Abstract
Mowat–Wilson syndrome (MWS) is a rare genetic neurodevelopmental congenital disorder associated with various defects of the zinc finger E-box binding homeobox 2 (ZEB2) gene. The ZEB2 gene is autosomal dominant and encodes six protein domains including the SMAD-binding protein, which functions as a transcriptional corepressor involved in the conversion of neuroepithelial cells in early brain development and as a mediator of trophoblast differentiation. This review summarizes reported ZEB2 gene variants, their types, and frequencies among the 10 exons of ZEB2. Additionally, we summarized their corresponding encoded protein defects including the most common variant, c.2083 C>T in exon 8, which directly impacts the homeodomain (HD) protein domain. This single defect was found in 11% of the 298 reported patients with MWS. This review demonstrates that exon 8 encodes at least three of the six protein domains and accounts for 66% (198/298) of the variants identified. More than 90% of the defects were due to nonsense or frameshift changes. We show examples of protein modeling changes that occurred as a result of ZEB2 gene defects. We also report a novel pathogenic variant in exon 8 in a 5-year-old female proband with MWS. This review further explores other genes predicted to be interacting with the ZEB2 gene and their predicted gene–gene molecular interactions with protein binding effects on embryonic multi-system development such as craniofacial, spine, brain, kidney, cardiovascular, and hematopoiesis.
Keywords: Mowat–Wilson syndrome (MWS), case report, review, ZEB2 gene variants, ZEB2 protein domains and defects, ZEB2 functional molecular interactions
1. Introduction
Rare genetic disorders have been estimated to affect up to 10% of the population [1]. Advances in genomic technologies such as exome sequencing are becoming more widely used to gain insight into the cause and diagnosis of these rare disorders by identifying specific molecular defects and outcomes in patients with Mendelian or non-Mendelian disorders. Exome sequencing studies have shown underlying causative genes in approximately 25% of cases [2]. A growing number of gene variants and types involved in protein production have not been well characterized in rare disorders, including Mowat–Wilson syndrome.
Mowat–Wilson syndrome (MWS) is an example of a rare genetic disorder with intellectual disabilities with multiple congenital anomalies and multi-system involvement. This disorder was first reported in 1998 [3] and now about 300 patients have been found in the medical literature or databases. Chromosome 2q22-2q23 deletions were reported in the first patients with this disorder and the zinc finger E-box binding homeobox 2 (ZEB2; NM_014795.4) gene was found in this region. Other patients with MWS with overlapping features having ZEB2 gene deletions or duplications of different sizes and intragenic variants have been identified [4,5,6].
Mowat–Wilson syndrome shows clinical variability and is recognized as a multiple congenital anomaly disorder involving several organ systems. Clinical facial features include a square-shaped face with a prominent and triangular chin, high forehead, large eyebrows with medial flaring, hypertelorism, deep-set and large eyes, broad and depressed nasal bridge, rounded nasal tip, prominent columella, open mouth, and M-shaped upper lip. The ears are posteriorly rotated with large, uplifted earlobes with a central depression reminiscent of a red blood corpuscle. The face lengthens with age and the chin becomes more prominent with the appearance of broad-appearing eyebrows. Individuals with this disorder often have severe intellectual disability with a mean age of walking at four years and a wide-based gait with a tendency for flexed arms in resting position [7,8,9,10]. Most individuals with MWS have seizures (84%) and an abnormal EEG. Short stature and microcephaly are often present with cerebral anomalies including corpus callosum and hippocampal defects, enlargement of cerebral ventricles, white matter abnormalities, large basal ganglia, and cortical and cerebellar malformations. Gastrointestinal problems such as chronic constipation are present and are most often related to lack of innervation causing Hirschsprung disease of either the short or long segment variety and are documented in about 50% of patients. Congenital heart disease is reported in 58% of patients including patent ductus arteriosus, atrial septal or ventricular septal defects, pulmonary stenosis, aortic coarctation, Tetralogy of Fallot, aortic valve abnormalities, and a pulmonary artery sling with or without tracheal stenosis/hypoplasia. Genitourinary and kidney anomalies are common including hypospadias, bifid scrotum, cryptorchidism, pelvic or duplex kidneys, and hydronephrosis [7,8]. The wide range of clinical findings may relate to different ZEB2 gene variants and therefore additional research is needed to identify the type and frequency of gene variants and their impact on protein structure and function.
The ZEB2 gene is located on chromosome 2q22.3 and expressed in the human nervous system throughout development, exemplifying its importance in gliogenic and neurogenic processes. ZEB2 has been documented to play roles in the induction of the neuroectoderm and neural crest. It acts to direct neural crest cells and regulates the development of cerebral regions, along with development of the spinal cord, cardiac, and enteric systems [8,9]. Hence, individuals with MWS can present with a combination of multi-system deficits with variable penetrance [10,11,12].
In searching the medical literature and unreported databases, we found a total of 298 patients with MWS and ZEB2 variants. We tabulated these variants in the ZEB2 gene and protein and summarized the frequency of gene variants within each of the 10 exons and their relationship to protein structure and function. We also obtained the predicted ZEB2 gene interactions with other genes implicated and molecular pathways pertaining to neurodevelopmental multi-system involvement. Additionally, we described a new patient with MWS having a novel pathogenic variant due to a heterozygous c.2471_2475del5 in exon 8 of the ZEB2 gene.
2. Detailed Case Description
2.1. Clinical Case Report
Our proband was prenatally diagnosed with a complex cardiovascular single ventricular disorder and subsequent amniocentesis genetic testing was normal. She was delivered at 37 weeks’ gestation with a double outlet right ventricle, subaortic and anterior muscular ventricular septal defects, and hypoplastic mitral and tricuspid valves with a hypoplastic left heart. The family initially took her home for palliative care, but upon further evaluation, underwent multiple cardiac and surgical procedures, including Fontan and Glenn procedures. These surgical interventions, which took place between one and six months of age, led to a partial recovery of cardiac function.
Our proband has two healthy siblings without cardiac or other disorders. The family history was also unremarkable for birth defects and no consanguinity was noted. The patient required G-tube feedings and close health monitoring with multiple evaluations and hospitalizations throughout infancy. She had a normal karyotype and chromosomal microarray studies during infancy.
Around one year of age, a comprehensive connective tissue genetic test including the autosomal dominant ZEB2 gene (NM_014795) was ordered via a commercially approved genetic testing laboratory (Connective Tissue Gene Tests (CTGT)). The DNA sequencing revealed a heterozygous c.2471_2475del5 in exon 8 of the ZEB2 gene. These five base pair deletions resulted in a frameshift, causing aberrant mRNA transcription and the defective protein causing MWS. All coding exons and exon boundaries of the gene were amplified by PCR and ABI 3730 sequencers, as standard genetic testing at the time. Additionally, coding exons and exon boundaries were analyzed for copy number variation using a high-density targeted array. The genetic defect was considered de novo in view of the negative family history, including two unaffected siblings for birth defects or features of MWS. ZEB2 is an autosomal dominant gene and parental DNA testing was not undertaken.
At about three years of age her height was 87.3 cm (6%ile), weight was 11.7 kg (7%ile), and body mass index was 15.35 kg/m2 (36%ile). At that age, her heart rate was 120, respiration was 20, oxygen saturation was 80%, blood pressure (right arm) was 85/33, and blood pressure (right leg) was 107/54. At five years and four months of age, her height was 102.1 cm (3%ile), weight was 14.3 kg (3%ile), and body mass index was 13.72 kg/m2 (15%ile). She was able to write, recognize a few written words, and perform several preschool-appropriate skills. She spoke at the level of a two-and-a-half-year-old and communicated her wants and needs, both verbally and with sign language. She had severe cardiac defects, including, but not limited to hypoplastic left heart syndrome (HLHS), ventricular septal defects (VSDs), left pulmonary artery (LPA) sling causing tracheomalacia, dysplastic tricuspid valve causing severe tricuspid regurgitation (TR), and partial anomalous pulmonary venous connection (PAPVR). She presented with typical MWS facial features, a duplex kidney and VUR, visual deficits, tooth abnormalities, hypotonia, global delays, secondary kidney and liver issues, high intracranial pressure (pseudotumor cerebri), and GI issues. She was not able to eat food, as she would develop severe abdominal pain, intractable vomiting, GI bleeding, and colitis, often requiring hospitalization for several weeks requiring IV fluids and/or TPN after ingestion of any amount of food. She was treated with diuretics. She was G-tube dependent and fed neonate infant formula (an elemental formula), with some GI bleeding, vomiting, and discomfort noted at baseline. Although Hirschsprung disease was ruled out based on rectal biopsy studies, she had gastroparesis and chronic constipation. Her EEG studies on more than one occasion were normal. She had no documented seizures/epilepsy (see Figure 1). Her overall health status, function, and quality of life continued to decline in spite of continuous care and monitoring. Her parents made the ultimate decision to enroll her in hospice care where she passed away at about 5 years of age.
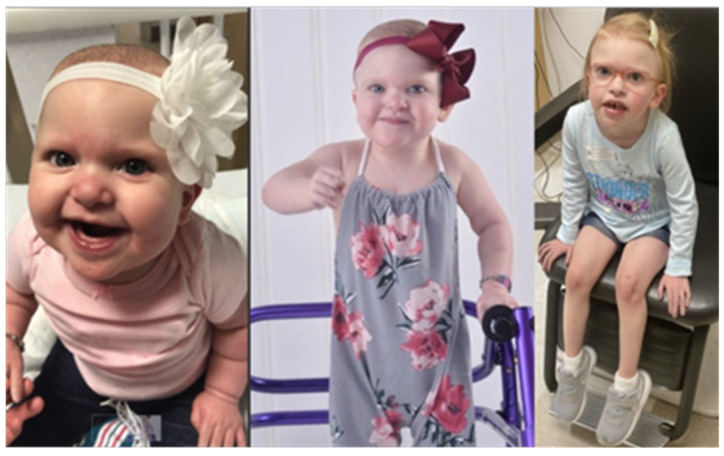
Figure 1.
Our proband with typical craniofacial features and phenotype of MWS. She had a pathogenic heterozygous c.2471_2475del5 in exon 8 of the ZEB2 gene. Photos were obtained with consent during infancy, early childhood, and before her death at about five years of age.
2.2. Genetic and Protein Domain Data Collection of Patients with Mowat-Wilson Syndrome
Computer literature and unreported databases were searched for keywords such as Mowat–Wilson, ZEB2 gene and protein defects or variants, or clinical features using PUBMED (www.pubmed.com; accessed on 1 October 2022) and performed from 2001 to the present (2023). About 180 published reports were found with the most useful data obtained from approximately 20 articles, as summarized in Table 1. From this study, we analyzed 266 cases of Mowat–Wilson syndrome and an additional 32 deidentified unpublished patients accessed from the Mowat–Wilson Syndrome Foundation for a total of 298 patients. These sources were used to collect data regarding ZEB2 gene variants, types, frequencies, and protein defects along with domain locations and functions. We found that exon 8 encodes at least three of the six protein domains of the ZEB2 gene and accounts for 66% (198/298) of the variants identified.
Table 1.
Review of Mowat–Wilson syndrome with ZEB2 gene and protein variants.
| Our Study ID |
Publication ID/
(Patient Number) |
ZEB2 Exon | ZEB2 Gene Variant or Defect | Protein Defect |
Type of Genetic
Defect |
ZEB2 Protein Domain |
|---|---|---|---|---|---|---|
| 1 | Mowat–Wilson Syndrome Foundation [MWSF]/(P1) | - | c.2180del | p.Leu727Tyrfs*7 | Frameshift | - |
| 2 | MWSF/(P2) | 8 | c.2083C>T | p.Arg695Ter | Nonsense | HD |
| 3 | MWSF/(P3) | 9 | c.3002del | p.Cys1001LeufsX74 | Frameshift | C-ZFa |
| 4 | MWSF/(P4) | 8 | c.2761C>T | p.Arg921* | Nonsense | - |
| 5 | MWSF/(P5) | 9 | c.3095G>A | p.Cys1032Tyr | Missense | C-ZFb |
| 6 | MWSF/(P6) | 10 | c.3213dup | p.Q1072AfsX52 | Frameshift | C-ZFb |
| 7 | MWSF/(P7) | 6 | c.696C>G | p.Tyr232* | Nonsense | N-ZFa |
| 8 | MWSF/(P8) | 6 | c.805C>T | p.Q269X | Nonsense | N-ZFb |
| 9 | MWSF/(P9) | 8 | c.2061del | p.Phe687Leufs*2 | Frameshift | HD |
| 10 | MWSF/(P10) | 10 | - | p.Tyr999* | Nonsense | C-ZFa |
| 11 | MWSF/(P11) | 3 | c.108del | p.E37fsX74 | Frameshift | - |
| 12 | MWSF/(P12) | 8 | c.2721del | p.Thr908LeufsTer22 | Frameshift | - |
| 13 | MWSF/(P13) | 2 | c.81_84dup | p.Asp29Leufs*2 | Frameshift | - |
| 14 | MWSF/(P14) | 8 | c.2094C>A | p.Y698X | Nonsense | HD |
| 15 | MWSF/(P15) | 6 | c.763C>T | p.Q255X | Nonsense | N-ZFa |
| 16 | MWSF/(P16) | intron 3 | c.331+1_331+2dup | - | Splicing | - |
| 17 | MWSF/(P17) | 10 | c.3533del | - | Deletion | - |
| 18 | MWSF/(P18) | 10 | c.3196C>T | p.His1066Tyr | Missense | C-ZFb |
| 19 | MWSF/(P19) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 20 | MWSF/(P20) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 21 | MWSF/(P21) | 8 | c.2367del | - | Frameshift | CID |
| 22 | MWSF/(P22) | 6 | c.674C>A | p.S225X | Nonsense | N-ZFa |
| 23 | MWSF/(P23) | 8 | c.909_910ins | p.H304Ffs*5 | Frameshift | N-ZFc |
| 24 | MWSF/(P24) | 8 | c.1795del | p.His599MetfsX8 | Frameshift | - |
| 25 | MWSF/(P25) | 2 | c.31del | p.Arg11Glyfs*16 | Frameshift | - |
| 26 | MWSF/(P26) | 8 | c.1571_1572ins | p.Ser524Argfs*4 | Frameshift | - |
| 27 | MWSF/(P27) | 1–10 | 6.2 Mb deletion Ch2q22.1-q22.3 | - | Chromosome deletion |
- |
| 28 | MWSF/(P28) | 8 | c.1956C>A | p.Y652X | Nonsense | HD |
| 29 | MWSF/(P29) | 1–10 | 6.9 Mb deletion Ch2q22.1-q22.3 | - | Chromosome deletion |
- |
| 30 | MWSF/(P30) | 4 | c.357_358del | p.Met120GlyfsX11 | Frameshift | - |
| 31 | MWSF/(P31) | - | 144 kb deletion Ch2q22.3 | - | Chromosome deletion |
- |
| 32 | MWSF/(P32) | 10 | c.3212_3215dup | p.Gln1072HisfsTer53 | Frameshift | C-ZFb |
| 33 | [13] Zou et al., 2020/(P1) |
3 | c.290G>A | p.Trp97X | Nonsense | - |
| 34 | Zou et al., 2020/(P2) |
8 | c.1067_1068ins | p.Val357Aspfs*15 | Frameshift | - |
| 35 | Zou et al., 2020/(P3) |
8 | c.2761C>T | p.Arg921X | Nonsense | - |
| 36 | Zou et al., 2020/(P4) |
8 | c.3214C>T | p.Gln1072X | Nonsense | C-ZFb |
| 37 | [14] Hu et al., 2020/(P1) | 3 | c.250G>T | p.E84* | Nonsense | - |
| 38 | [15] Ho et al., 2020/(P1) | 8&9 | ZEB2 gene Exons 8 and 9 deletion | - | Deletion | - |
| 39 | Ho et al., 2020/(P2) | 8 | c.1472_c.1473ins | p.Met491llefs*9 | Small insertion, Frameshift | - |
| 40 | Ho et al., 2020/(P3) | 8 | c.2083C>T | p.Arg695* | Nonsense | HD |
| 41 | Ho et al., 2020/(P4) | 8 | c.1387del | p.Val463Phefs*24 | Frameshift | SMD |
| 42 | Ho et al., 2020/(P5) | 8 | c.2646del | p.Val883Cysfs*4 | Small deletion, Frameshift | - |
| 43 | Ho et al., 2020/(P6) | 3 | c.189del | p.Ser64Valfs*11 | Small deletion, Frameshift | - |
| 44 | Ho et al., 2020/(P7) | 3 | c.189del | p.Ser64Valfs*11 | Small deletion, Frameshift | - |
| 45 | Ho et al., 2020/(P8) | 1–10 | ZEB2 gene Exons 1-10 deletion | - | Deletion | - |
| 46 | Ho et al., 2020/(P9) | 9 | c1297C>T | p.Gln433* | Nonsense | - |
| 47 | Ho et al., 2020/(P10) | 10 | c.3335del | p.Tyr1112Cysfs*128 | Small deletion, Frameshift | - |
| 48 | Ho et al., 2020/(P11) | 1–10 |
ZEB2 gene Exons 1-10 deletion |
- | Deletion | - |
| 49 | Ho et al., 2020/(P12) | 7 | c.857_858del | p.Glu286Valfs*8 | Small deletion, Frameshift | N-ZFb |
| 50 | Ho et al., 2020/(P13) | 3 | c.291G>A | p.Trp97* | Nonsense | - |
| 51 | Ho et al., 2020/(P14) | 8 | c.2865C>A | p.Tyr955* | Nonsense | - |
| 52 | Ho et al., 2020/(P15) | 9 | c.169delins | p.Leu565* | Small indel, Frameshift | - |
| 53 | [16] Wenger et al., 2014/(P1) | 8 | - | p.R695X | Nonsense | HD |
| 54 | Wenger et al., 2014/(P2) | 8 | - | p.Q384X | Nonsense | - |
| 55 | Wenger et al., 2014/(P3) | 8 | - | p.G1182KfsX59 | Frameshift | - |
| 56 | Wenger et al., 2014/(P4) | 5 | - | p.E181Rfs211X | Frameshift | - |
| 57 | Wenger et al., 2014/(P5) | 9 | - | p.F1008C | Missense | C-ZFa |
| 58 | Wenger et al., 2014/(P6) | 7 | c.808-1G>T | - | Splicing | N-ZF |
| 59 | Wenger et al., 2014/(P7) | 10 | - | p.Ser1071Pro | Missense | C-ZFb |
| 60 | Wenger et al., 2014/(P8) | 8 | - | p.L894FfsX53 | Frameshift | - |
| 61 | Wenger et al., 2014/(P9) | 8 | - | p.S434Vfs7X | Frameshift | - |
| 62 | Wenger et al., 2014/(P10) | 8 | - | p.R695X | Nonsense | HD |
| 63 | Wenger et al., 2014/(P11) | 8 | - | p.R695X | Nonsense | HD |
| 64 | Wenger et al., 2014/(P12) | 8 | - | p.R695X | Nonsense | HD |
| 65 | Wenger et al., 2014/(P13) | 8 | - | p.M476WfsX11 | Frameshift | - |
| 66 | Wenger et al., 2014/(P14) | 8 | - | p.P906LfsX24 | Frameshift | - |
| 67 | Wenger et al., 2014/(P15) | 6 | - | p.Q209X | Nonsense | - |
| 68 | Wenger et al., 2014/(P16) | 8 | - | p.V627SfsX4 | Frameshift | - |
| 69 | Wenger et al., 2014/(P17) | 9 | - | p.S1011AfsX53 | Frameshift | C-ZFa |
| 70 | Wenger et al., 2014/(P18) | 6 | - | p.R218RfsX21 | Frameshift | - |
| 71 | Wenger et al., 2014/(P19) | 8 | - | p.V621AfsX25 | Frameshift | - |
| 72 | Wenger et al., 2014/(P20) | 1–10 | - | - | Whole gene deletion | - |
| 73 | Wenger et al., 2014/(P21) | 8 | - | p.S359TfsX3 | Frameshift | - |
| 74 | Wenger et al., 2014/(P22) | 8 | - | p.R695X | Nonsense | HD |
| 75 | Wenger et al., 2014/(P23) | 8 | - | p.Q962X | Nonsense | - |
| 76 | Wenger et al., 2014/(P24) | 8 | - | p.A907LfsX23 | Frameshift | - |
| 77 | Wenger et al., 2014/(P25) | 8 | - | p.G351VfsX19 | Frameshift | - |
| 78 | Wenger et al., 2014/(P26) | 5 | - | p.E181RfsX211 | Frameshift | - |
| 79 | Wenger et al., 2014/(P27) | 8 | c.1224del | - | Deletion | - |
| 80 | Wenger et al., 2014/(P28) | 8 | - | p.I390LfsX6 & p.L388F | Frameshift | - |
| 81 | [6] Baxter et al., 2017/(P1) | 1&2 | - | - | Partial duplication | - |
| 82 | [17] Mundhofir et al., 2012/(P1) | 8 | c.1965C>G | p.Tyr652∗ | Nonsense | HD |
| 83 | [18] Murray et al., 2015/(P1) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 84 | [19] Wang et al., 2019/(P1) |
7 | c.904C>T | p.R302X | Nonsense | N-ZFc |
| 85 | Wang et al., 2019/(P2) |
6 | c.756C>A | p.Y252X | Nonsense | N-ZFa |
| 86 | Wang et al., 2019/(P3) |
8 | c.2761C>T | p.R921X | Nonsense | - |
| 87 | [20] Yamada et al., 2014/(P1) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 88 | Yamada et al., 2014/(P2) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 89 | Yamada et al., 2014/(P3) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 90 | Yamada et al., 2014 (P4) | 7 | c.811C>T | p.Q271X | Nonsense | N-ZFb |
| 91 | Yamada et al., 2014/(P5) | 7 | c.904C>T | p.R302X | Nonsense | N-ZFc |
| 92 | Yamada et al., 2014/(P6) | 8 | c.936C>A | p.C312X | Nonsense | N-ZFc |
| 93 | Yamada et al., 2014/(P7) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 94 | Yamada et al., 2014/(P8) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 95 | Yamada et al., 2014/(P9) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 96 | Yamada et al., 2014/(P10) | 8 | c.1298C>T | p.Q433X | Nonsense | - |
| 97 | Yamada et al., 2014/(P11) | 8 | c.1489C>T | p.Q497X | Nonsense | - |
| 98 | Yamada et al., 2014/(P12) | 8 | c.1645A>T | p.R549X | Nonsense | - |
| 99 | Yamada et al.,2014/(P13) | 8 | c.1825G>T | p.E609X | Nonsense | - |
| 100 | Yamada et al., 2014/(P14) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 101 | Yamada et al., 2014/(P15) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 102 | Yamada et al., 2014/(P16) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 103 | Yamada et al., 2014/(P17) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 104 | Yamada et al., 2014/(P18) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 105 | Yamada et al., 2014/(P19) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 106 | Yamada et al., 2014/(P20) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 107 | Yamada et al., 2014/(P21) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 108 | Yamada et al., 2014/(P22) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 109 | Yamada et al., 2014/(P23) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 110 | Yamada et al., 2014/(P24) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 111 | Yamada et al., 2014/(P25) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 112 | Yamada et al., 2014/(P26) | 8 | c.2399C>G | p.S800X | Nonsense | CID |
| 113 | Yamada et al., 2014/(P27) | 8 | c.2615C>G | p.S872X | Nonsense | - |
| 114 | Yamada et al., 2014/(P28) | 8 | c.2761C>T | p.R921X | Nonsense | - |
| 115 | Yamada et al., 2014/(P29) | 8 | c.2761C>T | p.R921X | Nonsense | - |
| 116 | Yamada et al., 2014/(P1) | 3 | c.162_164del | p.P55Lfs*20 | Frameshift | - |
| 117 | Yamada et al., 2014/(P2) | 3 | c.175_182del | p.T60Sfs*3 | Frameshift | - |
| 118 | Yamada et al., 2014/(P3) | 3 | c270_272del | p.G91Vfs*17 | Frameshift | - |
| 119 | Yamada et al., 2014/(P4) | 3 | c.311_312dup | p.A105Sfs*16 | Frameshift | - |
| 120 | Yamada et al., 2014/(P5) | 5 | c.459_460del | p.E154Rfs*58 | Frameshift | - |
| 121 | Yamada et al., 2014/(P6) | 6 | c.635_638dup | p.P214Lfs*26 | Frameshift | - |
| 122 | Yamada et al., 2014/(P7) | 6 | c.647del | p.C216Sfs*8 | Frameshift | - |
| 123 | Yamada et al., 2014/(P8) | 6 | c.759_760dup | p.Q255Pfs*8 | Frameshift | N-ZFa |
| 124 | Yamada et al., 2014/(P9) | 7 | c.852_855del | p.T285Rfs*9 | Frameshift | N-ZFb |
| 125 | Yamada et al., 2014/(P10) | 7 | c.855_858del | p.E286Vfs*8 | Frameshift | N-ZFb |
| 126 | Yamada et al., 2014/(P11) | 7 | c.862_863del | p.G288Afs*10 | Frameshift | N-ZFb |
| 127 | Yamada et al., 2014/(P12) | 8 | c.1169ins | p.I390Tfs*41 | Frameshift | - |
| 128 | Yamada et al., 2014/(P13) | 8 | c.1169_1170del | p.T392Qfs*4 | Frameshift | - |
| 129 | Yamada et al., 2014/(P14) | 8 | c.1174_1178del | p.T392Nfs*3 | Frameshift | - |
| 130 | Yamada et al., 2014/(P15) | 8 | c.1176del | p.E393Nfs*3 | Frameshift | - |
| 131 | Yamada et al., 2014/(P16) | 8 | c.1212_1213del | p.A405Lfs*12 | Frameshift | - |
| 132 | Yamada et al., 2014/(P17) | 8 | c.1268_1273del | p.S424Lfs*2 | Frameshift | - |
| 133 | Yamada et al., 2014/(P18) | 8 | c.1280_1286del7ins | p.G427Dfs*2 | Frameshift | - |
| 134 | Yamada et al., 2014/(P19) | 8 | c.1334_1337dup | p.L447Ffs*9 | Frameshift | SMD |
| 135 | Yamada et al., 2014/(P20) | 8 | c.1395_1408del14ins | p.Q465Hfs*9 | Frameshift | SMD |
| 136 | Yamada et al., 2014/(P21) | 8 | c.1417del | p.R473Gfs*14 | Frameshift | SMD |
| 137 | Yamada et al., 2014/(P22) | 8 | c.1417del | p.R473Gfs*14 | Frameshift | SMD |
| 138 | Yamada et al., 2014/(P23) | 8 | c.1421_1426dup | p.M476Nfs*6 | Frameshift | SMD |
| 139 | Yamada et al., 2014/(P24) | 8 | c.1492_1493del | p.P498Lfs*18 | Frameshift | - |
| 140 | Yamada et al., 2014/(P25) | 8 | c.1534_1535del | p.G512Vfs*4 | Frameshift | - |
| 141 | Yamada et al., 2014/(P26) | 8 | c.1822del | p.E608Kfs*13 | Frameshift | - |
| 142 | Yamada et al., 2014/(P27) | 8 | c.1966_1967del | p.M656Vfs*17 | Frameshift | HD |
| 143 | Yamada et al., 2014/(P28) | 8 | c.2178_2180del | p.L727Ifs*28 | Frameshift | - |
| 144 | Yamada et al., 2014/(P29) | 8 | c.2254dup | p.T752Nfs*4 | Frameshift | - |
| 145 | Yamada et al., 2014/(P30) | 8 | c. 2282del | p.T761Kfs*26 | Frameshift | CID |
| 146 | Yamada et al., 2014/(P31) | 8 | c.2349_2351dup | p.S784Ffs*11 | Frameshift | CID |
| 147 | Yamada et al., 2014/(P32) | 8 | c.2579del | p.L860Rfs*3 | Frameshift | CID |
| 148 | Yamada et al., 2014/(P33) | 8 | c.2740_2743dup | p.S916Dfs*34 | Frameshift | - |
| 149 | Yamada et al., 2014/(P34) | 10 | c.3608_3614del | p.D1204Rfs*29 | Frameshift | - |
| 150 | [21] Tronina et al., 2023/(P1) | 6 | - | p.Gln694Ter | Missense | HD |
| 151 | [22] Jakubiak et al., 2021/(P1) | 8 | c.1027C > T | p.R343* | Nonsense | - |
| 152 | Jakubiak et al., 2021/(P2) | 1–10 | - | - | Deletion | - |
| 153 | Jakubiak et al., 2021/(P3) | 6 | c.648C > A | p.C216* | Nonsense | - |
| 154 | Jakubiak et al., 2021/(P4) | 10 | ZEB2 gene Exon 10 deletion | - | Deletion | - |
| 155 | Jakubiak et al., 2021/(P5) | 8 | c.1946del | p.I649Tfs*17 | Frameshift | HD |
| 156 | Jakubiak et al., 2021/(P6) | 6 | c.607ins | p.Thr203IlefsTer37 | Frameshift | - |
| 157 | Jakubiak et al., 2021/(P7) | 4 | c.399_400dup | p.Thr134IlefsTer3 | Frameshift | - |
| 158 | Jakubiak et al., 2021/(P8) | 8 | c.1276T > A | p.Leu426Ile | Missense | - |
| 159 | Jakubiak et al., 2021/(P9) | 6 | c.696C > G | p.Y232* | Nonsense | N-ZFa |
| 160 | Jakubiak et al., 2021/(P10) | - | 8 Mb deletion 2q22.3q23.3 | - | Chromosome deletion |
- |
| 161 | Jakubiak et al., 2021/(P11) | 1–10 | - | - | Deletion | - |
| 162 | Jakubiak et al., 2021/(P12) | 3–10 | - | - | Deletion | - |
| 163 | Jakubiak et al., 2021/(P13) | 7 | c.857-858del | p.Glu286ValfsTer8 | Frameshift | N-ZFb |
| 164 | Jakubiak et al., 2021/(P14) | 6 | c.607ins | p.Thr203IlefsTer37 | Frameshift | - |
| 165 | Jakubiak et al., 2021/(P15) | 8 | c.1445T > G | p.Leu482* | Nonsense | SMD |
| 166 | Jakubiak et al., 2021/(P16) | 3 | c.84T > G | p.Tyr28* | Nonsense | - |
| 167 | Jakubiak et al., 2021/(P17) | 8 | c.1421-1426del | p.Gln474_Met476delins | Insertion, deletion | SMD |
| 168 | Jakubiak et al., 2021/(P18) | 8 | c.2230A > G | p.Ile744Val | Missense | - |
| 169 | Jakubiak et al., 2021/(P19) | 10 | c.3202G > T | p.Gly1068Cys | Missense | C-ZFb |
| 170 | Jakubiak et al., 2021/(P20) | 8 | c.2087_2088del | Lys696Serfs*24 | Frameshift | HD |
| 171 | Jakubiak et al., 2021/(P21) | 8 | c.2073G > A | p.Trp691Ter | Nonsense | HD |
| 172 | Jakubiak et al.,2021/(P22) | - | - | - | Deletion | - |
| 173 | Jakubiak et al., 2021/(P23) | 8 | c.2562_2564del | p.N855Lfs*3 | Frameshift | CID |
| 174 | Jakubiak et al., 2021/(P24) | 8 | c.1177dup | p.E393Gfs*7 | Frameshift | - |
| 175 | Jakubiak et al., 2021/(P25) | 8 | c.1437_1440del | p.H304Qfs*3 | Frameshift | N-ZFc |
| 176 | Jakubiak et al., 2021/(P26) | 8 | c.2083C > T | p.Arg695Ter | Nonsense | HD |
| 177 | Jakubiak et al., 2021/(P27) | 8 | c.2083C > T | p.Arg695Ter | Nonsense | HD |
| 178 | Jakubiak et al., 2021/(P28) | 3–10 | 258.2 kb deletion | - | Chromosome deletion |
- |
| 179 | [23] Refaat et al., 2021/(P1) | - | 2.27 Mb deletion | - | Chromosome deletion |
- |
| 180 | [24] Musaad et al., 2022/(P1) | - | Chr2:145161574del | - | Frameshift | - |
| 181 | [25] Pachajoa et al., 2022/(P1) | 8 | c.2761C>T | p.Arg921Ter | Nonsense | - |
| 182 | [26] Wu et al., 2022/(P1) | 8 | c.2417del | p.Phe807Serfs*11 | Frameshift | CID |
| 183 | Wu et al., 2022/(P2) | 8 | c.1200T>A | p.Tyr400X | Nonsense | - |
| 184 | Wu et al., 2022/(P3) | 8 | c.1027C>T | p.Arg343X | Nonsense | - |
| 185 | Wu et al., 2022/(P4) | 8 | c.2621del | p.Asn874Ilefs*12 | Frameshift | - |
| 186 | Wu et al., 2022/(P5) | 8 | c.2456C>G | p.Ser819X | Nonsense | CID |
| 187 | Wu et al., 2022/(P6) | 8 | c.2002del | p.Glu668Serfs*8 | Frameshift | HD |
| 188 | Wu et al., 2022/(P7) | 2–10 | Chr2:145147017-145274917del | - | Large deletion | - |
| 189 | Wu et al., 2022/(P8) | 5 | c.492_517del | p.Glu164Aspfs*9 | Frameshift | - |
| 190 | Wu et al., 2022/(P9) | 6 | c.779dup | p.Met260Ilefs*19 | Frameshift | N-ZFa |
| 191 | Wu et al., 2022/(P10) | 1–10 | chr2:141213978-148010654del | - | Large deletion | - |
| 192 | Wu et al., 2022/(P11) | 8 | c.2083C>T | p.Arg695X | Nonsense | HD |
| 193 | Wu et al., 2022/(P12) | 1–10 | chr2:145000000-145351228 del | - | Large deletion | - |
| 194 | Wu et al., 2022/(P13) | 7 | c.904C>T | p.Arg302X | Nonsense | N-ZFc |
| 195 | Wu et al., 2022/(P14) | 8 | c.2712del | p.Pro906Leufs*24 | Frameshift | - |
| 196 | Wu et al., 2022/(P15) | 8 | c.2670_2677del | p.Ala891Phefs*55 | Frameshift | - |
| 197 | Wu et al., 2022/(P16) | 8 | c.2177_2180del | p.Ser726Tyrfs*7 | Frameshift | - |
| 198 | Wu et al., 2022/(P17) | 1–10 | chr2:138434153-145285163 del | - | Large Deletion | - |
| 199 | Wu et al., 2022/(P18) | 8 | c.2851C>T | p.Gln951X | Nonsense | - |
| 200 | Wu et al., 2022/(P19) | 8 | c.1426dup | p.Met476Asnfs*5 | Frameshift | SMD |
| 201 | Wu et al., 2022/(P20) | 8 | c.2761C>T | p.Arg921X | Nonsense | - |
| 202 | Wu et al., 2022/(P21) | 8 | c.1027C>T | p.Arg343X | Nonsense | - |
| 203 | Wu et al., 2022/(P22) | 8 | c.1106_1115delins | p.Leu369X | Nonsense | - |
| 204 | [27] Fu et al., 2022/(P1) | 8 | c.2136del | p.Lys713Serfs*3 | Frameshift | - |
| 205 | Fu et al., 2022/(P2) | 8 | c.2740del | p. Gln914Argfs*16 | Frameshift | - |
| 206 | Fu et al., 2022/(P3) | 7 | c.808-2del | - | Splicing | N-ZFb |
| 207 | [28] Wei et al., 2021/(P1) | 8 | c.1137_1146del | p.S380Nfs*13 | Deletion | - |
| 208 | [29] Şenbil et al., 2021/(P1) | 6 | c.646dup | p.Cys216LeufsTer23 | Frameshift | - |
| 209 | [30] Ivanoski et al., 2018/(P1) | 6 | c.805C>T | p.Q269* | Nonsense | N-ZFb |
| 210 | Ivanoski et al., 2018/(P2) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 211 | Ivanoski et al., 2018/(P3) | 1–10 | 7.3 Mb deletion Ch2q22.1q22.3 | - | Large deletion | - |
| 212 | Ivanoski et al., 2018/(P4) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 213 | Ivanoski et al., 2018/(P5) | 8 | c.1052_1057delins | p.G351Vfs*19 | Small deletion, Frameshift | - |
| 214 | Ivanoski et al., 2018/(P6) | 6 | c.696C>G | p.Y232* | Nonsense | N-ZFa |
| 215 | Ivanoski et al., 2018/(P7) | 8 | c.1073_1122delins | p.S359Tfs*3 | Small indel, Frameshift | - |
| 216 | Ivanoski et al., 2018/(P8) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 217 | Ivanoski et al., 2018/(P9) | 1–10 | 2.6 Mb deletion Ch2q22.2q22.3 | - | Large deletion | - |
| 218 | Ivanoski et al., 2018/(P10) | 8 | c.2718del | p.A907Lfs*23 | Small deletion, Frameshift | - |
| 219 | Ivanoski et al., 2018/(P11) | 8 | c.2180T>A | p.L727* | Nonsense | - |
| 220 | Ivanoski et al., 2018/(P12) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 221 | Ivanoski et al., 2018/(P13) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 222 | Ivanoski et al., 2018/(P14) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 223 | Ivanoski et al., 2018/(P15) | 5 | c.460G>T | p.E154* | Nonsense | - |
| 224 | Ivanoski et al., 2018/(P16) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 225 | Ivanoski et al., 2018/(P17) | 8 | c.1426dup | p.M476Nfs*6 | Small insertion, Frameshift | SMD |
| 226 | Ivanoski et al., 2018/(P18) | 1–10 | 4.6 Mb deletion Ch2q22q22.3 | - | Large deletion | - |
| 227 | Ivanoski et al., 2018/(P19) | 8 | c.2682_2687delins | p.L894Ffs*36 | Small indel, Frameshift | - |
| 228 | Ivanoski et al., 2018/(P20) | 3 | c.274G>T | p.G92* | Nonsense | - |
| 229 | Ivanoski et al., 2018/(P21) | 8 | c.2053C>T | p.Q685* | Nonsense | HD |
| 230 | Ivanoski et al., 2018/(P23) | 9 | c.3031del | p.S1011Afs*64 | Small deletion, Frameshift | C-ZFa |
| 231 | Ivanoski et al., 2018/(P24) | 8 | c.2227del | p.S743Lfs*2 | Small deletion, Frameshift | - |
| 232 | Ivanoski et al., 2018/(P25) | 5 | c.460del | p.E154Rfs*58 | Small deletion, Frameshift | - |
| 233 | Ivanoski et al., 2018/(P26) | 6 | c.625C>T | p.Q209* | Nonsense | - |
| 234 | Ivanoski et al., 2018/(P27) | 7 | c.817del | p.L273* | Nonsense | N-ZFb |
| 235 | Ivanoski et al., 2018/(P28) | 8 | c.1635_1636ins | p.D546Lfs*11 | Small insertion, Frameshift | - |
| 236 | Ivanoski et al., 2018/(P29) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 237 | Ivanoski et al., 2018/(P30) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 238 | Ivanoski et al., 2018/(P31) | 8 | c.2701C>T | p.Q901* | Nonsense | - |
| 239 | Ivanoski et al., 2018/(P32) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 240 | Ivanoski et al., 2018/(P34) | 8 | c.2718del | p.A907Lfs*23 | Small deletion, Frameshift | - |
| 241 | Ivanoski et al., 2018/(P35) | 8 | c.2317_2318del | p.E773Kfs*8 | Small deletion, Frameshift | CID |
| 242 | Ivanoski et al., 2018/(P36) | 1–10 | 0.6 Mb deletion Ch2q22.2 | - | Large deletion | - |
| 243 | Ivanoski et al., 2018/(P37) | 3 | c.264_267del | p.I88Mfs*19 | Small deletion, Frameshift | - |
| 244 | Ivanoski et al., 2018/(P38) | 8 | c.1541del | p.P414Rfs*2 | Small deletion, Frameshift | - |
| 245 | Ivanoski et al., 2018/(P39) | 8 | c.2856del | p.R953Efs*24 | Small deletion, Frameshift | - |
| 246 | Ivanoski et al., 2018/(P40) | 8 | c.2254dup | p.Y752Nfs*4 | Small insertion, Frameshift | - |
| 247 | Ivanoski et al., 2018/(P41) | 8 | c.930C>A | p.Y310* | Nonsense | N-ZFc |
| 248 | Ivanoski et al., 2018/(P42) | 8&9 | c.917_3067del | p.E307_G1023del | Deletion, in-frame | N-ZFc C-ZFa |
| 249 | Ivanoski et al., 2018/(P43) | 8 | c.975C>A | p.Y325* | Nonsense | N-ZFc |
| 250 | Ivanoski et al., 2018/(P44) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 251 | Ivanoski et al., 2018/(P45) | 6 | c.691dup | p.Y232Vfs*7 | Frameshift | N-ZFa |
| 252 | Ivanoski et al., 2018/(P46) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 253 | Ivanoski et al., 2018/(P47) | 7 | c.901del | p.L301Cfs*37 | Small deletion, Frameshift | N-ZFc |
| 254 | Ivanoski et al., 2018/(P48) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 255 | Ivanoski et al., 2018/(P49) | 3 | c.81_84dup | p.D29Lfs*2 | Small insertion, Frameshift | - |
| 256 | Ivanoski et al., 2018/(P50) | 8 | c.1202dup | p.K401Ifs*17 | Small insertion, Frameshift | - |
| 257 | Ivanoski et al., 2018/(P51) | 6 | c.648C>A | p.C216* | Nonsense | - |
| 258 | Ivanoski et al., 2018/(P52) | 8 | c.1910C>G | p.S637* | Nonsense | - |
| 259 | Ivanoski et al., 2018/(P53) | 8 | c.2713del | p.P906Lfs*24 | Small deletion, Frameshift | - |
| 260 | Ivanoski et al., 2018/(P54) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 261 | Ivanoski et al., 2018/(P55) | 5 | c.540del | p.E181Rfs*31 | Small deletion, Frameshift | - |
| 262 | Ivanoski et al., 2018/(P56) | 6 | c.609del | p.P204Qfs*9 | Small deletion, Frameshift | - |
| 263 | Ivanoski et al., 2018/(P57) | 5 | c.477_484del | p.H159Qfs*10 | Small deletion, Frameshift | - |
| 264 | Ivanoski et al., 2018/(P58) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 265 | Ivanoski et al., 2018/(P59) | 5–8 | c.403_2887del | p.V135Gfs*5 | Frameshift | - |
| 266 | Ivanoski et al., 2018/(P60) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 267 | Ivanoski et al., 2018/(P61) | 1–10 | 16.7 Mb deletion Ch2q21.1q22.3 | - | Large deletion | - |
| 268 | Ivanoski et al., 2018/(P62) | 6 | c.653_654ins | p.G219Pfs*21 | Small insertion, Frameshift | - |
| 269 | Ivanoski et al., 2018/(P63) | 8 | c.1851del | p.H617Qfs*4 | Small deletion, Frameshift | - |
| 270 | Ivanoski et al., 2018/(P64) | 4 | c.389_390dup | p.T134Lfs*3 | Small insertion, Frameshift | - |
| 271 | Ivanoski et al., 2018/(P65) | 8 | c.2076del | p.F692Lfs*24 | Small deletion, Frameshift | HD |
| 272 | Ivanoski et al., 2018/(P66) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 273 | Ivanoski et al., 2018/(P67) | 7 | c.823C>T | p.Q275* | Nonsense | N-ZFb |
| 274 | Ivanoski et al., 2018/(P68) | 8 | c.1313del | p.H438Pfs*2 | Small deletion, Frameshift | SMD |
| 275 | Ivanoski et al., 2018/(P69) | 1&2 | ZEB2 gene Exons 1 and 2 deletion | - | Large deletion | - |
| 276 | Ivanoski et al., 2018/(P70) | 8 | c.1027C>T | p.R343* | Nonsense | - |
| 277 | Ivanoski et al., 2018/(P71) | 6 | c.648C>A | p.C216* | Nonsense | - |
| 278 | Ivanoski et al., 2018/(P72) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 279 | Ivanoski et al., 2018/(P73) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 280 | Ivanoski et al., 2018/(P74) | 10 |
ZEB2 gene Exon 10 deletion (3′UTR c.3642+750) |
- | Large deletion | - |
| 281 | Ivanoski et al., 2018/(P75) | 8 | c.1946del | p.I649Tfs*17 | Small deletion, Frameshift | HD |
| 282 | Ivanoski et al., 2018/(P76) | 2 | c.32dup | p.R11Pfs*9 | Small insertion, Frameshift | - |
| 283 | Ivanoski et al., 2018/(P77) | 6 | c.761del | p.Q255Sfs*7 | Small deletion, Frameshift | N-ZFa |
| 284 | Ivanoski et al., 2018/(P78) | 8 | c.1553del | p.H518Lfs*26 | Small deletion, Frameshift | - |
| 285 | Ivanoski et al., 2018/(P79) | 8 | c.936C>A | p.C312* | Nonsense | N-ZFc |
| 286 | Ivanoski et al., 2018/(P80) | 3 | c.187_228dup | p.A77Lfs*13 | Small insertion, Frameshift | - |
| 287 | Ivanoski et al., 2018/(P81) | 8 | c.1150C>T | p.Q384* | Nonsense | - |
| 288 | Ivanoski et al., 2018/(P82) | 5 | c.553_554ins | p.R185Lfs*28 | Small insertion, Frameshift | - |
| 289 | Ivanoski et al., 2018/(P83) | 7 | c.857_858del | p.E286Vfs*8 | Small deletion, Frameshift | N-ZFb |
| 290 | Ivanoski et al., 2018/(P84) | 10 | c.3567_3568ins | p.M1190Pfs*52 | Small insertion, Frameshift | - |
| 291 | Ivanoski et al., 2018/(P85) | 8 | c.2677del | p.P893Lfs*37 | Small deletion, Frameshift | - |
| 292 | Ivanoski et al., 2018/(P86) | 8 | c.2372del | p.T791Nfs*26 | Small deletion, Frameshift | CID |
| 293 | Ivanoski et al., 2018/(P87) | 6 | c.715del | p.E239Rfs*23 | Small deletion, Frameshift | N-ZFa |
| 294 | Ivanoski et al., 2018/(P88) | IVS1 | c.-69-2A>C | p.M1_N24delins | Splicing | - |
| 295 | Ivanoski et al., 2018/(P89) | 8 | c.1578_1579delins | p.D527Tfs*17 | Small indel, Frameshift | - |
| 296 | [12] Ghoumid et al., 2013/(P1) | 10 | c.3134A>G | p.His1045Arg | Missense | C-ZFb |
| 297 | Ghoumid et al., 2013/(P2) | 10 | c.3164A>G | p.Tyr1055Cys | Missense | C-ZFb |
| 298 | Ghoumid et al., 2013/(P3) | 10 | c.3211T>C | p.Ser1071Pro | Missense | C-ZFb |
Ter, X or * represent stop codons at the time of publication; N-ZF = N-terminal zinc finger clusters domain (221–334); SMD = SMAD-binding domain (437–482); IVS1 = intervening sequence; HD = homeodomain-like domain (644–703); CID = CtBP-interacting domain (757–868); C-ZF = C-terminal zinc finger clusters domain (998–1078). Protein domain regions coded by exons are represented based on codon location within the domains (e.g., exon 9 codes for C-ZFa and exon 10 codes for C-ZFb). References are listed in brackets [6,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30].
2.3. ZEB2 Gene Variant Types and Frequencies
ZEB2 gene variants, types, and frequencies were analyzed from 298 studied individuals with MWS and summarized in Figure 2. The most prevalent variant type was frameshift (134 patients; 45%) followed by nonsense (112 patients; 38%).
Figure 2.
Frequency of ZEB2 gene variants identified by variant type in 298 reported patients with MWS. The frequencies of ZEB2 gene variants were identified and grouped. Frameshift* represents a combination of frameshift alone (N = 88), and frameshift plus variants such as frameshift with small deletion (N = 29), frameshift with small insertion (N = 13), or frameshift with small indel (N = 4). Large deletion* represents a combination of large deletions (N = 15) and chromosome deletions (N = 6). Deletion* represents a combination of deletions involving DNA (N = 9) or whole exome based (N = 4). Other* represents a combination of in-frame defects with intragenic deletion (N = 1), partial duplication (N = 1) or insertion deletion (N = 1).
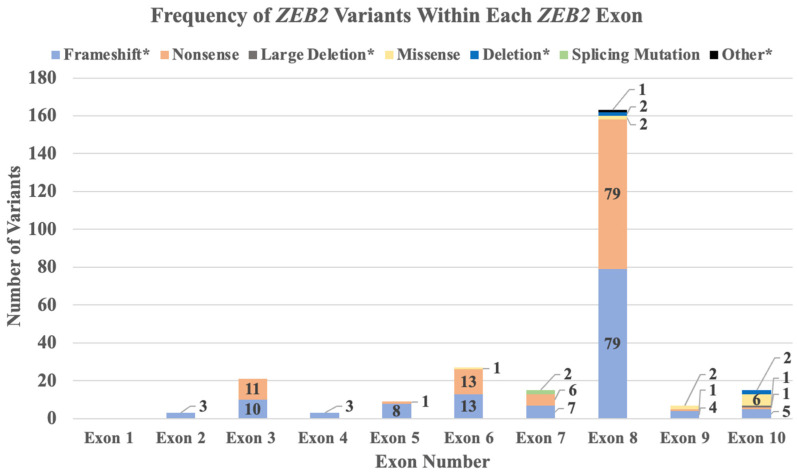
Of the 298 patients with MWS, 262 had sufficient information to determine the ZEB2 exon variant type and to analyze the distribution and frequency of ZEB2 variants within a specific exon. Figure 3 represents the exons within the gene, along with the frequency and type of variants within each exon. The most common variant site or location was c.2083C>T within exon 8 of the ZEB2 gene. This variant was found in 11 percent of all patients reported with MWS.
Figure 3.
Number of ZEB2 gene variants and types identified in each exon.
2.4. ZEB2 Gene–Gene or Protein Interactions
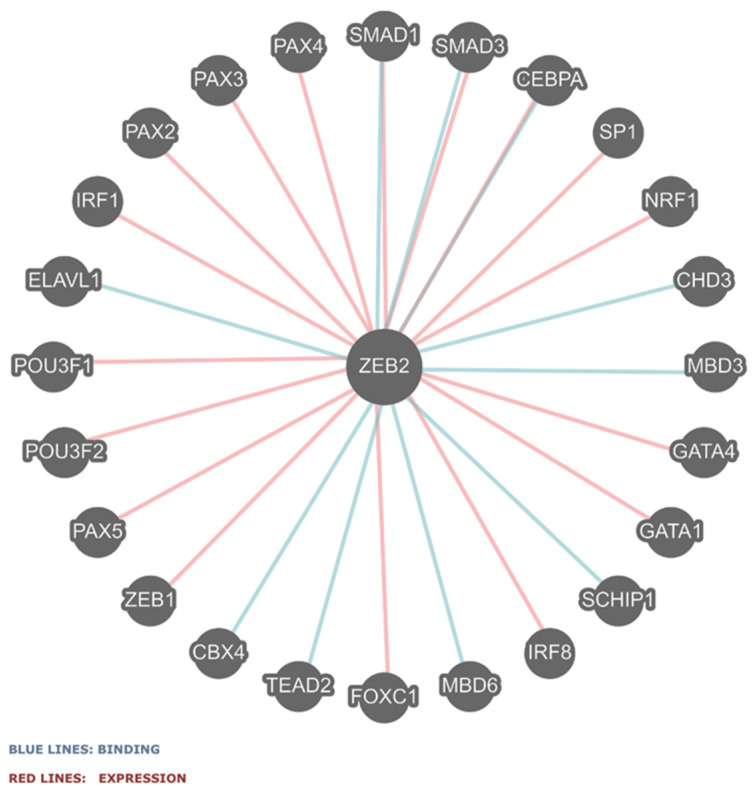
The protein–protein interaction networks for the ZEB2 gene were obtained as shown in Figure 4. ZEB2 interacts with 24 other proteins and these interactions are predicted to impact gene expression and the binding of encoded proteins (see Figure 4). Seventeen of the twenty-four genes are predicted to show co-expression with ZEB2 protein binding, while ten genes are predicted to interact via protein binding. The two SMAD proteins, SMAD1 and SMAD3, which are shown to interact with ZEB2 via co-expression and binding, are main signal transducers for receptors of the transforming growth factor beta superfamily, critical for regulating cell development and growth. The encoded ZEB2 protein contains a SMD protein domain. Additionally, the ZEB2 protein is predicted to interact with four PAX proteins, two POU3F proteins, and two GATA proteins. These interactions have the potential to impact the ZEB2 protein’s functional pathways and interactions related to the TGF receptor for which ZEB1, ZEB2, and SMAD are major players.
Figure 4.
ZEB2 gene–gene functional interactions are identified via binding (blue lines) and co-expression (red lines) [http://pathwaycommons.org/pc12/Pathway_751ce7ccec6e191682a33d7252aac8] (accessed on 1 November 2023). These gene interactions relate to TGF receptor pathways in which ZEB1, ZEB2, and SMAD are major players.
2.5. ZEB2 Structural Protein Domains, Functions and Models
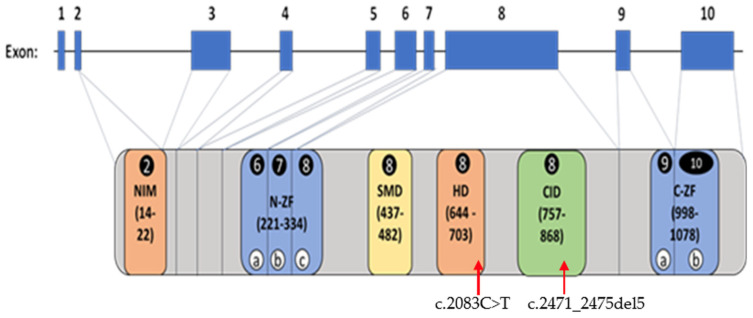
The ZEB2 protein domains and their corresponding exons are represented in Figure 5. As illustrated in this figure, the ZEB2 gene consists of 10 exons, each of varying size, and encodes for six protein domains with specific functions and domain relationships described below.
Figure 5.
Schematic representation of ZEB2 gene structure, exons, and codons listed and incorporated into each of the six protein domains. The protein domains and their corresponding exons are illustrated and modified from Zou et al. [13] such as NIM (nucleosome remodeling and deacetylase-interaction motif), N-ZF (N-terminal zinc finger cluster), SMD (SMAD-binding domain), HD (homeodomain), CID (CtBP-interacting domain), and C-ZF (C-terminal zinc finger cluster) modified from reference [13]. Exons coding the individual protein domains and domain regions are circled. The circled lowercase letters (a/b/c) found in both the N-ZF and C-ZF domains represent the regions coded by different exons (exon numbers circled). The most common gene variant (c.2083C>T) found in our study of 298 patients with MWS and the pathogenetic variant (c.2471_2475del5) found in our 5-year-old proband along with their locations in the HD and CID domains, respectively, are indicated by red arrows.
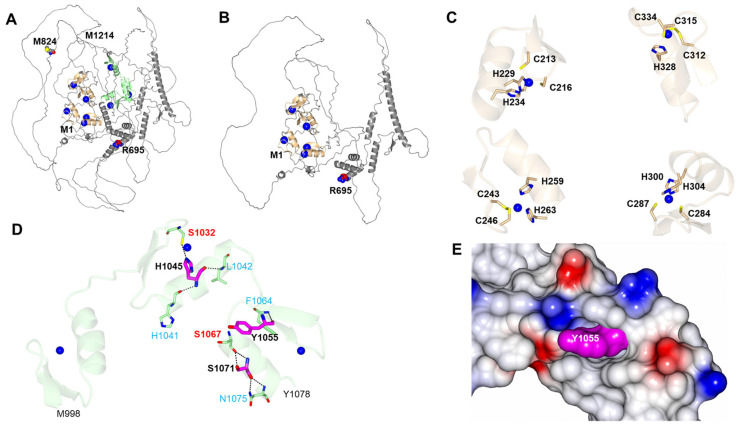
A predicted model from a computer-based Alphafold2 program (Uniprot:O60315) was used to generate an altered ZEB2 protein structure from a truncated protein, while haploinsufficiency leads to a lower amount of protein resulting from non-sense mediated decay (NMD) of mRNA containing the abnormal gene defect. As shown below in Figure 6A, the ZEB2 protein is composed of many flexible/disordered regions that contain well-structured ZF domains. Residues after R695, the most common protein defect/site found in our studied MWS population, generated a stop codon in exon 8, which may result in a polypeptide lacking the C-terminal zinc finger (C-ZF) domain due to a truncated protein as shown in Figure 6B. Additionally, the N-ZF contains ZF domains predicted to coordinate zinc ions by the residues shown in Figure 6C.
Figure 6.
Predicted AlphaFold2 structure of ZEB2 (www.uniprot.org/uniprotkb/O60315/entry; accessed on 29 September 2023). (A) Full-length structure showing the N-terminal zinc finger domain (tan, L211-C334) and the C-terminal zinc finger domain (green, M998-Y1078) with codon positions at R695 and M824 with their predicted zinc ion binding sites drawn as blue spheres. M824 is a codon that is deleted in our case report of a 5-year-old female with a pathogenic variant c.2472_2475del5 in exon 8, while the R695 protein variant was the most common protein defect found in the 298 reported patients with MWS summarized in Table 1. (B) Same view as panel A but with residues after R695 omitted, resulting in a polypeptide lacking the C-terminal zinc finger domain. (C) N-terminal zinc finger domain shows the predicted zinc binding sites and coordination with ZEB2 residues. (D) C-terminal zinc finger domain shows predicted side chain hydrogen bond interactions (dashed lines) between H1045, Y1055, and S1071 missense ZEB2 changes reported by Ghoumid et al. [12] and other ZEB2 residues (red). Y1055 does not form interactions with the side chain -OH but does form a backbone hydrogen bond interaction with the backbone N-atom of F1064. Other backbone interactions with H1045 and Y1055 are indicated with blue text. (E) Electrostatic surface representation shows packing of Y1055 within a cleft of the ZEB2 C-terminal zinc finger domain.
The depicted H1045R, Y1055C, and S1071P variants in three patients with MWS were reported in 2013 by Ghoumid et al. [12] as missense mutations within the C-ZF protein domain. Interestingly, the H1045R and S1071P variants would likely result in the breakage of hydrogen bonds between the side chains and residues of S1032 and S1067 which could destabilize the ZF domains (Figure 6D). Although the side chain of Y1055 does not form hydrogen bond interactions with its side chain, this residue is tightly packed within a cleft and mutations would likely affect this interaction (Figure 6E).
The five base pair ZEB2 deletion in exon 8 at c.2471_2475del5 at the M824 residue in our proband would potentially impact codons of four of the six domains (about one-third of the N-ZF domain and all of the SMD, HD, and CID domains). Hence, this novel ZEB2 deletion variant could not be predicted or modeled due to a lack of computer protein access data information for modeling purposes with the involvement of such a large proportion of the ZEB2 protein. The Met824 residue is predicted to be absent leading to disruption of the Lys825 residue upstream of the C-ZF protein domain. This predicted mistranslated string of residues would result in the removal of the C-ZF domain. Therefore, the ZEB2 protein would be altered and/or presumably degraded in our proband, potentially impacting 24 other interacting genes found in gene–gene interaction and protein binding studies, as noted in Figure 4.
3. Discussion
3.1. Mowat–Wilson Syndrome (MWS) and Clinical Findings
Mowat–Wilson syndrome (MWS) is a rare autosomal dominant disorder caused by pathogenic variants in the ZEB2 gene, located at chromosome 2q22.3. We identified about 300 known cases of MWS in the literature or unpublished databases. Most cases of MWS are sporadic heterozygous de novo occurrences with a low recurrence risk in siblings. We analyzed and summarized a wide range of ZEB2 defects including frameshift, nonsense, missense, and deletions or duplications. Those with the larger deletions tend to have more clinical severity.
Mowat–Wilson syndrome was first reported in 1998 [3,7] with multi-organ system involvement along with moderate-to-severe intellectual disability, speech delay, and microcephaly with brain anomalies. Seizures are often found with onset between several months to over 10 years of age. Specific facial dysmorphisms exist with a wide range of cardiac anomalies requiring intervention. Genitourinary and kidney anomalies are also present in about 50% of cases including undescended testicles, penile malformations, hydronephrosis, and kidney defects. Gastrointestinal problems include Hirschsprung disease in nearly one-half of reported patients accompanied by constipation. Other occasional abnormalities include long tapering toes and fingers, pes planus, nystagmus, cleft lip/palate, and pigmentary changes [3,7,8,9,10].
3.2. ZEB2 Gene Structure and Variants
The ZEB2 gene consists of ten exons of different lengths and codes for six protein domains of variable sizes and functions. Exon 8 showed the largest number of variants and comprised 162 of the cases in our study population, followed by exon 6 with 27 variants. The large exon 8 accounts for about one-half of the ZEB2 coding sequence and encodes the last zinc finger region of the N-ZF domain along with SMD, HD, and CID protein domains; hence, about 60% of the ZEB2 protein is coded by this exon, representing a similar proportion of the reported ZEB2 gene defects. Exon 8 is also involved in our 5-year-old proband with MWS.
Frameshift mutations or premature termination codons (PTCs) do lead to non-sense mediated decay (NMD), impacting protein production. NMD is a process that typically affects the mRNA of most genes with PTCs before the last exon, which leads to mRNA degradation with haploinsufficiency. It is likely that NMD does occur, affecting the amount of protein produced and causing the truncated protein to alter protein function in MWS as well as other genetic disorders. However, it is predicted that mutated transcripts of ZEB2 may not undergo non-sense mediated decay in MWS, but more research is needed [31].
We studied ZEB2 variant types and their locations within each exon. Nine of the ten exons, except exon 1, harbored damaging frameshift variants. Frameshift mutations occur when one or two bases are either inserted or deleted from a strand of DNA. This results in an aberrant protein impacting the protein function. This aligns with the fact that a frameshift defect was the most identified variant seen in our study, followed by nonsense mutations present in 6 of the 10 exons. A frameshift defect was seen in 134 of the 298 individuals [45%] reported with MWS. The second most frequent variant in our patient population was a nonsense defect seen in 112 patients [38%]. The third most frequent variant found was a large deletion in 21 individuals [7%]. These patients had a complete absence or deletion of the chromosome 2 region where the ZEB2 gene is located. This was followed by missense variants in 11 individuals [4%]. Collectively, MWS is caused by haploinsufficiency of the autosomal dominant ZEB2 gene function due to the heterozygous variants and copy number variations. Further studies to elucidate the precise molecular mechanisms for the pathogenicity of MWS are needed.
3.3. ZEB2 Gene Functions and Interactions
Recognized cellular components impacted by the ZEB2 gene include the chromatin, cytosol, nucleolus, nucleus, and nucleoplasm, and impacted processes include the molecular function and disturbances of DNA-binding transcription factor activity via RNA polymerase and transcription repression, metal ion binding for zinc, and phosphatase regulatory activity. These derangements of the ZEB 2 protein may play a role in clinical outcomes, variability, and severity with early multi-organ development in MWS (www.uniprot.org/uniprotkb/O60315/entry, accessed on 1 November 2023). As described in our review, the ZEB2 gene, when disturbed, could impact other interacting developmental genes by leading to a cascade of early perturbed biological processes and abnormal embryogenesis, requiring more research. The ZEB2 gene functions as a transcriptional inhibitor by binding to specific DNA structure (5′-CACCT-3′) at gene promoters. It represses the transcription of cadherin 1 (CDH1; NM_004360.5) gene and cell adhesion required for the establishment and maintenance of epithelial cells during embryogenesis and adulthood which are important for brain and other organ development and function [32]. The mesenchyme homeobox 2 (MEOX2; NM_005924.5) gene [33] also interacts with ZEB2 as a homeobox gene involved in the regulation of anatomical development such as morphogenesis. Moreover, homeodomain proteins regulate gene expression and cell differentiation during early embryonic development and organogenesis including the heart, brain, and gut [34], which are involved in Mowat–Wilson syndrome. Gene functions and outcomes may also be altered by gene–gene or protein interactions or epistasis, whereby a single gene function may be impacted by interactions with other genes and disease processes [35].
3.4. Genes That Interact with ZEB2 and Potential Relationship with Mowat–Wilson Syndrome
The 24 genes recognized to interact with ZEB2 are shown in Figure 4. These gene–gene or protein interactions may play important roles in the derangement of multi-system embryogenesis and organogenesis. Among the interactive genes are five gene families (SMAD, GATA, IRF, PAX, and POU3F) represented with the ZEB1 gene, presumably contributing to clinical findings seen in MWS. One of the interacting genes is a member of the SMAD gene family, SMAD1 (SMAD1; NM_005900.3). The SMAD-related protein domain is one of the six domains found in the ZEB2 protein. The SMAD1 encoded protein is Involved in the downstream signaling pathway for bone morphogenic protein (BMP) subfamily members [36]. BMPs are known to play vital roles during the formation and maintenance of various organs disturbed in MWS, are members of TGF-β receptor signaling and are involved in skeletal dysplasia, DNA-binding transcription factor activity, and protein kinase binding. SMAD3 (SMAD; NM_005902.4) targets genes involved in epithelial cells including cyclin-dependent kinase (CDK) inhibitors that generate a cytostatic response important for the regulation of muscle-specific genes [8]. The GATA-binding protein 1 (GATA1) interactive gene is X-linked and encodes the zinc finger DNA-binding transcription factor, which plays a critical role in the normal development of hematopoietic cells and is associated with thrombocytopenia, beta thalassemia, and dyserythropoietic anemia. The GATA4 (GATA4; NM_001308093.3) interactive gene is associated with congenital heart disease, as seen in our 5-year-old female, and cardiac conduction, testicular development, and chromatin binding [37,38].
Other interactive genes include interferon regulatory factor 1 (IRF1: NM_002198.3) which is highly regulated in human vascular lesions and exhibits a growth inhibitory function in coronary artery smooth muscle, nitric oxide production, endothelial tissue, vascular intimal growth with healing, and pathophysiology of primary atherosclerosis [39]. The IRF8 (IRF8; NM_002163.4) gene codes transcriptional agents of the interferon regulatory factor (IRF) family involved with conserved DNA-binding domains in the N-terminal regions and divergent C-terminal regions serving as regulatory sites, and in immunodeficiency [40].
The paired box 2 (PAX2; NM_000278.5) gene and other PAX family members are involved with mesenchyme to epithelium transition in renal development and related pathways for neural stem cells, lineage-specific markers, and Wnt/Hedgehog/Notch signaling [8]. For example, defects of the PAX3 (PAX3: NM_181458.4) gene cause Waardenburg syndrome, involved with chromatin organization and binding, ectoderm differentiation, and craniofacial-spinal development with myogenesis, which are possibly important in MWS, as well [41]. The PAX4 (PAX4; NM_001366110.1) gene encodes transcription factors that are essential for the formation of several tissues representing all germ layers and induced pluripotent stem cells with lineage-specific markers [42]. PAX5 (PAX5; NM_016734.3) is a transcription factor gene essential for B-cell differentiation and other hematopoietic lineages and diseases [43]. The POU domain, class 3, transcription factor 1 (POU3F1; NM_002699.4) gene is associated with related pathways involved with nervous system development, mammalian neurogenesis, and myelination [8,44,45]; POU3F2 (POU3F2; NM_005604.4) is involved with microphthalmia and melanoma, and MECP2 (MECP2; NM_001110792.2) activity that is disturbed in Rett syndrome [8].
The STRING database (STRING.org) was used to study predicted protein–protein associations, networks, and functional enrichment analysis [46]. In this database, there are 10 significantly associated proteins with ZEB2 including CTBP1, CHURC1, ARHGAP31, TWIST1, TWIST2, SMAD3, CDH1, CDH2, HDAC1, and ZEB1. Only ZEB1 and SMAD3 are in common between this database and the Pathwayscommons.org database [32] depicted in Figure 4.
The C-terminal binding protein 1 (CTBP1) which interacts with the ZEB2 protein in the STRING database involves dehydrogenase activity and functions in brown adipose tissue differentiation [47]. The Churchill domain-containing 1 (CHURC1) protein is involved in the positive regulation of transcription with ubiquitous expression in multiple tissues [8]. The Rho GTPase-activating protein 31 (ARHGAP31) functions as a GTPase-activating protein (GAP) for RAC1 and CDC42 required for cell spreading, polarized lamellipodia formation, and cell migration. The Twist family transcription factor 1 or twist-related protein 1 (TWIST1) also acts as a transcriptional regulator, inhibits myogenesis, and represses the expression of pro-inflammatory cytokines such as TNFA and IL1B. It also regulates cranial suture patterning and fusion. The Twist-related protein 2 (TWIST2) binds to the E-box consensus sequence 5′-CANNTG-3′ and represses the expression of proinflammatory cytokines involved with glycogen storage and energy metabolism as well as inhibiting the premature differentiation of pre-osteoblasts during osteogenesis. The Mothers against decapentaplegic homolog 3, receptor-regulated (SMAD3) binds to the TRE element of the promotor of many genes regulated by TGF-β, working with the SMAD4 (SMAD4: NM_005359.6) gene, playing a role in multiple organ development and wound healing. Both cadherin-1 (CDH1) and cadherin-2 (CDH2) are calcium-dependent cell adhesion proteins, specifically for neural stem cells, and mediate anchorage to ependymocytes during maturation. Histone deacetylase 1 (HDAC1) is responsible for the deacetylation of lysine residues on the N-terminal part of the core histones and is involved in epigenetic repression with an important role in transcriptional regulation, cell cycle progression, and developmental events. Lastly, the zinc finger E-box-binding homeobox 1 (ZEB1; NM_001174096.2) is directly related to the ZEB2 gene causing Mowat–Wilson syndrome and acts as a transcriptional repressor by inhibiting interleukin-2 (IL-2; NM_000586.4) gene expression. It also represses the E-cadherin promoter and induces an epithelial mesenchymal transition that positively regulates neuronal differentiation and plays a role in neurogenesis, as similarly seen with ZEB2 [47,48].
3.5. ZEB2 Protein Domains and Functions
There are six protein domains within the ZEB2 gene, and these negatively impact ZEB2 function, if altered by gene variants. The NIM (nucleosome remodeling and deacetylase interaction motif) protein domain functions as one of the major chromatin remodeling complexes. The N-ZF (N-terminal zinc finger cluster) and C-ZF (C-terminal zinc finger cluster) are significant players in gene regulation and function [49]. Moreover, in Xenopus, the N-ZF domain was also found to have an important role in early stages of neural induction [50]. These two zinc finger clusters are responsible for functions such as ZEB2 binding to DNA. These DNA-binding proteins often work to pack and modify DNA or regulate gene expression and are therefore crucial for proper functioning for DNA binding in vitro [51]. The SMD (SMAD-binding domain) functions to mediate TGF-β signaling in metazoan embryo development and adult tissue regeneration and homeostasis [52], while HD (homeodomain) regulates the expression of other genes in development [53]. The CID (CtBP-interacting domain) functions to regulate transcription, predominantly as a corepressor in the nucleus [54] impeding transcription and translation. The CtBP-interacting domain (CID) is responsible for the direct interaction of ZEB2 with CtBPs found at repeated PLDLS-like motifs thought to make ZEB2 more efficient at transcriptional suppression. CtBPs on their own do not necessarily have the ability to bind DNA in a gene/promoter specific context but rely on the recruitment of DNA-binding transcription factors such as ZEB2 to function [48]. Additional research is required to further understand the role of these protein domains and their function in causing clinical variability among patients diagnosed with MWS. This research would impact diagnosis, clinical care, treatment, and surveillance as well as genetic counseling of first-degree family members.
4. Conclusions
In our study, we report on a 5-year-old female with features commonly seen in MWS and with a novel pathogenic heterozygous c.2471_2475del5 in exon 8 of the ZEB2 gene. This frameshift defect presumably disrupts ZEB2 protein production, quantity, and quality, as the five base pair deletion in exon 8 would impact the coding of three protein domains (CID, HD, and SMD) and about one-third of the N-ZF domain. The novel defect directly affects the encoding of the CID domain. The CID domain plays a role as a transcriptional corepressor, the HD domain regulates the expression of developmental genes, the SMD domain regulates signaling protein and embryo development, while the N-ZF domain is involved in gene regulation.
Computer literature and unreported databases were searched for keywords such as Mowat–Wilson, ZEB2 gene and protein defects or variants, or clinical features and about 180 published reports were found including 298 patients. These sources were used to collect data regarding ZEB2 gene variants, types, frequencies, and protein defects along with domain locations and functions. Frameshift variants followed by nonsense variants accounted for more than 90% of the ZEB2 gene defects. We found that exon 8, as the largest exon, encodes at least three of the six protein domains of the ZEB2 gene and accounts for 66% (198/298) of the variants identified. ZEB2 gene-gene or protein interactions were studied, and 24 separate proteins were predicted to share molecular functions with protein binding effects on embryo development impacting craniofacial, spine, brain, cardiovascular, kidney and hematopoiesis. ZEB2 also plays a role in the conversion of neuroepithelial cells in early brain formation and as a mediator of trophoblast differentiation.
Acknowledgments
We thank the parents of the child with Mowat–Wilson syndrome for allowing us to describe the clinical findings in this report and thank the Mowat–Wilson Syndrome Foundation for their support and encouragement.
Author Contributions
Research design conceptualization, M.G.B. and W.A.H.; methodology, validation, investigation, formal analysis, data curation: M.G.B., W.A.H., C.S.P., S.L. and S.K.R.; writing—original draft preparation: C.S.P., W.A.H. and M.G.B.; writing—review and editing, C.S.P., W.A.H., M.G.B., S.L. and S.K.R.; and funding acquisition: M.G.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The participant consented under an IRB-approved protocol (IRB# 11608) for research at the University of Kansas Medical Center (KUMC). The IRB-approved form was signed by the guardian prior to the study.
Informed Consent Statement
Written informed consent and photo permit forms were obtained from the parents as the patient was a minor.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research is supported by the Mowat–Wilson Syndrome Foundation, grant number KU Endowment 41149 and the National Institute of Child Health and Human Development (NICHD), grant number U54 HD061222.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Haendel M., Vasilevsky N., Unni D., Bologa C., Harris N., Rehm H., Hamosh A., Baynam G., Groza T., McMurry J., et al. How many rare diseases are there? Nat. Rev. Drug Discov. 2020;19:77–78. doi: 10.1038/d41573-019-00180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z., et al. Clinical whole-exome sequencing for the diagnosis of Mendelian Disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mowat D.R., Croaker G.D., Cass D.T., Kerr B.A., Chaitow J., Adès L.C., Chia N.L., Wilson M.J. Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: Delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J. Med. Genet. 1998;8:617–623. doi: 10.1136/jmg.35.8.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakamatsu N., Yamada Y., Yamada K., Ono T., Nomura N., Taniguchi H., Kitoh H., Mutoh N., Yamanaka T., Mushiake K., et al. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat. Genet. 2001;4:369–370. doi: 10.1038/86860. [DOI] [PubMed] [Google Scholar]
- 5.Cacheux V., Dastot-Le Moal F., Kääriäinen H., Bondurand N., Rintala R., Boissier B., Wilson M., Mowat D., Goossens M. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum. Mol. Genet. 2001;14:1503–1510. doi: 10.1093/hmg/10.14.1503. [DOI] [PubMed] [Google Scholar]
- 6.Baxter A.L., Vivian J.L., Hagelstrom R.T., Hossain W.A., Golden W.L., Wassman E.R., Vanzo R.J., Butler M.G. A Novel Partial Duplication of ZEB2 and Review of ZEB2 Involvement in Mowat-Wilson Syndrome. Mol. Syndromol. 2017;4:211–218. doi: 10.1159/000473693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mowat D.R., Wilson M.J., Goossens M. Mowat-Wilson syndrome. J. Med. Genet. 2003;5:305–310. doi: 10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adam M.P., Feldman J., Mirzaa F.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. GeneReviews [Internet] University of Washington, Seattle; Seattle, WA, USA: 2023. [Google Scholar]
- 9.Hegarty S.V., Sullivan A.M., O’Keeffe G.W. Zeb2: A multifunctional regulator of nervous system development. Prog. Neurobiol. 2015;132:81–95. doi: 10.1016/j.pneurobio.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Zweier C., Thiel C.T., Dufke A., Crow Y.J., Meinecke P., Suri M., Ala-Mello S., Beemer F., Bernasconi S., Bianchi P., et al. Clinical and mutational spectrum of Mowat–Wilson syndrome. Eur. J. Med. Genet. 2005;2:97–111. doi: 10.1016/j.ejmg.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Saunders C.J., Zhao W., Ardinger H.H. Comprehensive ZEB2 gene analysis for Mowat-Wilson syndrome in a North American cohort: A suggested approach to molecular diagnostics. Am. J. Med. Genet. A. 2009;11:2527–2531. doi: 10.1002/ajmg.a.33067. [DOI] [PubMed] [Google Scholar]
- 12.Ghoumid J., Drevillon L., Alavi-Naini S.M., Bondurand N., Rio M., Briand-Suleau A., Nasser M., Goodwin L., Raymond P., Yanicostas C., et al. ZEB2 zinc-finger missense mutations lead to hypomorphic alleles and a mild Mowat–Wilson syndrome. Hum. Mol. Genet. 2013;22:2652–2661. doi: 10.1093/hmg/ddt114. [DOI] [PubMed] [Google Scholar]
- 13.Zou D., Wang L., Wen F., Xiao H., Duan J., Zhang T., Yin Z., Dong Q., Guo J., Liao J. Genotype-phenotype analysis in Mowat-Wilson syndrome associated with two novel and two recurrent ZEB2 variants. Exp. Ther. Med. 2020;6:263. doi: 10.3892/etm.2020.9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y., Peng Q., Ma K., Li S., Rao C., Zhong B., Lu X. A novel nonsense mutation of ZEB2 gene in a Chinese patient with Mowat-Wilson syndrome. J. Clin. Lab. Anal. 2020;9:e23413. doi: 10.1002/jcla.23413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho S., Luk H.M., Chung B.H., Fung J.L., Mak H.H., Lo I.F.M. Mowat-Wilson syndrome in a Chinese population: A case series. Am. J. Med. Genet. A. 2020;182:1336–1341. doi: 10.1002/ajmg.a.61557. [DOI] [PubMed] [Google Scholar]
- 16.Wenger T.L., Harr M., Ricciardi S., Bhoj E., Santani A., Adam M.P., Barnett S.S., Ganetzky R., McDonald-McGinn D.M., Battaglia D. CHARGE-like presentation, craniosynostosis and mild Mowat-Wilson Syndrome diagnosed by recognition of the distinctive facial gestalt in a cohort of 28 new cases. Am. J. Med. Genet. A. 2014;164A:2557–2566. doi: 10.1002/ajmg.a.36696. Erratum in Am. J. Med. Genet. A 2015, 167, 1682–1683. [DOI] [PubMed] [Google Scholar]
- 17.Mundhofir F.E., Yntema H.G., van der Burgt I., Hame B.C., Faradz S.M., van Bon B.W. Mowat-Wilson syndrome: The first clinical and molecular report of an indonesian patient. Case Rep. Genet. 2012;2012:949507. doi: 10.1155/2012/949507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray S.B., Spangler B.B., Helm B.M., Vergano S.S. Polymicrogyria in a 10-month-old boy with Mowat-Wilson syndrome. Am. J. Med. Genet. A. 2015;167A:2402–2405. doi: 10.1002/ajmg.a.37171. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Yan Y.C., Li Q., Zhang Z., Xiao P., Yuan X.Y., Li L., Jiang Q. Clinical and genetic features of Mowat-Wilson syndrome: An analysis of 3 cases. Zhongguo Dang Dai Er Ke Za Zhi. 2019;21:468–473. doi: 10.7499/j.issn.1008-8830.2019.05.014. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada Y., Nomura N., Yamada K., Matsuo M., Suzuki Y., Sameshima K., Kimura R., Yamamoto Y., Fukushi D., Fukuhara Y., et al. The spectrum of ZEB2 mutations causing the Mowat-Wilson syndrome in Japanese populations. Am. J. Med. Genet. A. 2014;164A:1899–1908. doi: 10.1002/ajmg.a.36551. Erratum in Am. J. Med. Genet. A 2015, 167, 1428. [DOI] [PubMed] [Google Scholar]
- 21.Tronina A., Świerczyńska M., Filipek E. First Case Report of Developmental Bilateral Cataract with a Novel Mutation in the ZEB2 Gene Observed in Mowat-Wilson Syndrome. Medicina. 2023;59:101. doi: 10.3390/medicina59010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakubiak A., Szczałuba K., Badura-Stronka M., Kutkowska-Kaźmierczak A., Jakubiuk-Tomaszuk A., Chilarska T., Pilch J., Braun-Walicka N., Castaneda J., Wołyńska K., et al. Clinical characteristics of Polish patients with molecularly confirmed Mowat-Wilson syndrome. J. Appl. Genet. 2021;62:477–485. doi: 10.1007/s13353-021-00636-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Refaat K., Helmy N., Elawady M., El Ruby M., Kamel A., Mekkawy M., Ashaat E., Eid O., Mohamed A., Rady M. Interstitial Deletion of 2q22.2q22.3 Involving the Entire ZEB2 Gene in a Case of Mowat-Wilson Syndrome. Mol. Syndromol. 2021;12:87–95. doi: 10.1159/000513313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musaad W., Lyons A., Allen N., Letshwiti J. Mowat-Wilson syndrome presenting with Shone’s complex cardiac anomaly. BMJ Case Rep. 2022;15:e246913. doi: 10.1136/bcr-2021-246913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pachajoa H., Gomez-Pineda E., Giraldo-Ocampo S., Lores J. Mowat-Wilson Syndrome as a Differential Diagnosis in Patients with Congenital Heart Defects and Dysmorphic Facies. Pharmgenom. Pers. Med. 2022;15:913–918. doi: 10.2147/PGPM.S380908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L., Wang J., Wang L., Xu Q., Zhou B., Zhang Z., Li Q., Wang H., Han L., Jiang Q., et al. Physical, language, neurodevelopment and phenotype-genotype correlation of Chinese patients with Mowat-Wilson syndrome. Front. Genet. 2022;13:1016677. doi: 10.3389/fgene.2022.1016677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y., Xu W., Wang Q., Lin Y., He P., Liu Y., Yuan H. Three Novel De Novo ZEB2 Variants Identified in Three Unrelated Chinese Patients With Mowat-Wilson Syndrome and A Systematic Review. Front. Genet. 2022;13:853183. doi: 10.3389/fgene.2022.853183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei L., Han X., Li X., Han B., Nie W. A Chinese Boy with Mowat-Wilson Syndrome Caused by a 10 bp Deletion in the ZEB2 Gene. Pharmgenom. Pers. Med. 2021;14:1041–1045. doi: 10.2147/PGPM.S320128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Şenbil N., Arslan Z., Sayın Kocakap D.B., Bilgili Y. A Case Report of a Prenatally Missed Mowat-Wilson Syndrome With Isolated Corpus Callosum Agenesis. Child. Neurol. Open. 2021;8:2329048X211006511. doi: 10.1177/2329048X211006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanovski I., Djuric O., Caraffi S.G., Santodirocco D., Pollazzon M., Rosato S., Cordelli D.M., Abdalla E., Accorsi P., Adam M.P., et al. Phenotype and genotype of 87 patients with Mowat-Wilson syndrome and recommendations for care. Genet. Med. 2018;20:965–975. doi: 10.1038/gim.2017.221. [DOI] [PubMed] [Google Scholar]
- 31. [(accessed on 1 December 2023)]. Available online: http://pathwaycommons.org/pc12/Pathway_751ce7c7ccec6e191682a33d7252aac8.
- 32.Long J., Zuo D., Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 2005;280:35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y., Banda M., Speyer C.L., Smith J.S., Rabson A.B., Gorski D.H. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol. Cell. Biol. 2010;15:3902–3913. doi: 10.1128/MCB.01237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., Wu X., Jiang R. Integrating human omics data to prioritize candidate genes. BMC Med. Genom. 2013;6:57. doi: 10.1186/1755-8794-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han C., Hong K.-H., Kim Y.H., Kim M.-J., Song C., Kim M.J., Kim S.-J., Raizada M.K., Oh S.P. SMAD1 deficiency in either endothelial or smooth muscle cells can predispose mice to pulmonary hypertension. Hypertension. 2013;61:1044–1052. doi: 10.1161/HYPERTENSIONAHA.111.199158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calligaris R., Bottardi S., Cogoi S., Apezteguia I., Santoro C. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc. Nat. Acad. Sci. USA. 1995;92:11598–11602. doi: 10.1073/pnas.92.25.11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3(FoxA) and GATA-4. Mol. Cell. 2002;2:279–289. doi: 10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Wang S., Wesley R.A., Danner R.L. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J. Biol. Chem. 2003;278:29192–29200. doi: 10.1074/jbc.M213043200. [DOI] [PubMed] [Google Scholar]
- 40.Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 41.Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 42.Mansouri A., St-Onge L., Gruss P. Role of Pax genes in endoderm-derived organs. Trends Endocr. Metab. 1999;10:164–167. doi: 10.1016/S1043-2760(98)00133-7. [DOI] [PubMed] [Google Scholar]
- 43.Kawamata N., Pennella M.A., Woo J.L., Berk A.J., Koffler H.P. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene. 2012;31:966–977. doi: 10.1038/onc.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber E., Tobler A., Malipiero U., Schaffner W., Fontana A. cDNA cloning of human N-Oct 3, a nervous-system specific POU domain transcription factor binding to the octamer DNA motif. Nucleic Acids Res. 1993;21:253–258. doi: 10.1093/nar/21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atanasoski S., Toldo S.S., Malipiero U., Schreiber E., Fries R., Fontana A. Isolation of the human genomic brain-2/N-Oct 3 gene (POUF3) and assignment to chromosome 6q16. Genomics. 1995;26:272–280. doi: 10.1016/0888-7543(95)80211-4. [DOI] [PubMed] [Google Scholar]
- 46.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Lee S., The C.E., Bunting K., Ma L., Shannon M.F. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int. Immunol. 2009;3:227–235. doi: 10.1093/intimm/dxn143. [DOI] [PubMed] [Google Scholar]
- 48.Wang H., Xiao Z., Zheng J., Wu J., Hu X.L., Yang X., Shen Q. ZEB1 Represses Neural Differentiation and Cooperates with CTBP2 to Dynamically Regulate Cell Migration during Neocortex Development. Cell Rep. 2019;27:2335–2353.e6. doi: 10.1016/j.celrep.2019.04.081. [DOI] [PubMed] [Google Scholar]
- 49.Güleray Lafcı N., Karaosmanoglu B., Taskiran E.Z., Simsek-Kiper P.O., Utine G.E. Mutated Transcripts of ZEB2 Do Not Undergo Nonsense-Mediated Decay in Mowat-Wilson Syndrome. Mol. Syndromol. 2023;14:258–265. doi: 10.1159/000528769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinnadurai G. Madame Curie Bioscience Database [Internet] Landes Bioscience; Austin, TX, USA: 2007. [(accessed on 1 November 2023)]. CtBP Family Proteins: Unique Transcriptional Regulators in the Nucleus with Diverse Cytosolic Functions. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6557/ [Google Scholar]
- 51.Li X., Han M., Zhang H., Liu F., Pan Y., Zhu J., Liao Z., Chen X., Zhang B. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark. Res. 2022;10:2. doi: 10.1186/s40364-021-00345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birkhoff J.C., Huylebroeck D., Conidi A. ZEB2, the Mowat-Wilson Syndrome Transcription Factor: Confirmations, Novel Functions, and Continuing Surprises. Genes. 2021;12:1037. doi: 10.3390/genes12071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macias M.J., Martin-Malpartida P., Massagué J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 2015;40:296–308. doi: 10.1016/j.tibs.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bürglin T.R. Homeodomain subtypes and functional diversity. Subcell. Biochem. 2011;52:95–122. doi: 10.1007/978-90-481-9069-0_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.