Abstract
Colorectal cancer (CRC) is one of the most prevalent cancers and the second leading cause of cancer deaths in developed countries. Early CRC may have no symptoms and symptoms usually appear with more advanced diseases. Regular screening can identify people who are at increased risk of CRC in order to offer earlier treatment. A cost-effective non-invasive platform for the screening and monitoring of CRC patients allows early detection and appropriate treatment of the disease, and the timely application of adjuvant therapy after surgical operation is needed. In this study, a cohort of 71 plasma samples that include 48 colonoscopy- and histopathology-confirmed CRC patients with TNM stages I to IV were recruited between 2017 and 2019. Plasma mRNA profiling was performed in CRC patients using NanoString nCounter. Normalized data were analyzed using a Mann–Whitney U test to determine statistically significant differences between samples from CRC patients and healthy subjects. A multiple-group comparison of clinical phenotypes was performed using the Kruskal–Wallis H test for statistically significant differences between multiple groups. Among the 27 selected circulating mRNA markers, all of them were found to be overexpressed (gene expression fold change > 2) in the plasma of patients from two or more CRC stages. In conclusion, NanoString-based targeted plasma CRC-associated mRNAs circulating the marker panel that can significantly distinguish CRC patients from a healthy population were developed for the non-invasive diagnosis of CRC using peripheral blood samples.
Keywords: NanoString, nCounter, non-invasive screening, plasma RNA, CRC, colorectal cancer
1. Introduction
CRC results from an abnormal growth of cells on the wall of the large bowel, which is the last portion of the digestive system. CRC is a complex disease, and it is believed to be formed by the accumulation of a cascade of genetic mutations in the body over time. Most CRC starts from polyps that are usually benign; however, some may develop into cancer over time after accumulating different mutations [1]. The development of a polyp into cancer may take more than 10 years. If it is not treated, cancer cells may spread to other parts of the body via the bloodstream and lymphatic system to invade and damage nearby organs. Adenoma–carcinoma sequence describes a gradual progression from normal epithelial mucosa to adenoma and then to carcinoma as a result of a series of genetic changes such as mutation and gene amplification [1]. Studies have shown that the risk of recurrence and subsequent death due to CRC is closely associated with the stage of the disease at the time of the first diagnosis [1,2]. Moreover, the risk of death from CRC could be reduced by detecting the disease at an earlier stage via mass screening and intervening [1,2].
With the growth of the aging population, it is expected that the number of cases, incidence, and death rates of CRC will increase further. Fortunately, CRC is highly curable if it is detected at an early stage. It is estimated that approximately 60% of all CRC deaths can be prevented by undergoing regular screening [3]. Early CRC may have no symptoms and symptoms usually appear with more advanced diseases. Common symptoms of CRC include a change in bowel habits (e.g., diarrhea or constipation) for unknown reasons that lasts for more than 2 weeks; persistent urge after passing stool; presence of blood or large amount of mucus in stool; abdominal discomfort (e.g., persistent pain, bloating, fullness, or cramps); weight loss and tiredness with unknown reason [1,2]. Direct visualization tests such as colonoscopy and sigmoidoscopy are currently the gold standards for examination of the lower digestive tract such as the colon and rectum. It allows clinicians to detect both cancerous and precancerous lesions by direct visualization. However, because colonoscopy involves invasive procedures, there is a risk of bowel perforation during colonoscopy and post-colonoscopy bleeding. Full bowel preparation and sedation are also required before the procedure. Moreover, the high examination cost as well as the risk of perforation during invasive procedures makes colonoscopy less suitable for a widespread population screening [4,5].
Human plasma contains RNA transcripts released by multiple cell types, including CRC cells. The potential application of serum circulating microRNA (miRNA) as biomarkers for CRC detection using the NanoString platform has been explored previously [6]. In this study, a NanoString-based targeted plasma circulating marker panel protocol for CRC-associated mRNAs in plasma was developed for the non-invasive diagnosis of CRC using peripheral blood samples. NanoString Technology utilizes novel digital color-coded barcode technology that is based on direct multiplexed measurement of gene expression and it offers high levels of precision and sensitivity. The use of our NanoString-based targeted plasma circulating marker panel allowed for the non-invasive detection of CRC, during which patients did not feel pain and embarrassment. Although our NanoString-based targeted plasma circulating marker panel cannot replace colonoscopy at this stage and colonoscopy is still the gold standard in the diagnosis of CRC, the results generated from this study will contribute to the development of a new targeted digital counting method without PCR amplification for early non-invasive CRC detection and monitoring of CRC patients.
2. Results
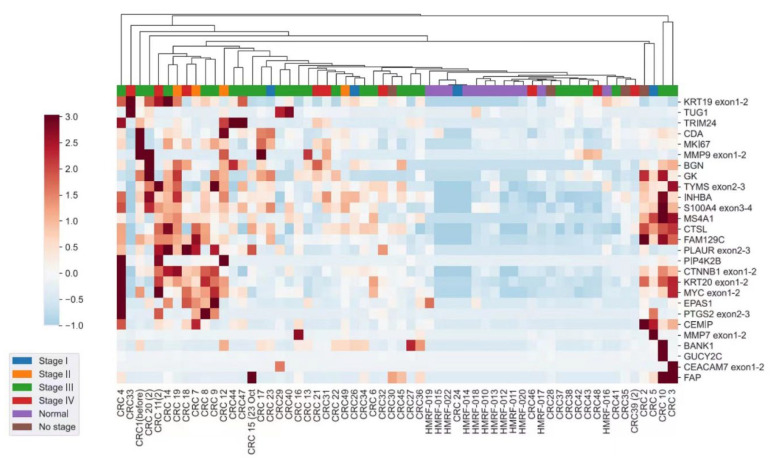
The CRC patients recruited in this study were aged between 50 and 88 (mean 71 ± 1) years old. The majority of the CRC patients were at stage III. Among the 27 selected candidate circulating mRNA markers, all of them were differentially expressed and able to distinguish CRC patients from healthy subjects significantly using plasma samples (p < 0.05). Those genes were found to be overexpressed (gene expression fold change >2) in the plasma of CRC patients in two or more CRC stages (Table 1). A heatmap showed a distinct gene expression pattern between CRC patients and healthy subjects without CRC (Figure 1). However, no significant difference in the expression of those genes from CRC TNM stages I to IV was found (Supplementary Table S2).
Table 1.
Demographic and clinical staging information of the study population.
| Category | Patient (n) | Age (Mean ± SD) | Male (%) |
|---|---|---|---|
| Healthy | 23 | 32.1 ± 10.7 | 48.7 |
| CRC | 48 | 71.1 ± 10.9 | 54.2 |
| Stage I | 4 | 69.5 ± 15.4 | 50.0 |
| Stage II | 4 | 64.5 ± 11.1 | 25.0 |
| Stage III | 27 | 71.2 ± 10.6 | 63.0 |
| Stage IV | 9 | 76.1 ± 9.1 | 55.6 |
| No stage | 4 | 66.8 ± 12.3 | 75.0 |
Figure 1.
Heatmap of the expression profile in CRC patients of each TNM stage independent of that of the healthy controls. The vertical axis shows the gene names and the horizontal axis shows the various TNM stages of CRC patients and healthy subjects. Hierarchical clustering was based on gene expression Z scores using the WPGMA algorithm. The red and blue colors denote high and low intensities of gene expression, respectively.
3. Discussion
Post-operative recurrence, metastasis, and spreading of tumor cells to lymph nodes are the major causes of cancer-related death in CRC patients. Prediction of the risk of tumor recurrence and metastasis is useful for guiding treatment decisions and disease management. Studies have shown that patients who receive early detection of disease recurrence and metastasis respond better to chemotherapy, and, thus, have better survival [7]. Hence, by identifying patients at high risk of recurrence and metastasis, the decision to adopt adjuvant treatment and close post-operative monitoring could be made immediately and appropriately in order to increase the survival rates of the patients. However, low sensitivity and low specificity for early-stage CRC detection using conventional screening methods such as the fecal occult blood test (FOBT) and fecal immunochemical test (FIT) [4,5], stool molecular tests [8,9], the blood-based carcinoembryonic antigen (CEA) [10] and SEPT9 tests [11] are still a major concern.
In this study, we analyzed a cohort of 71 plasma samples that included 48 colonoscopy- and histopathology-confirmed CRC patients with TNM stages I to IV as well as 23 colonoscopy-confirmed healthy subjects as controls. We aim to develop a non-invasive test utilizing a NanoString-based platform to detect and monitor CRC. Patients can benefit from the early detection and appropriate treatment of the disease, as well as the timely application of adjuvant therapy after surgical operation. From the heatmap produced in this study, all 27 selected circulating mRNA markers were differentially expressed and able to distinguish CRC patients from healthy subjects significantly using plasma samples. Those genes were found to be overexpressed (gene expression fold change >2) in the plasma of CRC patients in two or more CRC stages. However, no significant difference in the expression of those genes between CRC TNM stages I and IV was found. One of the possible reasons for this could be that the small sample size limited us from evaluating the sensitivity and accuracy of the panel in each TNM stage accurately in this study. Therefore, further validation is required.
To the best of our knowledge, this study is the first study using NanoString technologies to measure the expression of a cohort of RNA biomarkers in the plasma of CRC patients. Our results showed that this system can detect significant differences between colonoscopy-confirmed healthy people and CRC patients. The selected circulating mRNA markers in this study include important markers that have been shown to be involved and over-expressed during CRC development [5,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27]. Although potential CRC plasma circulating markers were identified in this study, this study has two limitations. First, the sample size was small. It prevented us from evaluating the sensitivity and accuracy of the panel in each TNM stage. An extensive evaluation with more CRC patients will be performed in the future. Second, the study was not designed to be age-matched between the CRC patients and healthy control subjects mainly because most CRC patients were diagnosed in the older age groups. An extensive evaluation with more age-matched patients and healthy subjects will be performed in the future to validate our results. In Hong Kong, the median age at diagnosis of CRC was 68 for males and 69 for females [28]. In order to focus on the over-expression of CRC-specific genes, we excluded patients who have been previously diagnosed and treated for CRC or the presence of synchronous malignancies. Therefore, it is not easy to find age-matched and colonoscopy-confirmed healthy control subjects for comparison in this study.
In conclusion, we have developed a NanoString-based targeted plasma circulating marker panel protocol for detecting CRC-associated mRNAs in plasma. This study is the first investigation to directly profile and analyze plasma mRNA markers in CRC patients utilizing a NanoString-based platform. The outcome of this study has also established a solid foundation for the non-invasive diagnosis of CRC using a peripheral blood sample. NanoString technology is a highly sensitive technology that can detect a scanty amount of RNA in plasma [29,30,31,32]. The results of this study have also shown that scanty amounts of RNA in plasma are detectable by NanoString technologies using random primers to amplify the RNA in plasma. This protocol can be applied in the development of non-invasive strategies or protocols for other cancers and diseases as well. Moreover, a panel of CRC-related markers was used for detection in this study. In order to translate this technology into routine clinical applications, a larger-scale study will be performed to include colorectal adenoma patients as well as patients with other common cancers such as breast cancer, liver cancer, and lung cancer to evaluate the specificity of this panel. Extensive evaluation of other factors such as logistics, regulation, and the cost of testing will also be needed to estimate the acceptance of this test in the future by the general public. Detection using multiple markers is more accurate than using individual single markers solely to predict the outcome of patients [33]. CRC detection using peripheral blood has long been an attractive approach because of its simplicity and non-invasive nature. The introduction of peripheral blood CRC screening has a potentially major impact on public health as well. For example, the Coronavirus Disease 2019 (COVID-19) pandemic has caused global changes in the delivery of healthcare services since 2019, including both outpatient community-based services and inpatient hospital-admission services [34]. During the COVID-19 pandemic, an accurate non-invasive CRC screening test could be an alternative to colonoscopy to reduce the risk of the transmission of infectious diseases and to relieve the workload in healthcare sectors [35,36].
However, in order to put this panel into clinical use, further clinical validation with a larger sample size is needed. Future work on this study can be focused on the follow-up on the plasma CRC-associated mRNA profiles in CRC patients (a) after surgical operation and before starting adjuvant therapy, (b) after completing adjuvant therapy, and through (c) tracing the change of plasma mRNA profiles for each patient to investigate if the observed changes are correlated with the patient’s clinical data such as metastasis, disease recurrence, and shorter survival to demonstrate the prognostic significance of the circulating marker panel.
4. Materials and Methods
4.1. Subjects Recruitment
In this study, a cohort of 71 plasma samples that include 48 colonoscopy- and histopathology-confirmed CRC patients with TNM stages from I to IV were recruited by the Departments of Surgery and Clinical Oncology, Queen Elizabeth Hospital between 1 September 2017 and 1 December 2019 with written informed consent. On the other hand, 23 colonoscopy-confirmed healthy subjects were recruited as healthy controls. The demographic and clinical staging information of the study population in this study are shown in Table 1. Ethics approval was obtained from the Kowloon Central Cluster Clinical Research Ethics Committee, Hong Kong (CREC Ref. No. 2019.542).
The inclusion criteria of this study were (1) patients who have been confirmed with CRC of various TNM stages, whereas the exclusion criteria of this study were (1) patients who have been previously diagnosed and treated for CRC, or (2) the presence of synchronous malignancies. Healthy volunteers with a family history of CRC, a history of colorectal polyps, cancer, inflammatory bowel, infectious diseases, or anemia were also excluded.
4.2. Customized CRC-Associated Targeted Plasma Circulating mRNA Markers Panel
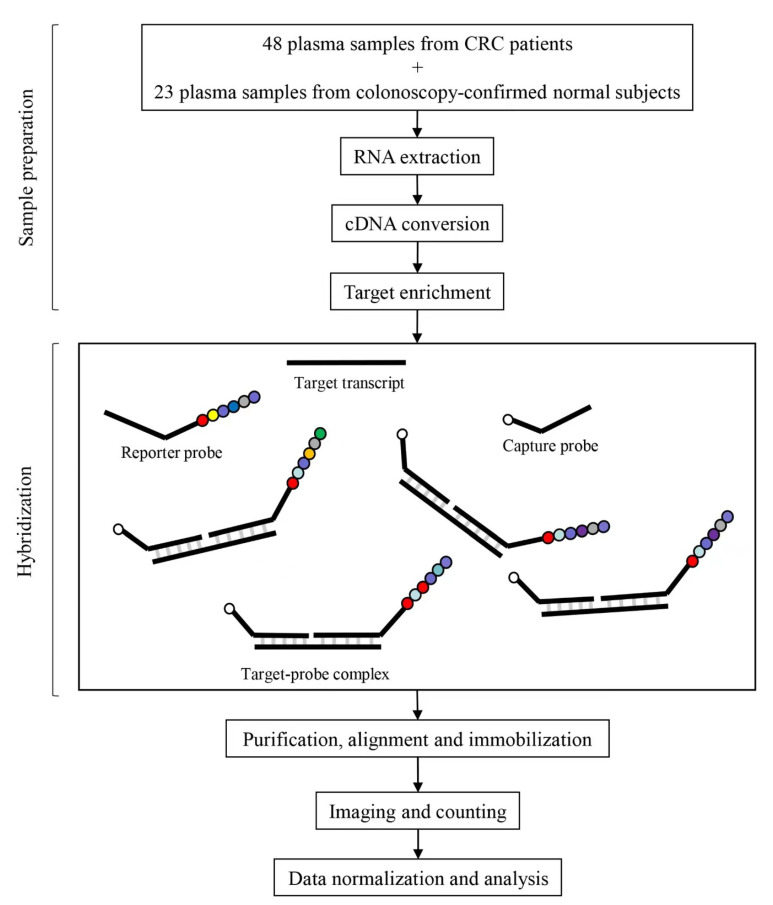
Literature review was performed and 27 published circulating mRNA markers that have diagnostic and prognostic potential for CRC were selected. They include CEA, CK19, GCC, KIAA1199, TUG1, TRIM24, PIP4K2B, GK, BANK1, CDA, CTSL1, MMP9, Ki-67, c-MYC, FAP, BGN, INHBA, BCNP1, MS4A1, CTNNB, CK20, CDX2, S100A4, EPAS1, TYMS, MMP7, COX2, and 2 house-keeping genes (GAPDH and TBP) (Table 2). The target sequences for the design of CodeSets are shown in Supplementary Table S1. Figure 2 illustrates the framework of this study.
Table 2.
Customized circulating mRNA markers panel composed of 27 published.
Figure 2.
A schematic illustration of the framework using NanoString technology to detect CRC-related circulating mRNA markers in plasma samples. A customized CodeSet for the circulating mRNA markers’ panel consists of sequence-specific capture and reporter probes, each of 35–50 bases long. The CodeSets are hybridized to the target transcript. Following hybridization, the target-probes complexes are purified, aligned, and immobilized in the sample cartridge using the automated nCounter Prep Station. Cartridges are then transferred to the nCounter Digital Analyzer for imaging and direct digital counting of the molecular barcodes on the reporter probes.
4.3. Blood Processing and Plasma RNA Extraction
For each subject, 9 mL of peripheral blood was collected in K3 EDTA tubes (Greiner Bio-one, Austria). Plasma was collected from peripheral blood samples using a double-centrifugation protocol described in our previous studies [45,46], which involves initial centrifugation at 1600× g followed by a second centrifugation at 16,000× g. Microfuge 22R centrifuge and an F301.5 rotor (Beckman Coulter) were used for centrifugation and plasma separation within 4 h after blood taking. After centrifugation, 4 mL of plasma was collected and preserved with 3.2 mL (0.8×) of Trizol (Life Technologies, Carsbad, CA, USA) before storage at −80 °C [45,46].
Cell-free RNA was extracted from plasma using our established protocol. In brief, 4.5 mL of preserved plasma with Trizol (Life Technologies, Carsbad, CA, USA) was used and the volume was topped up with Trizol to 5.5 mL. The plasma sample was then mixed with 2 mL of chloroform (Sigma–Aldrich, St. Louis, MO, USA), followed by centrifugation of 12,000× g for 15 min at 4 °C. The aqueous layer with RNA was collected and mixed with 1.5 volumes of absolute ethanol (Sigma–Aldrich, St. Louis, MO, USA) to achieve appropriate binding conditions. Total cell-free RNA was then extracted from the mixture using a miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol [47].
4.4. Targeted Genes Expression Profiling
The extracted RNA samples were then processed according to the manufacturer’s protocol for the nCounter low RNA input gene expression assay protocol.
4.5. cDNA Conversion
cDNA conversion was performed according to the manufacturer’s protocol of the nCounter Low RNA Input Amplification kit (NanoString Technologies, LOW-RNA-48, Seattle, WA, USA). In brief, the reverse transcription (RT) master mix was prepared by combining the RT Enzyme Mix and RT Primer Mix provided. For each sample, 1 μL of RT master mix was added to 4 μL of the extracted RNA sample. The cDNA conversion program was run on an Applied Biosystems 7500 Real-Time PCR System (Life Technologies) according to the manufacturer’s protocol. The concentrations of cDNA measured in this study are shown in Table 3.
Table 3.
The concentrations of cDNA measured in this study.
| Sample | Concentration (ng/µL) | Sample | Concentration (ng/µL) |
|---|---|---|---|
| 1 | 2635.037 | 37 | 681.436 |
| 2 | 2463.077 | 38 | 647.449 |
| 3 | 2475.2 | 39 | 624.455 |
| 4 | 2285.758 | 40 | 639.05 |
| 5 | 2368.351 | 41 | 605.24 |
| 6 | 2435.414 | 42 | 608.247 |
| 7 | 2307.185 | 43 | 643.833 |
| 8 | 2217.029 | 44 | 734.516 |
| 9 | 2253.341 | 45 | 614.606 |
| 10 | 2322.118 | 46 | 609.581 |
| 11 | 2398.608 | 47 | 624.743 |
| 12 | 2334.615 | 48 | 611.096 |
| 13 | 2571.032 | 49 | 323.357 |
| 14 | 2392.264 | 50 | 306.63 |
| 15 | 2557.589 | 51 | 299.239 |
| 16 | 2388.509 | 52 | 306.919 |
| 17 | 2900.842 | 53 | 302.598 |
| 18 | 2738.253 | 54 | 331.867 |
| 19 | 2737.108 | 55 | 325.641 |
| 20 | 2884.761 | 56 | 305.816 |
| 21 | 2799.384 | 57 | 312.801 |
| 22 | 2783.456 | 58 | 304.714 |
| 23 | 2860.305 | 59 | 316.975 |
| 24 | 2749.807 | 60 | 331.7 |
| 25 | 2753.355 | 61 | 153.338 |
| 26 | 2535.082 | 62 | 156.764 |
| 27 | 2573.109 | 63 | 150.78 |
| 28 | 2591.199 | 64 | 171.028 |
| 29 | 2521.445 | 65 | 162.975 |
| 30 | 2483.813 | 66 | 162.26 |
| 31 | 2557.493 | 67 | 158.024 |
| 32 | 2799.152 | 68 | 161.375 |
| 33 | 2453.478 | 69 | 151.518 |
| 34 | 2474.785 | 70 | 150.675 |
| 35 | 2432.49 | 71 | 179.454 |
| 36 | 2392.586 |
4.6. Multiplexed Target Enrichment
The amplification master mix was prepared by combining 5X dT Amp Master Mix and gene-specific primers at 500 nM per primer. For each sample, 2.5 µL of PCR master mix was added to the converted cDNA sample. The multiplexed target enrichment program was run on an Applied Biosystems 7500 Real-Time PCR System (Life Technologies) for 8 amplification cycles according to the manufacturer’s protocol.
4.7. NanoString Digital Profiling
Sequence-specific oligonucleotide probes tagged with fluorescent barcodes were used to bind to and digitally measure cDNA. Hybridization was conducted for 21 h at 65 °C. Subsequently, probes were purified and immobilized on the nCounter Prep Station (NanoString Technologies). Each sample was scanned for 600 FOV (fields of view) on the nCounter Digital Analyzer (NanoString Technologies). Data were extracted using the nCounter RCC Collector. The abundance of specific capture probe-bound cDNA molecules was measured using the nCounter digital analyzer to count individual fluorescent barcodes. The NanoString pre-designed panel simultaneously detects 27 CRC-related genes, including 2 housekeeping transcripts. Six positive and eight negative spike-in controls, hybridization controls, and ligation-specific controls were included to determine sample integrity, quality, and background.
4.8. Data Analysis
All data analyses were performed using nSolver 4.0 (NanoString Technologies). The raw data from NanoString nCounter were normalized for lane-to-lane variation with a dilution series of 6 spike-in positive controls. The sum of the 6 positive controls for a given lane was divided by the average sum across lanes to yield a normalization factor, which was then multiplied by the raw counts in each lane to give normalized values. Codeset normalization was performed using the 2 housekeeping genes. For each sample, the mean plus 2 times the standard deviation (threshold = mean + 2SD) of the 8 negative controls (probes without target sequences) was subtracted from each miRNA count in that sample.
Normalization of data was achieved by eliminating digital counts below 50. Comparison of gene expression profiles between (a) CRC patients and healthy controls (Table 4) and (b) CRC patients of different stages was performed (Supplementary Table S2). A heatmap with statistically significant differences was generated. Hierarchical clustering was based on gene expression Z scores using the weighted pair group method with an averaging (WPGMA) algorithm (Figure 1). Multiple group comparison of clinical phenotype was performed using the Kruskal–Wallis H test for statistically significant differences between multiple groups and the Mann–Whitney U test for statistically significant differences between two groups. The p-values were adjusted using the Benjamini–Hochberg procedure. p ≤ 0.05 was taken as statistically significant.
Table 4.
Results of Kruskal–Wallis H test for significant differences between multiple groups. Twenty-seven differentially expressed RNAs were found to be upregulated in the plasma of CRC patients.
| Gene Name | Refseq | Stage I | Stage II | Stage III | Stage IV | Normal | p-Value | Adjusted p-Value * |
|---|---|---|---|---|---|---|---|---|
| BANK1 | NM_001083907.1 | 3118.818 | 3773.453 | 3697.386 | 944.5667 | 169.1133 | 0.001758 | 0.004394 |
| BGN | NM_001711.3 | 103.0225 | 191.175 | 149.2763 | 123.5811 | 20 | 0.002165 | 0.004639 |
| CDA | NM_001785.2 | 82,333 | 133,665.2 | 79,813.29 | 30,877.19 | 41,917.4 | 0.03538 | 0.04746 |
| CEACAM7 | NM_006890.4 | 46.1025 | 51.425 | 13,521.77 | 28.64333 | 20 | 0.010153 | 0.016922 |
| CEMIP | NM_018689.1 | 47,200.59 | 59,372.89 | 23,834.92 | 18,151.22 | 4046.548 | 0.025741 | 0.035781 |
| CTNNB1 | NM_001098210.1 | 15,716.63 | 36,968.89 | 25,861.48 | 19,006.47 | 8143.892 | 0.003437 | 0.006751 |
| CTSL | NM_001912.4 | 26,001.42 | 36,041.48 | 22,480.74 | 15,920.57 | 3366.493 | 0.003889 | 0.00713 |
| EPAS1 | NM_001430.3 | 283.21 | 3203.148 | 3505.034 | 1865.092 | 1235.516 | 0.024221 | 0.035057 |
| FAM129C | NM_173544.4 | 58,697.87 | 99,605.14 | 59,054.28 | 47,203.52 | 16,909.75 | 0.000818 | 0.002248 |
| FAP | NM_004460.2 | 321.02 | 102.3725 | 869.8074 | 46.50889 | 20 | 0.002193 | 0.004639 |
| GK | NM_000167.3 | 19,534.83 | 29,865.46 | 20,696.59 | 10,550.17 | 3633.967 | 0.010934 | 0.017687 |
| GUCY2C | NM_004963.1 | 61.775 | 97.025 | 494.8544 | 68.26778 | 20 | 0.002057 | 0.004639 |
| INHBA | NM_002192.2 | 50,134.49 | 86,119.04 | 54,501 | 47,530.9 | 18,310.55 | 0.004839 | 0.008317 |
| KRT19 | NM_002276.4 | 12,402.46 | 32,193.63 | 26,178.84 | 29,384.17 | 4029.44 | 0.00057 | 0.001669 |
| KRT20 | NM_019010.2 | 46,344.5 | 11,6612.9 | 78,925.36 | 56,187.71 | 10,225.4 | 0.001336 | 0.003498 |
| MKI67 | NM_002417.2 | 56,987.24 | 74,004.38 | 77,026.36 | 52,881.03 | 22,054.26 | 0.026023 | 0.035781 |
| MMP7 | NM_002423.4 | 3649.235 | 277.8825 | 382.3 | 120.2722 | 20 | 0.00052 | 0.001669 |
| MMP9 | NM_004994.2 | 367.665 | 6548.393 | 7647.711 | 3684.182 | 20 | 0.000296 | 0.001086 |
| MS4A1 | NM_152866.2 | 18,589.93 | 22,468.3 | 17,110.81 | 7012.719 | 650.3958 | 0.000195 | 0.000893 |
| MYC | NM_002467.4 | 13,616.51 | 34,941.22 | 27,283.9 | 26,276.21 | 5521.895 | 0.003054 | 0.006222 |
| PIP4K2B | NM_003559.4 | 308.945 | 6175.323 | 1391.916 | 1687.758 | 20 | 0.002117 | 0.004639 |
| PLAUR | NM_002659.3 | 565.765 | 12,562.74 | 6378.578 | 9516.01 | 1035.063 | 0.003746 | 0.007105 |
| PTGS2 | NM_000963.3 | 27,295.66 | 20,618.94 | 25,963.25 | 16,800.94 | 20 | 0.004645 | 0.008242 |
| S100A4 | NM_019554.2 | 33,689 | 44,015.95 | 37,234.61 | 25,325.74 | 18,203.82 | 0.012145 | 0.019084 |
| TRIM24 | NM_015905.2 | 592.65 | 8514.685 | 7197.833 | 3787.37 | 20 | 0.0000283 | 0.000398 |
| TUG1 | NR_002323.1 | 975.32 | 5631.128 | 8363.993 | 21,629.69 | 2689.192 | 0.014668 | 0.021804 |
| TYMS | NM_001071.2 | 30,818.32 | 50,162.11 | 39,236.93 | 33,363.06 | 13,571.37 | 0.013251 | 0.020245 |
* adjusted p-value using the Benjamini–Hochberg procedure.
5. Conclusions
A NanoString-based targeted plasma CRC-associated mRNAs circulating marker panel that can significantly distinguish CRC patients from healthy populations was developed for the non-invasive diagnosis of CRC using peripheral blood samples. It allows for early detection as well as the timely application of adjuvant therapy after surgical operation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25053012/s1.
Author Contributions
H.F.T. and S.C.C.W. designed and performed the experiments and analyzed and interpreted the data; X.M.P. and Y.K.E.W. helped perform the experiments; X.M.P. and S.C.C.W. helped review and edit the manuscript; H.F.T. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Kowloon Central Cluster Clinical Research Ethics Committee, Hong Kong (CREC Ref. No. 2019.542).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that they have no competing interests.
Funding Statement
This study was supported by the Research Grants Council Hong Kong, Hong Kong Innovation and Technology Fund (Grant Number: RGCQ71P), University—Industry Collaborative Programme (Grant Number: UIM/354) and LimPeng Suan Charitable Trust Research Grant for S.C.C.W. (Grant Number: R-ZH5G).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Li J., Ma X., Chakravarti D., Shalapour S., DePinho R.A. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021;35:787–820. doi: 10.1101/gad.348226.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung J.J.Y., Chan F.K.L., Chiu H.M., Kim H.S., Matsuda T., Ng S.S.M., Lau J.Y.W., Zheng S., Adler S., Reddy N., et al. An updated Asia Pacific consensus recommendations on colorectal cancer screening. Gut. 2015;64:121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 3.Traverso G., Shuber A., Olsson L., Levin B., Johnson C., Hamilton S.R., Boynton K., Kinzler K.W., Vogelstein B. Detection of proximal colorectal cancers through analysis of faecal DNA. Lancet. 2002;359:403–404. doi: 10.1016/S0140-6736(02)07591-8. [DOI] [PubMed] [Google Scholar]
- 4.Schoen R.E. The case for population-based screening for colorectal cancer. Nat. Rev. Cancer. 2002;2:65–70. doi: 10.1038/nrc705. [DOI] [PubMed] [Google Scholar]
- 5.Wong S.C., Lo S.F., Cheung M.T., Ng K.O.E., Tse C.W., Lai B.S.P., Lee K.C., Lo Y.M.D. Quantification of plasma beta-catenin mRNA in colorectal cancer and adenoma patients. Clin. Cancer Res. 2004;10:1613–1617. doi: 10.1158/1078-0432.CCR-1168-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Raju G.S., Chang D.W., Lin S., Chen Z., Wu X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer. 2018;124:785–796. doi: 10.1002/cncr.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin B., Lieberman D.A., McFarland B., Andrews K.S., Brooks D., Bond J., Dash C., Giardiello F.M., Glick S., Johnson D., et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American cancer society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., Levin T.R., Lavin P., Lidgard G.P., Ahlquist D.A., Berger B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 9.Liang J.Q., Li T., Nakatsu G., Chen Y.-X., Yau T.O., Chu E., Wong S., Szeto C.H., Ng S.C., Chan F.K.L., et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2020;69:1248–1257. doi: 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moertel C.G., Fleming T.R., Macdonald J.S., Haller D.G., Laurie J.A., Tangen C. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA. 1993;270:943–947. doi: 10.1001/jama.1993.03510080047030. [DOI] [PubMed] [Google Scholar]
- 11.Nikolaou S., Qiu S., Fiorentino F., Rasheed S., Tekkis P., Kontovounisios C. Systematic review of blood diagnostic markers in colorectal cancer. Tech. Coloproctol. 2018;22:481–498. doi: 10.1007/s10151-018-1820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Pons M., Cruz-Correa M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed Res. Int. 2015;2015:149014. doi: 10.1155/2015/149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong S.C.C., Chan C.M.L., Ma B.B.Y., Hui E.P., Ng S.S.M., Lai P.B.S., Cheung M.T., Lo E.S.F., Chan A.K.C., Lam M.Y.Y., et al. Clinical significance of cytokeratin 20-positive circulating tumor cells detected by a refined immunomagnetic enrichment assay in colorectal cancer patients. Clin. Cancer Res. 2009;15:1005–1012. doi: 10.1158/1078-0432.CCR-08-1515. [DOI] [PubMed] [Google Scholar]
- 14.Wong S.C.C., Ng S.S.M., Cheung M.T., Luk L.Y., Chan C.M.L., Cheung A.H.K., Lee V.H.M., Lai P.B.S., Ma B.B.Y., Hui E.P., et al. Clinical significance of CDX2-positive circulating tumour cells in colorectal cancer patients. Br. J. Cancer. 2011;104:1000–1006. doi: 10.1038/bjc.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang C.-C., Yang S.-H., Chien C.-C., Chen S.-H., Pan S., Lee C.-L., Lin C.-M., Sun H.-L., Huang C.-C., Wu Y.-Y., et al. Clinical meaning of age-related expression of fecal cytokeratin 19 in colorectal malignancy. BMC Cancer. 2009;9:376. doi: 10.1186/1471-2407-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herszényi L., Sipos F., Galamb O., Solymosi N., Hritz I., Miheller P., Berczi L., Molnár B., Tulassay Z. Matrix metalloproteinase-9 expression in the normal mucosa–adenoma–dysplasia–adenocarcinoma sequence of the colon. Pathol. Oncol. Res. 2008;14:31–37. doi: 10.1007/s12253-008-9004-5. [DOI] [PubMed] [Google Scholar]
- 17.Kang J., Chen J., Lee C., Chang J., Shieh Y. Intratumoral macrophage counts correlate with tumor progression in colorectal cancer. J. Surg. Oncol. 2010;102:242–248. doi: 10.1002/jso.21617. [DOI] [PubMed] [Google Scholar]
- 18.Koskensalo S., Louhimo J., Nordling S., Hagström J., Haglund C. MMP-7 as a prognostic marker in colorectal cancer. Tumor Biol. 2011;32:259–264. doi: 10.1007/s13277-010-0080-2. [DOI] [PubMed] [Google Scholar]
- 19.Zaman K., Driscoll R., Hahn D., Werffeli P., Goodman S.L., Bauer J., Leyvraz S., Lejeune F., Stupp R., Rüegg C. Monitoring multiple angiogenesis-related molecules in the blood of cancer patients shows a correlation between VEGF-A and MMP-9 levels before treatment and divergent changes after surgical vs. conservative therapy. Int. J. Cancer. 2006;118:755–764. doi: 10.1002/ijc.21408. [DOI] [PubMed] [Google Scholar]
- 20.Lakatos G., Sipos F., Miheller P., Hritz I., Varga M.Z., Juhász M., Molnár B., Tulassay Z., Herszényi L. The behavior of matrix metalloproteinase-9 in lymphocytic colitis, collagenous colitis and ulcerative colitis. Pathol. Oncol. Res. 2012;18:85–91. doi: 10.1007/s12253-011-9420-9. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Sun L., Tang Z., Fu L., Xu Y., Li Z., Luo W., Qiu X., Wang E. Overexpression of TRIM24 correlates with tumor progression in non-small cell lung cancer. PLoS ONE. 2012;7:e37657. doi: 10.1371/journal.pone.0037657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambon M., Orsetti B., Berthe M.-L., Bascoul-Mollevi C., Rodriguez C., Duong V., Gleizes M., Thénot S., Bibeau F., Theillet C., et al. Prognostic significance of TRIM24/TIF-1α gene expression in breast cancer. Am. J. Pathol. 2011;178:1461–1469. doi: 10.1016/j.ajpath.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Z., Cao W., Li J., Song X., Mao L., Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS ONE. 2013;8:e63887. doi: 10.1371/journal.pone.0063887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuester D., Lippert H., Roessner A., Krueger S. The cathepsin family and their role in colorectal cancer. Pathol.—Res. Pr. 2008;204:491–500. doi: 10.1016/j.prp.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Han Y., Liu Y., Gui Y., Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J. Surg. Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 26.Mudd T.W., Lu C., Klement J.D., Liu K. MS4A1 expression and function in T cells in the colorectal cancer tumor microenvironment. Cell. Immunol. 2021;360:104260. doi: 10.1016/j.cellimm.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Jia S., Jiang W.G. KIAA1199 and its biological role in human cancer and cancer cells (Review) Oncol. Rep. 2014;31:1503–1508. doi: 10.3892/or.2014.3038. [DOI] [PubMed] [Google Scholar]
- 28.Hong Kong Cancer Registry, Hospital Authority. Colorectal Cancer in 2019. [(accessed on 1 October 2021)]. Available online: https://www3.ha.org.hk/cancereg/pdf/factsheet/2019/colorectum_2019.pdf.
- 29.Wallden B., Storhoff J., Nielsen T., Dowidar N., Schaper C., Ferree S., Liu S., Leung S., Geiss G., Snider J., et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med. Genom. 2015;8:54. doi: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Bioteechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 31.Veldman-Jones M.H., Brant R., Rooney C., Emery H., Harbron C.G., Wappett M., Sharpe A., Dymond M., Barrett J.C., Harrington E.A., et al. Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee A., Leichter A.L., Fan V., Tsai P., Purcell R.V., Sullivan M.J., Eccles M.R. A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients. Sci. Rep. 2015;5:srep10438. doi: 10.1038/srep10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichita C., Ciarloni L., Monnier-Benoit S., Hosseinian S., Dorta G., Rüegg C. A novel gene expression signature in peripheral blood mononuclear cells for early detection of colorectal cancer. Aliment. Pharmacol. Ther. 2014;39:507–517. doi: 10.1111/apt.12618. [DOI] [PubMed] [Google Scholar]
- 34.El Khoury C., Haro E., Alves M., O’Dwyer M.C., Meixner K., Albiac L.C., Capizzano J.N., Ramakrishnan M., Salada C., Gorin S.S., et al. Patient-Centered Home Cancer Screening Attitudes During COVID-19 Pandemic. J. Patient-Centered Res. Rev. 2021;8:340–346. doi: 10.17294/2330-0698.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falzone L., Gattuso G., Tsatsakis A., Spandidos D.A., Libra M. Current and innovative methods for the diagnosis of COVID-19 infection (Review) Int. J. Mol. Med. 2021;47:100. doi: 10.3892/ijmm.2021.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng Z., Guo S., Zhou Y., Li M., Wang M., Ying B. Applications of laboratory findings in the prevention, diagnosis, treatment, and monitoring of COVID-19. Signal Transduct. Target. Ther. 2021;6:316. doi: 10.1038/s41392-021-00731-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaPointe L.C., Pedersen S.K., Dunne R., Brown G.S., Pimlott L., Gaur S., McEvoy A., Thomas M., Wattchow D., Molloy P.L., et al. Discovery and validation of molecular biomarkers for colorectal adenomas and cancer with application to blood testing. PLoS ONE. 2012;7:e29059. doi: 10.1371/journal.pone.0029059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chee C.E., Meropol N.J. Current status of gene expression profiling to assist decision making in stage II colon cancer. Oncologist. 2014;19:704–711. doi: 10.1634/theoncologist.2013-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han M., Zhang H.W., Chao S., Zheng R., Yip K.T., Song Z.-Y., Li H.M., Geng X.P., Zhu L.X., Lin J.-J., et al. Novel blood-based, five-gene biomarker set for the detection of colorectal cancer. Clin. Cancer Res. 2008;14:455–460. doi: 10.1158/1078-0432.CCR-07-1801. [DOI] [PubMed] [Google Scholar]
- 40.Stein U., Burock S., Herrmann P., Wendler I., Niederstrasser M., Wernecke K.-D., Schlag P.M. Diagnostic and prognostic value of metastasis inducer S100A4 transcripts in plasma of colon, rectal, and gastric cancer patients. J. Mol. Diagn. 2011;13:189–198. doi: 10.1016/j.jmoldx.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collado M., Garcia V., Garcia J.M., Alonso I., Lombardia L., Diaz-Uriarte R., Fernández L.A.L., Zaballos A., Bonilla F., Serrano M. Genomic profiling of circulating plasma RNA for the analysis of cancer. Clin. Chem. 2007;53:1860–1863. doi: 10.1373/clinchem.2007.089201. [DOI] [PubMed] [Google Scholar]
- 42.Garcia V., García J.M., Peña C., Silva J., Domínguez G., Hurtado A., Alonso I., Rodriguez R., Provencio M., Bonilla F. Thymidylate synthase messenger RNA expression in plasma from patients with colon cancer: Prognostic potential. Clin. Cancer Res. 2006;12:2095–2100. doi: 10.1158/1078-0432.CCR-05-1644. [DOI] [PubMed] [Google Scholar]
- 43.Bujanda L., Sarasqueta C., Cosme A., Hijona E., Enríquez-Navascués J.M., Placer C., Villarreal E., Herreros-Villanueva M., Giraldez M.D., Gironella M., et al. Evaluation of alpha 1-antitrypsin and the levels of mRNA expression of matrix metalloproteinase 7, urokinase type plasminogen activator receptor and COX-2 for the diagnosis of colorectal cancer. PLoS ONE. 2013;8:e51810. doi: 10.1371/journal.pone.0051810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García J.M., Peña C., García V., Domínguez G., Muñoz C., Silva J., Millán I., Diaz R., Lorenzo Y., Rodriguez R., et al. Prognostic value of LISCH7 mRNA in plasma and tumor of colon cancer patients. Clin. Cancer Res. 2007;13:6351–6358. doi: 10.1158/1078-0432.CCR-07-0882. [DOI] [PubMed] [Google Scholar]
- 45.Wong S.C., Ma B.B., Lai P.B., Ng S.S., Lee J.F., Hui E.P., Lam M.Y., Chan C.M., Chan A.T. The effect of centrifugation on circulating mRNA quantitation opens up a new scenario in expression profiling from patients with metastatic colorectal cancer. Clin. Biochem. 2007;40:1277–1284. doi: 10.1016/j.clinbiochem.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Xue V.W., Ng S.S.M., Leung W.W., Ma B.B.Y., Cho W.C.S., Au T.C.C., Yu A.C.S., Tsang H.F.A., Wong S.C.C. The Effect of Centrifugal Force in Quantification of Colorectal Cancer-Related mRNA in Plasma Using Targeted Sequencing. Front. Genet. 2018;9:165. doi: 10.3389/fgene.2018.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong S.C.C., Lo E.S.F., Cheung M.T. An optimised protocol for the extraction of non-viral mRNA from human plasma frozen for three years. J. Clin. Pathol. 2004;57:766–768. doi: 10.1136/jcp.2003.007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.