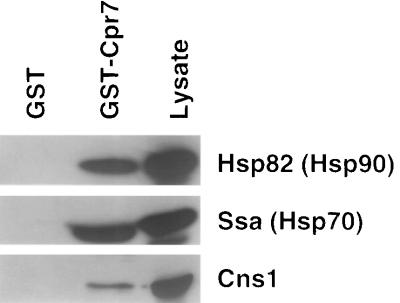
Because Cpr7 is a component of Hsp90 heterocomplexes (16), the ability of multicopy CNS1 plasmids to suppress the cpr7Δ phenotypes suggested that Cns1 might physically interact with the chaperone complex. Physical association between Cns1 and the Hsp90 machinery was tested in wild-type cells harboring a multicopy plasmid (pJM111) expressing HA epitope-tagged alleles of CNS1 (see Materials and Methods). Lysates from S. cerevisiae cells expressing Cns1-3HA fusion protein were mixed with glutathione beads coated with either GST or GST-Cpr7 fusion protein purified from bacterial extracts. The beads were then washed, and bound proteins were eluted and analyzed. Immunoblots of the eluates revealed the presence of Hsp90 and Ssa (an S. cerevisiae homolog of Hsp70), indicating that GST-Cpr7 precipitated Hsp90 heterocomplexes from the lysates (Fig. 5). Immunoblotting with anti-HA antibody revealed the presence of Cns1 among the precipitated proteins, demonstrating that Cns1 physically interacts with Hsp90 or its associated proteins. The fact that much less Cns1-3HA is obtained by precipitation with GST-Cpr7 than with Hsp90 (see below) suggests that only a minority of Cns1-containing complexes also contain Cpr7. GST-Cpr7 fusion protein was also able to precipitate Cns1-3HA from lysates expressing the epitope-tagged fusion protein from the low-copy-number plasmid pJM110 (not shown).
FIG. 5.
Precipitation of Cpr7 coprecipitates Cns1-3HA. Wild-type cells harboring the multicopy plasmid pJM111 expressing HA epitope-tagged Cns1 were cultured and lysates prepared as described in Materials and Methods. Cell lysates were incubated with glutathione beads charged with either GST or GST-Cpr7 fusion protein. Proteins precipitated by either GST or GST-Cpr7 or total cellular proteins (Lysate) were identified by immunoblotting with the appropriate antibodies.
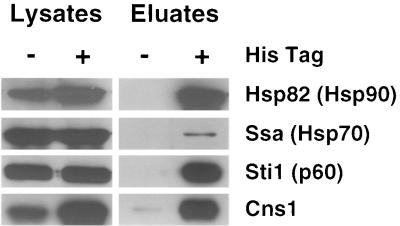
A second approach was used to test Hsp90 complexes for the presence of Cns1. Wild-type cells and cells expressing Hsp82FP, a histidine-tagged Hsp90 fusion protein, were transformed with a low-copy-number (pJM110) or multicopy (pJM107 or pJM111) CNS1::3HA plasmid. Hsp82FP but not Hsp82 was precipitated from lysates by a nickel matrix. Immunoblotting of the eluates showed that Ssa and Sti1 coprecipitated with Hsp82FP (Fig. 6). Immunoblotting with anti-HA antibody revealed that Cns1 also coprecipitated with the Hsp90 complex. Thus, two protein retention assays revealed that Cns1 is a component of the Hsp90 heterocomplexes.
FIG. 6.
Precipitation of Hsp90 coprecipitates Cns1-3HA. Cells expressing wild-type Hsp90 (Hsp82) or histidine-tagged Hsp90 (Hsp82FP) were transformed with a low-copy-number (pJM110) or multicopy (pJM111) CNS1::3HA plasmid. Total cellular protein (lanes 1 and 2) and proteins eluted from a nickel affinity matrix (lanes 3 and 4) were analyzed by immunoblotting with the appropriate antibodies. Coprecipitation of Cns1-3HA was also observed from lysates expressing the fusion protein from the low-copy-number plasmid pJM110 (not shown).
Finally, we tested for the ability of Cns1 to interact with Hsp90 by the yeast two-hybrid assay (26). Using a full-length Cns1-3HA fusion protein expressed from pJM123 as bait, we found that the carboxy-terminal 182 amino acids of Hsp82 were sufficient to strongly induce the HIS3, ADE2, and lacZ reporter genes (see Materials and Methods). Cells expressing GAL4BD-Cns1-3HA and GAL4AD-Hsp82C yielded 678 ± 91 U of β-galactosidase activity (n = 3), whereas cells harboring the GAL4AD-Hsp82C construct alone yielded only 26 ± 3.6 U of activity (n = 3). In addition, this interaction was not significantly diminished by deletion of CPR7 (631 ± 61 U; n = 3). Thus, physical association between Cns1 and the Hsp90 complex is independent of Cpr7.
DISCUSSION
CNS1 is an essential gene that we identified by a screen for multicopy suppressors of the severe slow-growth phenotype caused by deletion of the Hsp90-associated Cyp40-type cyclophilin Cpr7. Although isolated by its ability to suppress the severe growth defect of cpr7Δ hsc82Δ cells, overexpression of CNS1 also suppressed the slow-growth phenotype of cells containing only the cpr7Δ mutation. Increased CNS1 gene dosage also restored other Cpr7-dependent activities in cpr7Δ cells, including the activity of the heterologously expressed GR and negative regulation of HSF, both of which are Hsp90 dependent (6, 17). In addition, two-hybrid and in vitro protein-protein interaction analyses indicated that Cns1 can interact with one or more components of Hsp90 heterocomplexes. Thus, our results show that Cns1 is a previously unidentified component of the Hsp90 chaperone complex that is essential for viability.
The ability of increased CNS1 gene dosage to suppress distinct cpr7Δ phenotypes suggests that Cns1 broadly substitutes for Cpr7. While the precise function of Cpr7 in molecular chaperoning remains unknown, the cyclophilin clearly plays a major role in the Hsp90 activities, as it is required not only for rapid growth but also for full activity of GR and pp60v-src and for negative regulation of HSF (16–18). Cns1 appears to be the only TPR-containing component of Hsp90 heterocomplexes that can substitute functionally for Cpr7. The overexpression of CPR6 or STI1 does not increase the growth rate or restore the GR activity of cpr7Δ cells (reference 19 and data not shown). Despite an exhaustive search of a multicopy genomic library, CNS1 was the only extragenic suppressor obtained.
Two scenarios could explain the suppression of cpr7Δ phenotypes by Cns1. Since CNS1 acts as a suppressor of a null allele of CPR7, formally, Cns1 appears to act at or downstream of Cpr7. Thus, it is possible that the phenotypes that arise from the loss of Cpr7 might actually be due to decreased Cns1 activity and that the overexpression of CNS1 merely restores Cns1-dependent functions. Alternatively, the ability of Cns1 to suppress cpr7Δ phenotypes might reflect direct functional overlap between the two proteins. In this case, because both Cpr7 and Cns1 contain three-unit TPR domains and because both are components of Hsp90 heterocomplexes, a reasonable scenario is one in which Cns1 substitutes for Cpr7 in binding to the TPR-accepting pocket(s) of Hsp90 dimers of the appropriate heterocomplexes, imparting upon them Cpr7-like functions. If so, it is not surprising that the overexpression of Sti1 cannot suppress the cpr7Δ phenotypes because data from in vitro reconstitution studies with mammalian Hsp90 heterocomplexes indicate that p60 (Sti1) and the Cyp40-type cyclophilins are not found in the same heterocomplex (35). However, it is surprising that the closely related cyclophilin Cpr6 cannot substitute for Cpr7, even when overexpressed, as the region containing the TPR motifs of Cpr6 is 34% identical to that of Cpr7 whereas the TPR-containing regions of Cns1 and Cpr7 are only 9.6% identical. This is all the more surprising given that overexpression of the TPR-containing carboxy terminus of Cpr7 restores Cpr7-dependent functions to cpr7Δ cells (19). The inability of Cpr6 to replace Cpr7 does not appear to be due to an inhibitory function specific to the isomerase domain of Cpr6 since a Cpr6-Cpr7 chimera containing the amino-terminal half of Cpr6 and the carboxy-terminal half of Cpr7 can functionally substitute for Cpr7 in vivo (unpublished results). The hypothesis that Cns1 might occupy the TPR-accepting pocket(s) of the Hsp90 complex is also supported by recent work involving chimeric proteins between the Hsp90-associated immunophilins FKBP51 and FKBP52 (1). Individual chimeras displayed preferential binding to Hsp90-receptor complexes, depending on which TPR-containing carboxyl termini were present.
The repertoire of TPR proteins known to interact with Hsp90 heterocomplexes was broadened by the recent discovery that the mammalian protein phosphatase PP5, which harbors four TPR units, interacts with Hsp90 (11, 46). Although it can bind FK506 weakly, PP5 is not a member of the highly conserved FKBP family of proteins (46). This was the first evidence that a nonimmunophilin TPR-containing protein can associate with Hsp90 in mature receptor complexes. Our discovery (and that described in reference 14) that Cns1 is an essential nonimmunophilin TPR-containing component of the Hsp90 machinery suggests that these complexes are even more heterogeneous than previously thought. Whether Cns1 occupies specific niches among different types of heterocomplexes or even defines such complexes remains to be determined. The mammalian Hsp90-associated immunophilins do not precipitate with other Hsp90-associated immunophilins, indicating that TPR-containing immunophilins are not present in the same complex (see references 37 and 47 for reviews). Based on these and other studies, the stoichiometry of Hsp90-receptor complexes has been postulated to include one TPR-containing immunophilin per Hsp90 dimer (12, 39, 47). However, we found Cns1 among the proteins that could be removed from solution by GST-Cpr7. Thus, if Cns1 interacts directly with Hsp90 via its TPR domains, the one-TPR-accepting-pocket–one-TPR-partner hypothesis may be insufficient to explain the precipitation of Cns1 by Cpr7.
Although Hsp90-associated proteins are highly conserved among eukaryotes, in S. cerevisiae many, including Sti1 (p60), Cpr6 and Cpr7 (Cyp40), Ydj1 (DnaJ), Sba1 (p23), and Ppt1 (PP5), are not essential (16, 20, 28, 34, 46). In contrast, Cns1 is essential at all temperatures (2). Moreover, the lethality caused by the cns1Δ mutation cannot be suppressed by overexpression of other TPR-containing proteins, including Cpr6, Cpr7, or Sti1. Taken together, these observations suggest a key functional role for Cns1 in Hsp90 chaperone complexes.
ACKNOWLEDGMENTS
We thank S. Lindquist for plasmids, S. cerevisiae strains, and anti-Hsp82 antibodies, D. Toft for anti-Sti1 antibodies, and E. Craig for anti-Ssa antibodies. We thank D. Winge for the pHSE2-lacZ and pHSE12-lacZ reporter genes.
J.A.M. was supported in part by the ARCS Foundations and by predoctoral biotechnology training grant T32GM-08449. H.M.K. was supported by a National Science Foundation Research Experiences for Undergraduates supplement to R.F.G. This work was supported by National Science Foundation grant MCB-9724050 to R.F.G.
ADDENDUM IN PROOF
Our two-hybrid results with Cns1 are consistent with the findings of Young et al. (J. Biol. Chem. 273:18007–18010, 1998) published during review of this paper, in which the 104 carboxy-terminal residues of Hsp90 were sufficient to interact with TPR-containing Hsp90-associated proteins FKBP51, FKBP52, and hTom34p.
REFERENCES
- 1.Barent R L, Nair S C, Damon C C, Ruan Y, Rimerman R A, Fulton J, Zhang Y, Smith D F. Analysis of FKBP51/FKBP52 chimeras and mutants for Hsp90 binding and association with progesterone receptor complexes. Mol Endocrinol. 1998;12:342–354. doi: 10.1210/mend.12.3.0075. [DOI] [PubMed] [Google Scholar]
- 2.Baur A, Schaaff-Gerstenschlager I, Boles E, Miosga T, Rose M, Zimmermann F K. Sequence of a 4.8 kb fragment of Saccharomyces cerevisiae chromosome II including three essential open reading frames. Yeast. 1993;9:289–293. doi: 10.1002/yea.320090308. [DOI] [PubMed] [Google Scholar]
- 3.Bohen S P, Kralli A, Yamamoto K R. Hold’em and fold’em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- 4.Bohen S P, Yamamoto K R. Modulation of steroid receptor signal transduction by heat shock proteins. In: Morimoto R I, Tissiérres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- 5.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperature. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnick E H, Dalman F C, Sanchez E R, Pratt W B. Evidence that the 90-kDa heat shock protein is necessary for the steroid binding conformation of the L cell glucocorticoid receptor. J Biol Chem. 1989;264:4992–4997. [PubMed] [Google Scholar]
- 7.Brugge J S. Interaction of the Rous sarcoma virus protein pp60src with the cellular proteins pp50 and pp90. Curr Top Microbiol Immunol. 1986;123:1–22. doi: 10.1007/978-3-642-70810-7_1. [DOI] [PubMed] [Google Scholar]
- 8.Caplan A J. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrinol Metab. 1997;8:271–276. doi: 10.1016/s1043-2760(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 9.Chang H-C J, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 10.Chang H-C J, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M S, Silverstein A M, Pratt W B, Chinkers M. The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J Biol Chem. 1996;271:32315–32320. doi: 10.1074/jbc.271.50.32315. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Prapapanich V, Rimerman R A, Honore B, Smith D F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins Hsp90 and Hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 13.Cutforth T, Rubin G M. Mutations in Hsp83 and cdc37 impair signalling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 14.Dolinski K, Cardenas M E, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18:7344–7352. doi: 10.1128/mcb.18.12.7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 17.Duina A A, Kalton H M, Gaber R F. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974. [DOI] [PubMed] [Google Scholar]
- 18.Duina A A, Marsh J A, Gaber R F. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast. 1996;12:943–952. doi: 10.1002/(sici)1097-0061(199608)12:10<943::aid-yea997>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Duina A A, Marsh J A, Kurtz R B, Chang H-C J, Lindquist S, Gaber R F. The peptidyl-prolyl isomerase domain of the CyP-40 cyclophilin homolog Cpr7 is not required to support growth or glucocorticoid receptor activity in Saccharomyces cerevisiae. J Biol Chem. 1998;273:10819–10822. doi: 10.1074/jbc.273.18.10819. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Fliss A E, Rao J, Caplan A J. SBAI encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman B C, Toft D O, Morimoto R I. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 22.Galat A. Peptidylproline cis-trans-isomerases: immunophilins. Eur J Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa W C. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 24.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 25.Hutchison K A, Dittmar K D, Czar M J, Pratt W B. Proof that Hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with Hsp90. J Biol Chem. 1994;269:5043–5049. [PubMed] [Google Scholar]
- 26.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson J L, Toft D O. Novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269:24989–24993. [PubMed] [Google Scholar]
- 28.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone Ydj1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 29.Marks A R. Cellular functions of immunophilins. Physiol Rev. 1996;76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- 30.Miyata Y, Yahara I. The 90-kDa heat shock protein, Hsp90, binds and protects casein kinase II from self-aggregation and enhances its kinase activity. J Biol Chem. 1992;267:7042–7047. [PubMed] [Google Scholar]
- 31.Nair S C, Toran E J, Rimerman R A, Hjermstad S, Smithgall T E, Smith D F. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nathan D, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathan D F, Harju Vos M, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owens-Grillo J K, Czar M J, Hutchison K A, Hoffmann K, Perdew G H, Pratt W B. A model of protein targeting mediated by immunophilins and other proteins that bind to hsp90 via tetratricopeptide repeat domains. J Biol Chem. 1996;271:13468–13475. doi: 10.1074/jbc.271.23.13468. [DOI] [PubMed] [Google Scholar]
- 36.Owens-Grillo J K, Hoffmann K, Hutchison K A, Yem A W, Deibel M R, Jr, Handschumacher R E, Pratt W B. The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- 37.Pratt W B. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268:21455–21458. [PubMed] [Google Scholar]
- 38.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrine Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 39.Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 40.Redeuilh G, Moncharmont B, Secco C, Baulieu E E. Subunit composition of the molybdate-stabilized “8-9 S” nontransformed estradiol receptor purified from calf uterus. J Biol Chem. 1987;262:6969–6975. [PubMed] [Google Scholar]
- 41.Rose D W, Wettenhall R E, Kudlicki W, Kramer G, Hardesty B. The 90-kilodalton peptide of the heme-regulated eIF-2 alpha kinase has sequence similarity with the 90-kilodalton heat shock protein. Biochemistry. 1987;26:6583–6587. doi: 10.1021/bi00395a003. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford S L, Zuker C S. Protein folding and the regulation of signaling pathways. Cell. 1994;79:1129–1132. doi: 10.1016/0092-8674(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez E R, Toft D O, Schlesinger M J, Pratt W B. Evidence that the 90-kDa phosphoprotein associated with the untransformed L-cell glucorcorticoid receptor is a murine heat shock protein. J Biol Chem. 1985;86:1123–1127. [PubMed] [Google Scholar]
- 44.Schuh S, Yonemoto W, Brugge J, Bauer V J, Riehl R M, Sullivan W P, Toft D O. A 90,000-dalton binding protein common to both steroid receptors and the Rous sarcoma virus transforming protein, pp60v-src. J Biol Chem. 1985;260:14292–14296. [PubMed] [Google Scholar]
- 45.Shue G, Kohtz D S. Structural and functional aspects of basic helix-loop-helix protein folding by heat shock protein 90. J Biol Chem. 1994;269:2707–2711. [PubMed] [Google Scholar]
- 46.Silverstein A M, Galigniana M D, Chen M S, Owens-Grillo J K, Chinkers M, Pratt W B. Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J Biol Chem. 1997;272:16224–16230. doi: 10.1074/jbc.272.26.16224. [DOI] [PubMed] [Google Scholar]
- 47.Smith D F. Dynamics of heat shock protein 90-progesterone receptor binding and disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 48.Smith D F, Whitesell L, Nair S C, Chen S, Prapapanich V, Rimerman R A. Progesterone receptor structure and function altered by geldanamycin, and Hsp90-binding agent. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorger P K, Pelham H R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987;6:3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stancato L F, Chow Y-H, Hutchison K A, Perdew G H, Jove R, Pratt W B. Raf exists in a native heterocomplex with Hsp90 and p50 that can be reconstituted in a cell-free system. J Biol Chem. 1993;268:21711–21716. [PubMed] [Google Scholar]
- 50a.Stear, J., and R. F. Gaber. Unpublished data.
- 51.Wartmann M, Davis R J. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J Biol Chem. 1994;268:21711–21716. [PubMed] [Google Scholar]
- 52.Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperone protein folding in vitro. Nature. 1992;358:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]